Abstract
BACKGROUND
Variants in SCN2A that disrupt the encoded neuronal sodium channel NaV1.2 are important risk factors for autism spectrum disorder (ASD), developmental delay, and infantile seizures. Variants observed in infantile seizures are predominantly missense, leading to a gain of function and increased neuronal excitability. How variants associated with ASD affect NaV1.2 function and neuronal excitability are unclear.
METHODS
We examined the properties of 11 ASD-associated SCN2A variants in heterologous expression systems using whole-cell voltage-clamp electrophysiology and immunohistochemistry. Resultant data were incorporated into computational models of developing and mature cortical pyramidal cells that express NaV1.2.
RESULTS
In contrast to gain of function variants that contribute to seizure, we found that all ASD-associated variants dampened or eliminated channel function. Incorporating these electrophysiological results into a compartmental model of developing excitatory neurons demonstrated that all ASD variants, regardless of their mechanism of action, resulted in deficits in neuronal excitability. Corresponding analysis of mature neurons predicted minimal change in neuronal excitability.
CONCLUSIONS
This functional characterization thus identifies SCN2A mutation and NaV1.2 dysfunction as the most frequently observed ASD risk factor detectable by exome sequencing and suggests that associated changes in neuronal excitability, particularly in developing neurons, may contribute to ASD etiology.
Keywords: autism spectrum disorder, epilepsy, seizure, SCN2A, NaV1.2, electrophysiology
Introduction
Exome sequencing has transformed gene discovery in autism spectrum disorder (ASD). Observing multiple de novo protein truncating variants (PTVs, e.g. premature stop codons) in a gene demonstrates ASD association (1) and, to date, 65 such ASD genes have been identified (2–4). The gene SCN2A, which encodes the sodium channel NaV1.2, was one of the first ASD genes identified (1) and remains one of the genes with the strongest evidence for association with ASD (2) and, recently, developmental delay (5). SCN2A stands out from other ASD-associated genes in several ways. First, while most ASD-associated genes are related to either chromatin regulation or synapse structure (2), NaV1.2 channels are primarily expressed in the axon (6–10). Second, along with de novo PTVs, an excess of de novo missense mutations is also observed in SCN2A, a pattern seen in no other gene (2). Finally, genetic variants in SCN2A have previously been associated with infantile seizures, ranging from benign infantile familial seizures (BIFS) to epileptic encephalopathy (EE) with poor developmental outcome (11), however only 4 of 19 ASD patients for whom phenotypic data were available had a history of seizures and none had infantile seizures (Table S1 in Supplement 2).
NaV1.2 channels support central functions in developing and mature neurons, particularly in cortical glutamatergic pyramidal cells (6, 7) that are a locus of dysfunction in ASD (12–14). During early development, NaV1.2 is the only NaV isoform expressed at nodes of Ranvier and in the axon initial segment (AIS) (15–17), the site of action potential initiation in most neurons (18, 19). NaV1.2 is therefore critical for AP generation and propagation as these neurons integrate into circuits. Later in development, NaV1.2 is replaced by NaV1.6 (SCN8A), which has a lower voltage threshold for activation, at the majority of nodes and in the distal AIS (7, 10). Consequently, AP initiation occurs in this NaV1.6-rich region in mature neurons (19, 20), and NaV1.2, now restricted to the proximal AIS, takes on a new role, boosting rather than initiating APs (6). This developmental transition in AIS distribution may explain the resolution of seizures observed in BIFS before two years of age (13).
The majority of SCN2A missense variants associated with infantile seizures in BIFS and EE are gain of function (7, 21–25; but see 26). In contrast, mutations observed in ASD/developmental delay are either PTVs resulting in loss of NaV1.2 function or missense mutations of unknown effect. These observations raise the hypothesis that SCN2A variants exert opposing effects on NaV1.2 function in infantile seizures and ASD.
Here, we used bioinformatics, electrophysiology, and compartmental modeling to test how different SCN2A variants affect the function of NaV1.2 channels. We report that the majority of missense mutations observed in ASD completely blocked channel conductance, while the remainder altered NaV1.2 biophysical properties in ways that dampened channel function. Regardless of mechanism, all ASD-related variants impaired neuronal excitability in computational models of developing pyramidal cells. These impairments largely resolved as NaV1.6 replaced NaV1.2 in the distal AIS of mature models, supporting the notion that ASD is a consequence of disruption to early brain development leading to persistent neurodevelopmental dysfunction (27). Results from these de novo missense mutations in eight individuals, along with four individuals with de novo PTVs, identify SCN2A mutations in 0.3% of ASD cases (12 out of 4,109 cases with exome data). SCN2A therefore has the strongest evidence of ASD association of any gene discovered by exome sequencing, and is the most frequent single gene contributor to ASD, after Fragile X mutations.
Methods
Briefly, wild type and variants of SCN2A harboring an ASD-associated mutation were co-transfected with β1-β2 subunits in HEK293 cells. Sodium channel function was assessed with voltage-clamp techniques and immunohistochemistry, and then incorporated into a pyramidal cell computational model. Please see Supplemental Methods for details.
Results
A comprehensive literature review identified 148 SCN2A variants (117 unique) in 148 families (Table S1 in Supplement 2). Building on the description by Howell et al. (11), variants clustered in four phenotypic groups: 20 variants in families with BIFS; 8 variants in individuals with infantile seizures and mild developmental delay and/or episodic ataxia; 51 variants in individuals with EE characterized by infantile seizures and moderate to severe developmental delay, often accompanied by central hypotonia, cerebral atrophy, and microcephaly; and 39 variants in individuals with a developmental disorder characterized by ASD and/or intellectual disability. This latter group includes a few individuals with co-morbid seizures with onset after the age of 1.5 yrs, a frequent occurrence in both ASD and developmental delay. A further 16 variants lack sufficient phenotypic information to assign them to one of these four groups, while 15 variants do not clearly match these groups (e.g. febrile seizures, Herpes encephalitis, schizophrenia).
