Abstract
In silico transcriptome mining is a powerful tool for crustacean peptidome prediction. Using homology-based BLAST searches and a simple bioinformatics workflow, large peptidomes have recently been predicted for a variety of crustaceans, including the lobster, Homarus americanus. Interestingly, no in silico studies have been conducted on the eyestalk ganglia (lamina ganglionaris, medulla externa, medulla interna and medulla terminalis) of the lobster, although the eyestalk is the location of a major neuroendocrine complex, i.e., the X-organ-sinus gland system. Here, an H. americanus eyestalk ganglia-specific transcriptome was produced using the de novo assembler Trinity. This transcriptome was generated from 130,973,220 Illumina reads and consists of 147,542 unique contigs. Eighty-nine neuropeptide-encoding transcripts were identified from this dataset, allowing for the deduction of 62 distinct pre/preprohormones. Two hundred sixty-two neuropeptides were predicted from this set of precursors; the peptides include members of the adipokinetic hormone-corazonin-like peptide, allatostatin A, allatostatin B, allatostatin C, bursicon α, CCHamide, corazonin, crustacean cardioactive peptide, crustacean hyperglycemic hormone (CHH), CHH precursor-related peptide, diuretic hormone 31, diuretic hormone 44, eclosion hormone, elevenin, FMRFamide-like peptide, glycoprotein hormone α2, glycoprotein hormone β5, GSEFLamide, intocin, leucokinin, molt-inhibiting hormone, myosuppressin, neuroparsin, neuropeptide F, orcokinin, orcomyotropin, pigment dispersing hormone, proctolin, pyrokinin, red pigment concentrating hormone, RYamide, short neuropeptide F, SIFamide, sulfakinin, tachykinin-related peptide and trissin families. The predicted peptides expand the H. americanus eyestalk ganglia neuropeptidome approximately 7-fold, and include 78 peptides new to the lobster. The transcriptome and predicted neuropeptidome described here provide new resources for investigating peptidergic signaling within/from the lobster eyestalk ganglia.
Keywords: Crustacea, Decapoda, neurohormone, neuropeptide, transcriptomics, X-organ-sinus gland system
1. Introduction
The American lobster, Homarus americanus, is an iconic and economically important species due to its popularity as a luxury food item; the value of the lobster fishery in 2015 for the state of Maine alone was valued at approximately half a billion dollars (State of Maine Department of Marine Resources; http://www.maine.gov/dmr/commercial-fishing/landings/documents/11-15LandingsBySpecieswithBonus.Table.pdf). In addition to its commercial importance, H. americanus is one of several decapod crustaceans that have long been used to elucidate the basic principles governing the generation, maintenance and neuromodulation of rhythmically active motor behaviors, and as such, serve as models for understanding the control of walking, breathing and chewing in vertebrates (for review see: Blitz and Nusbaum, 2011; Christie et al., 2010a; Cooke, 2002; Fénelon et al., 2003; Hooper and DiCaprio, Marder and Bucher, 2007; Marder et al., 1995; Nusbaum et al., 2001; Selverston, 2005; Selverston and Ayers, 2006; Selverston et al., 1998; Skiebe, 2001; Stein, 2009). A major contribution to our understanding of rhythmic motor behavior that has come from work conducted using the lobster and other decapods is that numerically simple, “hard-wired” neural circuits are capable of producing a wide array of distinct motor outputs. This functional flexibility is due largely to the actions of locally-released and circulating chemicals that modify the intrinsic properties of circuit elements at the molecular, cellular and system levels. While a number of classes of chemical are known to serve as locally-released and/or hormonally-delivered neuromodulators, the peptides are by far the largest and most diverse group of these compounds (e.g., Christie et al., 2010a).
Much work has focused on identifying native neuropeptides in H. americanus. Early studies employing biochemical isolation and sequencing of peptides and/or the targeted molecular cloning of the genes/transcripts encoding them allowed for the identification of members of several peptide families from the lobster, e.g., crustacean hyperglycemic hormone (CHH)/molt-inhibiting hormone (MIH), FMRFamide-like peptides (FLPs) and proctolin (e.g., Chang et al., 1990; de Kleijn et al., 1994, 1995; Schwarz et al., 1984; Soyez et al., 1991; Tensen et al., 1991; Trimmer et al., 1987). However, large-scale peptide discovery in this species did not begin until the advent of biological mass spectrometry as a means for peptide identification. Via accurate mass matching and tandem mass spectrometric sequencing, a large peptidome, encompassing approximately 20 distinct families, was rapidly elucidated (e.g., Cape et al., 2008; Chen et al., 2010; Christie et al., 2006, 2008a; Dickinson et al., 2009a, 2009b; Fu et al., 2005; Jiang et al., 2012; Li et al., 2002; Ma et al., 2008, 2009a; Stemmler et al., 2005, 2006, 2007, 2010). While clearly a powerful means for peptidome discovery, peptides that are present in low abundance, are large, possess extensive post-translational modifications, and/or possess sequences that do not ionize well can be very difficult to identify via mass spectrometry (e.g., Christie et al., 2010a). In contrast, in silico genome/transcriptome mining with subsequent bioinformatics peptide prediction is not limited by these factors (e.g., Christie et al., 2010a), and thus can be used to complement and augment peptide discoveries made using mass spectral and other means (e.g., Christie, 2014a, Christie et al., 2011a; Torfs et al., 2002). In fact, in silico mining of a mixed tissue neural transcriptome (BioProject No. PRJNA300643; D. Schulz and E. Marder, unpublished direct GenBank submission) was recently employed for expansion of the H. americanus neuropeptidome (Christie et al., 2015).
The eyestalk ganglia of decapod species, which consist of the lamina ganglionaris, medulla externa, medulla interna and medulla terminalis, have long been known to be rich sources of neuropeptides (e.g., Christie, 2011). The sinus gland, a major neuroendocrine release site present in the eyestalk of decapods, is derived largely from neurons whose somata reside in the X-organ, a cluster of loosely associated cell bodies located within the medulla terminalis (e.g., Christie, 2011). Using mass spectrometry and other means, approximately 40 peptides encompassing about one dozen families (i.e., allatostatin C [AST-C], corazonin, CHH/MIH, crustacean hyperglycemic hormone precursor-related peptide [CPRP], myosuppressin, orcokinin, orcomyotropin, pigment dispersing hormone [PDH], red pigment concentrating hormone [RPCH], short neuropeptide F [sNPF], SIFamide and tachykinin-related peptide [TRP]) have been identified from the eyestalk ganglia/sinus gland of H. americanus (e.g., de Kleijn et al., 1994, 1995; Dickinson et al., 2009b; Fu et al., 2005; Ma et al., 2008; Stemmler et al., 2005, 2006, 2010). A number of these peptides have been shown (or implicated) to play critical roles in the control of key physiological processes in the lobster, including, but not limited to, molting, growth and reproduction (e.g., Christie et al., 2010a). Interestingly, and despite its clear importance in understanding physiological control in the lobster, no significant transcriptomic resources have been developed for the eyestalk ganglia of H. americanus. In the study presented here, an eyestalk ganglia-specific transcriptome for the lobster was assembled de novo and used to predict a peptidome for this portion of the H. americanus nervous system. The transcriptome, which consists of 147,542 transcripts, has been publicly deposited (BioProject No. PRJNA338672) to provide a resource for future molecular studies of physiological control within and by the eyestalk ganglia. The peptidome predicted using this resource expands the peptidome known for this portion of the lobster nervous system approximately 7-fold, and includes a number of neuropeptides previously unknown from the lobster. These new peptide discoveries provide an expanded foundation from which to initiate anatomical and physiological studies of peptidergic signaling within and from the eyestalk ganglia.
2. Materials and methods
2.1. De novo transcriptome assembly
2.1.1. Animals and tissue dissection
American lobsters, H. americanus, (N=4) were purchased from local (Brunswick, ME, USA) seafood retailers. All animals were housed in recirculating natural seawater aquaria at 10–12°C and were fed approximately weekly on a diet of chopped shrimp. For the isolation of the eyestalk ganglia, animals were cold-anesthetized by packing in ice for approximately 20–30 min. After anesthetization, the eyestalks were removed and the eyestalk ganglia were dissected from the overlying carapace and surrounding musculature in chilled (approximately 4°C) physiological saline (composition in mM/l: 479.12 NaCl, 12.74 KCl, 13.67 CaCl2, 20.00 MgSO4, 3.91 Na2SO4, 11.45 Trizma base, and 4.82 maleic acid [pH = 7.45]).
2.1.2. RNA isolation
Freshly dissected eyestalk ganglia pairs (N=4 pairs) were placed into sterile RNAase-free 1.5 ml microfuge tubes containing 300 μl of TRIzol Reagent (catalog no. 15596018; Thermo Fisher Scientific Inc., Waltham, MA, USA) and manually homogenized using a sterile RNAse-free disposable pestle (catalog no. 9950-901; Argos Technologies Inc., Elgin, IL, USA). RNA was isolated from the resulting homogenate using a Direct-zol RNA MiniPrep (catalog no. R2052; Zymo Research, Irvine, CA, USA) spin column system according to the manufacturer-supplied protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). All RNA samples were stored at −80°C until being shipped on dry ice to the Georgia Genomics Facility (University of Georgia, Athens, GA, USA) for library preparation and sequencing.
2.1.3. cDNA library production and Illumina sequencing
Double-stranded cDNA libraries were prepared from total RNA using a KAPA Stranded mRNA-seq kit (catalog No. KK8420; Kapa Biosystems, Wilmington, MA, USA) following the manufacturer’s instructions; 3 μg of total RNA/sample was used for library generation. In brief, total RNA samples were purified with two oligo-dT selection (poly(A) enrichment using oligo-dT beads). Samples were then fragmented and reverse transcribed into double-stranded complementary cDNA using random primers, with second strand synthesis marked using dUPT. Each eyestalk ganglia library was tagged with a unique indexed adapter. A Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), AATI Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA), and Kapa qPCR assays were used to determine the quality and quantity of the final pool of libraries. Paired-end Illumina sequencing (150 base pairs [bp]) was performed on a NextSeq 500 system (Illumina, San Diego, CA, USA) using the high output kit v2 with 300 cycles.
2.1.4. Transcriptome assembly
Prior to transcriptome assembly, raw sequencing reads were assessed for quality using FASTQC (v1.0.0) software (Illumina Basespace Labs). Specifically, each RNA-Seq eyestalk library was quality filtered using FASTQ Toolkit (v.2.0.0) by trimming the first 9 bp of each read, removing all Illumina adapters (TruSeqLT universal primer), culling all low quality reads (Phred cutoff score=30), and setting the minimum read length to 50 bp. This quality filtering resulted in the removal of <1% of the reads present in each library, leaving from ~27 to ~38 million filtered reads per eyestalk ganglion sample (Table 1).
Table 1.
Summary of Homarus americanus eyestalk ganglia samples and their Illumina sequencing
| Sample | Sex | Total RNA concentration (ng/μl) | Raw reads |
|---|---|---|---|
| E1 | M | 223 | 26,829,856 |
| E2 | M | 268 | 38,085,928 |
| E3 | F | 316 | 32,879,458 |
| E4 | M | 262 | 33,177,978 |
Filtered reads were de novo assembled using Trinity (v2.0.6) software (https://github.com/trinityrnaseq/trinityrnaseq/wiki; Grabherr et al., 2011) on the National Center for Genome Analysis Support’s (Indiana University, Bloomington, IN, USA) Mason Linux cluster. Each node of the computer system includes four Intel Xeon L7555 8-core processors running at 1.87 GHz with 512 GB of memory. For the assembly, reads from all eyestalk ganglia libraries were combined and the minimum sequence length in the assembly was set to 324 bp. For the de novo assembly, the initial parameters of Trinity were set as follows: maximum memory, 200GB; CPU, 32; normalize maximum read coverage, 50; minimum contig length, 324. A summary of the assembly statistics (Table 2) was obtained using the script TrinityStat.pl (v2.0.6). Quality filtered raw reads were mapped against the de novo assembled transcriptome using Bowtie2 (v2.1.0; Langmead et al., 2009) software (Table 3).
Table 2.
Homarus americanus eyestalk ganglia transcriptome assembly statistics
| Total number of bases assembled | 183,106,109 |
| Total number of reads assembled | 130,973,220 |
| Total number of assembled contigs | 147,542 |
| Total number of Trinity “genes” | 110,841 |
| Minimum contig length (bp) | 324 |
| Maximum contig length (bp) | 27,389 |
| Average contig length (bp) | 1,241 |
| Median contig length (bp) | 637 |
| N25 (bp) | 4,193 |
| N50 (bp) | 2,160 |
| N75 (bp) | 869 |
| Total GC count (bp) | 74,888,256 |
| GC content for the complete assembly (%) | 40.90 |
Abbreviations: bp, base pairs; GC, guanine-cytosine.
Transcriptome assembly statistics were generated using Trinity software. Reads used for the de novo assembly were trimmed for Illumina adapters and quality filtered (Phred score=30)
Read length ranged from 324-27389 bp.
Table 3.
Summary of the results of mapping RNA-Seq reads to the complete Homarus americanus eyestalk ganglia assembly
| Total reads used for mapping | 130,973,220 |
| Total mapped reads* | 119,120,860 |
| Overall alignment (%) | 91% |
| Reads mapped 1 time (#) | 68,454,575 |
| Reads mapped 1 time (%) | 52% |
| Reads mapped >1 time (#) | 50,666,285 |
| Reads mapped >1 time (%) | 39% |
97% of the mapped reads aligned as clusters (read pairs)
2.2. Peptidome prediction
2.2.1. Transcriptome mining
Searches of the H. americanus eyestalk ganglia transcriptome were conducted using BLAST software installed on an Intel-processor-based BEOWULF computer cluster (Pacific Biosciences Research Center, University of Hawaii at Manoa, Honolulu, HI, USA) using a protocol that has proven highly effective for peptide-encoding transcript discovery in a wide array of arthropod species (e.g., Christie, 2008a, 2008b, 2014a, 2014b, 2014c, 2014d, 2014e, 2014f, 2015a, 2015b, 2015c, 2015d, 2016a, 2016b; Christie and Chi, 2015a, 2015b, 2015c; Christie and Pascual, 2016; Christie et al., 2008b, 2010b, 2011b, 2013b, 2015; Gard et al., 2009; Ma et al., 2009b, 2010). Specifically, the H. americanus eyestalk transcriptome assembly was selected as the database to be searched using the tblastn algorithm, and a known neuropeptide precursor was input to the program as the protein query. The complete list of pre/preprohormones searched for, as well as the specific queries used, is provided in Table 4. All hits returned by a given search were translated using the “Translate” tool of ExPASy (http://web.expasy.org/translate/), and then checked manually for homology to the query sequence. The BLAST-generated maximum score and E-value for each of the transcripts identified as encoding a putative neuropeptide precursor are provided in Table 4.
Table 4.
