Abstract
Enzyme-instructed self-assembly (EISA) offers a facile approach to explore the supramolecular assemblies of small molecules in cellular milieu for a variety of biomedical applications. One of the commonly used enzymes is phosphatase, but the study of the substrates of phosphatases mainly focuses on the phosphotyrosine containing peptides. In this work, we examine the EISA of phosphoserine containing small peptides for the first time by designing and synthesizing a series of precursors containing only phosphoserine or both phosphoserine and phosphotyrosine. Conjugating a phosphoserine to the C-terminal of a well-established self-assembling peptide backbone, (naphthalene-2-ly)-acetyl-diphenylalanine (NapFF), affords a novel hydrogelation precursor for EISA. The incorporation of phosphotyrosine, another substrate of phosphatase, into the resulting precursor, provides one more enzymatic trigger on a single molecule, and meanwhile increases the precursors’ propensity to aggregate after being fully dephosphorylated. Exchanging the positions of phosphorylated serine and tyrosine in the peptide backbone provides insights on how the specific molecular structures influence self-assembling behaviors of small peptides and the subsequent cellular responses. Moreover, the utilization of D-amino acids largely enhances the biostability of the peptides, thus providing a unique soft material for potential biomedical applications.
Keywords: Enzyme-instructed self-assembly, phosphoserine, phosphatase, supramolecular hydrogel
Introduction
Although RNAs sometimes are capable of catalyzing some reactions,[1] most biological reactions in nature are under the control of enzymes. In the absence of enzymes, a great portion of biological reactions would not happen under the mild conditions (e.g., ambient condition) that are compatible with life. Cells contain thousands of different enzymes and chemical or biological transformations mediated by enzymes, which are ubiquitous in nature. For example, a cascade of enzymatic transformations constitutes the sophisticated cell death pathway and the process of metabolism is also a set of life-sustaining enzymatic transformations.[2–3] In biological system, another fundamental process is self-assembly[4–5]—the spontaneous generation of order in systems of components, which eliminates the tedious work in building up a set of covalent bonds at very specific positions. For example, nature effortless utilizes self-assembly to put together the phospholipids for building up the compartments of cells and subcellular organelles with great precision and flexibility.[6] Many biological processes, as a consequence of evolution, are a combination of enzymatic transformation and self-assembly—enzyme-instructed self-assembly (EISA)—for carrying out critical cellular functions. For example, the formation of microtubules, which serves as cytoskeleton and governs mitosis, is the integration of the enzymatic transformation of guanosine-5′-triphosphate (GTP) to guanosine-5′-diphosphate (GDP) and the self-assembly of tubulins.[7] The disease cascade of Alzheimer’s disease generally happens while enzymatic hyperphosphorylation of tau proteins, which eventually assemble to form neurofibrillary tangles inside nerve cells. The polymerization of actins, which governs the focal adhesion of cells, is essentially a self-assembly process that is regulated by enzymes.[8] These essential facts, as bioinspiration from cell biology,[9] support the notion that EISA promises a new paradigm for developing innovative biofunctional materials for a variety of applications.
As a powerful tool to control and direct supramolecular self-assembly by catalytically converting non-assembling precursors to self-assembling hydrogelators, EISA has been receiving increased attentions in the last decade due to its ability to control molecular self-assembly in cellular milieu and the advantages like high efficiency, selectivity, and mild conditions. Meanwhile, it is feasible to control the kinetics of the EISA process, giving rise to supramolecular hydrogels with tunable properties. The diversity of enzymes in nature offers many choices of the enzymes to catalyze the formation of supramolecular assemblies by EISA, such as phosphatas,[10–13] β-lactamase,[14] esterase,[15–18] metalloprotease-9/7 (MMP-9/7),[19–21] α-chymotrypsin,[22] thrombin,[23] chymotrypsin,[23] microbial transglutaminase (MTGase),[24] thermolysin,[15–18] tyrosinase,[25] etc. For example, our previous studies have demonstrated the feasibility of applying EISA, based on phosphatases, to modulate the self-assembly of small peptides,[26–30] cholesterol conjugates[31] and even nanoparticles[26–29, 32] for controlling the fates of cells. Pires and Ulijn recently also reported a simple carbohydrate amphiphile to self-assemble to form nanofibers upon enzymatic dephosphorylation for selectively inhibiting cancer cells.[33] Yang and coworkers have been building up a variety of biofunctional materials ranging from extracellular matrix mimics,[34] drug delivery carriers,[35] imaging reagents[36] to vaccine adjuvants[37–38] via the approach of EISA.