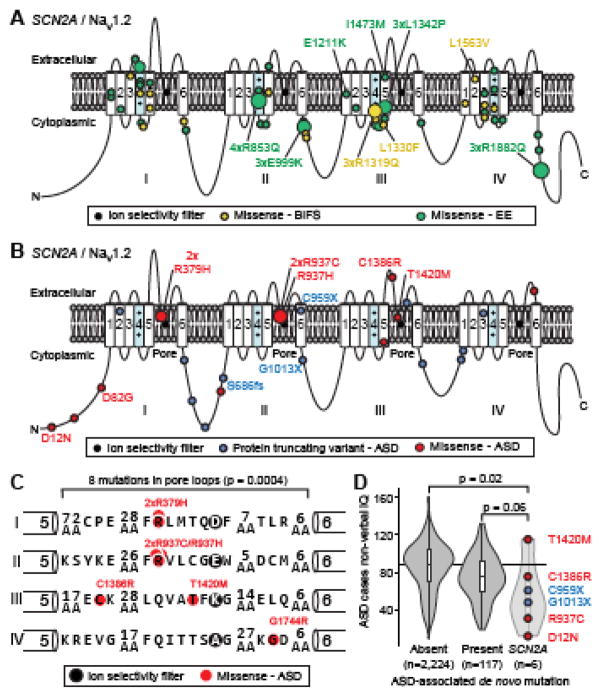
Of the 20 variants observed in BIFS, 19 (95%) are missense variants and one is a 507kbp duplication; 18 (90%) of the variants are inherited with the variant segregating with phenotype in multiple affected family members. BIFS missense variants cluster in the transmembrane segments and short connecting loops (16/19; 2.3-fold over expectation; p=0.0004, binomial test; Fig. 1A), particularly in and between transmembrane segments 4 and 5 that are proximal to the voltage sensor (11/19; 4.7-fold over expectation; p=4×10−5, binomial test; Fig. 1A).
Figure 1. SCN2A/NaV1.2 genotypes and phenotypes.
A. Location of 19 missense SCN2A variants in BIFS (yellow) and 49 missense SCN2A variants in EE (green) in the NaV1.2 sodium channel. The size of the circle corresponds to the number of individuals with a variant at a specific residue. Variants that are observed in three or more independent families are named, with the text color corresponding to the phenotype.
B. Location of 13 missense (red) and 10 PTV (blue) SCN2A variants in ASD cases in the NaV1.2 sodium channel. The size of the circle corresponds to the number of individuals with a variant at a specific residue. Variants that were functionally assessed are named, with the text color corresponding to the variant type.
C. A zoomed in view of the 8 missense SCN2A variants on the pore loop observed in ASD. Six of these variants are within five amino acid residues of the ion selectivity filter. Statistical significance was calculated using a two-sided Binomial Exact Test.
D. A violin plot of non-verbal IQ in 2,347 ASD cases from the Simons Simplex Collection; equivalent data are not available for the Autism Sequencing Consortium cases. The overlaid boxplot shows the median and interquartile range. The cases are divided into three groups: 2,224 with no known de novo PTV, deletion, or duplication mutations; 117 with a de novo PTV, deletion, or duplication in an ASD-associated gene or locus (2); 6 with a de novo PTV (blue) or missense (red) mutation in SCN2A. Statistical significance was calculated using a two-sided Wilcoxon Signed Rank Test.
Abbreviations: SCN2A: sodium channel, voltage-gated, type II, alpha subunit; PTV: protein truncating variant; ASD: Autism Spectrum Disorder; BIFS: Benign Infantile Familial Seizures; EE: Epileptic Encephalopathy; AA: Amino Acid; IQ: Intelligence Quotient.
Similarly, variants observed in EE are mainly missense variants (49/51, 96%) along with one 2.6Mbp duplication and one PTV; however, unlike BIFS variants, most are de novo (48 of 49 with inheritance data, 98%). EE missense variants also cluster in and between the transmembrane segments (39/49; 2-fold enrichment; p=2×10−6, binomial test; Fig. 1A), particularly segments 4 and 5 (24/49; 4-fold enrichment; p=4×10−10, binomial test; Fig. 1A). While the variants in BIFS and EE cluster in similar domains of the Nav1.2 protein, there are no examples of a variant contributing to both disorders, despite multiple recurrent variants in different families being observed in both BIFS (3xR1319Q) and EE (4xR853Q, 3xE999K, 3xL1342P, 3xR1882Q). Seven BIFS and three EE variants have been characterized electrophysiologically (Table S1, S2 in Supplement 2), largely revealing gain of function thought to result in neuronal hyperexcitability (7, 21–25) (but see 26).