Putative Homarus americanus neuropeptide-encoding transcripts identified via in silico transcriptome mining of a de novo assembled transcriptome for the eyestalk ganglia (lamina ganglionaris, medulla externa, medulla interna and medulla terminalis) and the proteins deduced from them
| Peptide family | Transcript/protein identifications | tblastn search statistics | |||||
|---|---|---|---|---|---|---|---|
| Transcript | Deduced protein | ||||||
| Identification No. | Length* | Name | Length† | Type | Score | E-value | |
| ACP | TR10790|c0_g1_i3 | 1033 | Prepro-ACP | 104 | F | 211 | 4e-55 |
| TR10790|c0_g1_i2 | 1077 | Prepro-ACP | 104 | F | 211 | 4e-55 | |
| TR10790|c0_g1_i1 | 1685 | Prepro-ACP | 104 | F | 211 | 4e-55 | |
| AST-A | TR19564|c0_g1_i1 | 1395 | Prepro-AST-A sv2 | 378 | N | 552 | 1e-156 |
| TR19564|c1_g1_i1 | 3284 | Prepro-AST-A sv2 | 189 | C | 357 | 7e-98 | |
| AST-B | TR68637|c1_g2_i2 | 647 | Prepro-AST-B | 157 | N | 120 | 1e-27 |
| TR68637|c1_g1_i2 | 950 | Prepro-AST-B sv1 | 153 | C | 69 | 3e-12 | |
| TR68637|c1_g1_i1 | 1058 | Prepro-AST-B sv2 | 189 | C | 69 | 3e-12 | |
| AST-C | TR39168|c1_g1_i1 | 1163 | Prepro-AST-C I | 65 | C | 97 | 1e-20 |
| TR48133|c0_g1_i1 | 1624 | Prepro-AST-C II | 105 | F | 174 | 4e-44 | |
| TR20160|c0_g1_i1 | 1609 | Prepro-AST-C III | 148 | F | 29 | 3.4 | |
| Allatotropin | |||||||
| Bursicon α | TR30087|c0_g1_i1 | 372 | Pre-bursicon α v2 | 100 | N | 209 | 2e-54 |
| Bursicon β | |||||||
| CAP2b | |||||||
| CCHamide | TR54993|c0_g1_i5 | 2717 | Prepro-CCHamide I | 116 | F | 237 | 6e-63 |
| TR54993|c0_g1_i4 | 3448 | Prepro-CCHamide I | 116 | F | 237 | 6e-63 | |
| TR54993|c0_g1_i3 | 3452 | Prepro-CCHamide I | 116 | F | 237 | 6e-63 | |
| TR54993|c0_g1_i2 | 3459 | Prepro-CCHamide I | 116 | F | 237 | 6e-63 | |
| TR54993|c0_g1_i1 | 3476 | Prepro-CCHamide I | 116 | F | 237 | 6e-63 | |
| TR75050|c0_g1_i1 | 3267 | Prepro-CCHamide II sv1 | 252 | F | 470 | 1e-132 | |
| TR75050|c0_g1_i3 | 3294 | Prepro-CCHamide II sv2 | 261 | F | 462 | 1e-130 | |
| TR75050|c0_g1_i2 | 1277 | Prepro-CCHamide II sv3 | 266 | F | 458 | 1e-129 | |
| TR75050|c0_g1_i4 | 1304 | Prepro-CCHamide II sv4 | 275 | F | 451 | 1e-126 | |
| Corazonin | TR17179|c1_g1_i6 | 1716 | Prepro-corazonin | 109 | F | 135 | 2e-32 |
| TR17179|c1_g1_i3 | 1716 | Prepro-corazonin | 109 | F | 135 | 2e-32 | |
| TR17179|c1_g1_i2 | 1715 | Prepro-corazonin | 109 | F | 135 | 2e-32 | |
| TR17179|c1_g1_i1 | 1715 | Prepro-corazonin | 109 | F | 135 | 2e-32 | |
| TR17179|c1_g1_i4 | 631 | Prepro-corazonin | 22 | C | 49 | 3e-06 | |
| CCAP | TR69144|c0_g1_i2 | 1098 | Prepro-CCAP | 140 | F | 254 | 5e-68 |
| TR69144|c0_g1_i1 | 1113 | Prepro-CCAP | 140 | F | 254 | 5e-68 | |
| CHH (CHH subgroup) | TR1098|c3_g6_i2 | 649 | Prepro-CHH Ia v1 | 134 | F | 260 | 6e-70 |
| TR1098|c3_g6_i3 | 497 | Prepro-CHH Ia v2 | 133 | F | 243 | 7e-65 | |
| TR1098|c3_g6_i4 | 589 | Prepro-CHH Ia v3 | 133 | F | 241 | 3e-64 | |
| TR1098|c3_g6_i1 | 747 | Prepro-CHH Ib v2 | 131 | F | 217 | 7e-57 | |
| TR25279|c0_g1_i1 | 673 | Prepro-CHH II sv2 | 111 | F | 87 | 1e-17 | |
| CHH (MIH subgroup) | TR12133|c0_g1_i1 | 1451 | Pre-MIH I v2 | 112 | F | 199 | 1e-51 |
| TR9706|c0_g1_i1 | 837 | Pre-MIH II | 111 | F | 87 | 1e-17 | |
| TR58586|c0_g1_i1 | 1736 | Pre-MIH III | 119 | F | 48 | 7e-06 | |
| DENamide | |||||||
| DH31 | TR60801|c0_g1_i2 | 2393 | Prepro-DH31 sv1 | 135 | F | 211 | 4e-55 |
| TR60801|c0_g1_i1 | 2371 | Prepro-DH31 sv2 | 128 | F | 190 | 1e-48 | |
| DH44 | TR48947|c0_g1_i2 | 1198 | Prepro-DH44 | 285 | F | 416 | 1e-116 |
| TR48947|c0_g1_i1 | 1551 | Prepro-DH44 | 285 | F | 416 | 1e-116 | |
| DXXRLamide | |||||||
| ETH | |||||||
| EH | TR8891|c0_g1_i1 | 555 | Pre-EH I | 88 | F | 120 | 1e-27 |
| TR33899|c2_g1_i1 | 2290 | Pre-EH II | 82 | F | 57 | 2e-08 | |
| Elevenin | TR73513|c0_g1_i1 | 884 | Prepro-elevenin | 127 | F | 39 | 0.003 |
| FLP | TR10371|c0_g1_i2 | 3112 | Prepro-FLP | 358 | F | 562 | 1e-160 |
| TR10371|c0_g1_i1 | 3101 | Prepro-FLP | 358 | F | 562 | 1e-160 | |
| FXGGXamide | |||||||
| GPα2 | TR7537|c0_g1_i1 | 2897 | Pre-GPα2 | 120 | F | 175 | 2e-44 |
| GPβ5 | TR31071|c0_g1_i2 | 2321 | Pre-GPβ5 | 163 | F | 170 | 8e-43 |
| TR31071|c0_g1_i1 | 2293 | Pre-GPβ5 | 163 | F | 170 | 8e-43 | |
| GSEFLamide | TR71156|c0_g1_i2 | 966 | Prepro-GSEFLamide sv1 | 268 | N | 39 | 0.003 |
| TR71156|c0_g1_i1 | 996 | Prepro-GSEFLamide sv2 | 278 | N | 39 | 0.003 | |
| TR33789|c3_g2_i1 | 1470 | Prepro-GSEFLamide | 35 | C | 75 | 3e-14 | |
| HIGSLYRamide | |||||||
| ILP | |||||||
| Intocin | TR17969|c0_g3_i1 | 1121 | Prepro-intocin | 154 | F | 309 | 2e-84 |
| TR17969|c0_g2_i1 | 1121 | Prepro-intocin | 154 | F | 309 | 2e-84 | |
| TR17969|c0_g1_i1 | 1121 | Prepro-intocin | 154 | F | 309 | 2e-84 | |
| Leucokinin | TR63890|c0_g1_i1 | 2739 | Prepro-leucokinin v1 | 606 | F | 920 | 0.0 |
| TR63890|c0_g1_i2 | 1230 | Prepro-leucokinin v2 | 130 | C | 64 | 1e-09 | |
| Myosuppressin | TR50807|c0_g1_i1 | 1600 | Prepro-myosuppressin | 100 | F | 169 | 2e-42 |
| Neuroparsin | TR33370|c0_g1_i1 | 726 | Pre-neuroparsin I | 98 | F | 207 | 5e-54 |
| TR49668|c0_g1_i1 | 1130 | Pre-neuroparsin II | 98 | F | 105 | 2e-23 | |
| TR70851|c4_g1_i1 | 1490 | Pre-neuroparsin III | 101 | F | 101 | 5e-22 | |
| TR43449|c0_g1_i2 | 1897 | Pre-neuroparsin IV | 103 | F | 100 | 2e-21 | |
| TR43449|c0_g1_i1 | 878 | Pre-neuroparsin IV | 103 | F | 100 | 2e-21 | |
| NPF | TR52169|c0_g1_i1 | 1470 | Prepro-NPF I sv2 | 132 | F | 161 | 4e-40 |
| TR47662|c0_g1_i1 | 1008 | Prepro-NPF II sv1 | 79 | F | 34 | 0.081 | |
| TR47662|c0_g1_i2 | 1119 | Prepro-NPF II sv2 | 116 | F | 28 | 4.4 | |
| NPLP1 | |||||||
| NPLP2 | |||||||
| NPLP3 | |||||||
| NPLP4 | |||||||
| Orcokinin | TR61279|c0_g2_i1 | 514 | Prepro-orcokinin II sv2 | 156 | N | 225 | 8e-59 |
| TR61279|c0_g1_i1 | 504 | Prepro-orcokinin II sv2 | 152 | N | 217 | 2e-56 | |
| TR4139|c0_g1_i1 | 1307 | Prepro-orcokinin I/II | 118 | C | 251 | 1e-66 | |
| TR4139|c0_g1_i2 | 1241 | Prepro-orcokinin IV | 96 | C | 203 | 3e-52 | |
| PDH | TR23970|c0_g1_i1 | 1058 | Prepro-PDH I | 79 | F | 122 | 2e-28 |
| TR9861|c0_g4_i1 | 481 | Prepro-PDH II | 79 | F | 101 | 4e-22 | |
| TR9861|c0_g3_i1 | 481 | Prepro-PDH II | 79 | F | 101 | 4e-22 | |
| TR9861|c0_g2_i1 | 481 | Prepro-PDH II | 79 | F | 101 | 4e-22 | |
| TR9861|c0_g1_i1 | 714 | Prepro-PDH II | 79 | F | 101 | 4e-22 | |
| Proctolin | TR11540|c0_g1_i2 | 962 | Prepro-proctolin | 88 | F | 111 | 4e-25 |
| TR11540|c0_g1_i1 | 951 | Prepro-proctolin | 88 | F | 111 | 4e-25 | |
| PTTH | |||||||
| Pyrokinin | TR21391|c0_g1_i1 | 1766 | Prepro-pyrokinin | 385 | F | 528 | 1e-150 |
| RPCH | TR11499|c0_g1_i1 | 770 | Prepro-RPCH | 98 | F | 101 | 4e-22 |
| RYamide | TR37059|c0_g1_i1 | 707 | Prepro-RYamide | 135 | F | 160 | 6e-40 |
| sNPF | TR46457|c1_g1_i2 | 677 | Prepro-sNPF sv1 | 96 | F | 137 | 9e-33 |
| TR46457|c1_g1_i1 | 663 | Prepro-sNPF sv1 | 96 | F | 137 | 9e-33 | |
| TR46457|c1_g1_i4 | 773 | Prepro-sNPF sv2 | 128 | F | 182 | 1e-46 | |
| TR46457|c1_g1_i3 | 759 | Prepro-sNPF sv2 | 128 | F | 182 | 1e-46 | |
| SIFamide | TR65061|c0_g1_i1 | 1734 | Prepro-SIFamide sv1 | 80 | F | 132 | 2e-31 |
| TR65061|c0_g1_i2 | 1302 | Prepro-SIFamide sv2 | 76 | F | 117 | 7e-27 | |
| Sulfakinin | TR6787|c0_g1_i1 | 1023 | Prepro-sulfakinin | 120 | F | 196 | 1e-50 |
| TRP | TR41108|c0_g1_i1 | 2211 | Prepro-TRP | 129 | F | 442 | 1e-124 |
| Trissin | TR50290|c0_g2_i1 | 943 | Prepro-trissin v1 | 187 | F | 58 | 7e-09 |
| TR50290|c0_g1_i1 | 940 | Prepro-trissin v2 | 186 | F | 56 | 2e-08 | |
Length in nucleotides.
Length in amino acids.
Peptide family abbreviations: ACP, adipokinetic-corazonin-like peptide; AST-A, allatostatin A; AST-B, allatostatin B; AST-C, allatostatin C; CAP2b, cardioacceleratory peptide 2b; CCAP, crustacean cardioactive peptide; CHH, crustacean hyperglycemic hormone; MIH, molt-inhibiting hormone; DH31, diuretic hormone 31; DH44, diuretic hormone 44; ETH, ecdysis-triggering hormone; EH, eclosion hormone; FLP, FMRFamide-like peptide; GPα2, glycoprotein hormone α2; GPβ5, glycoprotein hormone β5; ILP, insulin-like peptide; NPF, neuropeptide F; NPLP1, neuropeptide-like precursor 1; NPLP2, neuropeptide-like precursor 2; NPLP3, neuropeptide-like precursor 3; NPLP4, neuropeptide-like precursor 4; PDH, pigment dispersing hormone; PTTH, prothoracicotropic hormone; RPCH, red pigment concentrating hormone; sNPF, short neuropeptide F; TRP, tachykinin-related peptide.
Other abbreviations: C, carboxy-terminal partial protein; F, full-length protein; N, amino-terminal partial protein; sv, splice variant; v, variant.
Query proteins: ACP, Homarus americanus prepro-ACP (deduced from Accession No. GEBG01018127; Christie et al., 2015); AST-A, H. americanus prepro-AST-A (deduced from Accession No. GEBG01007827; Christie et al., 2015); AST-B, Procambarus clarkii prepro-allatostatin B (deduced from Accession No. GBEV01040422; Christie and Chi, 2015c); AST-C, H. americanus prepro-AST-C I (deduced from Accession No. GEBG01004053; Christie et al., 2015) and H. americanus prepro-AST-C II (deduced from Accession No. EY291152; Dickinson et al, 2009b); allatotropin, Tigriopus californicus prepro-allatotropin (deduced from Accession No. JW513825; Christie, 2014c); bursicon α, H. americanus pre-bursicon α (deduced from Accession No. GEBG01013055; Christie et al., 2015); bursicon β, H. americanus pre-bursicon β (deduced from Accession No. CN854188; Christie et al., 2010b); CAP2b, Nilaparvata lugens prepro-CAP2b splicing variant a (Accession No. BAO00940; Tanaka et al., 2014); CCHamide, H. americanus prepro-CCHamide I (deduced from Accession No. GEBG01016648; Christie et al., 2015) and H. americanus prepro-CCHamide II (deduced from Accession No. GEBG01015625; Christie et al., 2015); corazonin, H. americanus prepro-corazonin (deduced from Accession No. GEBG01047508; Christie et al., 2015); CCAP, H. americanus prepro-CCAP (deduced from Accession No. GEBG01001997; Christie et al., 2015); CHH, H. americanus prepro-CHH A (Accession No. P19806; de Kleijn et al., 1995); MIH, H. americanus pre-gonad-inhibiting hormone (Accession No. CAA60644; de Kleijn et al., 1994); DENamide, Daphnia pulex prepro-DENamide (Dircksen et al., 2011); DH31, H. americanus prepro-CLDH (Accession No. ACX46386; Christie et al., 2010c); DH44, H. americanus prepro-DH44 (deduced from Accession No. GEBG01010013; Christie et al., 2015); DXXRLamide, T. californicus prepro-DXXRLamide Ia (deduced from Accession No. JW528324; Christie, 2014c); ETH, D. pulex prepro-ETH (Dircksen et al., 2011); EH, pre-EH I (deduced from Accession No. GEBG01042722; Christie et al., 2015); elevenin, N. lugens prepro-elevenin (Accession No. BAO00952; Tanaka et al., 2014); FLP, H. americanus prepro-FLRFamide (deduced from Accession No. GEBG01004307; Christie et al., 2015); FXGGXamide, T. californicus prepro-FXGGXamide Ia (deduced from Accession No. JV193177; Christie, 2014c); GPα2, N. lugens pre-glycoprotein hormone α2 (Accession No. BAO00955; Tanaka et al., 2014); GPβ5, N. lugens pre-glycoprotein hormone β5 (Accession No. BAO00956; Tanaka et al., 2014); GSEFLamide, H. americanus prepro-GSEFLamide (deduced from Accession No. GEBG01035690; Christie et al., 2015); HIGSLYRamide, Carcinus maenas prepro-HIGSLYRamide (deduced from Accession No. DV111329; Christie et al., 2008b); ILP, H. americanus prepro-ILP (deduced from Accession No. GEBG01059205; Christie et al., 2015); intocin, H. americanus prepro-intocin (deduced from Accession No. GEBG01052869; Christie et al., 2015); leucokinin, H. americanus prepro-leucokinin (deduced from Accession No. GEBG01042414; Christie et al., 2015); myosuppressin, H. americanus prepro-myosuppressin (Accession No. ACX46385; Stevens et al., 2009); neuroparsin, H. americanus pre-neuroparsin (deduced from Accession No. GEBG01017677; Christie et al., 2015); NPF, H. americanus prepro-NPF (deduced from Accession No. GEBG01010211; Christie et al., 2015); NPLP1, N. lugens prepro-NPLP1 (Accession No. BAO00966; Tanaka et al., 2014); NPLP2, Drosophila melanogaster prepro-NPLP2, isoform A (Accession No. AAF49832; Adams et al., 2000); NPLP3, N. lugens prepro-NPLP3 (Accession No. BAO00967; Tanaka et al., 2014); NPLP4, N. lugens prepro-NPLP4 (Accession No. BAO00968; Tanaka et al., 2014); orcokinin, H. americanus prepro-orcokinin I (Accession No. ACB41787; Dickinson et al., 2009a); PDH, H. americanus prepro-PDH (deduced from Accession No. GEBG01005888; Christie et al., 2015); proctolin, H. americanus prepro-proctolin (deduced from Accession No. GEBG01005712; Christie et al., 2015); PTTH, N. lugens prepro-PTTH (Accession No. BAO00973; Tanaka et al., 2014); pyrokinin, H. americanus prepro-pyrokinin (deduced from Accession No. GEBG01039267; Christie et al., 2015); RPCH, Cherax quadricarinatus prepro-RPCH (Accession No. AAV80404; Martinez-Perez et al., 2007); RYamide, P. clarkii prepro-RYamide (deduced from Accession No. GBEV01010112; Christie and Chi, 2015c); sNPF, P. clarkii prepro-sNPF (deduced from Accession No. GBEV01004780; Christie and Chi, 2015c); SIFamide, H. americanus prepro-SIFamide (Accession No. ABV21807; Dickinson et al., 2008); sulfakinin, H. americanus prepro-sulfakinin (Accession No. ABQ95346; Dickinson et al., 2007); TRP, H. americanus prepro-TRP (Accession No. ACB41786; Christie et al., 2008a); trissin, D. melanogaster prepro-trissin (Accession No. AAF55203; Adams et al., 2000).
