Abstract
Candida albicans, one of the most common fungal pathogens, is responsible for several yeast infections in human hosts, being resistant to classically used antifungal drugs, such as azole drugs. Multifactorial and multistep alterations are involved in the azole resistance in Candida albicans. In this study, a FCZ-resistant C. albicans strain was obtained by serial cultures of a FCZ-susceptible C. albicans strain in incrementally increasing concentrations of FCZ. We performed an integrated profile of different classes of molecules related to azole resistance in C. albicans by combining several mass-spectrometry based methodologies. The comparative metabolomic study was performed with the sensitive and resistant strains of C.albicans to identify metabolites altered during the development of resistance to fluconazole, while the intervention strains and non-intervention strains of C.albicans to identify metabolites altered involved in cross-resistant to azole drugs. Our analysis of the different metabolites identified molecules mainly involved in metabolic processes such as amino acid metabolism, tricarboxylic acid cycle and phospholipid metabolism. We also compared the phospholipid composition of each group, revealing that the relative content of phospholipids significantly changed during the development of resistance to azole drugs. According with these results, we hypothesized that the metabolism shift might contribute to azole drugs resistance in C.albicans from multifactorial alterations. Our result paves the way to understand processes underlying the resistance to azole drugs in C. albicans, providing the basis for developing new antifungal drugs.
Introduction
Candida albicans is an asexual opportunistic fungus that causes infections in immunocompromised and debilitated individuals[1]. Azole drugs are widely used in clinical practice having a good antibacterial power and low side effects[2]. Unfortunately, widespread uses of azole drugs have led to the rapid development of drug resistance which hampers the efficacy of current treatments for invasive mycoses. The considerable number of side effects mainly related to high-concentration use of drugs and prolonged therapies. The increase of drug resistance still make a serious clinical problem for fungal infections[3]. A strong mobilization of the scientific community allowed elucidation of the molecular mechanisms underlying fluconazole resistance in the yeasts Candida, of which a number of them could be extrapolated to other azole drugs, such as mutations in the ERG11 gene, overexpression of efflux membrane transporters and overexpression of ERG11[4]. Metabolomics is a systems biology approach to study small molecules. It is a rapidly growing field that aims to profile as many low molecular weight metabolites as possible, rather than focus on single metabolites in cells, biofluids and tissue extracts[5, 6]. Almost all metabolomics approaches can be classified as targeted or untargeted. Targeted analyses always focus on a subset of known metabolites, while untargeted are global screening approaches[7]. Using bioinformatic and statistical tools, metabolomic profiles from different samples can be compared, identifying potential metabolite markers or patterns typical for a specific sample or condition. However, due to the wide range of metabolite concentrations and the diversity of their biochemical properties, no single analytical technique can provide a fully characterization of the metabolic profile of an organism. The best option is to use a combination of several analytical approaches. High-performance liquid chromatography mass spectrometry (HPLC-MS) and gas chromatography MS (GC-MS) are two widely used techniques for metabolomics analysis.
In this study, we designed to gain insight into alterations in metabolites associated with azole drugs resistance in laboratory C. albicans strains. Multiple metabolomic approaches were integrated for analyzing the metabolic fingerprinting and footprinting and profiling intracellular and extracellular metabolites associated with azole resistance in C. albicans. We adopted GC-MS and ultra-high performance liquid chromatography coupled with quadrupole time-of-flight MS (UHPLC-Q-TOF/MS) to obtain the metabolomic profiling. In addition, hydrophilic interaction liquid chromatography coupled with triple quadrupole MS (HILIC-QQQ/MS) method was used to profile phospholipid metabolism. Our results indicated that C. albicans responds to azole drugs by changing the expression of specific classes of metabolites. These metabolites were mainly related to anti-oxidative stress, and the function of cell membrane and mitochondrion. Although further studies are needed, our work intends to improve knowledge on the complex molecular networks involved in C. albicans resistance to azole drugs.
Materials and methods
Antifungal agents and chemicals
Fluconazole (FCZ), miconazole (MCZ), ketoconazole (KCZ) and the derivatizing reagent trimethylchlorosilane (TMCS), methoxyamine hydrochloride, pyridine and N-methyl-N-[trimethylsilyl]-trifluoroacetamide (MSTFA) were from Sigma Aldrich (St. Louis, MO, USA). Internal standards (IS) α-aminobutyric acid was from Dibo chemical and technology Co., Ltd (Shanghai, China). Ultrapure water was prepared with the Milli-Q water purification system (Millipore, Bedford, MA, USA) and solvents used for chromatographies were of HPLC-MS grade (Merck, Darmstadt, Germany). Other reagents were of analytical grade.
Induction of resistant strains
C. albicans strain SC5314 was kindly provided by Professor William A. Fonzi. A single colony of the strain SC5314 was first inoculated into 10 mL of YPD medium (1% w/v yeast extract, 2% w/v peptone, and 2% w/v dextrose) and cells were grown at 30°C with constant agitation (200 r/min). Every day, an aliquot of the overnight culture containing 106 cells was transferred into 10 mL of fresh medium containing twice their most recently measured minimal inhibitory concentration (MIC) of FCZ, and the cells were incubated overnight at 30°C with constant agitation. When the cultures reached a density of 108 cells/mL, anothor aliquot containing 106 cells was transferred into fresh medium and incubated as described above. Sensitive strains were grown without drug, while resistant strains were grown in twice their most recently measured MIC of fluconazole until they were grown steadily in 64 μg/mL of fluconazole. In order to study the developed cross-resistance to azole drugs, ketoconazole (KCZ) was set as an intervention drug. We divided these strains into 4 groups: sensitive strains (S group), resistant strains (R group), sensitive strains treated with ketoconazole (S+KCZ group), resistant strains treated with ketoconazole (R+KCZ group).
Determination of MICs
During the experiment, MIC of fluconazole, miconazole and ketoconazole were determined at each sampling time by the broth microdilution method using 96 well plates according to the National Committee for Clinical Laboratory Standards (M27-A2) protocol[8]. The lowest concentration of the extract that produced no visible growth (no turbidity) after 24h when compared with the control tubes was considered as initial MIC of this generation.
Culture conditions and determination of ketoconazole IC50
For the determination of the half maximal inhibitory concentration (IC50) of KCZ, C. albicans cells were suspended in YPD medium until reaching the optical density at 600 nm (OD600) of 0.1 and then allowed to reach the OD600 of 0.2 at 30°C. Several concentrations of KCZ were added and cells were incubated for 4 h at the same condition. Compared with control group, IC50 was 16 μg/mL and 32 μg/mL for sensitive and resistant strains, respectively. We thus set the concentration of KCZ for our experiments at 16 μg/mL.
Sample preparation for UHPLC-Q-TOF/MS experiments
Intracellular metabolites of C. albicans SC5314 were isolated as previously described with slight modifications[9, 10]. Briefly, following the cultivation, all samples were rapidly washed with ice-cold ultrapure sterile water. For quenching, samples were resuspended in 1 mL of ice-cold methanol:water (60:40) at -80°C for 5 min and centrifuged at 5,000 ×g for 5 min. For the extraction, pellets were resuspended in 1 mL of boiling water (containing 200 μg/mL IS), for 15 min. Samples were then subject to three repeated freeze-thaw cycles (15 min in a -80°C refrigerator and at 60°C in hot water, conducted alternatively). After centrifugation at 10,000 ×g for 5 min at 4°C, supernatants were collected, transferred into a filter cap and centrifuged again at the same condition. Supernatants were collected and dissolved in the initial mobile phase (water: acetonitrile = 95:5) after freeze-drying. The cells were dried at room temperature to constant weight. The dry fungus weight was measured at least three times, and the mean value was considered as the biomass of each sample.
Extracellular metabolites were obtained by the centrifugation of culture medium at 5,000 ×g for 5 min. The supernatant was collected and dissolved in the initial mobile phase after being freeze-dried.
Sample preparation for GC-MS experiments
For GC-MS analysis, intracellular metabolites were extracted as described above for UPLC-Q-TOF/MS, and derivatized after freeze-drying as follows: 75 μL of methoxyamine hydrochloride in pyridine (20 mg/mL) was added as first derivatization agent. The mixture was incubated at 70°C for 60 min. And 75 μL of MSTFA with 1% TMCS was added and incubated at 50°C for 60 min. Samples were then centrifuged at 10,000 ×g for 3 min at 4°C, supernatants were collected and mixed with 100 μL of heptane. The resulting solution (1 μL) was injected into the GC-MS system.