In spite that phosphatases are widely used for developing EISA, the study of the phosphatase substrates mainly focuses on the phosphotyrosine containing peptides. Another phosphorylated amino acid, phosphoserine, is less explored. Although Stupp et al. have reported the use of phosphoserine containing peptides as self-assembly amphiphiles and have used enzymatic reaction to generate phosphoserine residues in peptides,[39] there is no report of EISA based phosphoserine to create hydrogels or assemblies. To address this knowledge gap, in this work, we examine the EISA of phosphoserine-containing small peptides for the first time by designing and synthesizing a series of precursors (Figure 1) containing one phosphoserine or both one phosphoserine and one phosphotyrosine. Our study shows that it is feasible to use EISA to trigger the self-assembly of the serine-containing peptides and to form hydrogels (Scheme 1). The addition of a phosphotyrosine into the phosphoserine-containing peptides largely enhances the self-assembly propensity of peptides. As expected, the specific sequences of peptides also influences the self-assembly behaviors as well as the subsequent cellular responses. Moreover, the use of D-amino acids instead of L-amino acid renders the self-assembling molecules excellent biostability against proteolysis. This study not only provides a molecular validation of EISA of phosphoserine contain peptides, but also indicates that EISA of phosphoserine-containing peptides is relevant for developing cell compatible gels for a variety of applications.
Figure 1.
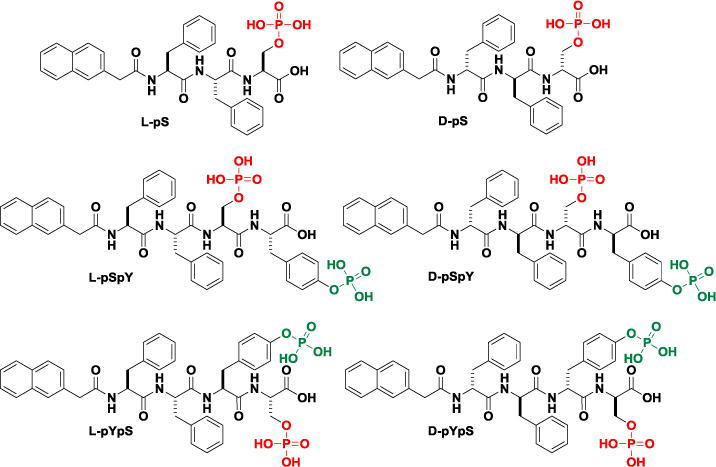
Chemical structures of the rationally designed precursors containing phosphoserine.
Scheme 1.
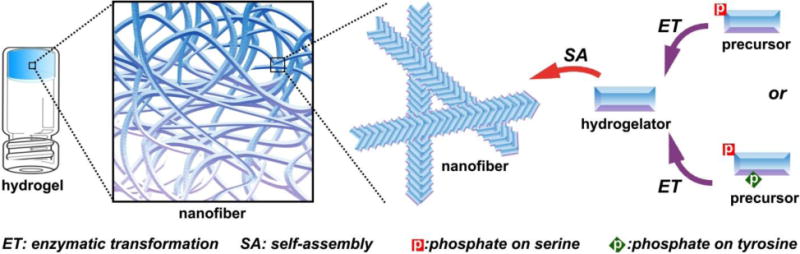
Schematic illustration of EISA of small molecules in water that usually results in supramolecular nanofibers/hydrogels.