2.2.2. Peptide prediction
The structures of mature peptides were predicted using a well-established workflow (e.g., Christie, 2008a, 2008, 2014a, 2014b, 2014c, 2014d, 2014e, 2014f, 2015a, 2015b, 2015c, 2015d, 2016a, 2016b; Christie and Chi, 2015a, 2015b, 2015c; Christie and Pascual, 2016; Christie et al., 2008b, 2010b, 2011b, 2011c, 2011d, 2013, 2015; Gard et al., 2009; Ma et al., 2009b, 2010). Specifically, each of the deduced precursor proteins was assessed for the presence of a signal peptide using the online program SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011); the D-cutoff values of SignalP 4.1 were set to “Sensitive” to better match the sensitivity of version 3.0 of this freeware program. Prohormone cleavage sites were identified based on the information presented in Veenstra (2000) and/or by homology to known arthropod pre/preprohormone processing schemes. When tyrosine residues were present, prediction of their sulfation state was conducted using the online program “Sulfinator” (http://www.expasy.org/tools/sulfinator/; Monigatti et al., 2002). Disulfide bonding between cysteine residues was predicted by homology to known peptide isoforms and/or by using the online program “DiANNA” (http://clavius.bc.edu/~clotelab/DiANNA/; Ferrè and Clote, 2005). Other post-translational modifications, i.e., cyclization of amino (N)-terminal glutamine/glutamic acid residues and carboxyl (C)-terminal amidation at glycine residues, were predicted by homology to known arthropod peptide isoforms. Figure 1 shows three examples of mature peptide structural prediction using the workflow just described; the mature structures of all peptides predicted in this study are provided in Table 5. All protein/peptide alignments were done using the online program MAFFT version 7 (http://mafft.cbrc.jp/alignment/software/; Katoh and Standley, 2013). To determine amino acid identity/similarity between peptides, the sequences in question were aligned using MAFFT version 7; amino acid identity/similarity was subsequently determined using the alignment output. Specifically, percent identity was calculated as the number of identical amino acids divided by the total number of residues in the longest sequence (x100). Amino acid similarity was calculated as the number of identical and similar amino acids divided by the total number of residues in longest sequence (x100).
Figure 1.
Two examples of the in silico workflow used for the prediction of putative mature Homarus americanus eyestalk ganglia peptide structures. (A) The predicted processing scheme for prepro-allatostatin C (AST-C) III. The structure of the putative mature AST-C isoform is shown in red, with the structures of three putative mature linker/precursor-related peptides shown in blue. In the AST-C isoform, the presence of a disulfide bridge between the two cysteine residues is indicated by an inverted red bracket. (B) The predicted processing scheme for prepro-sulfakinin. The structures of two putative mature sulfakinin isoforms are shown in red, with those of three putative mature linker/precursor-related peptides shown in blue. “Y(SO3H)” indicates the presence of sulfated tyrosine residues in the putative mature sulfakinins. In one of the two sulfakinins, the presence of pyroglutamic acid is indicated by “pE”.
Table 5.
Predicted neuropeptidome of the Homarus americanus eyestalk ganglia
| Peptide family | Predicted peptide structure |
|---|---|
| ACP | pQITFSRSWVPQa |
| ACP-PRP | SGGITGPLVTPGGGSDRGADPCKDVRLATLTQVASHLADLMDDTFDLPQDDAALALRLKHGLVA |
| MS | |
| AST-A | HSNYGFGLa |
| TPGYAFGLa | |
| SDLYSFGLa | |
| SGSYNFGLa | |
| AKYSFGIa | |
| SKLYGFGLa | |
| PRNYAFGLa | |
| SQMYSFGLa | |
| PRDYAFGLa | |
| PTAYSFGLa | |
| ATSYGFGLa | |
| SYDFGLa | |
| AGRYAFGLa | |
| TGPYAFGLa | |
| VGPYAFGLa | |
| AGHYAFGLa | |
| AGPYAFGLa | |
| SGPYAFGLa | |
| ADPYAFGLa | |
| AGQYSFGLa | |
| SGPYSFGLa | |
| SGVYSFGLa | |
| AGPYSFGLa | |
| AST-A-PRP | HDY(SO3H)LEDLDDPDTSRLLDVLQYYDTEPSYLYDYa |
| EGLYSLGLD | |
| SVGDLPEVSKVEDGASPRT | |
| DVSITEDTLED | |
| ESSKN | |
| DSGEE | |
| EDDDMENRTQQYSFGLGKQDPDMEIE | |
| ESDEDSD | |
| DPDMDMD | |
| ASSDEDDEERYYAYEQa | |
| AFSETDDY(SO3H)DNVNDNDDGDDELELSDLEQY(SO3H)SDDL | |
| SDAPDSGFa | |
| SDSDSDQYTLa | |
| EVSDDDHDEDEQDIGVEEEMSS | |
| AST-B | VGWSSMHGTWa |
| TNWNKFQGSWa | |
| NNWRSLQGSWa | |
| AWNKLQGAWa | |
| PDNTRVSPRSTNWSSLRGTWa | |
| SADWNKLRGAWa | |
| ASDWGQFRGSWa | |
| AST-B-PRP | SSSSPQQDDPASSPSHIEE |
| PHLEDAQLDAAEV | |
| GEELQAAED | |
| GDDLADAELQAAED | |
| SEDDNGDDLYDETNLEEDLAGNEEQVSPLALARLMAAAPQ | |
| GWTLWa | |
| PDNTRVSPRSTNWSSLRDMMSVAAPNQA | |
| APDMMSVAAPNQA | |
| AST-C | pQIRYHQCYFNPISCF |
| SYWKQCAFNAVSCFa | |
| GNGDGRLYWRCYFNAVSCF | |
| AST-C-PRP | +QQQQQQQQQQQQQQQQQQQQQGEEEV |
| MFVPLSGLPGELPTI | |
| KALPDQDPQVYGQMPHMLDPAGNHLIDDDGSLDAVLINYLFAKQMVERLRNNADIKDLQ | |
| ARVPQQAP | |
| PQYLEVVRPVLPNTALEPLGSLQDAPQQIAETVSTP | |
| AAIVLDKLMFALQKALDDSPAAPPGQPAPYSRPRTYAAGPMDLQ | |
| Bursicon α | 1 |
| CCHamide | SCSQFGHSCFGAHa |
| HRVLKGGCLNYGHSCLGAHa | |
| CCHamide-PRP | DGDQYARQEPSPLYPEANQLPEFEQRQEDRLSVDEAVTDREIVANA |
| NWLAVLSHRLRQRTSPQSSPSAQSLGYFQ | |
| AYVPVHPPVAPRPLLDVLLDALNTPTRSSHYSHARAANSVMGPRASYPEa | |
| VKSPPTSDQLSDMGLDLRGEDYASGTNDDLESVGAIGGVRGSLDDT | |
| DLAQDNVLYYGVLNDDY(SO3H)SDARY | |
| SAVSLPSRGRLGASPPLGVANNAVPQDRPHIL | |
| EEHTGKDEMDPKYLALASFPNWL | |
| YSIQHPFIRSAVSLPSRGRLGASPPLGVANNAVPQDRPHIL | |
| EEHTGKDEMDPKYLALASFPVSTILLASDPTLLPVVVLV | |
| Corazonin | pQTFQYSRGWTNa |
| Corazonin-PRP | SDPNVGVTELLADPP |
| LSAHSHPHPPTHTLPKNIEERLRALEAGLNAVLKANSINFSPGGDEEY(SO3H)YAEN | |
| CCAP | PFCNAFTGCa |
| CCAP-PRP | GPVA |
| DIGDLLEGKD | |
| SDPSMEGLASSSELDALAKHVLAEAKLWEQLQSKMEMMRSYAS | |
| MENHPVY | |
| STPHTQPRQHLTSTPQQKVETEKQ | |
| CHH (CHH subgroup) | pQVFDQACKGVYDRNLFKKLDRVCEDCYNLYRKPFVATTCRENCYSNWVFRQCLDDLLLSDVIDEYVSNVQMVGK |
| pQVFDQACKGVYDRNLFKKLNRVCEDCYNLYRKPFIVTTCRENCYSNRVFRQCLDDLLMIDVIDEYVSNVQMVGK | |
| pQVFDQACKGVYDRNLFKKLNRVCEDCYNLYRKPFIVTTCRQNCFEGDTFPRCVMDLGLDLELFLEFRDMIKa | |
| AVFDSACKGYYDREFWGKLSRVCWDCENLFRQPGYQDKCSEGCFVTTDFTQCVKALLLNVEEYNELAELVRa | |
| CPRP | RSVEGASRMEKLLSSSNSPSSTPLGFLSQDHSVN |
| RSVEGASRMEKLLSSISPSSTPLGFLSQDHSVN | |
| RSWLIDGDEDLQLSQYHSLN | |
| CHH (MIH subgroup) | WFTNDECPGVMGNRDLYEKVAWVCNDCANIFRNNDVGVMCKKDCFHTMDFLWCVYATERHGEIDQFRKWVSILRAa |
| IYIFNECPGRLGNGELHDKVDLVCDDCYNLFRDSALAVTCRGNCFTNHYFDMCIFATARRNQMQMYRRWVSILSAa | |
| 2 | |
| DH31 | GLDLGLGRGFSGSQAAKHLMGLAAANFAGGPa |
| DH31-PRP | AAFNREA |
| AVVQIEDPDY(SO3H)VLELLTRLGHSII | |
| ANELEKFVRSSGSA | |
| SSDDGLDLHHDDNLYAQDQAADLAESS | |
| ANELENA | |
| DH44 | ASGLSLSIDASMKVLREALYMEIIRKKQRQQMQRAQHNQKLLNSIa |
| DH44-PRP | 3 |
| NSNRSN | |
| SNSSSGISGSNTSSNSNTNNNSPDTISMA | |
| TWPNGFS | |
| DVTRQLQQEGIQGVYQRGQ | |
| EH | AANKVSVCIKNCAQCKIMYHDHFKGGLCADLCVQSGGKFIPDCGRPQTLIPFFLQRLE |
| ATFTSMCIRNCGQCKEMYGDYFHGQACAESCIMTQGISIPDCNNPATFNRFLKRFI | |
| Elevenin | VDCRKFVFAPVCRGIIA |
| Elevenin-PRP | MVSE |
| SSFRPTADTQWNSQYRAPTETEAENLLLASSYDDVMEPRPQEDMVVVRAGSDVVQVPAYVFGVIERSLQGE | |
| FLP | GYSDRNYLRFa |
| SGRNFLRFa | |
| NRNFLRFa | |
| DQNRNFLRFa | |
| GAHKNYLRFa | |
| GNRNFLRFa | |
| GDRNFLRFa | |
| FSHDRNFLRFa | |
| APSKNFLRFa | |
| FLP-PRP | APVPPVVAALDPPTDALLPAQSQEDDLFALPE |
| LLKYFLPASQAWGGDAYPIGQEGT | |
| SDDNS | |
| SDTNDY(SO3H)EGEEMPESPE | |
| SGSPMEFATDLQEDVELPVEE | |
| SVDRQLSSLSCEDCDEEQKAREFTSTPSPTTIQPLART | |
| DVSAVLSDDSIESSVLRQINAHRI | |
| AAAQNFYIPMAWASELQPEEDGIDVTSFEEPQVA | |
| DGSDDY(SO3H)PSSSSSAESPAPVVVVRPVEYPRYV | |
| GPα2 | 4 |
| GPβ5 | 5 |
| GSEFLamide | IGSEFLa |
| MGSEFLa | |
| AMGSEFLa | |
| ALGSEFLa | |
| VMGSEFLa | |
| AVGSEFLa | |
| GSEFLamide-PRP | ALPTHLPDELDDPVV |
| LAGTPHESMIRYFLMAMSNPAGRYKSPQLLNRGV | |
| SVGKLSDADNPRDFESENCSDDDGTEEEDL | |
| EQFSFTGQY(SO3H)DY(SO3H)DESAGENFGSQEDLFNTKP | |
| NIRSFHGGVNNDGLKNFFSMLMS | |
| QYEPEFAHTLDYDT | |
| Intocin | CFITNCPPGa |
| Intocin-PRP | SGPTAQLGRTRTCTACGPGLQGRCLGPEICCVLGIGCFLGTREA |
| MCHAENLVPVTCANRDLKSCa | |
| MQEGRCAAAGLCCTEMKCEFDSSCTVEGREE | |
| VGKQRAE | |
| QHLTFLSSLPEDQWNL | |
| Leucokinin | pQAFHPWGa |
| ASFNPWGa | |
| NTFAPWGa | |
| pESFSAWGa | |
| TRFSAWAa | |
| AFSAWAa | |
| TFSAWAa | |
| TFRAWAa | |
| TRFSPWAa | |
| PSFSAWAa | |
| PSFNAWAa | |
| pQGFSAWAa | |
| VPFSTWGa | |
| Leucokinin-PRP | LSSGAASVSFVTSEVMDVSPLALPHGRHPNLCTPDHVPSHPIIRCEVa |
| SSFKTAPGLPLSLREVYLALFQNARPRPPPPSEGEL | |
| SDPLLPASQHEPNT | |
| AAGYFTHDTNPLIIEEDLIPYIGVLSDDGEAEDVV | |
| GSFPADDWEEEEPTDLFVLDGSLPYPPVDRLRY | |
| EAETYTNTLVNSDDSGVKTEAENIKPDTKY(SO3H)DQTAEASSTTVA | |
| PDLQVIEDAVRKMAEHEPKTTE | |
| SSDVNLQDGEDDEPVSAWIa | |
| LQDANTDD | |
| SPSMDLSGNQD | |
| SSGDELDDHFLD | |
| AEGTLSRLSESTLKAALDENSPEDNVDNH | |
| SESNE | |
| SDSDE | |
| SDIDE | |
| SSETD | |
| NNGGSDDPTHSNNPQQISSILQQLQHQGLEFLH | |
| LPINDWGN | |
| ASPISEDSQLSDLYTSQL | |
| Myosuppressin | pQDLDHVFLRFa |
| Myosuppressin-PRP | VCVGVGETMPPPICLSQQVPLSPFA |
| LCSALINISEFSRAMEEY(SO3H)LGAQAIERSMPVNEPEV | |
| SQQ | |
| Neuroparsin | APRCNQGGNRLPANNCKYGTVVDWCGGSVCAKGPGEACGGEWSENGECGAGTYCSCGYCNGCSANLECWFGSYC |
| APSCDGHGTRTEPTDCDYGSFQDWCGNNVCAKGPGQRCGGEWWENDDCGHGMYCANCGNCAGCSVGIQCWFCDSGS | |
| TPLCPERNEIAPEDLSQCKYGVVLGWCGNAACGKGPDEPCGGRWEENGICGEGMYCVCGYCAGCTSTLECVLGRFC | |
| APRCDSHDSPAPTNCKYGTVRDWCRNGVCAKGPGESCGGYWYEYGKCGGGTFCLCGTCIGCSTIDGTCSQSSPAIIC | |
| NPF | ARPDNSAADTLQAIHEAAMAGILGSAEVQYPNRPSMFKSPVELRQYLDALNAYYAIAGRPRFa |
| KPDPNQLAAMADALKYLQELDKYYSQVSRPRFa | |
| KPDPNQLAAMADALKYLQELDKYYSQVSRPSLRSSPGPASQIQALEKALKFLQLQELGKMYSLRARPRFa | |
| NPF-PRP | GNHGAQRTEELYDY(SO3H) |
| SEYAMVPGDALVSYDGSE | |
| Orcokinin | NFDEIDRSGFGFH |
| NFDEIDRSGFGFN | |
| NFDEIDRSGFGFV | |
| Orcomyotropin | FDAFTTGFGHN |
| Orcokinin-PRP | GPIKAAPARSSPQQDAAAGYTDGAPV |
| SSEDMDRLGFGFN | |
| GDY(SO3H)DVYPE | |
| VYGPRDIANLY | |
| SAE | |
| PDH | NSELINSILGLPKVMNDAa |
| NSELINSLLGIPKVMNDAa | |
| PDH-PRP | QELKYPEREVVAELAAQILRVIQGPWGPMAAGPH |
| QELKYPEREVVADMAAQILRVALGPWGSVAAVP | |
| Proctolin | RYLPT |
| Proctolin-PRP | ADDTRLDEI |
| ELLREMLE | |
| TAEGANSRISGSGYD | |
| FMY | |
| SVPEEGAAEMVQPALNLPQ | |
| Pyrokinin | LYYSQRPa |
| SLFSPRLa | |
| GDDITNEELAY(SO3H)DDNLATSEYLRDDNNDYLPEELTEDVTEMSSPEMLSESAAALVGKNSVSFIPRLa | |
| GDGFAFSPRLa | |
| GADFAFSPRLa | |
| SEFVFSSRPa | |
| SDFAFSPRLa | |
| ADFAFSPRLa | |
| DSEDSSVESRNTKTQASIPRPa | |
| AYFSPRLa | |
| Pyrokinin-PRP | LEDEWAGLPQASFAQYPPALDDTSEAQPLSLLYNMYPSVTSADTVPPKSQELQYNSQDTP |
| SVDLY(SO3H)DDEDPERRM | |
| QTPQHDNEPTDDNDDSTHRWWWPFVAV | |
| RPCH | pQLNFSPGWa |
| RPCH-PRP | AAAASGTDPAAASLHPAPPAVLTAASGANAGDSCGTIPVSAVMHIYRLI |
| TEAARLIQCQEEEYMa | |
| RYamide | pQGFYTQRYa |
| FIGGSRYa | |
| RYamide-PRP | SDTGEVTVRSGFYAN |
| NGRSSPSQGLPEIKIRSS | |
| SGPAPAAEPEFTPVMNGEADDSDMPATLLVGDSVICLLVDVPDIYRCV | |
| STTDEASN | |
| sNPF | GPPSLRLRFa |
| DMGWQVAQRSMPSLRLRFa | |
| DTSTPALRLRFa | |
| sNPF-PRP | VPAPQDY(SO3H)DAVNEVYDWLVDHGLE |
| TVDQEEDLSSHEQ | |
| TVDQVKSLYDREVV | |
| DTTY(SO3H)GQEEDLSSHEQ | |
| SIFamide | VYRKPPFNGSIFa |
| SIFamide-PRP | AGADPREYTVFEPGKGLASVCQVAVEACAAWFPVQE |
| AGADPLFEPGKGLASVCQVAVEACAAWFPVQE | |
| Sulfakinin | pEFDEY(SO3H)GHMRFa |
| GGGEY(SO3H)DDY(SO3H)GHLRFa | |
| Sulfakinin-PRP | VSAPARPSSLARVLAPVV |
| QRLEESHLPPALVEELVQDFEDPELLDFHDAAa | |
| SLTHSDQHHHHDTTVN | |
| TRP | APSGFLGMRa |
| TPSGFLGMRa | |
| TRP-PRP | AGEGQDTPQDRE |
| DASTALDDNTAASEYSSLPDPYPLYGLRDNNLPMLFAVPWKT | |
| SDEEVFSDATADNDLEILL | |
| YYDDDSDMDAYIQALTAVVDGQQQQ | |
| AYYSENPDEEISMTGVD | |
| Trissin | WSSSEVSCTSCGSECQSACGTRNFRACCFNFQ |
| Trissin-PRP | 6 |
| PSPSLNQLQHQNLHQRYTPSPTSIKI | |
| 7 |
Peptides shown in bold black font are new discoveries for H. americanus, while those shown in normal black font are known from the lobster, but new discoveries for the eyestalk ganglia. Peptides shown in gray font are known lobster eyestalk ganglia peptides. Peptides highlighted in yellow are present in sinus gland.