Sample preparation for HILIC-QQQ/MS experiments
For the HILIC-QQQ/MS analysis, phospholipids were extracted according to the methyl tert-butyl ether (MTBE) method[11]. Briefly, each sample was resuspended in 1.5 mL of methanol and vortexed for 5 min. Then, 5 mL of MTBE was added, vortexed for other 30 min, and 1.25 mL of water was added. Samples were incubated for 10 min and centrifuged at 5,000 ×g for 5 min. The lower phase was extracted twice with 2 mL of solvent (MTBE: methanol: water = 10:3:2.5). Obtained samples were pooled and dried with N2.
UHPLC-Q-TOF/MS analysis
The UHPLC-Q-TOF/MS analysis was performed on an Agilent 1290 Infinity LC system equipped with Agilent 6530 Accurate Mass Quadrupole Time-of-Flight mass spectrometer (Agilent, USA). An ACQUITY UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm) keep at 40°C was employed for the peak separation. The mobile phase was a mixture of water with 0.1% formic acid (A) and acetonitrile (B) at a flow rate of 0.35 mL/min. The gradient elution was as follows: 5% B to 95% B for 15 min. The post time was set to 5 min for equilibrating the system and the injection volume was 4 μL. An electrospray ionization source (ESI) was used both in positive and negative mode. The optimized conditions were as follows: capillary voltage was 3.5 kV for both positive and negative mode; drying gas flow 11 L/min, gas temperature 350°C; nebulizer pressure 45 psi, fragmentor voltage 120 V, skimmer voltage 60 V. Data were collected in profile mode from m/z 50 to 1100. MS/MS analysis was carried out to study metabolite structures and the applied collision energy was from 10 to 30 eV.
GC-MS analysis
The GC-MS analysis was performed with a Thermo Finnigan Trace GC Ultra instrument equipped with a DSQ II single quadrupole MS (Thermo Fisher, City, USA). A DB-5ms capillary column (30 m × 0.25 mm, 0.25 μm film thickness, Agilent, Santa Clara, CA, USA) was used for separation, and helium was used as carrier gas at a constant flow rate of 1.0 mL/min. The temperature of injection and interface was set to 260°C and 280°C, respectively. The GC oven temperature was initially set to 70°C for 3 min. Upon injection, the temperature was firstly increased to 220°C at a rate of 4°C/min and then to 310°C at a rate of 8°C/min, holding for 10 min. The temperature of the ion source was maintained at 250°C for electron ionization (EI). The full scan mode with a mass range of m/z 50–600 was used.
HILIC-QQQ/MS analysis
Agilent 1290 Infinity LC system and 6460 triple quadrupole mass system equipped with HILIC Xbridge (3.0mm×100mm, 3.5 μm) keep at 3°C was employed to analyze phospholipid metabolites. The mobile phase was a mixture of water with 10 mM ammonium acetate (A) and 95% acetonitrile with 10 mM ammonium acetate (B) at a flow rate of 0.3 mL/min. The gradient elution was as follows: 100% B to 95% B for 2 min; 95% B for 7.5 min, changed to 75% B for 13 min. The post time was set to 2 min for equilibrating the system and the injection volume was 5 μL. An electrospray ionization source (ESI) was used both in positive and negative modes. The capillary voltage was 2500 V, drying gas flow 10 L/min, gas temperature 325°C and nebulizer pressure 50 psi. Other mass scan parameters are shown in Table 1.
Table 1. HILIC-QQQ/MS scan parameters.
| Start Time (min) |
Ionization mode | Scan Type | Mass range (m/z) |
Fragmentor voltage(V) | Collision energy(eV) |
|---|---|---|---|---|---|
| 0 | - | Pre 152.9 | 600–1000 | 200 | 20 |
| 5.0 | - | Pre 152.9 | 300–600 | 180 | 20 |
| 6.5 | + | NL 141 | 600–1000 | 200 | 10 |
| 9.0 | + | Pre 184.1 | 600–1000 | 180 | 20 |
| 11.5 | + | NL 141 | 300–600 | 120 | 5 |
| 12.5 | + | Pre 184.1 | 300–600 | 150 | 20 |
Pre: precursor-ion scanning; NL: neutral loss scanning
Data processing
The raw data from GC-MS and UHPLC-Q-TOF/MS were converted to common data format (.cdf and.mzData). In the R software environment, the XCMS program was used to process MS data, with peak identification, retention time correction, automatic integration pretreatment, alignment and annotation. Metabolites not present in 80% of the samples were filtered. The raw data of extracellular and intracellular metabolites from LC-Q-TOF/MS are reported in supplementary materials (S1 and S2 Tables, respectively). GC-MS data inherently contain apparent variability and complexity, an untargeted filtration of ion peaks was used. Indeed, the most abundant fragment ion with the respective retention time (the time bin is 0.01 min) was kept while other ions were excluded. By applying this untargeted filtration, the data set was simplified and 174 ion peaks were obtained (supplementary materials S3 Table). HILIC-QQQ/MS data were processed by using the Agilent software pack. Then, according to the m/z and characteristic fragment ions, we inferred the number of carbon atoms and the number of double bonds in the fatty acid chain by using the network database (supplementary materials S4 Table). All the output data were normalized using the internal standard (IS) and dry weight of each sample before being exported to perform multivariate data analysis.
The identification of metabolites from the GC-MS was performed by combining mass spectra and database consultation (NIST11). For LC-MS data, the measured accurate mass and isotopic pattern were matched with database such as Metlin (http://metlin.scripps.edu/), YMDB (http://www.ymdb.ca/) and Lipid Maps (www.lipidmaps.org). MS/MS analysis was carried out to elucidate the metabolite structure and further validation was achieved by using literature data and reference standards.
Statistical analysis
All the obtained data were analyzed by multivariate statistical data analysis. More in detail, the principal components analysis (PCA) and partial least-squares discriminant analysis (PLS-DA) were performed with SIMCA-P version 11.0 (Umetrics, Umea, Sweden)[12]. Unsupervised PCA was used to observe the separating trends and metabolic trajectories between each set of samples while PLS-DA was used to identify a metabolite candidate that could distinguish between different groups. Variable importance plot (VIP) was used to select interesting metabolite candidates with the threshold value of 1.0 (GC-MS) or 1.5 (UHPLC-Q-TOF/MS). The statistical significance of mean values was assessed by using the one-way ANOVA and the Tukey’s post hoc test by SPSS 17.0. Results were considered significant when p < 0.05.
Results
The sensitivity of the strain to azole drugs
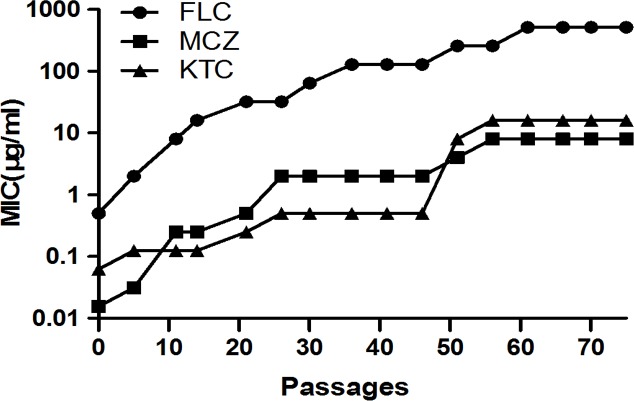
To obtain the resistant C. albicans strain in the laboratory, C. albicans SC5314 cells (FCZ MIC = 0.5 μg/mL) were cultivated in YPD medium with twice their most recently measured MIC of fluconazole. Reduced sensitivity to FCZ was detected after 5 passages, the high-level resistant strain (FCZ MIC = 256 μg/mL) was generated after about 51 passages and the next generations were cultivated in YPD medium with fluconazole (MIC = 512 μg/mL) and retained the resistant phenotype for more than 15 passages (Fig 1). The sensitivity testing showed that the increase in fluconazole MIC was accompanied by a corresponding increase in resistance to miconazole and ketoconazole (Fig 1). All the strains exposed to fluconazole adapted to the presence of drug, showing increased azole cross-resistance.