Result and Discussion
Molecule Design
In order to systematically investigate the enzyme-instructed assemblies based enzymatic transformation of phosphoserine peptides, we designed and synthesized a class of precursors containing phosphorylated serine with both L- and D-amino acids (Figure 1). We design the molecules according several principles: (i) Nap-Phe-Phe sequence, a well-documented self-assembling motif,[40] acts as the main self-assembling backbone of the six precursors to enhance aromatic-aromatic interactions; (ii) the incorporation of the phosphoserine is necessary for investigating the enzyme-instructed assemblies based on enzymatic reaction of phosphoserine, which not only facilitates the dissolution and dispersion of the precursors in water at physiological condition, but also provides a enzymatic trigger; (iii) while phosphorylated tyrosine, another well-established substrate of phosphatases, provides one more enzymatic trigger, the benzyl ring of tyrosine further increases the precursors’ propensity to aggregate after dephosphorylation; (iv) by replacing the L-amino acids with their D-enantiomers, we can evaluate and compare the biological responses of L- and D-precursors in the presence of proteases and cells; (v) the mutation of the position of phosphoserine in the precursors containing both phosphorylated serine and tyrosine permits evaluating the influence of the sequences of peptides on the self-assembling abilities and the subsequent biological behaviors of the assemblies of the serine-containing peptides.
Hydrogelation Preparation and TEM Characterization
We first use enzymatic hydrogelation[41] to assay the self-assembling ability of the serine-containing peptides. We dissolved each of the precursors (2 mg) into distilled water (300 μL), and adjusted pH of the solution carefully by adding 1 M NaOH (monitored by pH paper). After the pH reached 7.4, we added extra distilled water to make the final volume 400 μL so that the concentration of the precursor was 0.5 wt%. After adding alkaline phosphatase (ALP, final concentration 1 U/mL) into the solution, we briefly vortexed the vial containing the solution, kept it still on a bench, and monitored the hydrogelation by visual inspection. Since the dephosphorylation of phosphotyrosine is well-established, we used 31P-NMR to verify the dephosphorylation of phosphoserine (Figure S4).
The presence of phosphate(s) renders these six precursors good solubility in water. Though all of them completely dissolve in water at pH 7.4, the TEM image of the dried solution of each precursor shows amorphous aggregates (Figure 2), suggesting that the precursors likely are able to form oligomers in water. The solutions of precursors consisting of both serine and tyrosine phosphates (L-pSpY, D-pSpY, L-pYpS, D-pYpS) solidify gradually upon the addition of ALP and turn into rigid hydrogels within 4 hours. The L and D-enatiomeric precursors exhibit almost same behaviors in terms of gelation speed and microscopic morphologies. Based on our observation, L-pYpS and D-pYpS form hydrogels slightly faster than L-pSpY and D-pSpY, suggesting the better self-assembly abilities of L-YS and D-YS than those of L-SY and D-SY. This result implies that the stronger intermolecular aromatic-aromatic interactions offered by the sequence Nap-Phe-Phe-Tyr-Ser. TEM images reveals an entangled fibrous network in all of the four hydrogels and the length of the nanofibers to be more than several micrometers. The peptide sequence exhibits little influence on the morphologies of nanofibers since all of the four hydrogels contain the nanofibers with diameter of 8±2 nm (Figure 2). L-pS and D-pS only formed weak hydrogels, which are barely self-sustaining after the vials being inverted. Meanwhile, the nanofibers in the networks of these two hydrogels have diameters around 24 nm (Figure 2) and much longer persistence length, differing distinctively from those of the other four hydrogels. The morphologies of these nanofibers largely resemble the nanobelts reported by Cui and Stupp though they have dramatically different chemical structures.[42–43] These results indicate that the incorporation of tyrosine largely enhance self-assembly propensity of the peptides, which dictates the microscopic morphologies of the self-assembled structures.
Figure 2.