In contrast to BIFS and EE, over half of the ASD-associated variants are predicted to prevent translation of one SCN2A allele, with 12 PTVs and 2 deletions (14/27, 52%). ExAC (28) and RVIS (29), two methods that use large-scale population data to identify constrained genes that are intolerant of deleterious variation, both predict that SCN2A is highly intolerant of such haploinsufficiency, with 30-fold fewer PTVs observed in an adult population than expected. Consistent with a highly deleterious effect, all PTV and deletion variants are de novo (13/13 for which data are available, Table S1 in Supplement 2). The remaining 13 (48%) variants in ASD cases are missense (Table S1 in Supplement 2) and most of these are also de novo (12/13, 92%). Unlike the missense variants in BIFS and EE, ASD-missense variants cluster in the pore loop that forms the sodium ion selectivity filter (8/13; 3.8-fold enrichment; p=0.0004 binomial test; Fig. 1B and 1C), particularly in the five amino acid residues that line the pore upstream of the ion selectivity filter (3) (6/13; 33-fold enrichment; p=1×10−8 binomial test; Fig. 1C). Many individuals with ASD have co-existing intellectual disability (Fig. 1D) and developmental delay (Table S1 in Supplement 2). Accordingly, the 12 variants in SCN2A in individuals with developmental delay, but no known diagnosis of ASD, are mostly PTVs (8/12, 67%) and de novo (11/12, 92%).
This raises the hypothesis that SCN2A variants resulting in NaV1.2 gain of function and increased neuron excitability lead to infantile seizures, whereas SCN2A variants resulting in NaV1.2 loss of function and decreased neuron excitability lead to ASD and/or developmental delay. If this hypothesis were correct, we would expect the missense variants observed in ASD to reduce NaV1.2 channel function and decrease neuron excitability. We therefore performed functional analyses of all 12 de novo SCN2A mutations observed in the 4,109 ASD cases from the Simons Simplex Collection (SSC; 2,508 families with a single affected child) and Autism Sequencing Consortium (ASC; 1,601 families with one or more affected children), as these cases were identified as representing idiopathic ASD in clearly defined cohorts (4, 9, 22).
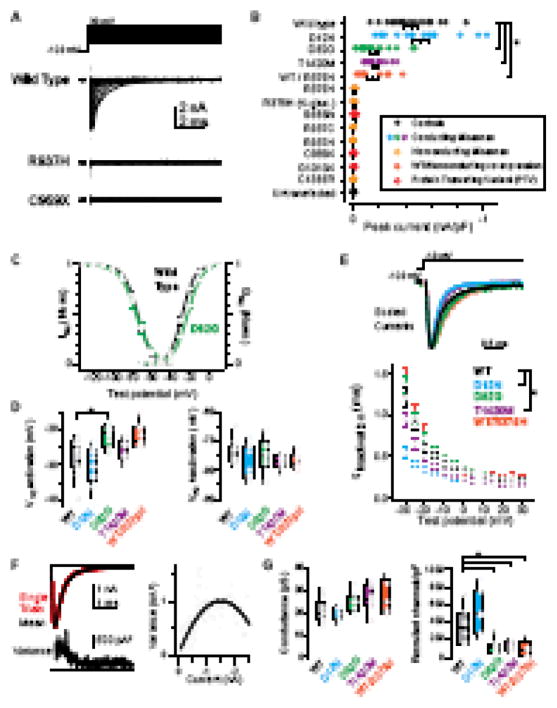
To confirm that PTV SCN2A mutations indeed impair NaV1.2 channel formation, and to determine whether missense mutations alter channel function, we made voltage-clamp recordings from HEK293 cells expressing wild type (WT) or mutated channels. β1 and β2 subunits were co-expressed in all experiments. As predicted, nonsense (p.C959X, p.G1013X) and frameshift (p.S686fs) PTVs lacked Na+ currents (Fig. 2A–B). Constructs recapitulating a fourth PTV that disrupts a canonical splice site in SCN2A (c.4821-1A>T) were not studied here, as it was unclear whether the mRNA recombines after this splice event. The missense mutations produced a range of results. Peak currents from D12N-mutated channels were comparable in size to wild type (WT: −447±45 pA/pF, n=18; D12N: −515±65 pA/pF, n=16; p=0.5), while currents from D82G and T1420M variants were smaller than wild type (D82G: −166±32 pA/pF, n=18, p<0.0001 vs WT; T1420M: −177±22 pA/pF, n=13, p<0.0001 vs WT). Remarkably, all other missense mutations, including pore-localized arginine mutations observed in multiple ASD cases (R379, R937), resulted in a loss of NaV1.2 function. Small residual currents were observed, but these were likely generated by endogenously expressed nonselective cation channels (30), as they were observed in all PTV cases (where no channel was formed but the plasmid vector still expressed GFP) and in untransfected cells (Fig. 2B). Furthermore, no current was observed when using a K+-based intracellular solution, indicating that these channels are not permeable to K+. Despite the lack of current, non-conducting missense mutations still allowed for NaV1.2 channel formation and trafficking, as membrane-associated NaV1.2 staining was evident using confocal and total internal reflectance fluorescence (TIRF) microscopy (Fig. S2 in Supplement 1). Thus, of the 12 ASD-variants tested here, 9 (75%) resulted in a complete loss of NaV1.2 conductance (Fig. 1B, 2B).
Figure 2. Electrophysiology of SCN2A variants.
A. NaV1.2 activation currents from HEK293 cells transfected with wild type, C959X, and R937H plasmids. Capacitance transient blanked for clarity. Note lack of current in either variant.
B. Peak current amplitudes observed during activation protocol for all mutations. Data are normalized to cell capacitance and color coded to match subsequent panels (WT, black; D12N, cyan; D82G, green; T1420M, magenta; other missense: yellow; all loss of function; red). WT/R379H denotes co-expression of the two variants. R379H (K-gluc) denotes only experiments performed with a K-based internal solution. Circles are individual cells, bars are mean±SEM. Data in yellow and red were not different than untransfected controls. Asterisk: D82G, T1420M, and WT/R379H currents were smaller than wild type (p<0.0006, Kruskal-Wallis).