Fig 1. Variations of MIC values of FCZ, MCZ and KCZ for the C. albicans strain SC5314 grown in medium containing twice their most recently measured MIC of FCZ.
The result showed that the increase in fluconazole MIC was accompanied by a corresponding increase in resistance to miconazole and ketoconazole.
Metabolic profiling analysis
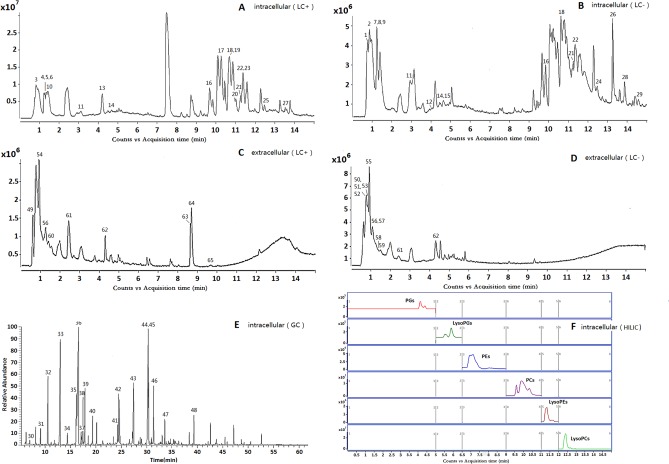
In this study, changes in intracellular and extracellular metabolites associate with azole resistance of C. albicans were evaluated. The typical total ion current chromatograms (TICs) are shown in Fig 2A–2E. In order to detect expression changes in metabolites due to external stimuli (i.e. KCZ treatment) or between strains themselves, four models were build for comparing: S group vs R group; S group vs S+KCZ group; R group vs R+KCZ group; S+KCZ group vs R+KCZ group (Fig 3, Fig 4, Fig 5). After merging redundant variables from the same metabolite, several molecules were selected as statistically significant (p<0.05; Table 2). According to this algorithm, we identified 48 and 17 differential molecules related to resistance among intracellular and extracellular metabolites, respectively. Moreover, by using the double factor analysis of variance, 25 significantly differential variables were identified as interacting (we set the C. albicans strain as resistant or not as factor A, and the presence of drug treatment or not as factor B, Table 3).
Fig 2. The typical total ions current chromatograms.
(A&B)Intracellular sample separated on UHPLC-Q-TOF/MS in positive and negative ionization mode; (C&D)Extracellular sample separated on UHPLC-Q-TOF/MS in positive and negative ionization mode; (E)Intracellular sample separated on GC-MS; (F)Extracted ion chromatography on HILIC-QQQ/MS.
Fig 3. PLS-DA scores plot of intracellular metabolites from GC-MS.
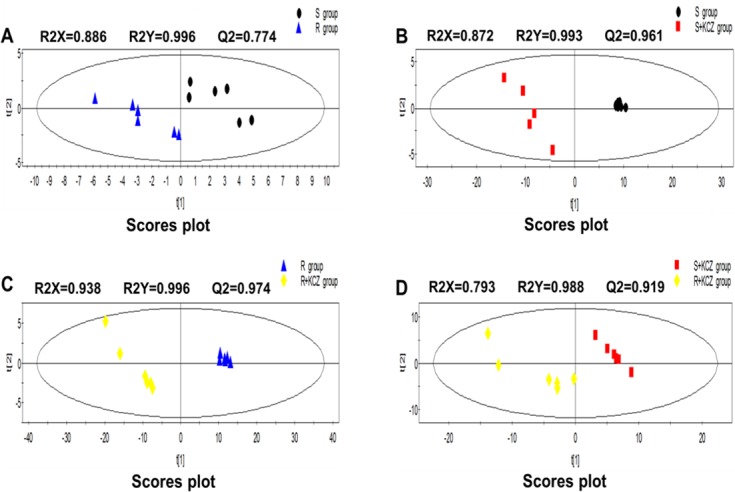
(A)S group vs R group; (B)S group vs S+KCZ group; (C)R group vs R+KCZ group; (D)S+KCZ group vs R+KCZ group.
Fig 4. PLS-DA scores plot of intracellular metabolites from LC-Q-TOF/MS.
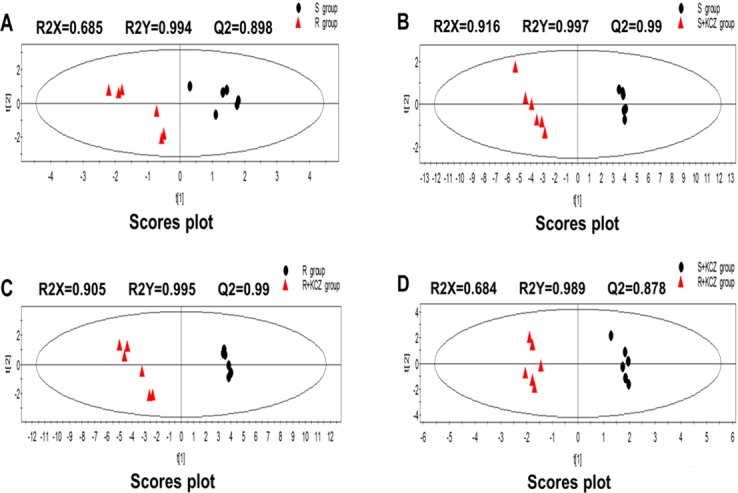
(A)S group vs R group; (B)S group vs S+KCZ group; (C)R group vs R+KCZ group; (D)S+KCZ group vs R+KCZ group.
Fig 5. PLS-DA scores plot of extracellular metabolites from LC-Q-TOF/MS.
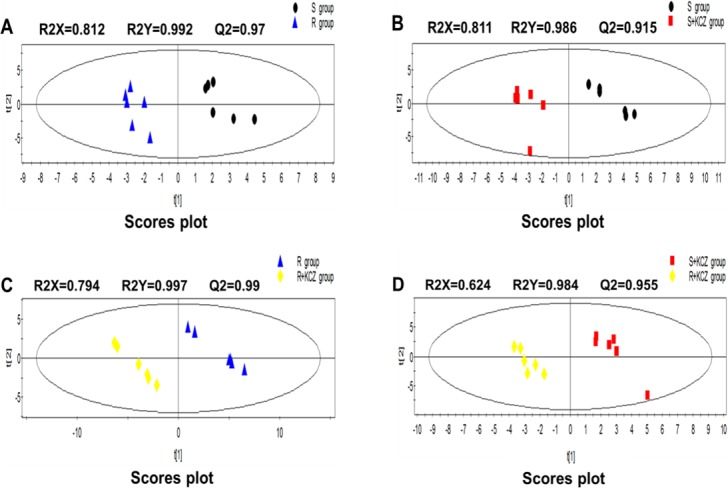
(A)S group vs R group; (B)S group vs S+KCZ group; (C)R group vs R+KCZ group; (D)S+KCZ group vs R+KCZ group.
