Abstract
Imaging gene expression non-invasively and deep into opaque tissues has been a long-standing goal of molecular science. Optical gene reporters such as green fluorescent protein and luciferase have revolutionized cellular and molecular biology, however their in vivo application is limited, due to poor tissue penetration of visible light. The iron storage protein ferritin forms a paramagnetic ferrihydrite core that affects the relaxation rate of surrounding nuclear spins. Ferritin has recently emerged as an MRI gene reporter for molecular applications, however its detection with MRI still has relatively low sensitivity. In this work we present an improved ferritin chimera, genetically engineered to exhibit stronger paramagnetic properties.
Keywords: MRI gene reporter, ferritin, transverse relaxation rate
Introduction
Preclinical development of gene therapy and basic molecular research on systems level may require methods for non-invasive visualization of gene expression [1]. Recently, active research in MRI/MRS has resulted in the development of gene reporters whose expression can be coupled to genes of interest [2]. The iron storage protein ferritin has been successfully used as an MR reporter gene in both vector mediated gene delivery and transgenic animal models [3, 4]. Ferritin is built of heavy H and light L subunits, which have a cooperative role in iron loading and core formation [5]. The ratio L:H varies throughout the tissues and is closely related to iron loading efficacy [5].
We constructed a single chain ferritin chimera, L*H ferritin (L*H Ft), where the two chains, heavy and light, are connected with a linker and expressed as a single transcript. The resulting protein has a 1:1 subunit ratio and has a slightly larger protein cage than the wild type. Our present work characterizes the relaxation properties of L*H Ft chimera and compares them to wild type ferritin. We targeted the central nervous system because it is particularly challenging to study due to its inherent tissue opacity and the blood-brain barrier. We established that the engineered chimera has higher transverse relaxation rates than the wild type ferritin and can be used as an improved MRI reporter.
Materials and Methods
We used HSV1 vector [6] with titer 2 × 1010 pfu/ml coding for L*H Ft and green fluorescent protein (GFP) under the Human Cytomegalovirus (CMV) promoter. First we transduced U2OS cell line and incubated with 1mM ferric citrate in DMEM for 24 hours. We then fixed the cells 78 hours post-transduction in 4% paraformaldehyde. We measured the transverse relaxation rate R2 of the cell pellet using a 500 MHz Bruker NMR spectrometer. We used conventional CPMG sequence, NEX = 4, delay time of 10 sec, 14 echoes ranging from 10 ms to 250 ms. All data was fit to monoexponential decay curve y = Ae′l*m + C using Bruker software.
For the in vivo work we injected 5 \i\ L*H Ft inoculum unilaterally into the cortex and striatum of C57BL/6J male mice, age 4–6 weeks (n=6). The contra-lateral side was injected with the same vector expressing control gene LacZ and no L*H Ft. All imaging was done on 11.7 T Bruker micro-imaging system. At 4 days post-injection, mice were imaged in vivo and then perfused with 4% paraformaldehyde. The brains were then embedded in paraffin and sectioned for histological study. We acquired spin echo (SE) images with TE = 15, TR = 1200ms, and NEX = 2. We used 9 slices covering the injection site with lmm thickness, no gap, 256 × 128 matrix size and 0.059 × 0.117 in plane resolution. In addition we acquired gradient echo images (GE), TE = 7 ms, TR = 500ms, and NEX = 8. For GE we used the same geometry and resolution.
Results and conclusions
The transgene vector mediated expression of L*H Ft resulted in an increase of the R2 relaxation rate of cell pellet, which was significantly higher than control LacZ, wild type ferritin and homopolymers (Figure 1). The R2 relaxation rates of L*H Ft chimera showed a pronounced increase of contrast in vivo at the site of vector inoculation (Figure 2). The histological stain using hematoxylin and eosin showed no apparent local tissue toxicity of the vector inoculation. In addition immunoreactivity showed colocali-zation of GFP with the flag epitope on the L*H chimera.
Figure 1.
