Abstract
Random forest (RF) missing data algorithms are an attractive approach for imputing missing data. They have the desirable properties of being able to handle mixed types of missing data, they are adaptive to interactions and nonlinearity, and they have the potential to scale to big data settings. Currently there are many different RF imputation algorithms, but relatively little guidance about their efficacy. Using a large, diverse collection of data sets, imputation performance of various RF algorithms was assessed under different missing data mechanisms. Algorithms included proximity imputation, on the fly imputation, and imputation utilizing multivariate unsupervised and supervised splitting—the latter class representing a generalization of a new promising imputation algorithm called missForest. Our findings reveal RF imputation to be generally robust with performance improving with increasing correlation. Performance was good under moderate to high missingness, and even (in certain cases) when data was missing not at random.
Keywords: Imputation, Correlation, Missingness, Splitting (random, univariate, multivariate, unsupervised), Machine Learning
1 Introduction
Missing data is a real world problem often encountered in scientific settings. Data that is missing is problematic as many statistical analyses require complete data. This forces researchers who want to use a statistical analysis that requires complete data to choose between imputing data or discarding missing values. But to simply discard missing data is not a reasonable practice, as valuable information may be lost and inferential power compromised [1]. Thus, imputing missing data in such settings is a more reasonable and practical way to proceed.
While many statistical methods have been developed for imputing missing data, many of these perform poorly in high dimensional and large scale data settings; for example, in genomic, proteomic, neuroimaging, and other high-throughput problems. In particular, it is generally recommended that all variables be included in multiple imputation to make it proper in general and in order to not create bias in the estimate of the correlations [2]. But this can lead to overparameterization when there are a large number of variables and the sample size is moderate. Computational issues may also arise. An example is the occurrence of non-convexity due to missing data when maximizing the log-likelihood. This creates problems for traditional optimization methods such as the EM algorithm [3]. Missing data methods are also often designed only for continuous data (for example, gene expression data [4]), and for methods applicable to mixed data (i.e., data having both nominal and categorical variables), implementation can often break down in challenging data settings [5]. Another concern is the inability to deal with complex interactions and nonlinearity of variables. Standard multiple imputation approaches do not automatically incorporate interaction effects, which leads to biased parameter estimates when interactions are present [6]. Although some techniques, such as fully conditional specification of covariates can be used to try to resolve this problem [7], these techniques can be difficult and inefficient to implement in settings where interactions are expected to be complicated.
For these reasons there has been much interest in using machine learning methods for missing data imputation. A promising approach can be based on Breiman’s random forests [8] (abbreviated hereafter as RF). RF have the desired characteristic that they: (1) handle mixed types of missing data; (2) address interactions and nonlinearity; (3) scale to high-dimensions while avoiding overfitting; and (4) yield measures of variable importance useful for variable selection. Currently there are several different RF missing data algorithms. This includes the original RF proximity algorithm proposed by Breiman [9] implemented in the randomForestR-package [10]. A different class of algorithms are the “on-the-fly-imputation” algorithms implemented in the randomSurvivalForestR-package [11], which allow data to be imputed while simultaneously growing a survival tree. These algorithms have been unified within the randomForestSRCR-package (abbreviated as RF-SRC) to include not only survival, but classification and regression among other settings [12]. A third approach is missForest, a method recently introduced in [13]. Missforest takes a different approach by recasting the missing data problem as a prediction problem. Data is imputed by regressing each variable in turn against all other variables and then predicting missing data for the dependent variable using the fitted forest. MissForest has been shown [14] to outperform well known methods such as k-nearest neighbors [15] and parametric MICE [16] (multivariate imputation using chained equation).