C. Activation and inactivation curves in wild type and D82G. D82G has depolarized activation. Circles and bars are mean±SEM at each test potential.
D. Voltage at which half of current was activated or inactivated for each conducting variant, compared to wild type. Data shown as box plots, with median and quartiles within the box and 10/90th percentile as tails. Individual data points are overlaid as circles. Asterisk: p=0.0043, Kruskal-Wallis.
E. Top, Exemplar currents from WT and conducting variants, scaled to peak. Bottom: inactivation tau determined from single exponential fit (10–90% of peak). Circles and bars are mean±SEM. Asterisk: p<0.01, Repeated measures ANOVA. n=13, 13, 10, 9, 6 cells for WT, D12N, D82G, and T1420N, WT/R379H respectively.
F. Top, overlay of 10 consecutive trials (black) and their average (red). Bottom, average variance2 between individual trials and the average. Right, nonstationary fluctuation analysis was performed by comparing variance with current amplitude. Dots are individual time points, curve is parabolic fit for nonstationary fluctuation analysis.
G. Single channel conductance and the number of channels contributing to the macroscopic current, as determined in F. Data and statistics presented as in D.
We next characterized the electrophysiological properties of the 3 conducting missense variants. Compared to WT, D82G was the only variant in which the voltage-dependence of channel activation was depolarized (Fig. 2C; WT: −26.9±1.3 mV, n=14; D12N: −30.2±1.5 mV, n=13; D82G: −22.0±0.9 mV, n=12, T1420M: −25.5±1.0 mV, n=9; p=0.004, WT vs D82G only). The voltage-dependence of inactivation was not altered in any variant (Fig. 2D, WT: −74.3±1.2 mV, n=12; D12N: −78.0±1.7 mV, n=13; D82G: −74.8±2.0 mV, n=10, T1420M: −77.0±0.8 mV, n=9; p>0.1, all variants vs WT), though both D12N and T1420M currents inactivated more quickly at any given voltage (Fig. 2E, e.g., tau at −30 mV; WT: 1.2±0.1 ms, n=13; D12N: 0.7±0.1 ms, n=13, p<0.001 vs WT; D82G: 1.4±0.1 ms, n=10, p=0.5 vs WT; T1420M: 0.9±0.09 ms, n=9; p<0.01 vs WT; repeated measures ANOVA).
Lastly, we used fluctuation analysis to estimate single channel conductance and functional channel density (Fig. 2F–G). Single channel conductance estimates were comparable to those obtained from recordings of single channels (WT: 20.8±1.8 pS, n=9) (31), and were not affected in any variant (D12N: 20.1±1.2 pS, n=7; D82G: 24.8±1.6 pS, n=8; T1420M: 26.9±1.9 pS, n=9; p>0.05, across all vs WT); however, the number of channels contributing to currents was reduced in D82G and T1420M variants (channels/pF; WT: 345±70, n=8; D12N: 512±91, n=7, p=0.15; D82G: 111±28, n=9, p=0.003; T1420M: 118±18, n=8, p=0.003, all vs WT). Surface NaV staining, assessed with TIRF, was similarly reduced in T1420M variants (Fig. S2C–D in Supplement 1; NaV/GFP mean intensity, WT: 0.45±0.05, n=13; T1420M: 0.31±0.03, n=13, p=0.006); however, no reduction was observed for D82G (0.40±0.03, n=10, p=0.3364). This discrepancy between fluctuation analysis and surface expression for D82G may be due in part to the observed shift in voltage dependent activation. Because D82G channels are less effectively activated at voltages used for fluctuation analysis (see Methods), a smaller fraction of surface-localized channels would contribute to the current. Overall, these results indicate that ASD-associated mutations can result in a wide range of dysfunction in NaV1.2 and that all three of the conducting missense mutations tested reduced NaV1.2 function (Fig. 2).
Dominant negative effects of PTVs on WT channels have been reported when SCN2A α subunits are expressed in isolation; however, these effects are not present when channels are co-expressed with β subunits (32). Some have hypothesized that NaVs form functional complexes in the AIS whereby the activation of one channel hyperpolarizes voltage-dependent activation of its neighbors (33). Non-conducting variants may interfere with coupling between neighboring functional channels. Therefore, we co-expressed WT with the non-conducting variant R379H. While peak currents were smaller than WT alone (−160±41 pA, n=9, p=0.0006), no change in voltage dependence, kinetics, or conductance were observed [V1/2 activation: −22±1.1 mV, n=9, p=0.0472 (post-correction α required for significance: <0.0167); V1/2 inactivation: −76.8±1.0 mV, n=7, p=0.227; tau at −30 mV: 1.4±0.2 ms, n=6, p=0.350; single channel conductance: 27.2±3.4 pS, n=5, p=0.112; channels/pF: 94±41, n=5, p=0.011, all vs WT]. This indicates that non-conducting variants lack dominant negative effects on WT channels and suggests that NaV1.2 channels do not couple functionally (6).