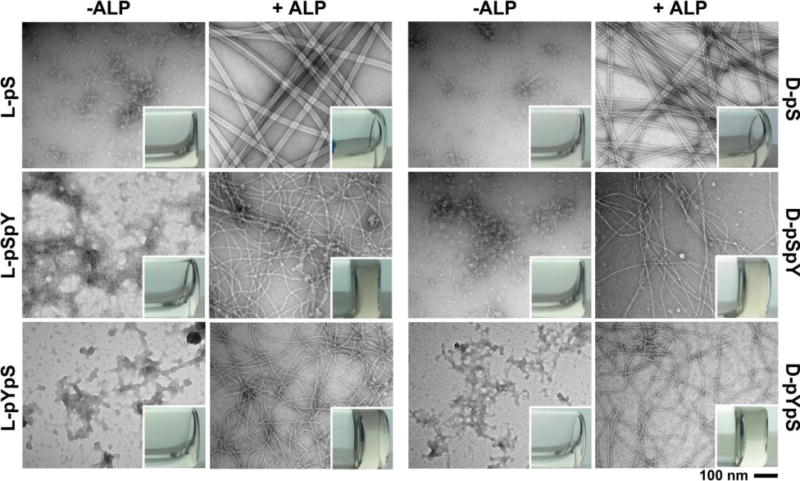
Transmission electron microscopy (TEM) images of the aggregates/nanofibers in the solutions of different precursors (L-pS, D-pS, L-pSpY, D-pSpY, L-pYpS, D-pYpS) or nanofibers in the hydrogels formed by treating the solutions of these precursors with alkaline phosphatase (ALP). C = 0.5 wt%, pH = 7.4, [ALP] = 1 U/ml. Insets are optical images of the solutions of the precursors and the hydrogels formed after enzymatic dephosphorylation. The scale bar is 100 nm.
Rheological Characterization
Rheological characterization of six hydrogles largely matches with the observations in hydrogelation and the TEM examination. The storage modulus (G′) of each of these hydrogels is greater than the loss modulus (G”), agreeing with the fact that they are hydrogels with certain viscosity (Figure 3). The storage modulus values of L- and D-enantiomeric hydrogels are almost identical, suggesting that the chirality exerts little affect on the enzyme-instructed assembly behaviors of these precursors. The storage modulus of the hydrogel formed by the dephosphorylation of L-pYpS (or D-pYpS) is greater than the dephosphorylation of L-pSpY (or D-pSpY). The storage modulus of the hydrogel formed by the dephosphorylation of L-pS (or D-pS) is the lowest among the three pairs. These results are consistent with that L-YS and D-YS form solid gels faster than L-SY and D-SY, while L-S and D-S only form weak hydrogels. We summarized all the properties related to EISA and hydrogelation in Table 1.
Figure 3.
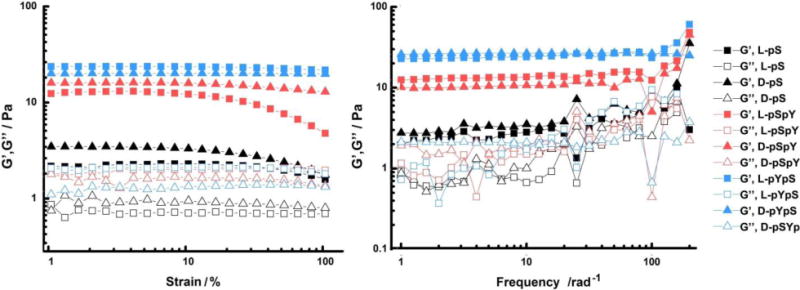
Rheological characterization of the six hydrogels fromed by treating the solutions of each of the precursors (0.5 wt%) with ALP (1 U/mL). The strain dependence of the dynamic storage (G′) and loss storage (G″) is taken at a frequency equal to 6.28 rad/s, and the frequency dependence is taken at a strain equal to 1.00 %.
Table 1.
Summary of the EISA of the Phosphoserine-Containing Precursors
| compounda | L-pS | L-pSpY | L-pYpS | D-pS | D-pSpY | D-pYpS |
|---|---|---|---|---|---|---|
| in pbs (pH 7.4) | solution | solution | Solution | solution | solution | solution |
| + ALP /(1U/mL) | gel | gel | gel | gel | gel | gel |
| storage modulus G′b /Pa | 2.3 | 12.5 | 23.7 | 3.4 | 16.0 | 20.0 |
| loss modulus G″b /Pa | 0.8 | 1.8 | 2.0 | 1.5 | 39 | 1.2 |
| morphology before ALP treatment (dc /nm) |
aggregate | aggregate | aggregate | aggregate | aggregate | aggregate |
| morphology after ALP treatment (dc /nm) |
nanofiber (24±2) |
nanofiber (8±2) |
nanofiber (8±2) |
nanofiber (24±2) |
nanofiber (8±2) |
nanofiber (8±2) |
The concentration is 0.5 wt%.
The modulus is taken at the frequency of 6.28 rad/s.