Table 2. The number of differential metabolites from each model.
| Models | UHPLC-Q-TOF/MS | GC-MS | |
|---|---|---|---|
| intracellular extracellular | intracellular | ||
| S group vs R group | 8 | 9 | 8 |
| S group vs S+KCZ group | 19 | 7 | 17 |
| R group vs R+KCZ group | 21 | 9 | 15 |
| S+KCZ group vs R+KCZ group | 8 | 6 | 6 |
| Interaction | 15 | 2 | 8 |
Table 3. Related metabolites and their metabolic pathway.
| No. | Mass | RT | Column | Formula | Metabolite | Related pathway | S vs Ra |
S vs S+KCZb |
R vs R+KCZc |
S+KCZ vs R+KCZd |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trend | FC | Trend | FC | Trend | FC | Trend | FC | |||||||
| intracellular | ||||||||||||||
| 1 | 155.0695 | 0.74 | LC | C6H9N3O2 | Histidinee | Amino acid metabolism | —— | ↓*** | 0.4 | ↓*** | 0.5 | —— | ||
| 2 | 117.0426 | 0.89 | LC | C4H7NO3 | L-Aspartate-4-semi aldehydefg | Amino acid metabolism | ↑*** | 2.0 | —— | —— | —— | |||
| 3 | 257.1028 | 0.96 | LC | C8H20NO6P | Glycerophosphocholineeg | Phospholipid metabolism | ↑** | 2.0 | ↑*** | 4.3 | ↑*** | 3.9 | ↑*** | 1.8 |
| 4 | 149.0510 | 1.24 | LC | C5H11NO2S | Methionineeg | Amino acid metabolism | —— | ↓*** | 0.4 | ↓*** | 0.2 | —— | ||
| 5 | 307.0838 | 1.24 | LC | C10H17N3O6S | Glutathioneeg | Glutathione metabolism | —— | —— | ↑*** | 2.8 | ↑*** | 2.2 | ||
| 6 | 136.0385 | 1.24 | LC | C5H4N4O | Hypoxanthineeg | Purine metabolism | —— | —— | —— | ↑*** | 2.4 | |||
| 7 | 192.0270 | 1.25 | LC | C6H8O7 | Citratee | Citrate cycle | —— | ↓*** | 0.1 | ↓*** | 0.2 | ↑*** | 2.7 | |
| 8 | 146.0215 | 1.25 | LC | C5H6O5 | 2-Oxoglutaratee | Citrate cycle | ↑** | 1.6 | ↑*** | 3.0 | ↑*** | 2.0 | —— | |
| 9 | 244.0695 | 1.25 | LC | C9H12N2O6 | Uridinee | Pyrimidine metabolism | —— | —— | ↓*** | 0.2 | —— | |||
| 10 | 137.0477 | 1.66 | LC | C7H7NO2 | N-Methyl nicotinatefg | Nicotinate metabolism | ↓*** | 0.2 | —— | —— | —— | |||
| 11 | 219.1107 | 3.01 | LC | C9H17NO5 | Pantothenic acideg | Vitamin metabolism | —— | ↑*** | 3.2 | ↑*** | 2.0 | —— | ||
| 12 | 177.0460 | 4.02 | LC | C6H11NO3S | N-Formylmethioninefg | Amino acid metabolism | —— | ↓*** | 0.2 | ↓*** | 0.2 | —— | ||
| 13 | 135.0545 | 4.19 | LC | C5H5N5 | Adeninee | Purine metabolism | ↑** | 1.7 | —— | —— | ↑** | 2.1 | ||
| 14 | 224.0797 | 4.61 | LC | C10H12N2O4 | 3-Hydroxy-kynu reninefg | Tryptophan metabolism | —— | —— | ↑*** | 5.5 | ↑*** | 3.7 | ||
| 15 | 180.0899 | 4.62 | LC | C9H12N2O2 | 5-Hydroxy-kynu renaminefg | Tryptophan metabolism | —— | —— | —— | ↑*** | 2.0 | |||
| 16 | 397.2593 | 9.85 | LC | C18H40NO6P | Phytosphingosine-1-Pe | Sphingolipid metabolism | ↑*** | 1.5 | —— | —— | ↑* | 1.5 | ||
| 17 | 493.3168 | 10.36 | LC | C24H48NO7P | LysoPC(16:1)e | Phospholipid metabolism | ↑** | 1.7 | ↑** | 2.6 | ↑*** | 2.1 | —— | |
| 18 | 477.2855 | 10.71 | LC | C23H44NO7P | LysoPE(18:2)f | Phospholipid metabolism | —— | ↑*** | 3.6 | ↑*** | 2.8 | —— | ||
| 19 | 519.3325 | 10.78 | LC | C26H50NO7P | LysoPC(18:2)f | Phospholipid metabolism | —— | ↑*** | 2.5 | ↑*** | 2.0 | —— | ||
| 20 | 495.3325 | 11.03 | LC | C24H50NO7P | LysoPC(16:0)eg | Phospholipid metabolism | —— | ↑*** | 7.8 | ↑*** | 4.5 | —— | ||
| 21 | 453.2855 | 11.20 | LC | C21H44NO7P | LysoPE(16:0)eg | Phospholipid metabolism | —— | ↑*** | 10.2 | ↑*** | 8.3 | —— | ||
| 22 | 479.3012 | 11.43 | LC | C23H46NO7P | LysoPE(18:1)f | Phospholipid metabolism | —— | ↑*** | 3.5 | ↑*** | 3.4 | —— | ||
| 23 | 521.3481 | 11.49 | LC | C26H52NO7P | LysoPC(18:1)f | Phospholipid metabolism | —— | ↑*** | 2.6 | ↑*** | 1.9 | —— | ||
| 24 | 481.3168 | 12.42 | LC | C23H48NO7P | LysoPE(18:0)eg | Phospholipid metabolism | —— | ↑*** | 17.1 | —— | —— | |||
| 25 | 523.3638 | 12.48 | LC | C26H54NO7P | LysoPC(18:0)e | Phospholipid metabolism | —— | ↑*** | 4.3 | ↑*** | 4.1 | —— | ||
| 26 | 278.2246 | 13.28 | LC | C18H30O2 | γ-Linoleic acidfg | Fatty acid metabolism | —— | ↑*** | 9.6 | ↑*** | 6.2 | —— | ||
| 27 | 511.4964 | 13.50 | LC | C32H65NO3 | Cer(d18:0/14:0)f | Sphingolipid metabolism | ↑* | 1.9 | —— | —— | —— | |||
| 28 | 280.2402 | 13.88 | LC | C18H32O2 | Linoleic acideg | Fatty acid metabolism | —— | ↑*** | 4.0 | ↑*** | 3.5 | —— | ||
| 29 | 282.2559 | 14.58 | LC | C18H34O2 | Oleatee | Fatty acid metabolism | —— | ↑*** | 4.3 | ↑*** | 4.3 | —— | ||
| 30 | 103.0633 | 7.08 | GC | C4H9NO2 | N,N-Dimethyl glycine | Amino acid metabolism | ↓* | 0.7 | ↑*** | 4.4 | ↑** | 5.6 | —— | |
| 31 | 90.0317 | 9.23 | GC | C3H6O3 | Lactate | Citrate cycle | ↑* | 1.4 | —— | —— | ↓** | 0.6 | ||
| 32 | 90.0550 | 10.63 | GC | C3H7NO2 | Alanine | Amino acid metabolism | —— | ↑* | 2.4 | ↑*** | 3.3 | —— | ||
| 33 | 131.0946 | 12.43 | GC | C6H13NO2 | Leucine | Amino acid metabolism | —— | ↑*** | 13.2 | —— | —— | |||
| 34 | 117.0790 | 14.43 | GC | C5H11NO2 | Valine | Amino acid metabolism | —— | ↑*** | 6.9 | ↑*** | 7.2 | —— | ||
| 35 | 97.9769 | 16.50 | GC | H3PO4 | Phosphateg | Energy metabolism | —— | ↑** | 2.9 | —— | —— | |||
| 36 | 92.0473 | 16.57 | GC | C3H8O3 | Glycerolg | Stress response | ↓* | 0.7 | ↑*** | 3.2 | ↑*** | 4.7 | ↑* | 1.4 |
| 37 | 115.0633 | 17.20 | GC | C5H9NO2 | Proline | Amino acid metabolism | ↓* | 0.8 | ↑*** | 4.6 | ↑*** | 1.8 | —— | |
| 38 | 75.0320 | 17.51 | GC | C2H5NO2 | Glycine | Amino acid metabolism | —— | ↑** | 1.5 | ↑*** | 1.5 | —— | ||
| 39 | 90.0317 | 17.89 | GC | C3H6O3 | Succinateg | Citrate cycle | —— | ↑*** | 2.5 | ↑*** | 3.8 | —— | ||
| 40 | 105.0426 | 19.36 | GC | C3H7NO3 | Serine | Amino acid metabolism | ↑* | 1.4 | ↑** | 3.8 | —— | —— | ||
| 41 | 133.0375 | 24.43 | GC | C4H7NO4 | Aspartate | Amino acid metabolism | ↓** | 0.7 | ↑*** | 11.2 | ↑*** | 13.5 | —— | |
| 42 | 103.0633 | 24.64 | GC | C4H9NO2 | γ-Aminobutryic acidg | Citrate cycle | —— | ↑*** | 23.5 | ↑*** | 47.7 | ↑*** | 2.1 | |
| 43 | 146.0691 | 27.43 | GC | C5H10N2O3 | Glutamineg | Amino acid metabolism | ↓*** | 0.6 | ↑*** | 1.9 | ↑*** | 3.9 | ↑* | 1.3 |
| 44 | 152.0685 | 30.29 | GC | C5H12O5 | Arabitol | Stress response | —— | ↑*** | 3.5 | ↑*** | 5.5 | —— | ||
| 45 | 152.0685 | 30.29 | GC | C5H12O5 | Ribitolg | Pentose phosphate metabolism | ↓** | 0.6 | ↑*** | 3.2 | ↑*** | 5.5 | —— | |