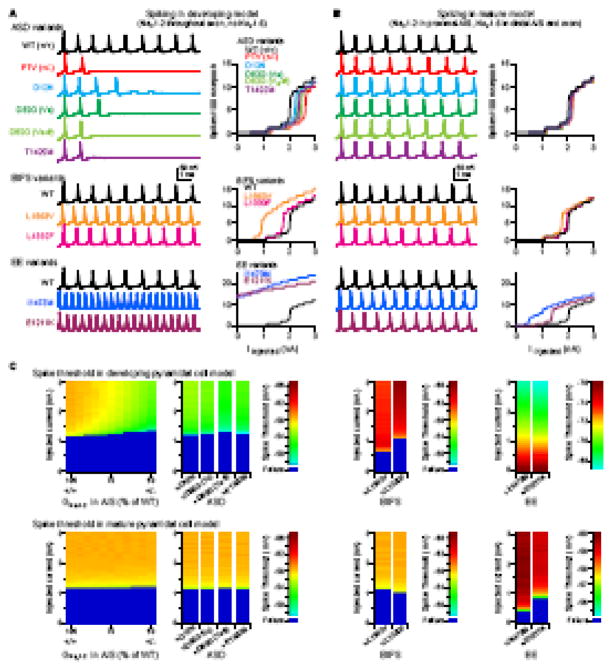
Multiple lines of evidence suggest that disruption in neocortical circuits can contribute to infantile seizures and ASD (3, 30, 32, 34). To test how dysfunction in NaV1.2 affects neuronal excitability, we incorporated each type of NaV1.2 mutation observed into an established computational model of a cortical pyramidal cell (35). Because the mutations observed in ASD are heterozygous, we incorporated equal amounts of wild type and mutated channels into the model and incorporated changes to the affected allele based on results in Fig. 2 (see Methods). To validate that these models accurately predict changes in neuronal excitability, and to compare ASD mutations with other conditions (Fig. 1), BIFS and EE variants were also modeled. These include L1563V and L1330F, which are associated with BIFS and produce modest (~5 mV) shifts in steady state voltage dependence (24), and E1211K and I1473M, which are associated with EE and produce larger (14–22 mV) changes in voltage dependence (23).
As pyramidal neurons are incorporating into cortical networks during early development, they express only NaV1.2 in the AIS (15–17), and it is thought that SCN2A variants associated with BIFS produce seizures because NaV1.2 plays the dominant role in neuronal excitability at this age (7). This time period may also be critical for ASD, as the early development of prefrontal layer 5/6 pyramidal cells has been implicated in the disorder (13). Therefore, we created a developmental model in which NaV1.2 was expressed throughout the AIS and axon and a mature model in which NaV1.2 was replaced by NaV1.6 in the distal AIS and axon. In control conditions, spike threshold was depolarized in the developmental model relative to the mature model, consistent with the differences in voltage-dependent activation between the two isoforms (Fig. 3; Fig. S3 in Supplement 1). Intermediary developmental timepoints with different densities or distributions of NaV1.2 vs NaV1.6 were also modeled (Fig. S4 in Supplement 1).
Figure 3. Neuronal excitability in a cortical pyramidal cell model.
A. Spiking generated with 2.2 nA somatic current in ASD, BIFS, and EE variants. Data are modeled in a developing neuron expressing only NaV1.2 in the AIS and axon. Top, all conducting ASD variants, compared to WT and PTV/non-conducting missense modeled as a 50% reduction in overall NaV1.2 conductance. D82G was modeled as either an activation shift alone (D82G Va) or with a reduction in channel density (D82G Va-#). Bottom: Two BIFS and two EE variants, each compared to WT. Data are vertically offset in 100 mV increments; all traces begin at −78 mV. Plots detail the number of spikes evoked in 100 ms epoch with varying somatic current injections. PTV is from 50% reduction model. Data are color coded as in legend.
B. Same as A, but in a mature pyramidal cell model, with a mix of NaV1.2 and NaV1.6 in the AIS, and NaV1.6 in the axon (see Fig. S3 in Supplement 1 for distributions).
C. Spike threshold with varying levels of NaV1.2 in the AIS, from WT (100%) through different levels of compensation in cells heterozygous for PTV/non-conducting missense variants (to 50%), conducting missense ASD, BIFS, and EE variants. Colored scale bar corresponds to spike threshold of the first spike observed in each trace; dark blue are conditions in which no spikes were generated. Note different scaling for EE conditions, as these variants were excitable at/near rest.
In the developmental model, BIFS variants hyperpolarized spike threshold and enhanced in spike rate, consistent with seizure phenotypes at this early age. EE variants were more excitable, firing spikes even at rest. In contrast, ASD variants were far less excitable than wild type. Full PTV/non-conducting missense models had a rheobase spike threshold 7 mV more depolarized than WT conditions. Similarly, spike threshold in models of—D12N, D82G, and T1420M—variants was depolarized to levels comparable to a 40–50% reduction in overall NaV1.2 conductance. In all cases, spike rate was suppressed, suggesting that even though missense mutations have different effects on NaV1.2 channels, they produce comparable deficits in neuronal excitability. Therefore, all 12 of the observed ASD variants were predicted to reduce the excitability of cortical pyramidal neurons during early development.
In the mature model, BIFS variants had only modest effects on threshold or spike rate, whereas EE variants still resulted in marked hyperexcitability. This is consistent with seizure resolution in BIFS, but not in EE. Interestingly, the ASD variants also resulted in minimal changes in neuronal excitability. Mature excitability developed gradually as more NaV1.6 was incorporated into the AIS (Fig. S4 in Supplement 1). Overall, this suggests that the critical period for SCN2A mutations to produce an ASD phenotype through changes in neuronal excitability occurs during early development.
Discussion
Mutations in SCN2A are strongly associated with both ASD and infantile seizures. Here, we characterized the functional impact of all 11 de novo mutations in SCN2A from ASD cases in the Simons Simplex Collection and Autism Sequencing Consortium. All three PTVs (i.e. nonsense and frameshift) and the majority of missense mutations resulted in a complete loss of NaV1.2 function, despite normal trafficking to the membrane (Fig. 2; Fig S2 in Supplement 1). Three missense mutations D12N, D82G, and T1420M altered channel function in different ways. Remarkably, all 11 alterations in channel function evoked comparable deficits in neuronal excitability in pyramidal cell simulations of the developing brain (Fig. 3) and were consistent with excitability changes observed in recordings from dissociated neurons from heterozygous SCN2A deficient mice (36). Thus, these results indicate that functional haploinsufficiency of SCN2A, generated by a variety of PTV and missense mutations, leads to substantial ASD risk.