Diameter of nanofibers.
Biostability
The addition of proteinase K, a powerful endopeptidase, into the solutions of all six precursors (500 μM, pH 7.4, in PBS) reveals that the use of D-amino acids in the peptide backbone completely eliminates the proteolysis catalyzed by proteinase K (Figure 4). This result agrees with that D-peptides rarely interact with endogenous proteins and exhibit proteolytic resistance. However, proteinase K digests three L-peptides efficiently and almost no start compounds remain after 24 hour. According to digestion curve in Figure 4, L-pS is more prone to proteolysis catalyzed by proteinase K, followed by L-pSpY and then L-pYpS. This result indicates that the presence of pY next to NapFF increases the proteolytic resistance of the peptides against proteinase K, an observation may be useful for enhance the biostability of peptidic hydrogelators and precursors.
Figure 4.
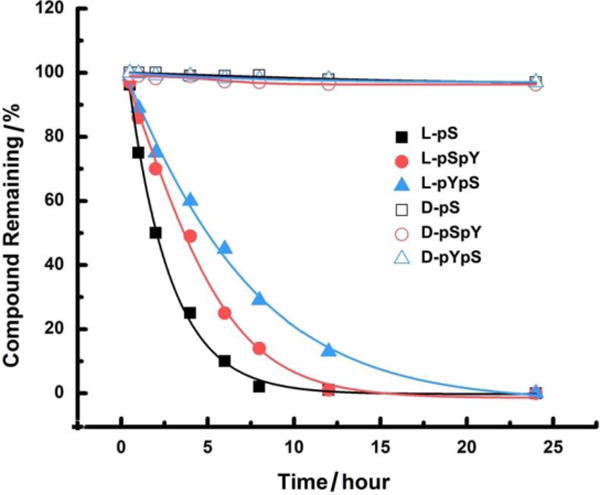
Digestion curve of all six precursors (500 μM) in the presence of proteinase K (2 U/mL) at 37 ℃. The digestion curve was determined on analytical HPLC.
Cell Compatibility Evaluation
To investigate how EISA based on serine and tyrosine phosphate affects the cellular response to the small molecular precursors, we utilized MTT cell viability assay to examine the cell viability of a cancer cell line (HeLa) and a stromal cell line (HS-5) treated by the six precursors. As shown in Figure 5, the precursors are overall more cytotoxic to HeLa cells than to HS-5, indicating that the cell inhibitory behaviors are largely associated with phosphatase mediated self-assembly, which we have observed in previous studies.[27–30] Because HeLa cells have a higher phosphatase level on their surfaces than on HS-5 cells,[44] which leads to faster enzyme-instructed assembly on cell surface to induce cell death. The D-peptides are overall more potent for killing the cancer cells due to their biostability. The stronger hydrogel (D/L-pYpS) induces less toxicity against HeLa cells than weaker ones (D/L-pSpY), indicating that the cellular responses may also be related to chemical structures and dephosphorylation rates in addition to the mechanical strength. Moreover, L-hydrogels are more prone to proteolysis in cell culture, resulting in lower cytotoxicity than D-hydrogels. Both L-pS and D-pS remain innocuous to HeLa and HS-5 cells, suggesting the hydrogels of L-S or D-S may serve as a hydrogel medium for 3D cell culture.
Figure 5.
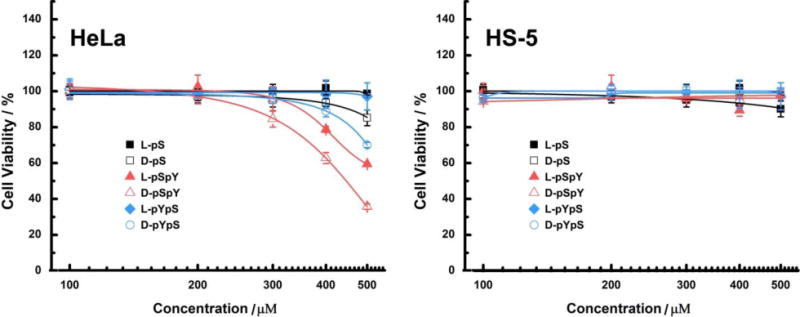
48-hour cell viability (determined by MTT assay) of HeLa (cancer) and HS-5 (stromal) cells, incubated with six precursors at the concentrations of 100, 200, 300, 400 and 500 μM in cell growth medium. The initial cell numbers are 1 × 104 cells/well.