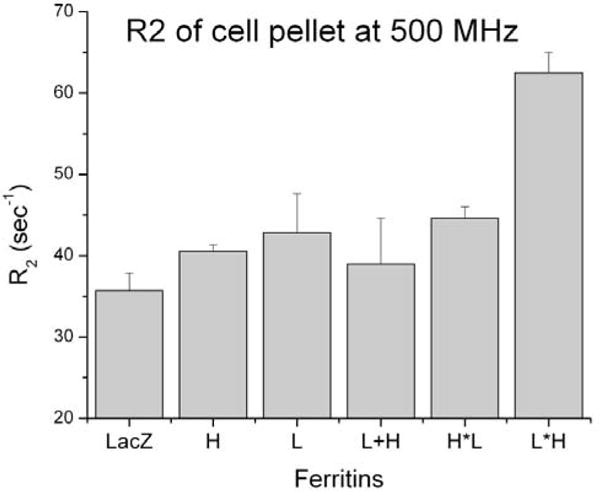
Relaxation rates at 500 MHz of U2OS cell pellet expressing different ferritin constructs. L*H ferritin chimera has the highest R2
Figure 2.
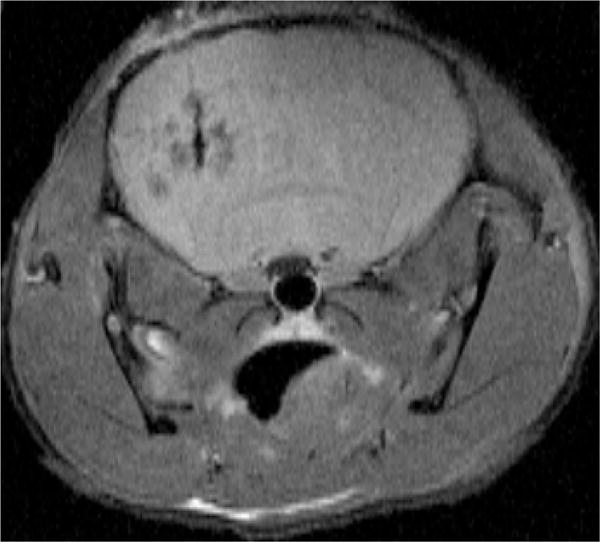
In vivo T2-weighted spin echo coronal image of mouse brain (TE = 15 ms, TR = 1200 ms) showing the site inoculated with the MRI transgene L*H Ft on the left and Lac Z control on the right
Both gene reporters, optical and MR, were expressed in the same population of transduced pyramidal neurons in the mouse striatum (data not shown).
Our study shows that the single chain L*H Ft chimera improves the MR imaging of gene expression in cells and intact animals. We are currently quantifying the level of gene expression in the mouse brain using 3D R2 relaxation maps. Our goal is to obtain numerical transverse relaxation values in vivo for the different ferritins. Our study offers a new MRI reporter with higher sensitivity for future research into molecular events in the central nervous system of living animals.
Acknowledgments
This work was funded by National Science Foundation Graduate Research Fellowship 2007053507 and NIH grants R01-EB005740 & P41-EB001977.
References
- 1.Massoud TF, Gambhir SS. Genes Dev. 2003 Mar 1;17(5):545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Gilad AA, Winnard PT, Jr, van Zijl PC. Bulte JW NMR in Biomedicine. 2007 May;20(3):275–90. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 3.Genove G, DeMarco U, Xu H, Goins W, Ahrens ET. Nature Medicine. 2005 Apr;11(4):450–4. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 4.Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benjamin LE, Neeman M. Nature Medicine. 2007 Apr;13(4):498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- 5.Levi S, Yewdall SJ, Harrison PM, Santambrogio P, Cozzi A, Rovida E, Albertini A, Arosio P. Biochem J. 1992 Dec 1;288(Pt 2):591–6. doi: 10.1042/bj2880591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frampton AR, Jr, Goins WF, Nakano K, Burton EA. Glorioso. Gene Therapy. 2005 Jun;12(11):891–901. doi: 10.1038/sj.gt.3302545. [DOI] [PubMed] [Google Scholar]