The most common result of ASD-related missense mutation was a loss of channel conductance without dominant negative interactions. One of these mutations occurred in the outer vestibule (C1386R), an area important for channel conductance (Cervenka et al., 2010). Several more were observed within the pore at conserved arginine residues (R379, R937). These residues are notable, as mutations here are the only instance of two de novo missense mutations occurring at the same residue in two different families among 4,109 cases with exome data (2). Two missense mutations located on the N-terminal domain altered inactivation kinetics (D12N) and the voltage dependence of steady-state activation (D82), demonstrating, to our knowledge, the first examples of pathological N-terminal mutations in NaV1.2 (37). T1420M, which is in close proximity to the pore selectivity filter (Fig. 1), also had deleterious effects on channel function, increasing the speed of inactivation and reducing functional (Fig. 2G) and physical channel surface density (Fig. S2C–D). Mutation of a homologous residue in rat NaV1.4 to cysteine had modest effects on channel function (38), suggesting that the observed change in pore hydrophobicity is critical at this residue.
Since all SCN2A mutations analyzed led to haploinsufficiency, including all missense variants characterized here, we can revise our estimate of the contribution of SCN2A mutations to ASD risk. Across the 4,109 ASD cases, 12 (0.29%) have mutations resulting in SCN2A haploinsufficiency, a figure marginally higher than that for CHD8 (0.24% if equivalent functional analyses validated three non-PTV mutations along with the seven PTV mutations), making SCN2A the gene with the strongest evidence for ASD-association based on exome analysis and second only to Fragile X Syndrome as a single gene cause of ASD. Therefore, examination of the neurobiology consequent to SCN2A haploinsufficiency, alongside parallel analysis of genes related to chromatin regulation, synaptic structure, and Fragile X Syndrome, is likely to provide critical insights into the etiology of ASD.
Modeling ASD-associated SCN2A mutations in pyramidal neurons predicted a reduction in neuronal excitability in the developing brain in which NaV1.2 is present throughout the AIS and axon, but minimal changes in neuronal excitability in the mature brain in which NaV1.2 is restricted to the proximal AIS (Fig. 3). If neuronal excitability were the main physiological consequence of reduced NaV1.2 function, this would imply that the contribution of SCN2A mutations to the ASD phenotype occurs primarily during early development. Of note, pyramidal cortical neurons during mid-fetal development have previously been implicated in ASD through co-expression networks (13). Since the ASD phenotype persists in the mature brain, the decrease in neuronal excitability would need to result in a downstream change that persists after neuronal excitability returns to physiological levels. Examples of such a downstream effect include the formation and maturation of cortical circuits and the balance of excitatory and inhibitory inputs within these circuits (39, 40).
Alternatively, NaV1.2 dysfunction may continue to affect circuits even after these early developmental time periods in a manner not simulated in our model. APs initiated in the distal AIS propagate forwards, towards axonal release sites, and backwards, throughout the soma and dendrites (backpropagation). In mature pyramidal cells, NaV1.2 channels in the proximal AIS are thought to provide an important electrical boost to promote effective backpropagation of APs into the somatodendritic compartment (6) and this function may still be impaired in the mature cell. Backpropagating spikes are critical signals for activity-dependent transcription, dendritic integration, and synaptic plasticity (41, 42). In this way, mutations in SCN2A may interact with other ASD-associated mutations at the synapse (43, 44), or at the transcriptional level. Thus, even though NaV1.2 channels are restricted to the axon, SCN2A haploinsufficiency may result in cellular and circuit dysfunctions that may be common to other causes of ASD.
In contrast to ASD, the SCN2A variants observed in infantile seizures have a gain of function effect on NaV1.2. Computational models showed that variants observed in BIFS result in increased neuronal excitability during early development, but not in the mature brain (Fig. 3), consistent with the observed resolution of infantile seizures without further apparent neurological impairment (Table S1 in Supplement 2). The variants characterized in EE are predicted to result in a greater degree of neuronal excitability that persists in the mature brain (Fig. 3). This is consistent with the severe seizure phenotype that often persists beyond infancy and is accompanied by profound neurological impairment from an early age (Table S1 in Supplement 2).
Numerous genetic loci associated with neuropsychiatric disorders are observed across a range of diagnoses including developmental delay, ASD, and schizophrenia. Previously, correlation between phenotype and genotype has been described for some large-scale recurrent deletions and duplications (2, 45). Here, we show that functional analysis of apparently similar SCN2A missense mutations (Fig. S1 in Supplement 1) can distinguish two neuropsychiatric phenotypes: infantile seizures and ASD/developmental delay, with further correlation between functional severity and phenotypic severity for infantile seizures (Fig. 3). Thus, the specific functional effect of each SCN2A mutation is directly affecting the observed neuropsychiatric phenotype, presumably with genetic background, environmental influences, and stochastic effects also playing a modifying role. It remains to be seen if further functional characterization can distinguish more closely related phenotypes such as ASD and intellectual disability (Fig. 1D).