Conclusion
This communication, for the first time, describes a systematic investigation of enzyme-instructed self-assembly of peptides containing phosphoserine to form supramolecular hydrogels as potential soft biomaterials. For example, being biocompatible, this type of assemblies/hydrogels maybe more suitable candidates for closely mimicking both the structural and functional aspects of native extracellular matrix (ECM) for cell culture and tissue engineering. We also validated that phosphotyrosine, as another enzymatic trigger, would leverage not only the self-assembly behaviors, but also the subsequent cellular response of the peptides. In addition, D-peptides, made exclusively of unnatural amino acids, are good alternatives to native peptides to significantly enhance proteinase inhibitors. The results in this work, coupled with expanding efforts in material science, nanotechnology, cell biology, will ultimately contribute to the development of more innovative biomaterials and therapies.
Supplementary Material
Acknowledgments
This work was partially supported by NIH (CA142746), NSF (MRSEC-1420382) and the W. M. Keck Foundation. We thank Brandeis EM and Optical Imaging facilities for TEM. JZ is an HHMI student fellow.
References
- 1.Strobel SA, Cochrane JC. Rna Catalysis: Ribozymes, Ribosomes, and Riboswitches. Current Opinion in Chemical Biology. 2007;11(6):636–643. doi: 10.1016/j.cbpa.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green DR, Reed JC. Mitochondria and Apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The Ubiquitin System. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Mitchison T, Kirschner M. Dynamic Instability of Microtubule Growth. nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 5.Schiff PB, Fant J, Horwitz SB. Promotion of Microtubule Assembly in Vitro by Taxol. nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 6.Meyers MA, Chen PY, Lin AYM, Seki Y. Biological Materials: Structure and Mechanical Properties. Progress in Materials Science. 2008;53(1):1–206. [Google Scholar]
- 7.Kirschner M, Mitchison T. Beyond Self-Assembly - from Microtubules to Morphogenesis. Cell. 1986;45(3):329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 8.Korn ED, Carlier MF, Pantaloni D. Actin Polymerization and Atp Hydrolysis. Science. 1987;238(4827):638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- 9.Whitesides GM. Bioinspiration: Something for Everyone. Interface Focus. 2015;5(4):20150031. doi: 10.1098/rsfs.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y, Shi J, Yuan D, Xu B. Imaging Enzyme-Triggered Self-Assembly of Small Molecules inside Live Cells. Nature Communication. 2012;3:1033. doi: 10.1038/ncomms2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Kuang Y, Shi J, Yuan G, Zhou J, Xu B. The Conjugation of Nonsteroidal Anti-Inflammatory Drugs (Nsaid) to Small Peptides for Generating Multifunctional Supramolecular Nanofibers/Hydrogels. Beilstein Journal of Organic Chemistry. 2013;9:908–917. doi: 10.3762/bjoc.9.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thornton K, Smith AM, Merry CLR, Ulijn RV. Controlling Stiffness in Nanostructured Hydrogels Produced by Enzymatic Dephosphorylation. Biochemical Society Transactions. 2009;37:660–664. doi: 10.1042/BST0370660. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Qian J, Tang A, An L, Zhong K, Liang G. Using Magnetic Resonance Imaging to Study Enzymatic Hydrogelation. Analytical Chemistry. 2014;86(12):5955–5961. doi: 10.1021/ac500967x. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Ho P-L, Liang G, Chow KH, Wang Q, Cao Y, Guo Z, Xu B. Using Beta-Lactamase to Trigger Supramolecular Hydrogelation. Journal of the American Chemical Society. 2007;129(2):266–267. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- 15.Guilbaud J-B, Vey E, Boothroyd S, Smith AM, Ulijn RV, Saiani A, Miller AF. Enzymatic Catalyzed Synthesis and Triggered Gelation of Ionic Peptides. Langmuir. 2010;26(13):11297–11303. doi: 10.1021/la100623y. [DOI] [PubMed] [Google Scholar]