| 46 | 172.0137 | 31.35 | GC | C3H9O6P | Glycerophosphate | Phospholipid metabolism | —— | ↑*** | 50.8 | ↑*** | 68.9 | —— | ||
| 47 | 188.1161 | 33.56 | GC | C8H16N2O3 | N-Acetyl-L-lysineg | Amino acid metabolism | —— | ↑*** | 8.2 | ↑*** | 18.1 | ↑* | 1.5 | |
| 48 | 180.0634 | 39.33 | GC | C6H12O6 | Inositolg | Inositol phosphate metabolism | —— | —— | —— | ↓* | 0.7 | |||
| extracellular | ||||||||||||||
| 49 | 202.2157 | 0.59 | LC | C10H26N4 | Sperminee | Spermine metabolism | ↑** | 1.4 | ↓*** | 0.8 | —— | ↓* | 0.7 | |
| 50 | 152.0685 | 0.78 | LC | C5H12O5 | Ribitole | Pentose phosphate metabolism | ↓** | 0.8 | —— | —— | ↓* | 0.8 | ||
| 51 | 180.0634 | 0.78 | LC | C6H12O6 | Glucosee | Energy metabolism | —— | ↑** | 1.1 | —— | —— | |||
| 52 | 120.0436 | 0.79 | LC | C5H4N4 | Purinee | Purine metabolism | —— | ↓** | 1.3 | —— | —— | |||
| 53 | 268.0808 | 0.85 | LC | C10H12N4O5 | Inosinee | Purine metabolism | ↑* | 1.2 | ↑** | 1.2 | ↑*** | 1.1 | —— | |
| 54 | 117.0790 | 0.86 | LC | C5H11NO2 | Valineeg | Amino acid metabolism | ↓*** | 0.6 | —— | —— | ↓*** | 0.6 | ||
| 55 | 175.0481 | 0.95 | LC | C6H9NO5 | N-Acetyl-aspartic acidf | Amino acid metabolism | ↓* | 0.8 | —— | —— | —— | |||
| 56 | 192.0270 | 1.10 | LC | C6H8O7 | Citratee | Citrate cycle | ↑** | 1.6 | ↓** | 0.6 | ↓*** | 0.5 | —— | |
| 57 | 134.0579 | 1.15 | LC | C5H10O4 | 2-Deoxy-Ribosef | Pentose phosphate metabolism | ↑** | 1.3 | —— | —— | ↑* | 1.8 | ||
| 58 | 181.0739 | 1.27 | LC | C9H11NO3 | Tyrosinee | Amino acid metabolism | —— | ↑** | 1.2 | ↑*** | 1.1 | —— | ||
| 59 | 152.0797 | 1.34 | LC | C4H12N2O4 | succinatee | Citrate cycle | ↑*** | 1.3 | —— | —— | ↑*** | 1.2 | ||
| 60 | 131.0946 | 1.40 | LC | C6H13NO2 | Leucineeg | Amino acid metabolism | —— | —— | ↓*** | 0.9 | ↓* | 0.8 | ||
| 61 | 165.0790 | 2.44 | LC | C9H11NO2 | Phenylalaninee | Amino acid metabolism | —— | —— | ↑* | 1.2 | —— | |||
| 62 | 204.0899 | 4.29 | LC | C11H12N2O2 | Tryptophane | Tryptophan metabolism | —— | —— | ↑** | 1.2 | —— | |||
| 63 | 317.2930 | 8.69 | LC | C18H39NO3 | Phytosphingosinee | Sphingolipid metabolism | —— | ↑* | 1.9 | —— | —— | |||
| 64 | 271.2511 | 8.91 | LC | C16H33NO2 | Sphingosinef | Sphingolipid metabolism | ↑* | 1.6 | ↑* | 2.5 | —— | —— | ||
| 65 | 301.2981 | 9.66 | LC | C18H39NO2 | Sphinganinee | Sphingolipid metabolism | —— | ↑** | 2.7 | —— | —— | |||
Abbreviations: RT, retention time; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine; Cer, ceramide
a The trend and fold change (FC) of relative amounts of the R group compared to the S group;(↑): up-regulated. (↓): down-regulated. (* p<0.05; ** p <0.01; ***p< 0.001).
b The trend and fold change of relative amounts of the S+KCZ group compared to the S group
c The trend and fold change of relative amounts of the R+KCZ group compared to the R group
d The trend and fold change of relative amounts of the R+KCZ group compared to the S+KCZ group.
e Metabolites validated with standard sample
f Metabolites putatively annotated by library searching
g Metabolites of the interaction.
Differential metabolites identified among different strains
Partial least squares discriminant analysis (PLS-DA) were performed to screen the differences of metabolites between sensitive and resistant strains. According to the VIP value and one-way analysis of variance (ANOVA, p<0.05), 25 variables were selected as candidates of potential biomarkers between sensitive and resistant strains. These potential biomarkers were mapped onto 12 principal pathways by using the KEGG PATHWAY Database (http://www.genome.jp/kegg/). Compared with the sensitive strain of C. albicans, the levels of N-Methyl nicotinate, N, N-dimethyl glycine, Glycerol, proline, Ribitol, L-aspartate, glutamine, valine, and N-Acetyl-aspartic acid were significantly decreased in the resistant strains, while the other metabolites including L-Aspartate-4-semi aldehyde, Glycerophosphocholine, 2-Oxoglutarate, Adenine, Phytosphingosine-1-P, LysoPC(16:1), Cer(d18:0/14:0), Lactate, Serine, Spermine, Inosine, Citrate, 2-Deoxy-Ribose, succinate and sphingosine were increased.
Differential metabolites associate with drug stress
Among the sensitive and resistant strains tretment with KCZ compared with no drug intervention, 41 differential metabolites were identified as potential biomarkers related to drug stress. Four of them (i.e. serine, sphingosine, dihydrosphingosine and phytosphingosine) were detected only in the sensitive strain, while 7 (i.e. glutathione, tryptophan, uridine, 3-hydroxy-kynurenine, phenylalanine, purine and glucose) only in the resistant strains. These compounds are mainly involved in processes such as amino acid metabolism, stress response, sphingolipid metabolism, and phospholipid metabolism. In our results, most of the amino acid levels (i.e. alanine aspartate, glutamine, glycine, N, N-dimethyl glycine, N-acetyl lysine, proline, valine, and tyrosine) were increased both in sensitive and resistant strains after the treatment with KCZ.
Differential metabolites related to drug resistance level
Twenty significantly differential metabolites were identified as potential biomarkers related to drug resistance level. Compared with S+KCZ group, it was found that fourteen of 20 metabolites, including glycerophosphocholine, glutathione, hypoxanthine, citrate, adenine, 3-hydroxy-kynurenine, 5-hydroxy-kynurenamine, phytosphingosine-1-P, glycerol, γ-aminobutryic acid, glutamine, 2-deoxy-ribose, N-acetyl-L-lysine and succinate, were up-regulated in the R+KCZ group, while the other six metabolites including lactate, inositol, spermine, ribitol, valine and leucine were reduced. These differentially expressed compounds are mainly related to the antioxidant activity or the action of drugs. For example, C. albicans regulates the content of VB6 by adjusting valine metabolism in order to assess the antioxidant activity. Besides, regulation of glutathione, polyamine and 3-Hydroxy-kynurenine may reduce intracellular reactive oxygen species, thus reducing the fungal sensitivity to drugs.