DECSLTPVIHILSYPGCVSKPIPSFACQGRCTSYVQVSGSKLWQTERSCMCCQESGEREASVVLNCPKVRKGEPTRRKVSV+
YFYKIRSGTQKEFELINCKQFNKTYYTELSRVCDDCQNIYRKYYNVGVDCKKDCFDNEWFPKCVTYIEHDHLLEEYKKMKEYLNLRDL
LSLGGGRADTSSLLSLPHPQELSQDDLQPFLSRQGNTDSAGAPSSVADYTGYDKSEVLRGLEDPTSSSAYRLQEALSEAVAAAAAAAEGAEGVRDGAAALSPTANEGVTLEDLVPYDPGYYLYPAFLNRGDEAMTGGSSGINSLRKV
TSFKHAWQNPGCHKVGHTRRISIPECLEFDITTNACRGFCESWSVPSAWQTLASNPHQVVTSIGQCCNIMDTEDVKVKVMCIQGPRELVFKSASTCDCFHCKKY
INPQSTLECHRRQYTYKVHKTDDEGRICWDFINVMSCWGRCDSNEIADWKFPYKRSHHPVCMHEETQLTVVTLGNCEDNAAPGTETYSYHEATRCACSVCKTSEASCEGLRYRGARRAPRAEVPRa
ADPRSHSSAGVNEADSMALEGLLKPDVSGTKDSHLSQILHLLSRALAESKTDPAFYKDPPSLSSVLSPLVQESTEEETDDTSDLLPSSGGDSDDPPLDNVIYLAF
ADPRSHSAGVNEADSMALEGLLKPDVSGTKDSHLSQILHLLSRALAESKTDPAFYKDPPSLSSVLSPLVQESTEEETDDTSDLLPSSGGDSDDPPLDNVIYLAF
Peptide family abbreviations: ACP, adipokinetic hormone-corazonin-like peptide; AST-A, allatostatin A; AST-B, allatostatin B; AST-C, allatostatin C; CCAP, crustacean cardioactive peptide; CHH, crustacean hyperglycemic hormone; CPRP, crustacean hyperglycemic hormone precursor-related peptide; DH31, diuretic hormone 31; DH44, diuretic hormone 44; EH, eclosion hormone; FLP, FMRFamide-like peptide; NPF, neuropeptide F; PDH, pigment dispersing hormone; RPCH, red pigment concentrating hormone; sNPF, short neuropeptide F; TRP, tachykinin-related peptide; PRP, precursor-related peptide.
Abbreviations in peptide structures: a, carboxyl-terminal amide group; C, one of a pair of cysteines that are linked by a disulfide bridge; pE/pQ, pyroglutamic acid; Y(SO3H), sulfated tyrosine.
See text for descriptions of the disulfide bridging patterns for all peptides with more than one pair of bridged cysteine residues.
3. Results
3.1. De novo assembly of an eyestalk ganglia-specific transcriptome
Four samples, each consisting of the two eyestalk ganglia from a single lobster (three males and one female), were used as the source of RNA for transcriptome development (Table 1). RNASeq yielded approximately 33 million paired-end reads per library (Table 1), which were collectively assembled using Trinity; a total of 130,973,220 trimmed and quality filtered paired-end reads ranging in length from 50 to 150 bp were input into Trinity for de novo assembly (Table 2). The transcriptome assembled by Trinity consists of 147,542 transcripts and 110,841 “Trinity predicted genes” (Table 2). Of the “Trinity predicted genes”, 96,484 are singletons (87%), with the remaining genes (14,357 in total) possessing from two to 55 “Trinity predicted isoforms”. The average transcript length is 1,241 bp (Table 2); half of the contigs (N50 in Table 2) were at least 2,160 bp long, with the shortest and longest assembled sequences being 324 and 27,389 bp, respectively (Table 2). Mapping for the Illumina-generated reads against the complete 147,542-transcript assembly using Bowtie yielded an overall alignment rate of 91%, with the majority of reads (52%) mapping just once (Table 3). The transcriptome just described, as well as all associated data, have been deposited in GenBank under BioProject No. PRJNA338672.
3.2. Prediction of an eyestalk ganglia-specific neuropeptidome
Transcripts encoding precursors for 48 distinct peptide families/subfamilies were searched for within the de novo assembled H. americanus eyestalk ganglia-specific transcriptome described in the previous section (Table 4). In the interest of space, only those searches that resulted in the identification of protein-encoding transcripts (33 families/subfamilies) are described in the following subsections, with the data presented in alphabetical order based on peptide family name. All precursor proteins listed as “full-length” exhibit a functional signal sequence (including a “start” methionine) and are flanked on their C-terminal end by a stop codon. Proteins described here as “partial” lacked a start methionine (referred to as C-terminal partial proteins), a stop codon (referred to as N-terminal partial proteins), or both of these features (referred to as internal fragment proteins).
3.2.1. Adipokinetic hormone-corazonin-like peptide (ACP)
Three transcripts were identified as encoding putative ACP precursors (Table 4). Translation of these sequences revealed each to encode an identical 104 amino acid protein (Homam-prepro-ACP; Table 4 and Fig. 2A); this protein is identical in sequence to the query protein used to identify the transcripts encoding it (Christie et al., 2015). Three distinct peptides were predicted from Homam-prepro-ACP (Table 5 and Fig. 2A): the ACP isoform pQITFSRSWVPQamide and two linker/precursor-related sequences. While all three of the peptides predicted from Homam-prepro-ACP are known H. americanus peptides (Christie et al., 2015), they are identified from the lobster eyestalk ganglia for the first time here.
Figure 2.
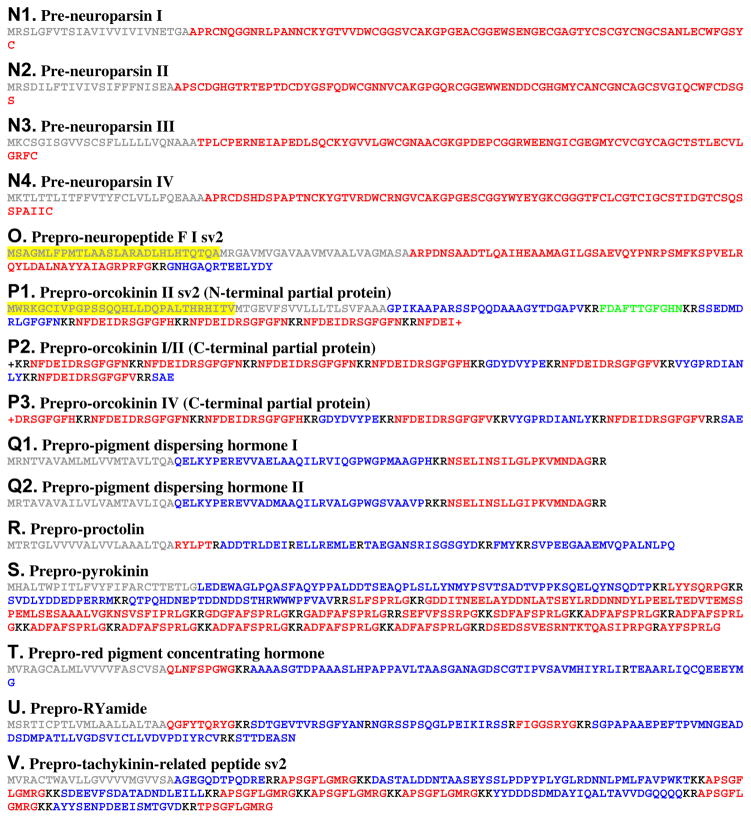
Putative Homarus americanus pre/preprohormones deduced from transcriptome shotgun assembly sequence data. This figure does not include predicted preprohormones for which multiple putative splice variants were identified in the eyestalk ganglia transcriptome. (A) Prepro-adipokinetic hormone-corazonin-like peptide. (B) Prepro-allatostatin A. (C1) The carboxyl (C)-terminal portion of prepro-allatostatin C I. (C2) Prepro-allatostatin C II. (D) The amino (N)-terminal portion of pre-bursicon α variant (v) 2. (E). Prepro-CCHamide I. (F) Prepro-corazonin. (G) Prepro-crustacean cardioactive peptide. (H). Prepro-diuretic hormone 44. (I1) Pre-eclosion hormone I. (I2) Pre-eclosion hormone II. (J) Prepro-FMRFamide-like peptide. (K) Prepro-intocin. (L1) Prepro-leucokinin v1. (L2) The C-terminal portion of prepro-leucokinin v2. (M) Prepro-myosuppressin. (N1) Pre-neuroparsin I. (N2) Pre-neuroparsin II. (N3) Pre-neuroparsin III. (N4) Pre-neuroparsin IV. (O) Prepro-neuropeptide F splice variant (sv) 2. (P1) The N-terminal portion of prepro-orcokinin II sv2. (P2) The C-terminal portion of prepro-orcokinin I/II. (P3) The C-terminal portion of prepro-orcokinin IV. (Q1) Prepro-pigment dispersing hormone I. (Q2) Prepro-pigment dispersing hormone II. (R) Prepro-proctolin. (S) Prepro-pyrokinin. (T) Prepro-red pigment concentrating hormone. (U) Prepro-RYamide. (V) Prepro-tachykinin-related peptide sv2. In this figure, signal peptides are shown in gray, while all mono/dibasic cleavage loci are shown in black. For each sequence, the isoform(s) of the peptide for which the precursor is named is/are shown in red, with all linker/precursor related peptides shown in blue. The “+” symbol indicate the presence of additional, unknown, amino acid residues at the N- and/or C-termini of the protein in question. It should be noted that there is an N-terminal extension prior to theorized start of the signal peptide in the neuropeptide F and orcokinin precursors shown in O and P1 (highlighted in yellow in each panel). These extensions may be the result of an artifact in the process of assembling the transcript encoding each of these proteins, or may have true biological significance, e.g., they may function as potential regulatory elements. In P1, an isoform of orcomyotropin has been colored green.
3.2.2. Allatostatin A (AST-A)
Two transcripts were identified as encoding putative AST-A precursors (Table 4). Translation of these sequences revealed one to encode the N-terminus and the other the C-terminus of a 561 amino acid full-length preprohormone (Table 4 and Fig. 2B). This protein appears to be a splice variant (sv) of the Homarus AST-A precursor used for identifying the transcripts encoding it (i.e., Homam-prepro-AST-A [termed here sv1] of Christie et al. [2015]; alignment not shown); Homam-prepro-AST-A sv2 (the protein identified here) possesses two copies of the AST-A isoform TGPYAFGLamide that are missing in the sv1 protein. Twenty-three distinct isoforms of AST-A and 14 distinct linker/precursor-related sequences were predicted from Homam-prepro-AST-A sv2 (Table 5 and Fig. 2B), all of which, while known H. americanus peptides (e.g., Christie et al., 2015; Ma et al., 2008), are identified for the first time from the eyestalk ganglia in this study.
3.2.3. Allatostatin B (AST-B)
Three transcripts were identified as encoding putative AST-B precursors (Table 4). Translation of these transcripts revealed that one encodes the N-terminal partial protein, and the two others encode distinct C-terminal partial sequences. Given a common region of overlap between the N-terminal partial protein and two C-terminal partial precursors, two distinct full-length preprohormones can be generated by combining the partial sequences. One full-length precursor is 274 amino acids long, while the other preprohormone is 310 amino acids in length. As the longer protein differs from the shorter only in that it possesses a single 36 amino acid insertion (Fig. 3A), the two precursors appear to be splice variants of a common gene; these precursors are consequently named Homam-prepro-AST-B sv1 (the shorter protein; Table 4 and Fig. 3A) and Homam-prepro-AST-B sv2 (the longer protein; Table 4 and Fig. 3A). Fifteen distinct peptides were predicted from this pair of preprohormones collectively (Table 5 and Fig. 3A); seven of these possess the C-terminal motif −WX6Wamide, the hallmark of the AST-B family (e.g., Christie et al., 2010a). All of the peptides predicted from Homam-prepro-AST-B sv1/sv2 except TNWNKFQGSWamide, an AST-B isoform identified previously via mass spectrometry from the lobster brain, pericardial organ and stomatogastric ganglion (Ma et al., 2008), are novel discoveries for this species. All of these peptides are new discoveries for the H. americanus eyestalk ganglia.
Figure 3.
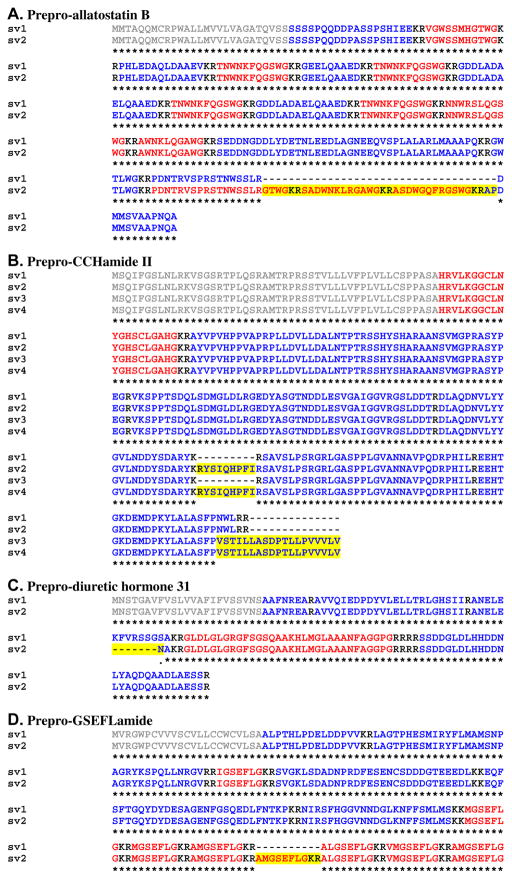
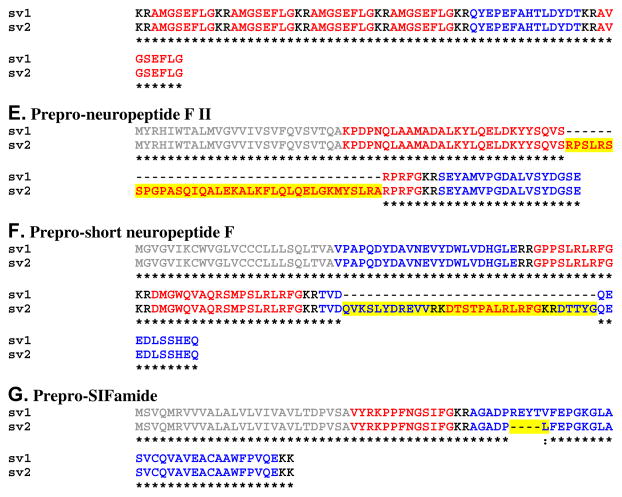
Alignment of Homarus americanus preprohormones for which putative splice variants were identified from eyestalk ganglia transcriptome shotgun assembly data. (A) Prepro-allatostatin B. (B) Prepro-CCHamide II. (C) Prepro-diuretic hormone 31. (D) Prepro-GSEFLamide. (E) Prepro-neuropeptide F II. (F) Prepro-short neuropeptide F. (G) Prepro-SIFamide. In this figure, signal peptides are shown in gray, while all mono/dibasic cleavage loci are shown in black. For each sequence, the isoform(s) of the peptide for which the precursor is named is/are shown in red, with all linker/precursor related peptides shown in blue. Residues (or gaps) that vary from the first splice variant in each alignment are highlighted in yellow. In the line below each sequence grouping, amino acids that are identically conserved are indicated by “*”, while conservative amino acid substitutions are marked by “:” or “.”.