- 16.Das AK, Collins R, Ulijn RV. Exploiting Enzymatic (Reversed) Hydrolysis in Directed Self-Assembly of Peptide Nanostructures. Small. 2008;4(2):279–287. doi: 10.1002/smll.200700889. [DOI] [PubMed] [Google Scholar]
- 17.Williams RJ, Gardiner J, Sorensen AB, Marchesan S, Mulder RJ, McLean KM, Hartley PG. Monitoring the Early Stage Self-Assembly of Enzyme-Assisted Peptide Hydrogels. Australian Journal of Chemistry. 2013;66(5):572–578. [Google Scholar]
- 18.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. Enzyme-Triggered Self-Assembly of Peptide Hydrogels Via Reversed Hydrolysis. Journal of the American Chemical Society. 2006;128(4):1070–1071. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Ma M, Xu B. Using Matrix Metalloprotease-9 (Mmp-9) to Trigger Supramolecular Hydrogelation. Soft Matter. 2009;5(13):2546–2548. [Google Scholar]
- 20.Bremmer SC, McNeil AJ, Soellner MB. Enzyme-Triggered Gelation: Targeting Proteases with Internal Cleavage Sites. Chemical Communications. 2014;50(14):1691–1693. doi: 10.1039/c3cc48132h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalafatovic D, Nobis M, Son J, Anderson KI, Ulijn RV. Mmp-9 Triggered Self-Assembly of Doxorubicin Nanofiber Depots Halts Tumor Growth. Biomaterials. 2016;98:192–202. doi: 10.1016/j.biomaterials.2016.04.039. [DOI] [PubMed] [Google Scholar]
- 22.Qin X, Xie W, Tian S, Cai J, Yuan H, Yu Z, Butterfoss GL, Khuong AC, Gross RA. Enzyme-Triggered Hydrogelation Via Self-Assembly of Alternating Peptides. Chemical Communications. 2013;49(42):4839–4841. doi: 10.1039/c3cc41794h. [DOI] [PubMed] [Google Scholar]
- 23.Bremmer SC, Chen J, McNeil AJ, Soellner MB. A General Method for Detecting Protease Activity Via Gelation and Its Application to Artificial Clotting. Chemical Communications. 2012;48(44):5482–5484. doi: 10.1039/c2cc31537h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song F, Zhang L-M. Enzyme-Catalyzed Formation and Structure Characteristics of a Protein- Based Hydrogel. Journal of Physical Chemistry B. 2008;112(44):13749–13755. doi: 10.1021/jp8041389. [DOI] [PubMed] [Google Scholar]
- 25.Choi YC, Choi JS, Jung YJ, Cho YW. Human Gelatin Tissue-Adhesive Hydrogels Prepared by Enzyme-Mediated Biosynthesis of Dopa and Fe3+ Ion Crosslinking. Journal of Materials Chemistry B. 2014;2(2):201–209. doi: 10.1039/c3tb20696c. [DOI] [PubMed] [Google Scholar]
- 26.Zhou R, Kuang Y, Zhou J, Du XW, Li J, Shi JF, Haburcak R, Xu B. Nanonets Collect Cancer Secretome from Pericellular Space. Plos One. 2016;11(4):e0154126. doi: 10.1371/journal.pone.0154126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Xu B. Enzyme-Instructed Self-Assembly: A Multistep Process for Potential Cancer Therapy. Bioconjugate Chemistry. 2015;26(6):987–999. doi: 10.1021/acs.bioconjchem.5b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Du XW, Yamagata N, Xu B. Enzyme-Instructed Self-Assembly of Small D-Peptides as a Multiple-Step Process for Selectively Killing Cancer Cells. Journal of the American Chemical Society. 2016;138(11):3813–3823. doi: 10.1021/jacs.5b13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Du XW, Xu B. Regulating the Rate of Molecular Self-Assembly for Targeting Cancer Cells. Angewandte Chemie International Edition. 2016;55(19):5770–5775. doi: 10.1002/anie.201600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi JF, Du XW, Yuan D, Zhou J, Zhou N, Huang YB, Xu B. D-Amino Acids Modulate the Cellular Response of Enzymatic-Instructed Supramolecular Nanofibers of Small Peptides. Biomacromolecules. 