While we focused on understanding NaV1.2 dysfunction in cortical pyramidal cells, SCN2A expression in other cell types may also contribute to ASD. Though the majority of cortical interneurons express SCN1A/NaV1.1 at the AIS (46, 47), with loss of function resulting in Dravet Syndrome with EE (48), somatostatin positive interneurons, which play a role in cortical circuit maturation (49, 50), express a mix of both NaV1.1 and 1.2 in the proximal AIS (9). Further, NaV1.2 channels are expressed throughout cerebellar granule cell axons (8), potentially contributing to episodic ataxia (25). Consequently, further functional studies, including in vivo models expressing specific SCN2A variants, will be required to fully understand the pathophysiological consequences of deleterious SCN2A variants. Nevertheless, the comprehensive functional assessment of ASD-associated mutations alongside the existing literature on SCN2A variation and electrophysiology (Table S1 in Supplement 2) provides an initial insight into the genotype-phenotype correlations for this gene. The variants associated with infantile seizures lead to gain of function in NaV1.2, resulting in increased neuronal excitability and the extent of the effect on neuronal excitability may explain the varying severity of this phenotype. These gain of function variants are usually missense variants in proximity to the voltage sensing 4th transmembrane segment and possibly in the C-terminus (Fig. 1A). In contrast, our analysis shows that variants observed in ASD are usually de novo and lead to loss of function by introducing a premature stop codon (PTV) or by disrupting the pore loop and possibly the N-terminus (Fig. 1B). These variants lead to loss of function in NaV1.2 (Fig. 2), resulting in reduced neuronal excitability during development only (Fig. 3). We predict that this defect in neuronal excitability would result in a persistent change in neuronal circuitry or activity that would converge with neurobiological changes observed following disruption of ASD-associated chromatin regulating or synaptic genes.
Supplementary Material
Acknowledgments
We are grateful to all the families participating in this research, including the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC) and the Autism Sequencing Consortium (ASC). We thank members of the Bender and Sanders labs, as well as Drs. A. Brumback, J. Rubenstein, V. Sohal, and M. State for comments. This work was supported by the Simons Foundation (SFARI 362242, KJB) and the NIH (F32NS095580, RBS and R01 MH110928, SJS).
Footnotes
Author Contributions:
Conceptualization, RB, SJS, and KJB; Acquisition of data, RB, CMK, KNB, and JYA; Data Analysis, RB, KNB, JYA, SJS, and KJB; Writing, RB, SJS, and KJB.
Financial Disclosure:
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald TW, Gerety SS, Jones WD, van Kogelenberg M, King DA, McRae J, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu W, Tian C, Li T, Yang M, Hou H, Shu Y. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nature neuroscience. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- 7.Liao Y, Deprez L, Maljevic S, Pitsch J, Claes L, Hristova D, et al. Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain: a journal of neurology. 2010;133:1403–1414. doi: 10.1093/brain/awq057. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Hernandez J, Ballesteros-Merino C, Fernandez-Alacid L, Nicolau JC, Aguado C, Lujan R. Polarised localisation of the voltage-gated sodium channel Na(v)1.2 in cerebellar granule cells. Cerebellum. 2013;12:16–26. doi: 10.1007/s12311-012-0387-1. [DOI] [PubMed] [Google Scholar]
- 9.Li T, Tian C, Scalmani P, Frassoni C, Mantegazza M, Wang Y, et al. Action potential initiation in neocortical inhibitory interneurons. PLoS biology. 2014;12:e1001944. doi: 10.1371/journal.pbio.1001944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian C, Wang K, Ke W, Guo H, Shu Y. Molecular identity of axonal sodium channels in human cortical pyramidal cells. Frontiers in cellular neuroscience. 2014;8:297. doi: 10.3389/fncel.2014.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell KB, McMahon JM, Carvill GL, Tambunan D, Mackay MT, Rodriguez-Casero V, et al. SCN2A encephalopathy: A major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85:958–966. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nature neuroscience. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boiko T, Van Wart A, Caldwell JH, Levinson SR, Trimmer JS, Matthews G. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:2306–2313. doi: 10.1523/JNEUROSCI.23-06-02306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazina EV, Leaw BT, Richards KL, Wimmer VC, Kim TH, Aumann TD, et al. ‘Neonatal’ Nav1.2 reduces neuronal excitability and affects seizure susceptibility and behaviour. Human molecular genetics. 2015;24:1457–1468. doi: 10.1093/hmg/ddu562. [DOI] [PubMed] [Google Scholar]
- 17.Osorio N, Alcaraz G, Padilla F, Couraud F, Delmas P, Crest M. Differential targeting and functional specialization of sodium channels in cultured cerebellar granule cells. The Journal of physiology. 2005;569:801–816. doi: 10.1113/jphysiol.2005.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender KJ, Trussell LO. The physiology of the axon initial segment. Annual review of neuroscience. 2012;35:249–265. doi: 10.1146/annurev-neuro-062111-150339. [DOI] [PubMed] [Google Scholar]
- 19.Kole MH, Stuart GJ. Signal processing in the axon initial segment. Neuron. 2012;73:235–247. doi: 10.1016/j.neuron.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Kole MH, Ilschner SU, Kampa BM, Williams SR, Ruben PC, Stuart GJ. Action potential generation requires a high sodium channel density in the axon initial segment. Nature neuroscience. 2008;11:178–186. doi: 10.1038/nn2040. [DOI] [PubMed] [Google Scholar]