3.2.4. Allatostatin C (AST-C)
Three transcripts were identified as encoding putative AST-C precursors (Table 4). Translation of one of these transcripts revealed a 65 amino acid C-terminal partial protein (Table 4 and Fig. 2C1) that is identical in sequence to a previously identified H. americanus partial precursor (i.e., Homam-prepro-AST-C I of Christie et al. [2015]). Three distinct peptides (one partial and two full-length) were predicted from the extant portion of Homam-prepro-AST-C I (Table 5 and Fig. 2C1), one of which, pQIRYHQCYFNPISCF (disulfide bridging between the two cysteine residues), is a known lobster eyestalk AST-C isoform (Stemmler et al., 2010). The partial and full-length linker/precursor related peptides derived from Homam-prepro-AST-C I, while previously known H. americanus peptides (Christie et al., 2015), are new discoveries for the lobster eyestalk ganglia.
Translation of the second transcript revealed a 105 amino acid full-length preprohormone (Table 4 and Fig. 2C2) that is identical in sequence to a known H. americanus precursor protein, i.e., Homam-prepro-AST-C II (Christie et al., 2015; Dickinson et al., 2009b). Two peptides were predicted from Homam-prepro-AST-C II (Table 5 and Fig. 2C2), one of which, SYWKQCAFNAVSCFamide (disulfide bridging between the two cysteine residues), is a known Homarus eyestalk AST-C isoform (Dickinson et al., 2009b); while previously identified from the lobster, this is the first report of the linker/precursor-related peptide derived from Homam-prepro-AST-C II in the H. americanus eyestalk ganglia.
Translation of the third transcript revealed a 148 amino acid full-length precursor protein that is distinct in sequence from Homam-prepro-AST-C I and II, and for convenience of future discussion has been named Homam-prepro-AST-C III (Table 4 and Fig. 1A). Four distinct peptides were predicted from Homam-prepro-AST-C III (Table 5 and Fig. 1A), one of which, GNGDGRLYWRCYFNAVSCF (disulfide bridging between the two cysteine residues), is a novel H. americanus isoform of AST-C; the three linker/precursor-related peptides derived from Homam-prepro-AST-C III are also new peptide discoveries for the lobster.
3.2.5. Bursicon α
A single transcript was identified as encoding a putative bursicon α precursor (Table 4). Translation of this sequence revealed a 100 amino acid N-terminal partial protein (Homam-pre-bursicon α v2; Table 4 and Fig. 2D). A single partial isoform of bursicon α (81 amino acids in length) was predicted from Homam-pre-bursicon α (Table 5 and Fig. 2D); except for its last three residues, this partial peptide is identical in amino acid sequence to an isoform of bursicon α predicted from a prehormone described in an earlier study (Christie et al., 2015; for convenience of later discussion this earlier identified variant is termed here Homam-pre-bursicon α v1).
3.2.6. CCHamide
Nine transcripts were identified as encoding putative CCHamide precursors (Table 4). Five of the transcripts encode an identical 116 amino acid preprohormone (Homam-prepro-CCHamide I; Table 4 and Fig. 2E); this precursor is identical in sequence to that of a CCHamide I preprohormone identified in a previous study (Christie et al., 2015). Three peptides, including the CCHamide isoform SCSQFGHSCFGAHamide (disulfide bridging between the two cysteine residues), were predicted from Homam-prepro-CCHamide I (Table 5 and Fig. 2E). Although all three peptides were previously identified from the lobster, this is their first identification from the eyestalk ganglia of H. americanus.
Translation of the remaining four transcripts revealed four distinct putative splice variants of a second CCHamide gene (Homam-prepro-CCHamide II sv1–sv4; Table 4 and Fig. 3B). The four CCHamide II precursors differ from one another in two alternatively spliced regions, one involving the presence/absence of a nine amino acid insertion within the preprohormone, and the other involving one of two alternative C-termini, one of which is five amino acids long (the short C-terminus) and the other of which is a 19 amino acid segment (the long C-terminus). Homam-prepro-CCHamide II sv1, which is identical in sequence to a previously identified H. americanus CCHamide II precursor (the tblastn query sequence used for the identification the CCHamide II transcripts identified here from the eyestalk ganglia; Christie et al., 2015), lacks the insertion and possesses the short C-terminus (first row of sequence in Fig. 3B). Homam-prepro-CCHamide II sv2 possesses the nine amino acid insertion and the short C-terminus (second row of sequence in Fig. 3B). Homam-prepro-CCHamide II sv3 lacks the nine amino acid insertion and possesses the long C-terminus (third row of sequence in Fig. 3B). Homam-prepro-CCHamide II sv4 possesses both the nine amino acid insertion and the long C-terminus (fourth row of sequence in Fig. 3B). Eight distinct peptides were predicted from the collective set of Homam-CCHamide II variants (Table 5 and Fig. 3B), six of which, including the CCHamide isoform HRVLKGGCLNYGHSCLGAHamide (disulfide bridging between the two cysteine residues) are known H. americanus peptides (Christie et al., 2015), though identified here for the first time from the eyestalk ganglia. The remaining two peptides, both linker/precursor-related sequences, are new discoveries for H. americanus.
3.2.7. Corazonin
Five transcripts were identified as encoding putative corazonin precursors (Table 4). Translation of four of these revealed identical 109 amino acid full-length precursors (Homam-prepro-corazonin), with the fifth transcript encoding the C-terminus of the full-length protein (Table 4 and Fig. 2F). Homam-prepro-corazonin is identical in sequence to a previously identified H. americanus corazonin precursor (Christie et al., 2015). Three distinct peptides were predicted from Homam-prepro-corazonin (Table 5 and Fig. 2F): pQTFQYSRGWTNamide (authentic corazonin) and two linker/precursor-related sequences. Corazonin is a known H. americanus eyestalk ganglia peptide (Ma et al., 2008); the two linker/precursor-related peptides, while known from the lobster (Christie et al., 2015), are new discoveries for the eyestalk system.
3.2.8. Crustacean cardioactive peptide (CCAP)
Two transcripts were identified as encoding putative CCAP precursors (Table 4). Translation of these transcripts revealed that each encodes an identical 140 amino acid full-length protein (Homam-prepro-CCAP; Table 4 and Fig. 2G); Homam-prepro-CCAP possesses the same amino acid sequences as a known H. americanus CCAP precursor (Christie et al., 2015). Six peptides were predicted from Homam-prepro-CCAP (Table 5 and Fig. 2G), one being authentic CCAP, i.e., PFCNAFTGCamide (disulfide bridging between the two cysteine residues). While all of the peptides derived from Homam-prepro-CCAP have been identified previously from the lobster (e.g., Christie et al., 2015; Ma et al., 2008), this is their first identification from the H. americanus eyestalk ganglia.
3.2.9. Crustacean hyperglycemic hormone (CHH) superfamily
The CHH superfamily of peptides consists of two distinct subgroups, the CHH proper subfamily and the molt-inhibiting hormone (MIH) subfamily (e.g., Böcking et al., 2002; Chan et al., 2003; Chung et al., 2010; Fanjul-Moles, 2006). Members of the CHH subgroup are typically 70 or so amino acids long and possess a stereotypical pattern of disulfide bridges between six identically conserved cysteine residues (namely bridging between the first and fifth, the second and fourth, and the third and sixth cysteines). The preprohormones that give rise to CHH subfamily members also contain a linker/precursor-related peptide between the signal sequence and the CHH, a peptide commonly referred to as crustacean hyperglycemic hormone precursor-related peptide or CPRP. In contrast, members of MIH subgroup are typically longer than the CHHs, their arrangements of disulfide bridging between cysteine residues are more variable than that seen in the CHHs, and the precursors from which they are cleaved lack a CPRP.
3.2.9.1. CHH subgroup
Five transcripts were identified as encoding putative CHH precursors (Table 4). Four of the transcripts encode full-length preprohormones that possess distinct but highly similar amino acid sequences, and have been named here Type I CHH precursors (Table 4 and Fig. 4A). One of the deduced proteins is 134 amino acids in overall length and is identical in sequence to a known H. americanus X-organ-sinus gland system CHH precursor (Accession No. P19806; de Kleijn et al., 1995); for ease of discussion, this preprohormone has been named Homam-prepro-CHH Ia v1 (Table 4 and Fig. 4A). One isoform of CHH and one isoform of CPRP were predicted from Homam-prepro-CHH Ia v1 (Table 5 and Fig. 4A), both known eyestalk system peptides (e.g., de Kleijn et al., 1995). Two 133 amino acid preprohormones are also members of the Type I grouping. The sequences of these two proteins are identical except for a single substituted residue (position 2) in their signal peptides, with that being a methionine in one protein (Homam-prepro-CHH Ia v2; Table 4 and Fig. 4A) and a phenylalanine in the other (Homam-prepro-CHH Ia v3; Table 4 and Fig. 4A); to the best of our knowledge, both of these precursors, while similar to known lobster CHH preprohormones (e.g., Accession No. 2105187B; de Kleijn et al., 1995), are described here for the first time. The CHH and CPRP derived from Homam-prepro-CHH Ia v2 and v3 possess identical structures (Table 5 and Fig. 4A), with the CPRP being one amino acid shorter than that derived from Homam-prepro-CHH Ia v1 (i.e., missing the position 17 asparagine) and the CHH having five substituted positions relative to its Homam-prepro-CHH Ia v1 counterpart. CHH derived from Homam-prepro-CHH Ia v2 and v3 is a known eyestalk system peptide (de Kleijn et al., 1995); the CPRP is a new lobster variant. The final Type I protein deduced here is 131 amino acids long and, while its N-terminus is similar to those of the previously described preprohormones, its C-terminus is quite distinct, and hence it has been named Homam-prepro-CHH Ib (Table 4 and Fig. 4A); with the exception of a 12 amino acid N-terminal extension and a single substituted residue (in the CPRP), this protein is identical to a previously known Homarus CHH precursor and hence has been named Homam-prepro-CHH Ib v2 (Table 4 and Fig. 4A). The CPRP derived from Homam-prepro-CHH Ib v2 is identical to that predicted from Homam-prepro-CHH Ia v2 and v3, while its CHH is distinct from all of those described earlier (Table 5 and Fig. 4A) and is described here from the lobster eyestalk ganglia for the first time here. Regardless of amino acid sequence, each of the CHHs identified in this study was predicted by DiANNA to possess a stereotypical pattern of disulfide bonding, namely bonds between the first and fifth, the second and fourth, and the third and sixth cysteines.
Figure 4.
Alignment of crustacean hyperglycemic hormone (CHH)/molt inhibiting hormone (MIH) precursors. (A) Alignment of prepro-CHH Type I variants. (B) Alignment of CHH Type I and Type II precursors. (C) Alignment of putative MIH prehormones. In this figure, signal peptides are shown in gray, while all mono/dibasic cleavage loci are shown in black. For each sequence, the isoform of CHH/MIH is shown in red, with isoforms of CHH precursor-related peptide shown in pink. Residues (or gaps) that vary between from the top protein in each alignment are highlighted in yellow. In the line below each sequence grouping, amino acids that are identically conserved are indicated by “*”, while conservative amino acid substitutions are marked by “:” or “.”.
The protein deduced from the fifth and final CHH-encoding transcript identified here is 111 amino acids long and, like those described above, is a full-length precursor; this protein is identical in sequence to a recently predicted CHH preprohormone (Christie et al., 2015), with the exception that it is missing a 16 amino acid N-terminal extension. Given its sequence identity to the known precursor (both are likely splice variants of a common gene), but its variation from the other proteins in the Type I series, we have named this preprohormone Homam-prepro-CHH II sv2 (Table 4 and Fig. 4B). One isoform of CPRP and one isoform of CHH were predicted from Homam-prepro-CHH II v2 (Table 5 and Fig. 4B), both distinct from those derived from the members of the Type I precursor series. While known lobster peptides, this is the first description of the CHH and CPRP derived from Homam-prepro-CHH II v2 from the H. americanus eyestalk system. Analysis of the CHH derived from Homam-prepro-CHH II by DiANNA suggest it possesses disulfide bridging between its first and fifth, the second and fourth, and the third and sixth cysteines.
3.2.9.2. Molt-inhibiting hormone (MIH) subgroup
Three transcripts were identified as encoding putative MIH precursors (Table 4). Translation of one of these sequences revealed a 112 amino acid full-length prehormone that is nearly identical in sequence to a known lobster MIH precursor (Accession No. CAA60644; de Kleijn et al., 1994); it differs by a single substituted residue, namely one in the signal peptide portion of the prehormone (an alanine for glycine substitution at position 6). The protein identified here has been named Homam-prepro-MIH I v2 (Table 4 and Fig. 4C). The MIH predicted from Homam-prepro-MIH I v2 (Table 5 and Fig. 4C) is a known H. americanus eyestalk system peptide (de Kleijn et al., 1994). The remaining two transcripts encode novel full-length MIH prehormones of 111 (Homam-pre-MIH II; Table 4 and Fig. 4C) and 119 (Homam-pre-MIH III; Table 4 and Fig. 4C) amino acids. A single isoform of MIH was predicted from Homam-pre-MIH II and Homam-pre-MIH III, each possessing a distinct sequence (Table 5 and Fig. 4C); these two MIHs are new discoveries for H. americanus. While DiANNA analyses suggest that the MIHs derived from Homam-pre-MIH I v2 and Homam-pre-MIH II possess disulfide bonding between their first and fifth, the second and fourth, and the third and sixth cysteines, a different bridging pattern was predicted for the MIH produced from Homam-pre-MIH III, i.e., bridges between its first and third, second and fourth, and fifth and sixth cysteines.
3.2.10. Diuretic hormone 31 (DH31)
Two transcripts were identified as encoding putative DH31 precursors (Table 4). Translation of these transcripts revealed one to encode a 135 amino acid full-length preprohormone and the other a 128 amino acid full-length precursor (Table 4 and Fig. 3C). These two proteins appear to be splice variants of a common gene and hence have been named Homam-prepro-DH31 sv1 and Homam-prepro-DH31 sv2, respectively; the former precursor is identical in sequence to a previously identified H. americanus DH31 preprohormone (Christie et al., 2010c). Six distinct peptides were predicted from Homam-prepro-DH31 sv1/2 (Table 5 and Fig. 3C), including the DH31 isoform GLDLGLGRGFSGSQAAKHLMGLAAANFAGGPamide. This DH31 isoform, and four of the five linker/precursor-related sequences, are previously known lobster peptides (Christie et al., 2010c). However, they are identified here from the H. americanus eyestalk ganglia for the first time. The fifth linker/precursor-related peptide is a new discovery for H. americanus.
3.2.11. Diuretic hormone 44 (DH44)
Two transcripts were identified as encoding putative DH44 precursors (Table 4). Both transcripts encode the same 285 amino acid protein (Table 4 and Fig. 2H), which is identical in sequence to a previously described H. americanus DH44 precursor (Christie et al., 2015). Six distinct peptides were predicted from Homam-prepro-DH44 (Table 5 and Fig. 2H): the DH44 isoform ASGLSLSIDASMKVLREALYMEIIRKKQRQQMQRAQHNQKLLNSIamide and five linker/precursor-related sequences. While all of the peptides predicted from Homam-prepro-DH44 are known lobster peptides (Christie et al., 2015), this is their first identification from the eyestalk ganglia.
3.2.12. Eclosion hormone (EH)
Two transcripts were identified as encoding putative EH precursors (Table 4). Translation of these transcripts revealed one to encode an 88 amino acid full-length protein (Homam-pre-EH I; Table 4 and Fig. I1), and the other, an 82 amino acid prehormone (Homam-pre-EH II; Table 4 and Fig. I2); these proteins are identical in sequence to a pair of EH precursors identified previously from H. americanus (Christie et al., 2015). A single EH isoform was predicted from each protein (Table 5 and Fig. 2I1–I2); these EHs possess distinct structures. Analysis of the EH isoforms derived from both Homam-pre-EH I and Homam-pre-EH II using DiANNA suggests disulfide bridges between the first and second, third and fourth, and fifth and sixth cysteines in it. While both of the predicted EH isoforms are known H. americanus peptides (Christie et al., 2015), this is the first identification of each EH from the eyestalk ganglia.
3.2.13. Elevenin
A single transcript was identified as encoding a putative elevenin precursor (Table 4). This transcript encodes a 127 amino acid full-length protein (Homam-prepro-elevenin; Table 4 and Fig. 5A). Three distinct peptides were predicted from Homam-prepro-elevenin (Table 5 and Fig. 5A). One of these peptides, VDCRKFVFAPVCRGIIA (disulfide bridging between the two cysteines), possesses structural homology to known arthropod elevenin isoforms; it is approximately 72% identical/89% similar in amino acid composition to the elevenin predicted from the Nilaparvata lugens precursor used to identify the transcript encoding Homam-prepro-elevenin. This is the first report of an elevenin from H. americanus.
Figure 5.
Alignments of selected Homarus americanus precursor proteins and the insect queries used for their identifications. (A) Alignment of Nilaparvata lugens (Nillu) and H. americanus (Homam) prepro-elevenins. (B) Alignment of N. lugens and H. americanus glycoprotein hormone α2 (GPα2) precursors. (C) Alignment of N. lugens and H. americanus glycoprotein hormone β5 (GPβ5) precursors. (D) Alignment of Drosophila melanogaster (Drome) and H. americanus prepro-trissins (lobster variant 1 shown). In this figure, signal peptides are shown in gray, while all mono/dibasic cleavage loci are shown in black. For each sequence, the isoform(s) of the peptide for which the precursor is named is/are shown in red, with all linker/precursor related peptides shown in blue. In the line below each sequence grouping, amino acids that are identically conserved are indicated by “*”, while conservative amino acid substitutions are marked by “:” or “.”.