Phospholipid metabolism analysis
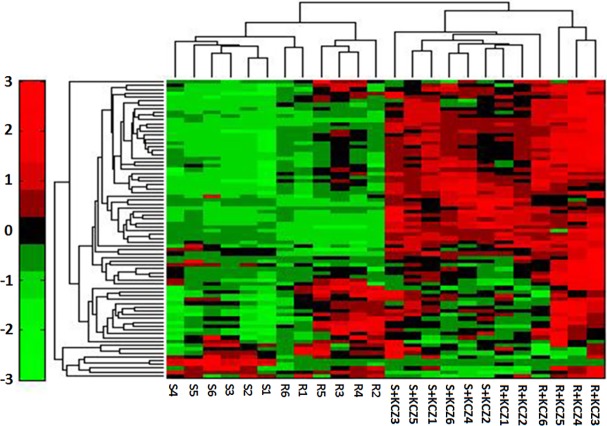
Lipids, in addition to being structural and metabolic components of yeast cells, also appear to be responsible for drug resistance in C. albicans. Considering the central role of phospholipid metabolism in several biochemical processes, we decided to perform the phospholipid metabolomic analysis. 79 phospholipidic compounds were identified by HILIC-QQQ/MS (Fig 2F). All these metabolites are reported in a heatmap,including 28 phosphatidylcholines (PC), 10 lysophosphatidylcholines (LysoPC), 20 phosphatidyl ethanolamines (PE), 6 lysophosphatidyl ethanolamines (LysoPE), 12 phosphatidylglycerols (PG) and 3 lysophosphatidylglycerols (LysoPG) (Fig 6). We also compared the percentage of glycerol phospholipid contained in each group, observing that phosphatidylcholine was the most abundant compound. Finally, glycerol-phospholipid fatty acids analysis showed that resistant strains of C. albicans presented the highest content of glyceryl phosphatide fatty acids, which contain more than 34-C and an odd number of C atoms (Table 4, Table 5).
Fig 6. Heatmap of glycerophospholipid from HILIC-QQQ/MS.
Colors represent fold change in the metabolite concentration in different groups. Red indicates increased concentration levels of metabolites; green indicates decreased concentration levels of metabolites.
Table 4. The result of total number of carbons in the fatty acid chains of phosphoglyceride.
| No. of carbons | S group | R group | S+KCZ group | R+KCZ group | ||||
|---|---|---|---|---|---|---|---|---|
| mean | S.D. | mean | S.D. | mean | S.D. | mean | S.D. | |
| <34-C | 5.72 | 0.27 | 5.45 | 0.23 | 5.47 | 0.11 | 5.27 | 0.13 |
| 34-C | 30.23 | 0.78 | 34.97 | 0.78 | 28.65 | 0.68 | 28.97 | 1.10 |
| 36-C | 42.99 | 1.03 | 47.65 | 0.91 | 39.21 | 1.01 | 43.48 | 0.82 |
| 38-C | 0.58 | 0.01 | 0.75 | 0.08 | 0.69 | 0.04 | 0.74 | 0.05 |
The data is represented as % of total PGLs mass spectral signal(n = 6)
Table 5. The result of odd chain fatty acid of phosphoglyceride.
| No. of carbons | S group | R group | S+KCZ group | R+KCZ group | ||||
|---|---|---|---|---|---|---|---|---|
| mean | S.D. | mean | S.D. | mean | S.D. | mean | S.D. | |
| 33-C | 1.25 | 0.06 | 1.20 | 0.12 | 0.82 | 0.02 | 0.76 | 0.05 |
| 35-C | 1.38 | 0.10 | 1.36 | 0.05 | 1.28 | 0.07 | 1.27 | 0.06 |
The data is represented as % of total PGLs mass spectral signal(n = 6)
Discussion
To investigate biological mechanisms of azole resistance in C. albicans, a laboratory azole resistant C. albicans strain was obtained by serial cultures of a FCZ susceptible C. albicans strain in inhibitory concentrations of FCZ. This resistant strain possessed high-level and stable resistant characteristic, as well as cross-resistance to two other azole antifungal agents. Comparative analysis of metabolomics in groups showed that the differentially expressed metabolites were found to be involved in multiple biochemical functions. It is reasonable to assume that many of the observed alternations are somewhat related to azole resistance in C. albicans.
Amino acids are key precursors for the synthesis of important low molecular weight nitrogenous compounds[13]. It has been reported that pyridoxal phosphate is not only able to regulated valine metabolism by combining with the pyridoxal protein, but also to activate VB6 as a catalyst[14, 15]. VB6, as a singlet oxygen scavenger, can protect fungi from the oxidative damage, and it has been considered as a potential antioxidant[16]. Therefore, the observed changes of valine may be associated with a stimulation of the activity of VB6. Previous studies have reported that proline, being a kind of universal antioxidant, could protect filamentous fungi and yeast against oxidative stress[17, 18]. Proline is also able to reduce the production of reactive oxygen species in the mitochondria[19, 20]. In our study, on the one hand, the down-regulation of proline in the R group may suggest the idea that resistant strains had a wider oxidative stress tolerance than sensitive strains. On the other hand, proline was found at higher levels both in resistant and sensitive strains after tretment with ketoconazole. We suspected that proline is induced for protective purposes under stress conditions, such as drug stimulation. The redox system plays an important role in the occurrence and progress of several diseases, and glutathione is an indispensable compound that maintains redox homeostasis responding to oxidative stress[21]. In our study, glutathione was significant up-regulation in resistant strains after ketoconazole intervention. We speculated that such alternations might be associated with endogenous reactive oxygen species (ROS) production. Resistant strains synthesized a large amount of glutathione in response to endogenous ROS production in cells, thus reducing the sensitivity of antifungal agents. It is well known that yeasts activate a series of protective defense measures in response to external environment stimulations, such as the rapid accumulation of polyols (e.g. glycerin, arabitol, sorbitol and mannitol). For example, C. albicans secrets a large amount of glycerin and arabitol in response to osmotic stress, temperature and oxidative stress[22, 23]. Therefore, the up-regulation of glycerin and arabitol both in resistant and sensitive strains after tretment with ketoconazole suggested that C. albicans cells againsted the oxidative stress through the accumulation of polyols (Fig 7).
Fig 7. Schematic overview of the main differences in the metabolite change associated with drug resistance of Candida albicans.
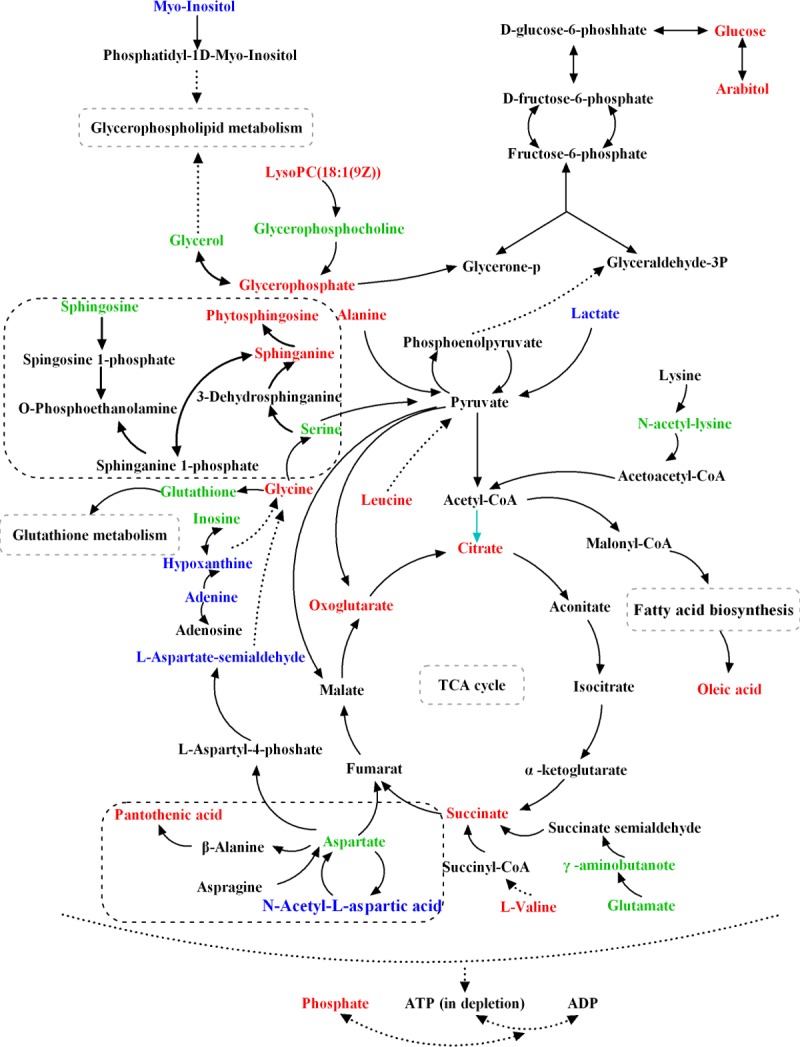
The differentially produced metabolites are mainly involved in amino metabolism, sphingolipid metabolism, TCA cycle, oxidative stress, glutathione metabolism and lipid metabolism. Metabolites in red marks are only related to drug stress; metabolites in blue marks are only related to strain type or drug resistance level; metabolites marked green are associated with the three described above.