2014;15(10):3559–3568. doi: 10.1021/bm5010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Feng Z, Wu D, Fritzsching KJ, Rigney M, Zhou J, Jiang Y, Schmidt-Rohr K, Xu B. Enzyme-Regulated Supramolecular Assemblies of Cholesterol Conjugates against Drug-Resistant Ovarian Cancer Cells. Journal of the American Chemical Society. 2016;138(34):10758–10761. doi: 10.1021/jacs.6b06075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du XW, Zhou J, Xu B. Ectoenzyme Switches the Surface of Magnetic Nanoparticles for Selective Binding of Cancer Cells. Journal of Colloid and Interface Science. 2015;447:273–277. doi: 10.1016/j.jcis.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I. Controlling Cancer Cell Fate Using Localized Biocatalytic Self-Assembly of an Aromatic Carbohydrate Amphiphile. Journal of the American Chemical Society. 2015;137(2):576–579. doi: 10.1021/ja5111893. [DOI] [PubMed] [Google Scholar]
- 34.Lv L, Liu H, Chen X, Yang Z. Glutathione-Triggered Formation of Molecular Hydrogels for 3d Cell Culture. Colloids and Surfaces B: Biointerfaces. 2013;108:352–357. doi: 10.1016/j.colsurfb.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Wang HM, Yang ZM. Short-Peptide-Based Molecular Hydrogels: Novel Gelation Strategies and Applications for Tissue Engineering and Drug Delivery. Nanoscale. 2012;4(17):5259–5267. doi: 10.1039/c2nr31149f. [DOI] [PubMed] [Google Scholar]
- 36.Cai Y, Shi Y, Wang H, Wang J, Ding D, Wang L, Yang Z. Environment-Sensitive Fluorescent Supramolecular Nanofibers for Imaging Applications. Analytical Chemistry. 2014;86(4):2193–2199. doi: 10.1021/ac4038653. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Luo Z, Wang Y, He T, Yang C, Ren C, Ma L, Gong C, Li X, Yang Z. Enzyme-Catalyzed Formation of Supramolecular Hydrogels as Promising Vaccine Adjuvants. Advanced Functional Materials. 2016;26(11):1822–1829. [Google Scholar]
- 38.Tian Y, Wang H, Liu Y, Mao L, Chen W, Zhu Z, Liu W, Zheng W, Zhao Y, Kong D, Yang Z, Zhang W, Shao Y, Jiang X. A Peptide-Based Nanofibrous Hydrogel as a Promising DNA Nanovector for Optimizing the Efficacy of Hiv Vaccine. Nano Letter. 2014;14(3):1439–1445. doi: 10.1021/nl404560v. [DOI] [PubMed] [Google Scholar]
- 39.Sargeant TD, Aparicio C, Goldberger JE, Cui HG, Stupp SI. Mineralization of Peptide Amphiphile Nanofibers and Its Effect on the Differentiation of Human Mesenchymal Stem Cells. Acta Biomaterialia. 2012;8(7):2456–2465. doi: 10.1016/j.actbio.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Kuang Y, Gao YA, Xu B. Versatile Small-Molecule Motifs for Self-Assembly in Water and the Formation of Biofunctional Supramolecular Hydrogels. Langmuir. 2011;27(2):529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, Liang G, Xu B. Enzymatic Hydrogelation of Small Molecules. Accounts of Chemical Research. 2008;41(2):315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 42.Cui H, Cheetham AG, Pashuck ET, Stupp SI. Amino Acid Sequence in Constitutionally Isomeric Tetrapeptide Amphiphiles Dictates Architecture of One-Dimensional Nanostructures. Journal of the American Chemical Society. 2014;136(35):12461–12468. doi: 10.1021/ja507051w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui H, Muraoka T, Cheetham AG, Stupp SI. Self-Assembly of Giant Peptide Nanobelts. Nano Letter. 2009;9(3):945–951. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, Du X, Berciu C, He H, Shi J, Nicastro D, Xu B. Enzyme-Instructed Self-Assembly for Spatiotemporal Profiling of the Activities of Alkaline Phosphatases on Live Cells. Chem. 1(2):246–263. doi: 10.1016/j.chempr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