3.2.14. FMRFamide-like peptide (FLP)
Two transcripts were identified as encoding putative FLP precursors (Table 4). Both transcripts encode identical 358 amino acid full-length proteins (Homam-prepro-FLP; Table 4 and Fig. 2J); this protein was identified in a previous study as an H. americanus FLP precursor (Christie et al., 2015). Eighteen distinct peptides were predicted from Homam-prepro-FLP (Table 5 and Fig. 2J), nine of which are FLP isoforms. While all 18 of the peptides predicted from Homam-prepro-FLP have been reported previously from the lobster, this is the first identification of them from the H. americanus eyestalk ganglia.
3.2.15. Glycoprotein hormone
3.2.15.1. α-subunit 2 (GPα2)
One transcript was identified as encoding a putative GPα2 precursor (Table 4). Translation of this transcript revealed a 120 amino acid full-length prehormone (Homam-pre-GPα2; Table 4 and Fig. 5B). A single 104 amino acid isoform of GPα2 was predicted from Homam-pre-GPα2 (Table 5 and Fig. 5B); this peptide shares extensive amino acid conservation with other members of this peptide family, e.g., it is 69% identical/94% similar in sequence to the GPα2 predicted from the N. lugens precursor (Accession No. BAO00955; Tanaka et al., 2014) used to identify the transcript encoding it. Analysis of the H. americanus GPα2 by DiANNA suggests disulfide bridging between the peptide’s first and tenth, second and eighth, third and fifth, fourth and ninth, and sixth and seventh cysteine residues; homology to known GPα2 isoforms (e.g., Sudo et al., 2005) suggest that peptide is also glycosylated, potentially at its position 34 asparagine. This is the first identification of a GPα2 from H. americanus.
3.2.15.2. β-subunit 5 (GPβ5)
Two transcripts were identified as encoding putative GPβ5 precursors (Table 4). Translation of these transcripts revealed each to encode an identical 163 amino acid full-length prehormone (Homam-pre-GPβ5; Table 4 and Fig. 5C). A single 125 amino acid isoform of GPβ5 was predicted from Homam-pre-GPβ5 (Table 5 and Fig. 5C), this GPβ5 is 54% identical/81% similar in sequence to the GPβ5 predicted from the N. lugens precursor (Accession No. BAO00956; Tanaka et al., 2014) used to identify the transcript encoding it. Analysis of this GPβ5 isoform by DiANNA suggests disulfide bridging between its first and tenth, second and fifth, third and fourth, sixth and eighth, and seventh and ninth cysteines. This is the first identification of a GPβ5 from H. americanus.
3.2.16. GSEFLamide
Three transcripts were identified as encoding putative GSEFLamide precursors (Table 4). Two of the transcripts encode N-terminal partial preprohormones of 268 and 278 amino acids, while the third transcript encodes a 35 amino acid C-terminal partial protein, which is identical to the query sequence (Christie et al., 2015) used to identify the transcripts encoding these partial proteins. Two full-length preprohormones can be generated by combining each of the N-terminal fragments with the C-terminal partial sequence. As these two proteins differ from one another only by the presence/absence of a 10 amino acid insertion, they are likely splice variants of a common gene. Given this hypothesis, they have been named here Homam-prepro-GSEFLamide sv1 (Fig. 3D; the shorter precursor), and Homam-prepro-GSEFLamide sv2 (Fig. 3D, the longer precursor). Twelve distinct peptides were predicted from these two proteins (Table 5 and Fig. 3D), six of which are isoforms of GSEFLamide. Two of the GSEFLamides, AMGSEFLamide AVGSEFLamide, and one of the linker/precursor-related peptides, QYEPEFAHTLDYDT, while known lobster peptides (Christie et al., 2015), are new discoveries for the eyestalk ganglia. The remaining nine peptides are described here for H. americanus for the first time.
3.2.17. Intocin
Three transcripts were identified as encoding putative intocin precursors (Table 4). All three transcripts encode a common 154 amino acid protein (Table 4 and Fig. 2K) that is identical in sequence to a previously described H. americanus intocin precursor (Christie et al., 2015). Six distinct peptides were predicted from Homam-intocin (Table 5 and Fig. 2K): CFITNCPPGamide (disulfide bridging between its two cysteine residues) and five linker/precursor-related sequences. All six of the peptides predicted from Homam-intocin are known H. americanus peptides (Christie et al., 2015). However, their descriptions here are the first identifications of them from the lobster eyestalk ganglia.
3.2.18. Leucokinin
Two transcripts were identified as encoding putative leucokinin precursors (Table 4). Translation of one transcript revealed a 606 amino acid full-length preprohormone (Table 4 and Fig. 2L1), while translation of the other revealed a 130 amino acid C-terminal partial protein (Table 4 and Fig. 2L2). The N-terminus of the full-length preprohormone (named here Homam-prepro-leucokinin v2) is identical in sequence, save a single substituted residue, to that of a previously identified H. americanus N-terminal partial leucokinin precursor (Christie et al., 2015; referred to here prepro-leucokinin v1). The C-terminal partial protein discovered here differs from the C-terminus of Homam-prepro-leucokinin v2 at three positions, and has been named Homam-prepro-leucokinin v3. Thirty-two distinct peptides were predicted from the combination of Homam-prepro-leucokinin v2 and v3 (Table 5 and Fig. 2L1–2), 13 of which are isoforms of leucokinin. Of this collective set of peptides, all are new discoveries for the lobster eyestalk ganglia, with 10 being described here for H. americanus for the first time.
3.2.19. Myosuppressin
One transcript was identified as encoding putative myosuppressin precursors (Table 4). Translation of this transcript revealed a 100 amino acid preprohormone (Homam-prepro-myosuppressin; Table 4 and Fig. 2M); this protein is identical in sequence to a previously described H. americanus myosuppressin precursor (Accession No. ACX46385; Stevens et al., 2009), the query sequence used to identify the transcript encoding it. Four peptides were predicted from Homam-prepro-myosuppressin (Table 5 and Fig. 2M): the myosuppressin isoform pQDLDHVFLRFamide and three distinct linker/precursor-related sequences. While all four of the peptides predicted from Homam-prepro-myosuppressin are known lobster peptides (e.g., Christie et al., 2015; Ma et al., 2008), and pQDLDHVFLRFamide was identified from the lobster eyestalk ganglia via mass spectrometry in a previous study (Ma et al., 2008), this is the first identification of the three linker/precursor-related sequences from the eyestalk ganglia.
3.2.20. Neuroparsin
Five transcripts were identified as encoding putative neuroparsin precursors (Table 4). Translation of one transcript revealed a 98 amino acid full-length prehormone (Homam-pre-neuroparsin I; Table 4 and Fig. 2N1); this protein is identical in sequence to a known H. americanus pre-neuroparsin (Christie et al., 2015), which was the query protein used for the identification of neuroparsin-encoding transcripts reported here. A single 74 amino acid neuroparsin isoform was predicted from Homam-pre-neuroparsin I (Table 5 and Fig. 2N1). Analysis of this neuroparsin isoform by DiANNA suggests that disulfide bridges are present between its first and seventh, second and third, fourth and tenth, fifth and eleventh, sixth and eighth, and ninth and twelfth cysteine residues (Table 5). While this is a previously predicted lobster peptide (Christie et al., 2015), this is the first description of the neuroparsin derived from Homam-pre-neuroparsin I from the H. americanus eyestalk ganglia.
Translation of another transcript also revealed a 98 amino acid full-length prehormone (Table 4 and Fig. 2N2); this protein is distinct in amino acid sequence from Homam-pre-neuroparsin I, and thus was named Homam-pre-neuroparsin II. A 76 amino acid isoform of neuroparsin was predicted from this prehormone (Table 5 and Fig. 2N2), with disulfide bridges predicted between its first and eleventh, second and seventh, third and fourth, fifth and twelfth, sixth and eighth, and ninth and tenth cysteines by DiANNA (Table 5); this is the first description of this peptide from H. americanus.
The third neuroparsin transcript encodes a 101 amino acid full-length prehormone (Homam-pre-neuroparsin III; Table 4 and Fig. 2N3). Like Homam-pre-neuroparsin II, a 76 amino acid isoform of neuroparsin was predicted from Homam-pre-neuroparsin III (Table 5 and Fig. 2N3); this peptide was predicted by DiANNA to possess disulfide bridges between its first and twelfth, second and tenth, third and sixth, fourth and fifth, seventh and ninth, and eighth and eleventh cysteines (Table 5). It is a novel H. americanus neuroparsin isoform.
The remaining two transcripts encode identical 103 amino acid full-length prehormones (Homam-pre-neuroparsin IV; Table 4 and Fig. 2N4). Like the neuroparsin precursors described earlier, a single neuroparsin isoform was predicted from Homam-pre-neuroparsin IV (Table 5 and Fig. 2N4). This peptide, which is 77 amino acids long, was predicted by DiANNA to possess disulfide bonding between its first and fourth, second and eighth, third and sixth, fifth and seventh, ninth and twelfth, and tenth and eleventh cysteines (Table 5). The neuroparsin predicted from Homam-pre-neuroparsin IV is a new discovery for H. americanus.
3.2.21. Neuropeptide F (NPF)
Three transcripts were identified as encoding putative NPF precursors (Table 4). Translation of one transcript revealed a 132 amino acid full-length preprohormone (Homam-prepro-NPF I sv2; Table 4 and Fig. 2O). With the exception of a 28 amino acid N-terminal extension, Homam-prepro-NPF I sv2 is identical in sequence to a known H. americanus NPF precursor (Christie et al., 2015), and the two are likely splice variants of a common gene (alignment not shown). For ease of discussion, the precursor described in Christie et al. (2015) has been renamed here Homam-prepro-NPF I sv1; Homam-prepro-NPF I sv1 was the protein query used for the discovery of the NPF-encoding transcripts discovered in this study. Two peptides were predicted from Homam-prepro-NPF I sv2 (Table 5 and Fig. 2O): the NPF isoform ARPDNSAADTLQAIHEAAMAGILGSAEVQYPNRPSMFKSPVELRQYLDALNAYYAIAGRPRFamide and a linker/precursor related sequence, both of which, while previously identified from H. americanus (Christie et al., 2015) are new discoveries for the lobster eyestalk ganglia.
Translation of the remaining two transcripts revealed full-length proteins of 79 and 116 amino acids, which appear to be splice variants of a second NPF gene, and have been named here Homam-prepro-NPF II sv1 (the shorter protein; Table 4 and Fig. 3E) and Homam-prepro-NPF II sv2 (the longer protein; Table 4 and Fig. 3E). Two peptides were predicted from each preprohormone (Table 5 and Fig. 3E): an isoform of NPF and a linker/precursor-related sequence. While the linker/precursor-related peptide is shared by the two splice variants, the NPF isoform derived from Homam-prepro-NPF II sv2 possesses a 37 amino acid insertion relative to the NPF predicted from Homam-prepro-NPF II sv1, i.e., KPDPNQLAAMADALKYLQELDKYYSQVSRPSLRSSPGPASQIQALEKALKFLQLQELGKMYSLRARPRFamide vs. KPDPNQLAAMADALKYLQELDKYYSQVSRPRFamide. All of the peptides predicted from Homam-prepro-NPF II sv1 and sv2 are new discoveries for H. americanus.
3.2.22. Orcokinin
Four transcripts were identified as encoding putative orcokinin precursors (Table 4). Translation of one transcript revealed a 156 amino acid N-terminal partial protein, with a second transcript encoding a portion (152 amino acids) of the same sequence. With the exception of a 30 amino acid N-terminal extension, this partial protein is identical in amino acid sequence to the N-terminus of a known H. americanus orcokinin precursor, namely Homam-prepro-orcokinin II (Accession No. ACD13197; Dickinson et al., 2009a), and likely represents a splice variant of this gene; it was named here Homam-prepro-orcokinin II sv2 (Table 4 and Fig. 2P1). The remaining two transcripts encode C-terminal partial proteins of 118 and 96 amino acids, which also differ from one another at a single residue. The longer C-terminal partial preprohormone is identical in amino acid sequence to the C-termini of two known H. americanus orcokinin precursors, namely Homam-prepro-orcokinin I (Accession No. ACB41787; Dickinson et al., 2009a) and II (see above), and is missing the portion of the protein that allows for differentiating between the two sequences; this partial protein is referred to here as Homam-prepro-orcokinin I/II in Table 4 and Figure 2P2. The other C-terminal partial protein appears to represent the C-terminus of a novel orcokinin precursor and has been named Homam-prepro-orcokinin IV (Table 4 and Fig. 2P3). It should be noted that a protein named Homam-prepro-orcokinin III currently exists in GenBank (Accession No. ACD13198; Dickinson et al., 2009a); although the nucleotide sequence from which Homam-prepro-orcokinin III was deduced is distinct from that encoding Homam-prepro-orcokinin II, (Accession No. ACD13197; Dickinson et al., 2009a), the proteins encoded by these two nucleotide sequences are identical. Nine distinct peptides were predicted from the collective set of orcokinin precursors derived from the eyestalk ganglia (Table 5 and Fig. 2P1–3): three orcokinins, NFDEIDRSGFGFH, NFDEIDRSGFGFN and NFDEIDRSGFGFV, the orcomyotropin FDAFTTGFGHN, and five linker/precursor-related sequences, all of which have been previously described from H. americanus (e.g., Dickinson et al., 2009a; Ma et al., 2008). With the exception of the linker/precursor-related peptides GPIKAAPARSSPQQDAAAGYTDGAPV and SAE, all have also been reported from the lobster eyestalk system (e.g., Dickinson et al., 2009a; Ma et al., 2008).
3.2.23. Pigment dispersing hormone (PDH)
Five transcripts were identified as encoding putative PDH precursors (Table 4). Translation of one transcript revealed a 79 amino acid full-length preprohormone (Homam-prepro-PDH I; Table 4 and Fig. 2Q1). This protein is identical in sequence to a known H. americanus PDH precursor (Christie et al., 2015), which was the query sequence used for the discovery of the PDH-encoding transcripts reported here (alignment not shown). Two peptides were predicted from Homam-prepro-PDH I (Table 5 and Fig. 2Q1): a linker/precursor-related sequence and the PDH isoform NSELINSILGLPKVMNDAamide. Both of the peptides predicted from Homam-prepro-PDH I have been described from the lobster previously (e.g., Christie et al., 2015; Ma et al., 2008), with NSELINSILGLPKVMNDAamide identified from the eyestalk system via mass spectrometry (e.g., Ma et al., 2008).
The remaining four transcripts encode identical 79 amino acid full-length proteins (Homam-prepro-PDH II; Table 4 and Fig. 2Q2); Homam-prepro-PDH II, while similar to Homam-prepro-PDH I in amino acid sequence (alignment not shown), is distinct from it, and likely represents the product of a different gene. Two peptides were predicted from Homam-prepro-PDH II (Table 5 and Fig. 2Q2): a linker/precursor-related sequence and the PDH isoform NSELINSLLGIPKVMNDAamide. Both peptides are distinct in structure from their counterparts derived from Homam-prepro-PDH I, and are new discoveries for H. americanus.
3.2.24. Proctolin
Two transcripts were identified as encoding putative proctolin precursors (Table 4). Translation of these transcripts revealed that they encode identical 88 amino acid full-length preprohormones (Homam-prepro-proctolin; Table 4 and Fig. 2R), which are identical in sequence to the H. americanus prepro-proctolin that was used as the query sequence for the identification of the transcripts encoding them (Christie et al., 2015). Six distinct peptides, including RYLPT, i.e., authentic proctolin, were predicted from Homam-prepro-proctolin (Table 5 and Fig. 2R). While all of the peptides derived Homam-prepro-proctolin were identified previously from H. americanus (e.g., Christie et al., 2015; Ma et al., 2008), they are new discoveries from the lobster eyestalk ganglia.
3.2.25. Pyrokinin
One transcript was identified as encoding a putative pyrokinin precursor (Table 4). This transcript encodes a 385 amino acid full-length preprohormone (Homam-prepro-pyrokinin; Table 4 and Fig. 2S). The partial H. americanus pyrokinin precursor used as the query sequence in the search that identified the transcript encoding Homam-prepro-pyrokinin (Christie et al., 2015) is identical in sequence to the corresponding portion of the full-length protein (alignment not shown), and likely represents an internal fragment of it. Thirteen distinct peptides were predicted from Homam-prepro-pyrokinin, including 10 isoforms of pyrokinin and three linker/precursor-related sequences (Table 5 and Fig. 2S). Seven of the pyrokinins and all three linker/precursor-related peptides have been described previously from H. americanus (Christie et al., 2015), though all are new discoveries for the lobster eyestalk ganglia. The remaining three pyrokinins, ADFAFSPRLamide, DSEDSSVESRNTKTQASIPRPamide and AYFSPRLamide, are new discoveries for H. americanus.
3.2.26. Red pigment concentrating hormone
One transcript was identified as encoding a putative RPCH precursor (Table 4), translation of which revealed a 98 amino acid full-length preprohormone (Homam-prepro-RPCH; Table 4 and Fig. 2T). Three distinct peptides were predicted from Homam-prepro-RPCH (Table 5 and Fig. 2T): pQLNFSPGWamide, authentic RPCH, and two linker/precursor-related sequences. While RPCH has been identified previously from the lobster eyestalk system (e.g., Ma et al., 2008), the two linker/precursor-related peptides predicted from Homam-prepro-RPCH are new discoveries for H. americanus.