Sphingosine, phytosphingosine and sphinganine were observed up-regulation only in sensitive strains after KCZ intervention (Fig 7). Sushma et al. have reported that the deletion of the sphingolipid biosynthetic pathway genes FEN1 and SUR4 of C. albicans resulted in a major sensitivity to amphotericin B (AmB) than parent strains[24]. Vinay K. et al. have demonstrated that the modulation of AmB resistance by PMP3 was dependent on sphingolipid biosynthetic pathway, since AmB sensitivity of PMP3 deletants was suppressed by phytosphingosine, a sphingolipid pathway intermediate[25]. As a signal molecule, phytosphingosine-1-P could enhance the miconazole drug efflux to reduce the sensitivity. Several studies have showed that the overexpression of drug transporters is one of the main resistant mechanisms for several pathogens[26, 27]. As a consequence, we may speculate that the up-regulation of phytosphingosine-1-P in C. albicans can reduce its sensitivity to antifungal drugs by increasing the drug efflux. These results all indicated that sphingolipid biosynthetic pathway is essential for the achievement of drug resistance in C. albicans, highlighting that cell membrane is a main target for antibacterial action in C. albicans.
Furthermore, phospholipid metabolomics showed that resistant strains of C. albicans had high content of PC, PE, PG and low content of LysoPC, LysoPE, LysoPG. These results imply that phospholipase B is strongly expressed and the resistant strains probably enhanced its pathogenicity by this high expression[28]. Intracellular drug content is strictly related with the expression level of membrane transporters, and the rate of transported drugs depends on the fluidity of cell membrane[29]. This parameter is linked to the length of the lipid C-chain: the longer is the C-chain, the weaker is the fluidity of the cell membrane. We found that resistant strains of C. albicans had the highest content of glyceryl phosphatide fatty acids with more than 34-C. This content was significantly reduced after the treatment with ketoconazole in both resistant and sensitive groups. This suggests that strains can reduce the drug uptake by enhancing the cell membrane fluidity. At the same time, we also observed that phosphoglycerides containing an odd number of C atoms were down-regulated in resistant strains group. This is in good agreement with a previous study showing that phospholipid fatty acid with an odd number of C atoms affected the sensitivity of C. albicans against drugs[30].
Mitochondria play an important role in multiple processes, such as hyphal development, resistance to stress, virulence, apoptosis and sensitivity of yeast cells to drugs[31–33]. Cardiolipin and its precursor PG, the principal anionic phospholipid in mitochondrial membranes, are key elements in the response conferring resistance to osmotic and thermal stresses[34, 35]. We found that PG was increased in treatment groups, thus maybe contributing to mitochondrial defects, which probably reduce the content of antifungal drugs through the regulation of the expression of transporters[36]. Shingu-Vazquez et al. have found that mitochondrial dysfunction in some fungi, such as S. cerevisiae and C. glabrata, could lead to drug resistance, especially in reducing susceptibility to azole and polyene drugs[31]. Phosphatidylcholine is a precursor of lysophosphatidylcholine, which beyond the control of mitochondrial functions regulates intracellular vesicle trafficking[37, 38]. LysoPC was demonstrated to be able to inhibit the transition of C. albicans from yeast to hyphae via the MAP kinase pathway, but did not affect the growth of either yeast or hyphae[39]. Compared to the sensitive strains, we observed that lysoPC was up-regulated in the resistant group, thus indicating that lysoPC may be associated with C. albicans resistance.
Conclusions
In this study, different MS-based approaches were used to investigate the metabolic profile associated with azole resistance in C. albicans. Our results allowed, as a model containing potential biomarkers changing in azole resistance in C. albicans. that the majority of these potential biomarkers were involved in metabolic processes related to amino acid metabolism, sphingolipid metabolism and phospholipid metabolism. It is a complex process probably involving, reducing the endogenous ROS production, strengthening the expression of drug transporters, changing the function of cell membranes and mitochondria. Overall, our study explores global changes in metabolites involved in resistance to azole drugs, providing a more detailed understanding of the evolution of drug resistance in C. albicans.
Supporting information
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank Professor William A. Fonzi for kindly providing the C. albicans strains SC5314. This work was financially supported by two National Nature Science Foundation of China (81273474 and 81671991). In addition, the authors would like to thank the referees and editor for their useful comments and suggestions, which have helped us to improve our manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by two National Nature Science Foundation of China (81273474 and 81671991). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nakamura K, Niimi M, Niimi K, Holmes AR, Yates JE, Decottignies A, et al. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrobial agents and chemotherapy. 2001;45(12):3366–74. Epub 2001/11/16 doi: 10.1128/AAC.45.12.3366-3374.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tagliaferri E, Menichetti F. Treatment of invasive candidiasis: between guidelines and daily clinical practice. Expert review of anti-infective therapy. 2015;13(6):685–9. Epub 2015/03/31 doi: 10.1586/14787210.2015.1029916 [DOI] [PubMed] [Google Scholar]
- 3.Bona E, Cantamessa S, Pavan M, Novello G, Massa N, Rocchetti A, et al. Sensitivity of Candida albicans to essential oils: are they an alternative to antifungal agents? Journal of applied microbiology. 2016;121(6):1530–45. Epub 2016/10/25. doi: 10.1111/jam.13282 [DOI] [PubMed] [Google Scholar]
- 4.White TC; Marr KA; Bowden RA Clinical, Cellular, and Molecular Factors That Contribute to Antifungal Drug Resistance. Clinical microbiology reviews 1998, 11:382–402. Epub 1998/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebeke M, Lalk M. Staphylococcus aureus metabolic response to changing environmental conditions—a metabolomics perspective. International journal of medical microbiology: IJMM. 2014;304(3–4):222–9. Epub 2014/01/21. doi: 10.1016/j.ijmm.2013.11.017 [DOI] [PubMed] [Google Scholar]
- 6.Teoh ST, Kitamura M, Nakayama Y, Putri S, Mukai Y, Fukusaki E. Random sample consensus combined with partial least squares regression (RANSAC-PLS) for microbial metabolomics data mining and phenotype improvement. Journal of bioscience and bioengineering. 2016;122(2):168–75. Epub 2016/02/11. doi: 10.1016/j.jbiosc.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 7.Madji Hounoum B, Blasco H, Nadal-Desbarats L, Dieme B, Montigny F, Andres CR, et al. Analytical methodology for metabolomics study of adherent mammalian cells using NMR, GC-MS and LC-HRMS. Analytical and bioanalytical chemistry. 2015;407(29):8861–72. Epub 2015/10/09. doi: 10.1007/s00216-015-9047-x [DOI] [PubMed] [Google Scholar]
- 8.Van EJ; Joosten L; Verhaeghe V; Surmont I, Fluconazole and Amphotericin B Antifungal Susceptibility Testing by National Committee for Clinical Laboratory Standards Broth Macrodilution Method Compared with E-test and Semiautomated Broth Microdilution Test. Journal of clinical microbiology 1996, 34:842–847. Epub 1996/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolten CJ, Wittmann C. Appropriate sampling for intracellular amino acid analysis in five phylogenetically different yeasts. Biotechnology letters. 2008;30(11):1993–2000. Epub 2008/07/08. doi: 10.1007/s10529-008-9789-z [DOI] [PubMed] [Google Scholar]