- 21.Xu R, Thomas EA, Jenkins M, Gazina EV, Chiu C, Heron SE, et al. A childhood epilepsy mutation reveals a role for developmentally regulated splicing of a sodium channel. Molecular and cellular neurosciences. 2007;35:292–301. doi: 10.1016/j.mcn.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Lauxmann S, Boutry-Kryza N, Rivier C, Mueller S, Hedrich UB, Maljevic S, et al. An SCN2A mutation in a family with infantile seizures from Madagascar reveals an increased subthreshold Na(+) current. Epilepsia. 2013;54:e117–121. doi: 10.1111/epi.12241. [DOI] [PubMed] [Google Scholar]
- 23.Ogiwara I, Ito K, Sawaishi Y, Osaka H, Mazaki E, Inoue I, et al. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–1053. doi: 10.1212/WNL.0b013e3181b9cebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scalmani P, Rusconi R, Armatura E, Zara F, Avanzini G, Franceschetti S, et al. Effects in neocortical neurons of mutations of the Na(v)1.2 Na+ channel causing benign familial neonatal-infantile seizures. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:10100–10109. doi: 10.1523/JNEUROSCI.2476-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz N, Hahn A, Bast T, Muller S, Loffler H, Maljevic S, et al. Mutations in the sodium channel gene SCN2A cause neonatal epilepsy with late-onset episodic ataxia. Journal of neurology. 2016;263:334–343. doi: 10.1007/s00415-015-7984-0. [DOI] [PubMed] [Google Scholar]
- 26.Misra SN, Kahlig KM, George AL., Jr Impaired NaV1.2 function and reduced cell surface expression in benign familial neonatal-infantile seizures. Epilepsia. 2008;49:1535–1545. doi: 10.1111/j.1528-1167.2008.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders SJ. First glimpses of the neurobiology of autism spectrum disorder. Current opinion in genetics & development. 2015;33:80–92. doi: 10.1016/j.gde.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrovski S, Gussow AB, Wang Q, Halvorsen M, Han Y, Weir WH, et al. The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity. PLoS genetics. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu G, Zhang Y, Xu H, Jiang C. Identification of endogenous outward currents in the human embryonic kidney (HEK 293) cell line. Journal of neuroscience methods. 1998;81:73–83. doi: 10.1016/s0165-0270(98)00019-3. [DOI] [PubMed] [Google Scholar]
- 31.Stuhmer W, Methfessel C, Sakmann B, Noda M, Numa S. Patch clamp characterization of sodium channels expressed from rat brain cDNA. European biophysics journal: EBJ. 1987;14:131–138. doi: 10.1007/BF00253837. [DOI] [PubMed] [Google Scholar]
- 32.Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naundorf B, Wolf F, Volgushev M. Unique features of action potential initiation in cortical neurons. Nature. 2006;440:1060–1063. doi: 10.1038/nature04610. [DOI] [PubMed] [Google Scholar]
- 34.Noebels J. Pathway-driven discovery of epilepsy genes. Nature neuroscience. 2015;18:344–350. doi: 10.1038/nn.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallermann S, de Kock CP, Stuart GJ, Kole MH. State and location dependence of action potential metabolic cost in cortical pyramidal neurons. Nature neuroscience. 2012;15:1007–1014. doi: 10.1038/nn.3132. [DOI] [PubMed] [Google Scholar]
- 36.Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, et al. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophysical journal. 2000;78:2878–2891. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugawara T, Tsurubuchi Y, Agarwala KL, Ito M, Fukuma G, Mazaki-Miyazaki E, et al. A missense mutation of the Na+ channel alpha II subunit gene Na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6384–6389. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamagishi T, Xiong W, Kondratiev A, Velez P, Mendez-Fitzwilliam A, Balser JR, et al. Novel Molecular Determinants in the Pore Region of Sodium Channels Regulate Local Anesthetic Binding. Molecular pharmacology. 2009;76:861–871. doi: 10.1124/mol.109.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepherd GM, Katz DM. Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and Met. Current opinion in neurobiology. 2011;21:827–833. doi: 10.1016/j.conb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldman DE. The Spike-Timing Dependence of Plasticity. Neuron. 2012;75:556–571. doi: 10.1016/j.neuron.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha RN, Dudek SM. Action potentials: to the nucleus and beyond. Exp Biol Med (Maywood) 2008;233:385–393. doi: 10.3181/0709-MR-241. [DOI] [PubMed] [Google Scholar]
- 43.Aceti M, Creson TK, Vaissiere T, Rojas C, Huang WC, Wang YX, et al. Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biological psychiatry. 2015;77:805–815. doi: 10.1016/j.biopsych.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philpot BD, Thompson CE, Franco L, Williams CA. Angelman syndrome: advancing the research frontier of neurodevelopmental disorders. Journal of neurodevelopmental disorders. 2011;3:50–56. doi: 10.1007/s11689-010-9066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. The British journal of psychiatry: the journal of mental science. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Catterall WA, Kalume F, Oakley JC. NaV1.1 channels and epilepsy. The Journal of physiology. 2010;588:1849–1859. doi: 10.1113/jphysiol.2010.187484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Depienne C, Trouillard O, Saint-Martin C, Gourfinkel-An I, Bouteiller D, Carpentier W, et al. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. Journal of medical genetics. 2009;46:183–191. doi: 10.1136/jmg.2008.062323. [DOI] [PubMed] [Google Scholar]
- 49.Anastasiades PG, Marques-Smith A, Lyngholm D, Lickiss T, Raffiq S, Katzel D, et al. GABAergic interneurons form transient layer-specific circuits in early postnatal neocortex. Nature communications. 2016;7:10584. doi: 10.1038/ncomms10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuncdemir SN, Wamsley B, Stam FJ, Osakada F, Goulding M, Callaway EM, et al. Early Somatostatin Interneuron Connectivity Mediates the Maturation of Deep Layer Cortical Circuits. Neuron. 2016;89:521–535. doi: 10.1016/j.neuron.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.