3.2.27. RYamide
One transcript was identified as encoding a putative RYamide precursor (Table 4). Translation of this transcript revealed a 135 amino acid full-length preprohormone (Homam-prepro-RYamide; Table 4 and Fig. 2U). Six distinct peptides were predicted from Homam-prepro-RYamide (Table 5 and Fig. 2U), two of which, pQGFYTQRYamide and FIGGSRYamide, possess −RYamide C-termini, the hallmark of the RYamide family (Christie et al., 2010a). All of the peptides derived from Homam-prepro-RYamide are new discoveries for H. americanus.
3.2.28. Short neuropeptide F (sNPF)
Four transcripts were identified as encoding putative sNPF precursors (Table 4). Two of these transcripts encode identical 96 amino acid full-length preprohormones, with the remaining pair encoding identical 128 amino acid full-length precursor proteins. As the two proteins are identical in sequence except for a 32 amino acid insertion/deletion, they likely represent splice variants of a common gene, and hence have been named Homam-prepro-sNPF sv1 (the shorter variant; Table 4 and Fig. 3F) and Homam-prepro-sNPF sv2 (the longer precursor; Table 4 and Fig. 3F). Four distinct peptides were predicted from Homam-prepro-sNPF sv1 (Table 5 and Fig. 3F), two of which possess the structural hallmarks of the sNPF family, i.e., the C-terminal motif −RLRFamide and an overall length of approximately 10 amino acids (Christie et al., 2010a). Six peptides were predicted from Homam-prepro-sNPF v2 (Table 5 and Fig. 3F), three of which, the sNPFs GPPSLRLRFamide and DMGWQVAQRSMPSLRLRFamide and the linker/precursor-related peptide VPAPQDY(SO3H)DAVNEVYDWLVDHGLE, are shared with Homam-prepro-sNPF sv1. The three remaining peptides derived from Homam-prepro-sNPF v2 include the sNPF isoform DTSTPALRLRFamide and two distinct linker/precursor-related sequences. Two of the sNPFs, GPPSLRLRFamide and DTSTPALRLRFamide, while known lobster peptides (Ma et al., 2008), are new discoveries for the eyestalk ganglia. The remaining sNPF and all of the linker/precursor-related peptides are new discoveries for H. americanus in a general sense.
3.2.29. SIFamide
Two transcripts were identified as encoding putative SIFamide precursors (Table 4). Translation of one transcript revealed an 80 amino acid full-length preprohormone, while translation of the other sequence revealed a 76 amino acid full-length precursor protein (Table 4 and Fig. 3G). The longer preprohormone is identical in sequence to a known SIFamide precursor (Accession No. ABV21807; Dickinson et al., 2008), while the shorter protein differs from it by a four amino acid deletion and one substituted residue (Fig. 3G); both are considered here to be splice variants of a common gene. For ease of discussion, the longer protein has been named Homam-prepro-SIFamide sv1 and the shorter, Homam-prepro-SIFamide sv2. Two peptides were predicted from each of the two Homarus SIFamide precursors (Table 5 and Fig. 3G); both preprohormones share the SIFamide isoform VYRKPPFNGSIFamide, a known peptide from the lobster eyestalk ganglia (e.g., Ma et al., 2008). However, they possess distinct linker/precursor-related peptides, i.e., AGADPREYTVFEPGKGLASVCQVAVEACAAWFPVQE, a known H. americanus peptide (Dickinson et al., 2008), which is described here for the first time from the eyestalk ganglia, and AGADPLFEPGKGLASVCQVAVEACAAWFPVQE, which has not previously been identified in the lobster.
3.2.30. Sulfakinin
One transcript was identified as encoding a putative sulfakinin precursor (Table 4). Translation of this transcript revealed a 120 amino acid full-length precursor protein (Homam-prepro-sulfakinin; Table 4 and Fig. 1B); Homam-prepro-sulfakinin is identical in sequence to a known lobster sulfakinin precursor (Accession No. ABQ95346; Dickinson et al., 2007). Five distinct peptides were predicted from Homam-prepro-sulfakinin (Table 5 and Fig. 1B), two of which, pEFDEY(SO3H)GHMRFamide and GGGEY(SO3H)DDY(SO3H)GHLRFamide, are isoforms of sulfakinin. While all of the peptides predicted from Homam-prepro-sulfakinin have been described previously from the lobster (Dickinson et al., 2007), they are new discoveries for the eyestalk ganglia.
3.2.31. Tachykinin-related peptide (TRP)
One transcript was identified as encoding a putative TRP precursor (Table 4). Translation of this transcript revealed a 229 amino acid full-length preprohormone (Table 4). With the exception of a missing 20 amino acid N-terminal extension in its signal peptide, this protein is identical in amino acid sequence to a known lobster TRP precursor (Accession No. ACB41786; Christie et al., 2008a), and the two precursors are likely splice variants of a common gene. For ease of later discussion, the previously known preprohormone has been termed Homam-prepro-TRP sv1, while the precursor predicted here has been termed Homam-prepro-TRP sv2 (Fig. 2V). Seven distinct peptides were predicted from Homam-prepro-TRP sv2 (Table 5 and Fig. 2V), two of which, APSGFLGMRamide and TPSGFLGMRamide, are isoforms of TRP. The TRP isoform APSGFLGMRamide has been identified previously from the eyestalk ganglia of H. americanus (Ma et al., 2008); the remaining peptides derived from Homam-prepro-TRP sv2, while known from the lobster (Christie et al., 2008a), are new discoveries from the eyestalk system.
3.2.32. Trissin
Two transcripts were identified as encoding putative trissin precursors (Table 4). Translation of these transcripts revealed each to encode a full-length preprohormone, with one protein being 187 amino acids in length, and the other 186 amino acids long. The two precursors are identical in amino acid sequences except that the shorter protein is missing the position 61 serine of the longer sequence. The two proteins have been named Homam-prepro-trissin v1 (Table 4 and Fig. 5D; the longer sequence) and Homam-prepro-trissin v2 (Table 4 and Fig. 5D; the shorter sequence). Three peptides were predicted from each trissin precursor (Table 5 and Fig. 5D), two of which, the putative trissin isoform WSSSEVSCTSCGSECQSACGTRNFRACCFNFQ (disulfide bridges predicted by DiANNA between the first and sixth, second and fourth, and third and fifth cysteine residues) and the linker/precursor-related peptide PSPSLNQLQHQNLHQRYTPSPTSIKI, are present in both preprohormones. The third peptide derived from Homam-prepro-trissin v1 and v2 is a linker/precursor-related sequence; the peptides derived from Homam-prepro-trissin v1 and v2 differ from one another in the presence/absence of the serine residue discussed earlier. The H. americanus trissin isoform is approximately 50% identical/75% similar in amino acid composition to the trissin derived from the Drosophila melanogaster precursor (Accession No. AAF55203; Adams et al., 2000) used to identify the transcripts encoding the two lobster trissin preprohormones. To the best of our knowledge, this is the first report of trissin (and trissin linker/precursor-related peptides) from H. americanus.
4. Discussion
4.1. Development of new molecular resources for investigations of peptidergic signaling in Homarus americanus
In the study presented here, high-throughput nucleotide sequencing was conducted on the eyestalk ganglia of the lobster H. americanus using the Illumina NextSeq platform. This sequencing resulted in the generation of approximately 131,000,000 high-quality reads, which were de novo assembled into a transcriptome consisting of ~150,000 unique contigs. While other nucleotide data sets have been generated for H. americanus (e.g., McGrath et al., 2016; Stepanyan et al., 2006; Towle and Smith, 2006), including neural-specific ones (e.g., BioProject No. PRJNA300643; D. Schulz and E. Marder, unpublished direct GenBank submission), the transcriptome generated here is by far the largest currently extant for this species, and is the only one that includes the eyestalk ganglia, the location of X organ-sinus gland complex, a major neuroendocrine center in this commercially and biomedically important decapod (e.g., Christie, 2011). The public deposition of this eyestalk transcriptome, as well as the raw reads from which it was generated, should provide a powerful resource for investigations of neural control in the lobster, including, but certainly not limited to, the peptidergic modulation of physiology and behavior that was the impetus for this study.
4.2. The peptidome of the lobster eyestalk ganglia contains members of most known crustacean peptide families
Via the mining of the eyestalk ganglia-specific transcriptome developed here, ~90 neuropeptide encoding transcripts were identified. Analyses of the proteins deduced from these transcripts allowed for the prediction of 262 distinct neuropeptides. This eyestalk peptidome includes isoforms of ACP, AST-A, AST-B, AST-C, bursicon α, CCHamide, corazonin, CCAP, CHH, CPRP, DH31, DH44, EH, elevenin, FLP, GPα2, GPβ5, GSEFLamide, intocin, leucokinin, MIH, myosuppressin, neuroparsin, NPF, orcokinin, orcomyotropin, PDH, proctolin, pyrokinin, RPCH, RYamide, sNPF, SIFamide, sulfakinin, TRP and trissin, as well as a large number of linker/precursor-related peptides. Of the identified peptides, 19 are reidentifications of known H. americanus eyestalk peptides, 165 are ones previously described from the lobster, but new discoveries from the eyestalk ganglia, and 78 are new to the lobster in a general sense (see Table 5). All members of the elevenin, GPα2, GPβ5, RYamide and trissin families are included in the latter grouping, being the first isoforms of these peptide families identified from H. americanus.
Although they were searched for, no transcripts encoding members of 13 other peptide families were found in the eyestalk transcriptome (see Table 4). Of these 13 families, isoforms of only two, bursicon β and insulin-like peptide (ILP), have been identified in the lobster (e.g., Christie et al., 2015). It is possible that our lack of detection of bursicon β- and ILP-encoding transcripts (as well as those for members of the other 11 families), is due to the eyestalk system not possessing members of these peptide families. Alternatively, our lack of detection of transcripts for some or all of these peptide groups could be a result of incomplete coverage of the transcriptome that was mined. As additional molecular data are generated for the H. americanus eyestalk system, it will be interesting to see if members of these peptide families can be identified from this portion of the nervous system or if these groups are truly absent from the eyestalk ganglia.
4.3. Local modulatory vs. hormonal roles for Homarus americanus eyestalk peptides
The eyestalks of lobsters are complex structures; in addition to their functions in vision, they are involved in the control of reproduction and are the site of the X-organ-sinus gland complex, a major neuroendocrine organ (e.g., Christie, 2011). Because the eyestalk transcriptomes used in this study included all parts of the eyestalk ganglia (lamina ganglionaris, medulla externa, medulla interna and medulla terminalis), the roles played by the peptides we have identified are likely diverse. First, they could exert their effects within the eyestalk ganglia themselves, modulating the neuronal output of these ganglia and the interactions that take place within them. Future research examining the peptide receptors located within the eyestalk ganglia transcriptome could help shed light on which neuropeptides are likely to exert such local modulatory effects.
In addition to local regulation, the neuropeptides identified here could serve as neurohormones, exerting their effects on other parts of the nervous system or on other tissues. Peptides synthesized within the X-organ are released from the sinus gland into the hemolymph, allowing them to exert effects throughout the body. The present study does not distinguish those peptides that are synthesized in the X-organ-sinus gland complex from those that are present in other parts of the eyestalk ganglia, and so cannot determine which peptides might be released from the sinus gland. However, previous mass spectral studies (e.g., Fu et al., 2005; Ma et al., 2008) have shown that a number of peptides identified here (see peptides highlighted in yellow in Table 5), for example, the orcokinins, orcomyotropin and many of the orcokinin linker/precursor-related peptides, are present in the lobster sinus gland. As additional studies using mass spectrometry are conducted, it will be interesting to see what additional peptides discovered here are also present in the sinus gland of H. americanus and which are absent from this neuroendocrine organ; such studies will help clarify which peptides are local modulators, which are neurohormones, and which are likely to serve both roles.
4.4. Conclusions and future directions
This study has significantly expanded both the number of individual peptides and the number of families of peptides known to be present in the H. americanus eyestalk ganglia. The peptides identified here provide a rich resource for examining local control within the eyestalk ganglia as well as hormonal control of other tissues, including the highly modulated neuronal networks that control rhythmic movements in the lobster. These include the stomatogastric nervous system, which controls the foregut, and the cardiac ganglion, which controls the rhythmic contractions of the heart. Both of these neuronal networks are bathed in hemolymph, the cardiac ganglion due to its location within the lumen of the heart and the stomatogastric system due to the location of the stomatogastric ganglion within the ophthalmic artery. A number of the peptides identified here from the eyestalk ganglion (e.g., RPCH) are known to modulate one or both of these networks in members of the Decapoda (e.g., Cruz-Bermúdez and Marder, 2007; Thirumalai et al., 2006); the present study identifies others whose modulatory effects are presently unknown, but could be examined to increase our understanding of neurohormonal modulatory systems.