- 10.Canelas AB, ten Pierick A, Ras C, Seifar RM, van Dam JC, van Gulik WM, et al. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Analytical chemistry. 2009;81(17):7379–89. Epub 2009/08/06. doi: 10.1021/ac900999t [DOI] [PubMed] [Google Scholar]
- 11.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. Journal of lipid research. 2008;49(5):1137–46. Epub 2008/02/19. doi: 10.1194/jlr.D700041-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, et al. A tutorial review: Metabolomics and partial least squares-discriminant analysis—a marriage of convenience or a shotgun wedding. Analytica chimica acta. 2015;879:10–23. Epub 2015/05/24. doi: 10.1016/j.aca.2015.02.012 [DOI] [PubMed] [Google Scholar]
- 13.Wu G. Amino acids: metabolism, functions, and nutrition. Amino acids. 2009;37(1):1–17. Epub 2009/03/21. doi: 10.1007/s00726-009-0269-0 [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee T, Hanes J, Tews I, Ealick SE, Begley TP. Pyridoxal phosphate: biosynthesis and catabolism. Biochimica et biophysica acta. 2011;1814(11):1585–96. Epub 2011/07/20. doi: 10.1016/j.bbapap.2011.06.018 [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Iimori J, Takayama S, Moriyama A, Yamauchi A, Hemmi H, et al. Conserved pyridoxal protein that regulates Ile and Val metabolism. Journal of bacteriology. 2013;195(24):5439–49. Epub 2013/10/08. doi: 10.1128/JB.00593-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bilski P., Li MY., Ehrenshaft M., Daub ME., Chignell CF. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol. 2000; 71(2): 129–34. Epub 1999/10/19. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Alfano JR, Becker DF. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. Journal of bacteriology. 2015;197(3):431–40. Epub 2014/11/12. doi: 10.1128/JB.02282-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan SK, Zhu W, Liang X, Zhang L, Demers AJ, Zimmerman MC, et al. Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free radical biology & medicine. 2012;53(5):1181–91. Epub 2012/07/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saye M, Miranda MR, di Girolamo F, de los Milagros Camara M, Pereira CA. Proline modulates the Trypanosoma cruzi resistance to reactive oxygen species and drugs through a novel D, L-proline transporter. PloS one. 2014;9(3):e92028 Epub 2014/03/19. doi: 10.1371/journal.pone.0092028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan N., Dickman MB., Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med. 2008; 44(4):671–81. Epub 2009/02/15. doi: 10.1016/j.freeradbiomed.2007.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G. Maturation of cytosolic iron-sulfur proteins requires glutathione. The Journal of biological chemistry. 2002;277(30):26944–9. Epub 2002/05/16. doi: 10.1074/jbc.M200677200 [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Fresneda R, Guirao-Abad JP, Arguelles A, Gonzalez-Parraga P, Valentin E, Arguelles JC. Specific stress-induced storage of trehalose, glycerol and D-arabitol in response to oxidative and osmotic stress in Candida albicans. Biochemical and biophysical research communications. 2013;430(4):1334–9. Epub 2012/12/25. doi: 10.1016/j.bbrc.2012.10.118 [DOI] [PubMed] [Google Scholar]
- 23.Li L, Ye Y, Pan L, Zhu Y, Zheng S, Lin Y. The induction of trehalose and glycerol in Saccharomyces cerevisiae in response to various stresses. Biochemical and biophysical research communications. 2009;387(4):778–83. Epub 2009/07/29. doi: 10.1016/j.bbrc.2009.07.113 [DOI] [PubMed] [Google Scholar]
- 24.Sharma S., Alfatah Md., Bari V K., Rawal Y., Paul S., Ganesan K. Sphingolipid Biosynthetic Pathway Genes FEN1 and SUR4 Modulate Amphotericin B Resistance. Antimicrob Agents Chemother. 2014; 58(4): 2409–14. Epub 2013/12/08. doi: 10.1128/AAC.02130-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bari VK, Sharma S, Alfatah M, Mondal AK, Ganesan K. Plasma Membrane Proteolipid 3 Protein Modulates Amphotericin B Resistance through Sphingolipid Biosynthetic Pathway. Scientific reports. 2015;5:9685 Epub 2015/05/13. doi: 10.1038/srep09685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuentes M., Hermosilla G., Alburquenque C., Falconer MA., Amaro J., Tapia C. Characterization of azole resistance mechanisms in Chilean clinical isolates of Candida albicans. Rev Chilena Infectol. 2014; 31(5): 511–517. Epub 2014/07/08. [DOI] [PubMed] [Google Scholar]
- 27.Luo L, Jiang X, Wu Q, Wei L, Li J, Ying C. Efflux pump overexpression in conjunction with alternation of outer membrane protein may induce Acinetobacter baumannii resistant to imipenem. Chemotherapy. 2011;57(1):77–84. Epub 2011/02/25. doi: 10.1159/000323620 [DOI] [PubMed] [Google Scholar]
- 28.Theiss S, Ishdorj G, Brenot A, Kretschmar M, Lan CY, Nichterlein T, et al. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int J Med Microbiol. 2006; 296(6): 405–420. Epub 2008/07/23. doi: 10.1016/j.ijmm.2006.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peetla C, Vijayaraghavalu S, Labhasetwar V. Biophysics of cell membrane lipids in cancer drug resistance: Implications for drug transport and drug delivery with nanoparticles. Advanced drug delivery reviews. 2013;65(13–14):1686–98. Epub 2013/09/24. doi: 10.1016/j.addr.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh A, Yadav V, Prasad R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PloS one. 2012;7(6):e39812 Epub 2012/07/05. doi: 10.1371/journal.pone.0039812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryotic cell. 2011;10(11):1376–83. Epub 2011/09/20. doi: 10.1128/EC.05184-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, et al. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS pathogens. 2009;5(1):e1000268 Epub 2009/01/17. doi: 10.1371/journal.ppat.1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan N. Two-component phosphorelays in fungal mitochondria and beyond. Mitochondrion. 2015;22:60–5. Epub 2015/04/11. doi: 10.1016/j.mito.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 34.Romantsov T, Guan Z, Wood JM. Cardiolipin and the osmotic stress responses of bacteria. Biochimica et biophysica acta. 2009;1788(10):2092–100. Epub 2009/06/23. doi: 10.1016/j.bbamem.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luevano-Martinez LA, Forni MF, dos Santos VT, Souza-Pinto NC, Kowaltowski AJ. Cardiolipin is a key determinant for mtDNA stability and segregation during mitochondrial stress. Biochimica et biophysica acta. 2015;1847(6–7):587–98. Epub 2015/04/07. doi: 10.1016/j.bbabio.2015.03.007 [DOI] [PubMed] [Google Scholar]
- 36.Pokorna L, Cermakova P, Horvath A, Baile MG, Claypool SM, Griac P, et al. Specific degradation of phosphatidylglycerol is necessary for proper mitochondrial morphology and function. Biochimica et biophysica acta. 2016;1857(1):34–45. Epub 2015/10/21. doi: 10.1016/j.bbabio.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe A.G., McMaster C.R. Regulation of vesicle trafficking, transcription, and meiosis: lessons learned from yeast regarding the disparate biologies of phosphatidylcholine. Biochim Biophys Acta. 2001;1534:65–77. Epub 2001/10/04. [DOI] [PubMed] [Google Scholar]
- 38.Flis VV., Fankl A., Ramprecht C, Zellnig G, Leitner E, Hermetter A, et al. Phosphatidylcholine Supply to Peroxisomes of the Yeast Saccharomyces cerevisiae. PLoS ONE. 2015; 10(8):e0135084 Epub 2015/07/17. doi: 10.1371/journal.pone.0135084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min J. Lee YJ. Kim YA, Park HS, Han SY, Jhon GH,et al. Lysophosphatidylcholine derived from deer antler extract suppresses hyphal transition in Candida albicans through MAP kinase pathway. Biochim Biophys Acta. 2001;1531:77–89. Epub 2001/01/16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.