Acknowledgments
Financial support for this study was provided by the National Science Foundation (IOS-1353023, IOS-1354567 and OCE-1459235), the National Institutes of Health (5P20RR016463-12 and 8P20GM103423-12), the Cades Foundation of Honolulu, Hawaii, the University of Hawaii at Manoa's Undergraduate Research Opportunities Program, the American Physiological Society Undergraduate Research Fellowship Program, and a gift from the Henry L. and Grace Doherty Charitable Foundation to Bowdoin College.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Blitz DM, Nusbaum MP. Neural circuit flexibility in a small sensorimotor system. Curr Opin Neurobiol. 2011;21:544–552. doi: 10.1016/j.conb.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking D, Dircksen H, Keller R. The crustacean neuropeptide of the CHH/MIH/GIH family: structures and biological activities. In: Wiese K, editor. The Crustacean Nervous System. Springer; Heidelberg: 2002. pp. 84–97. [Google Scholar]
- Cape SS, Rehm KJ, Ma M, Marder E, Li L. Mass spectral comparison of the neuropeptide complement of the stomatogastric ganglion and brain in the adult and embryonic lobster, Homarus americanus. J Neurochem. 2008;105:690–702. doi: 10.1111/j.1471-4159.2007.05154.x. [DOI] [PubMed] [Google Scholar]
- Chan SM, Gu PL, Chu KH, Tobe SS. Crustacean neuropeptide genes of the CHH/MIH/GIH family: implications from molecular studies. Gen Comp Endocrinol. 2003;134:214–219. doi: 10.1016/s0016-6480(03)00263-6. [DOI] [PubMed] [Google Scholar]
- Chang ES, Prestwich GD, Bruce MJ. Amino acid sequence of a peptide with both molt-inhibiting and hyperglycemic activities in the lobster, Homarus americanus. Biochem Biophys Res Commun. 1990;171:818–826. doi: 10.1016/0006-291x(90)91219-i. [DOI] [PubMed] [Google Scholar]
- Chen R, Jiang X, Conaway MC, Mohtashemi I, Hui L, Viner R, Li L. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J Proteome Res. 2010;9:818–832. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE. Neuropeptide discovery in Ixodoidea: an in silico investigation using publicly accessible expressed sequence tags. Gen Comp Endocrinol. 2008a;157:174–185. doi: 10.1016/j.ygcen.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Christie AE. In silico analyses of peptide paracrines/hormones in Aphidoidea. Gen Comp Endocrinol. 2008;159:67–79. doi: 10.1016/j.ygcen.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Christie AE. Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res. 2011;345:41–67. doi: 10.1007/s00441-011-1183-9. [DOI] [PubMed] [Google Scholar]
- Christie AE. Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen Comp Endocrinol. 2014a;206:235–254. doi: 10.1016/j.ygcen.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Christie AE. Prediction of the first neuropeptides from a member of the Remipedia (Arthropoda, Crustacea) Gen Comp Endocrinol. 2014b;201:74–86. doi: 10.1016/j.ygcen.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Christie AE. Prediction of the peptidomes of Tigriopus californicus and Lepeophtheirus salmonis (Copepoda, Crustacea) Gen Comp Endocrinol. 2014c;201:87–106. doi: 10.1016/j.ygcen.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Christie AE. Peptide discovery in the ectoparasitic crustacean Argulus siamensis: identification of the first neuropeptides from a member of the Branchiura. Gen Comp Endocrinol. 2014d;204:114–125. doi: 10.1016/j.ygcen.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Christie AE. In silico characterization of the peptidome of the sea louse Caligus rogercresseyi (Crustacea, Copepoda) Gen Comp Endocrinol. 2014e;204:248–260. doi: 10.1016/j.ygcen.2014.05.031. [DOI] [PubMed] [Google Scholar]
- Christie AE. Identification of the first neuropeptides from the Amphipoda (Arthropoda, Crustacea) Gen Comp Endocrinol. 2014f;206:96–110. doi: 10.1016/j.ygcen.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Christie AE. In silico characterization of the neuropeptidome of the Western black widow spider Latrodectus hesperus. Gen Comp Endocrinol. 2015a;210:63–80. doi: 10.1016/j.ygcen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Christie AE. Neuropeptide discovery in Eucyclops serrulatus (Crustacea, Copepoda): in silico prediction of the first peptidome for a member of the Cyclopoida. Gen Comp Endocrinol. 2015b;211:92–105. doi: 10.1016/j.ygcen.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Christie AE. Neuropeptide discovery in Symphylella vulgaris (Myriapoda, Symphyla): In silico prediction of the first myriapod peptidome. Gen Comp Endocrinol. 2015c;223:73–86. doi: 10.1016/j.ygcen.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Christie AE. In silico prediction of a neuropeptidome for the eusocial insect Mastotermes darwiniensis. Gen Comp Endocrinol. 2015d;224:69–83. doi: 10.1016/j.ygcen.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Christie AE. Prediction of Scylla olivacea (Crustacea; Brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen Comp Endocrinol. 2016a;230–231:1–16. doi: 10.1016/j.ygcen.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Christie AE. Expansion of the neuropeptidome of the globally invasive marine crab Carcinus maenas. Gen Comp Endocrinol. 2016b;235:150–169. doi: 10.1016/j.ygcen.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Christie AE, Cashman CR, Brennan HR, Ma M, Sousa GL, Li L, Stemmler EA, Dickinson PS. Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen Comp Endocrinol. 2008b;156:246–264. doi: 10.1016/j.ygcen.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Christie AE, Cashman CR, Stevens JS, Smith CM, Beale KM, Stemmler EA, Greenwood SJ, Towle DW, Dickinson PS. Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus. Peptides. 2008a;29:1909–1918. doi: 10.1016/j.peptides.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chapline MC, Jackson JM, Dowda JK, Hartline N, Malecha SR, Lenz PH. Identification, tissue distribution and orexigenic activity of neuropeptide F (NPF) in penaeid shrimp. J Exp Biol. 2011a;214:1386–1396. doi: 10.1242/jeb.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Chi M. Neuropeptide discovery in the Araneae (Arthropoda, Chelicerata, Arachnida): elucidation of true spider peptidomes using that of the Western black widow as a reference. Gen Comp Endocrinol. 2015a;213:90–109. doi: 10.1016/j.ygcen.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M. Identification of the first neuropeptides from the enigmatic hexapod order Protura. Gen Comp Endocrinol. 2015b;224:18–37. doi: 10.1016/j.ygcen.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M. Prediction of the neuropeptidomes of members of the Astacidea (Crustacea, Decapoda) using publicly accessible transcriptome shotgun assembly (TSA) sequence data. Gen Comp Endocrinol. 2015c;224:38–60. doi: 10.1016/j.ygcen.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M, Lameyer TJ, Pascual MG, Shea DN, Stanhope ME, Schulz DJ, Dickinson PS. Neuropeptidergic signaling in the American lobster Homarus americanus: new insights from high-throughput nucleotide sequencing. PLoS One. 2015;10:e0145964. doi: 10.1371/journal.pone.0145964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Durkin CS, Hartline N, Ohno P, Lenz PH. Bioinformatic analyses of the publicly accessible crustacean expressed sequence tags (ESTs) reveal numerous novel neuropeptide-encoding precursor proteins, including ones from members of several little studied taxa. Gen Comp Endocrinol. 2010b;167:164–178. doi: 10.1016/j.ygcen.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Christie AE, McCoole MD, Harmon SM, Baer KN, Lenz PH. Genomic analyses of the Daphnia pulex peptidome. Gen Comp Endocrinol. 2011d;171:131–150. doi: 10.1016/j.ygcen.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Christie AE, Nolan DH, Garcia ZA, McCoole MD, Harmon SM, Congdon-Jones B, Ohno P, Hartline N, Congdon CB, Baer KN, Lenz PH. Bioinformatic prediction of arthropod/nematode-like peptides in non-arthropod, non-nematode members of the Ecdysozoa. Gen Comp Endocrinol. 2011c;170:480–486. doi: 10.1016/j.ygcen.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Christie AE, Nolan DH, Ohno P, Hartline N, Lenz PH. Identification of chelicerate neuropeptides using bioinformatics of publicly accessible expressed sequence tags. Gen Comp Endocrinol. 2011b;170:144–155. doi: 10.1016/j.ygcen.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Christie AE, Pascual MG. Peptidergic signaling in the crab Cancer borealis: tapping the power of transcriptomics for neuropeptidome expansion. Gen Comp Endocrinol. 2016 doi: 10.1016/j.ygcen.2016.08.002. In press. [DOI] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Wu LS, Ganote CL, Doak T, Lenz PH. Peptidergic signaling in Calanus finmarchicus (Crustacea, Copepoda): in silico identification of putative peptide hormones and their receptors using a de novo assembled transcriptome. Gen Comp Endocrinol. 2013;187:117–135. doi: 10.1016/j.ygcen.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Christie AE, Stemmler EA, Dickinson PS. Crustacean neuropeptides. Cell Mol Life Sci. 2010a;67:4135–4169. doi: 10.1007/s00018-010-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Stemmler EA, Peguero B, Messinger DI, Provencher HL, Scheerlinck P, Hsu YW, Guiney ME, de la Iglesia HO, Dickinson PS. Identification, physiological actions, and distribution of VYRKPPFNGSIFamide (Val1)-SIFamide) in the stomatogastric nervous system of the American lobster Homarus americanus. J Comp Neurol. 2006;496:406–421. doi: 10.1002/cne.20932. [DOI] [PubMed] [Google Scholar]
- Christie AE, Stevens JS, Bowers MR, Chapline MC, Jensen DA, Schegg KM, Goldwaser J, Kwiatkowski MA, Pleasant TK, Jr, Shoenfeld L, Tempest LK, Williams CR, Wiwatpanit T, Smith CM, Beale KM, Towle DW, Schooley DA, Dickinson PS. Identification of a calcitonin-like diuretic hormone that functions as an intrinsic modulator of the American lobster, Homarus americanus, cardiac neuromuscular system. J Exp Biol. 2010c;213:118–127. doi: 10.1242/jeb.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JS, Zmora N, Katayama H, Tsutsui N. Crustacean hyperglycemic hormone (CHH) neuropeptides family: functions, titer, and binding to target tissues. Gen Comp Endocrinol. 2010;166:447–454. doi: 10.1016/j.ygcen.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Cooke IM. Reliable, responsive pacemaking and pattern generation with minimal cell numbers: the crustacean cardiac ganglion. Biol Bull. 2002;202:108–136. doi: 10.2307/1543649. [DOI] [PubMed] [Google Scholar]
- Cruz-Bermúdez ND, Marder E. Multiple modulators act on the cardiac ganglion of the crab, Cancer borealis. J Exp Biol. 2007;210:2873–2884. doi: 10.1242/jeb.002949. [DOI] [PubMed] [Google Scholar]
- de Kleijn DP, de Leeuw EP, van den Berg MC, Martens GJ, van Herp F. Cloning and expression of two mRNAs encoding structurally different crustacean hyperglycemic hormone precursors in the lobster Homarus americanus. Biochim Biophys Acta. 1995;1260:62–66. doi: 10.1016/0167-4781(94)00173-z. [DOI] [PubMed] [Google Scholar]
- de Kleijn DP, Sleutels FJ, Martens GJ, Van Herp F. Cloning and expression of mRNA encoding prepro-gonad-inhibiting hormone (GIH) in the lobster Homarus americanus. FEBS Lett. 1994;353:255–258. doi: 10.1016/0014-5793(94)01055-2. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Stemmler EA, Barton EE, Cashman CR, Gardner NP, Rus S, Brennan HR, McClintock TS, Christie AE. Molecular, mass spectral, and physiological analyses of orcokinins and orcokinin precursor-related peptides in the lobster Homarus americanus and the crayfish Procambarus clarkii. Peptides. 2009a;30:297–317. doi: 10.1016/j.peptides.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Stemmler EA, Cashman CR, Brennan HR, Dennison B, Huber KE, Peguero B, Rabacal W, Goiney CC, Smith CM, Towle DW, Christie AE. SIFamide peptides in clawed lobsters and freshwater crayfish (Crustacea, Decapoda, Astacidea): a combined molecular, mass spectrometric and electrophysiological investigation. Gen Comp Endocrinol. 2008;156:347–360. doi: 10.1016/j.ygcen.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Stevens JS, Rus S, Brennan HR, Goiney CC, Smith CM, Li L, Towle DW, Christie AE. Identification and cardiotropic actions of sulfakinin peptides in the American lobster Homarus americanus. J Exp Biol. 2007;210:2278–2289. doi: 10.1242/jeb.004770. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Wiwatpanit T, Gabranski ER, Ackerman RJ, Stevens JS, Cashman CR, Stemmler EA, Christie AE. Identification of SYWKQCAFNAVSCFamide: a broadly conserved crustacean C-type allatostatin-like peptide with both neuromodulatory and cardioactive properties. J Exp Biol. 2009b;212:1140–1152. doi: 10.1242/jeb.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dircksen H, Neupert S, Predel R, Verleyen P, Huybrechts J, Strauss J, Hauser F, Stafflinger E, Schneider M, Pauwels K, Schoofs L, Grimmelikhuijzen CJ. Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J Proteome Res. 2011;10:4478–4504. doi: 10.1021/pr200284e. [DOI] [PubMed] [Google Scholar]
- Fanjul-Moles ML. Biochemical and functional aspects of crustacean hyperglycemic hormone in decapod crustaceans: review and update. Comp Biochem Physiol C Toxicol Pharmacol. 2006;142:390–400. doi: 10.1016/j.cbpc.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Fénelon V, Le Feuvre Y, Bem T, Meyrand P. Maturation of rhythmic neural network: role of central modulatory inputs. J Physiol Paris. 2003;97:59–68. doi: 10.1016/j.jphysparis.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Ferrè F, Clote P. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 2005;33:W230–W232. doi: 10.1093/nar/gki412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Goy MF, Li L. Identification of neuropeptides from the decapod crustacean sinus glands using nanoscale liquid chromatography tandem mass spectrometry. Biochem Biophys Res Commun. 2005;337:765–778. doi: 10.1016/j.bbrc.2005.09.111. [DOI] [PubMed] [Google Scholar]
- Gard AL, Lenz PH, Shaw JR, Christie AE. Identification of putative peptide paracrines/hormones in the water flea Daphnia pulex (Crustacea; Branchiopoda; Cladocera) using transcriptomics and immunohistochemistry. Gen Comp Endocrinol. 2009;160:271–287. doi: 10.1016/j.ygcen.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SL, DiCaprio RA. Crustacean motor pattern generator networks. Neurosignals. 2004;13:50–69. doi: 10.1159/000076158. [DOI] [PubMed] [Google Scholar]
- Jiang X, Chen R, Wang J, Metzler A, Tlusty M, Li L. Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. ACS Chem Neurosci. 2012;3:439–450. doi: 10.1021/cn200107v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E. Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J Comp Neurol. 2002;444:227–244. doi: 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- Ma M, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, Henry RP, Smith CM, Towle DW, Christie AE, Li L. Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen Comp Endocrinol. 2009b;161:320–334. doi: 10.1016/j.ygcen.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen Comp Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Gard AL, Xiang F, Wang J, Davoodian N, Lenz PH, Malecha SR, Christie AE, Li L. Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides. 2010;31:27–43. doi: 10.1016/j.peptides.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Szabo TM, Jia C, Marder E, Li L. Mass spectrometric characterization and physiological actions of novel crustacean C-type allatostatins. Peptides. 2009a;30:1660–1668. doi: 10.1016/j.peptides.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- Marder E, Christie AE, Kilman VL. Functional organization of cotransmission systems: lessons from small nervous systems. Invert Neurosci. 1995;1:105–112. doi: 10.1007/BF02331908. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez F, Durán-Gutiérrez D, Delaye L, Becerra A, Aguilar G, Zinker S. Loss of DNA: a plausible molecular level explanation for crustacean neuropeptide gene evolution. Peptides. 2007;28:76–82. doi: 10.1016/j.peptides.2006.09.021. [DOI] [PubMed] [Google Scholar]
- McGrath LL, Vollmer SV, Kaluziak ST, Ayers J. De novo transcriptome assembly for the lobster Homarus americanus and characterization of differential gene expression across nervous system tissues. BMC Genomics. 2016;17:63. doi: 10.1186/s12864-016-2373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E. The roles of co-transmission in neural network modulation. Trends Neurosci. 2001;24:146–154. doi: 10.1016/s0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Schwarz TL, Lee GM, Siwicki KK, Standaert DG, Kravitz EA. Proctolin in the lobster: the distribution, release, and chemical characterization of a likely neurohormone. J Neurosci. 1984;4:1300–1311. doi: 10.1523/JNEUROSCI.04-05-01300.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selverston AI. A neural infrastructure for rhythmic motor patterns. Cell Mol Neurobiol. 2005;25:223–244. doi: 10.1007/s10571-005-3154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selverston AI, Ayers J. Oscillations and oscillatory behavior in small neural circuits. Biol Cybern. 2006;95:537–554. doi: 10.1007/s00422-006-0125-1. [DOI] [PubMed] [Google Scholar]
- Selverston A, Elson R, Rabinovich M, Huerta R, Abarbanel H. Basic principles for generating motor output in the stomatogastric ganglion. Ann NY Acad Sci. 1998;860:35–50. doi: 10.1111/j.1749-6632.1998.tb09037.x. [DOI] [PubMed] [Google Scholar]
- Skiebe P. Neuropeptides are ubiquitous chemical mediators: Using the stomatogastric nervous system as a model system. J Exp Biol. 2001;204:2035–2048. doi: 10.1242/jeb.204.12.2035. [DOI] [PubMed] [Google Scholar]
- Soyez D, Le Caer JP, Noel PY, Rossier J. Primary structure of two isoforms of the vitellogenesis inhibiting hormone from the lobster Homarus americanus. Neuropeptides. 1991;20:25–32. doi: 10.1016/0143-4179(91)90036-i. [DOI] [PubMed] [Google Scholar]
- Stein W. Modulation of stomatogastric rhythms. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195:989–1009. doi: 10.1007/s00359-009-0483-y. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Bruns EA, Cashman CR, Dickinson PS, Christie AE. Molecular and mass spectral identification of the broadly conserved decapod crustacean neuropeptide pQIRYHQCYFNPISCF: the first PISCF-allatostatin (Manduca sexta- or C-type allatostatin) from a non-insect. Gen Comp Endocrinol. 2010;165:1–10. doi: 10.1016/j.ygcen.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, Christie AE. High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides. 2007;28:2104–2115. doi: 10.1016/j.peptides.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Gardner NP, Guiney ME, Bruns EA, Dickinson PS. The detection of red pigment-concentrating hormone (RPCH) in crustacean eyestalk tissues using matrix-assisted laser desorption/ionization-Fourier transform mass spectrometry: [M + Na]+ ion formation in dried droplet tissue preparations. J Mass Spectrom. 2006;41:295–311. doi: 10.1002/jms.989. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Provencher HL, Guiney ME, Gardner NP, Dickinson PS. Matrix-assisted laser desorption/ionization fourier transform mass spectrometry for the identification of orcokinin neuropeptides in crustaceans using metastable decay and sustained off-resonance irradiation. Anal Chem. 2005;77:3594–3606. doi: 10.1021/ac0502347. [DOI] [PubMed] [Google Scholar]
- Stepanyan R, Day K, Urban J, Hardin DL, Shetty RS, Derby CD, Ache BW, McClintock TS. Gene expression and specificity in the mature zone of the lobster olfactory organ. Physiol Genomics. 2006;25:224–233. doi: 10.1152/physiolgenomics.00276.2005. [DOI] [PubMed] [Google Scholar]
- Stevens JS, Cashman CR, Smith CM, Beale KM, Towle DW, Christie AE, Dickinson PS. The peptide hormone pQDLDHVFLRFamide (crustacean myosuppressin) modulates the Homarus americanus cardiac neuromuscular system at multiple sites. J Exp Biol. 2009;212:3961–3976. doi: 10.1242/jeb.035741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo S, Kuwabara Y, Park JI, Hsu SY, Hsueh AJ. Heterodimeric fly glycoprotein hormone-α2 (GPA2) and glycoprotein hormone-β5 (GPB5) activate fly leucine-rich repeat-containing G protein-coupled receptor-1 (DLGR1) and stimulation of human thyrotropin receptors by chimeric fly GPA2 and human GPB5. Endocrinology. 2005;146:3596–3604. doi: 10.1210/en.2005-0317. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Suetsugu Y, Yamamoto K, Noda H, Shinoda T. Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides. 2014;53:125–133. doi: 10.1016/j.peptides.2013.07.027. [DOI] [PubMed] [Google Scholar]
- Tensen CP, De Kleijn DP, Van Herp F. Cloning and sequence analysis of cDNA encoding two crustacean hyperglycemic hormones from the lobster Homarus americanus. Eur J Biochem. 1991;200:103–106. doi: 10.1111/j.1432-1033.1991.tb21054.x. [DOI] [PubMed] [Google Scholar]
- Thirumalai V, Prinz AA, Johnson CD, Marder E. Red pigment concentrating hormone strongly enhances the strength of the feedback to the pyloric rhythm oscillator but has little effect on pyloric rhythm period. J Neurophysiol. 2006;95:1762–1770. doi: 10.1152/jn.00764.2005. [DOI] [PubMed] [Google Scholar]
- Torfs P, Baggerman G, Meeusen T, Nieto J, Nachman RJ, Calderon J, De Loof A, Schoofs L. Isolation, identification, and synthesis of a disulfated sulfakinin from the central nervous system of an arthropods the white shrimp Litopenaeus vannamei. Biochem Biophys Res Commun. 2002;299:312–320. doi: 10.1016/s0006-291x(02)02624-4. [DOI] [PubMed] [Google Scholar]
- Towle DW, Smith CM. Gene discovery in Carcinus maenas and Homarus americanus via expressed sequence tags. Integr Comp Biol. 2006;46:912–918. doi: 10.1093/icb/icl002. [DOI] [PubMed] [Google Scholar]
- Trimmer BA, Kobierski LA, Kravitz EA. Purification and characterization of FMRFamidelike immunoreactive substances from the lobster nervous system: isolation and sequence analysis of two closely related peptides. J Comp Neurol. 1987;266:16–26. doi: 10.1002/cne.902660103. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch Insect Biochem Physiol. 2000;43:49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]