Given that RF meets all the characteristics for handling missing data, it seems desirable to use RF for imputing data. However, practitioners have very little guidance about which of these algorithms to use, as there have been no systematic studies looking at the comparative behavior of RF missing data algorithms. Comparisons of RF imputation to other procedures have been considered by [13, 14], and there have been studies looking at effectiveness of RF imputation when combined with other methods (for instance [17] showed that parameter estimates were less biased when using MICE with random forest based imputation), but no systematic study of performance between RF algorithms has been attempted. To address this gap in the literature, we therefore sought to study various RF algorithms and systematically document their performance using a large empirical study involving 60 data sets. Performance was assessed by imputation accuracy and computational speed. Different missing data mechanisms (missing at random and not missing at random) were used to assess robustness. In addition to the RF missing data algorithms described above, we also considered several new algorithms, including a multivariate version of missForest, referred to as mForest. Despite the superior performance of missForest (a finding confirmed in our experiments), the algorithm is computationally expensive to implement in high-dimensions as a separate forest must be calculated for each variable and the algorithm run until convergence is achieved. The mForest algorithm helps to alleviate this problem by grouping variables and using multivariate forests with each group used in turn as the set of dependent variables. This replaces p regressions, where p is the number of variables, with ≈ 1/α regressions, where 0 < α < 1 is a user specified group fraction size. Computational savings were found to be substantial for mForest without overly compromising accuracy even for relatively large α. Other RF algorithms studied included a new multivariate unsupervised algorithm and algorithms utilizing random splitting.
All forests constructed in the manuscript followed the RF methodology of [8]. Trees were grown using independently sampled bootstrap data. For univariate regression, continuous variables were split using squared-error splitting; categorical variables by the Gini index [18]. More general splitting rules, such as unsupervised and multivariate splitting, were also employed, and are described later in the manuscript. Random feature selection was used, with mtryvariables selected at random prior to splitting a tree node, and trees grown as deeply as possible subject to the constraint of a lower bound of nodesizeunique data values within a terminal node. RF missing data algorithms were implemented using the randomForestSRCR-package [12], which has been extended to include a new impute.rfsrcfunction optimized specifically for data imputation. The package implements openMP parallel processing, which allows for parallel processing on user desktops as well as large scale computing clusters; thus greatly reducing computational times.
2 RF approaches to imputation
Three general strategies have been used for RF missing data imputation:
Preimpute the data; grow the forest; update the original missing values using proximity of the data. Iterate for improved results.
Simultaneously impute data while growing the forest; iterate for improved results.
Preimpute the data; grow a forest using in turn each variable that has missing values; predict the missing values using the grown forest. Iterate for improved results.
Proximity imputation [9] uses strategy A, on-the-fly-imputation [11] (OTFI) uses strategy B, and missforest [13] uses strategy C. The way these algorithms impute data is very different. Even the manner in which they model outcome data (i.e. “Y” variables) is different. Strategy C does not use outcome data at all and is solely a technique for imputing features (X variables), while strategies A and B utilize outcome information in their model building. Below we detail each of these strategies and describe various algorithms which utilize one of these three approaches. These new algorithms take advantage of new splitting rules, including random splitting, unsupervised splitting, and multivariate splitting [12].
2.1 Strawman imputation
We first begin by describing a “strawman imputation” which will be used throughout as our baseline reference value. While this algorithm is rough, it is also very rapid, and for this reason it was also used to initialize some of our RF procedures. Strawman imputation is defined as follows. Missing values for continuous variables are imputed using the median of non-missing values, and for missing categorical variables, the most frequently occurring non-missing value is used (ties are broken at random).
2.2 Proximity imputation: RFprx and RFprxR
Here we describe proximity imputation (strategy A). In this procedure the data is first roughly imputed using strawman imputation. A RF is fit using this imputed data. Using the resulting forest, the n × n symmetric proximity matrix (n equals the sample size) is determined where the (i, j) entry records the inbag frequency that case i and j share the same terminal node. The proximity matrix is used to impute the original missing values. For continuous variables, the proximity weighted average of non-missing data is used; for categorical variables, the largest average proximity over non-missing data is used. The updated data are used to grow a new RF, and the procedure is iterated.
We use RFprx to refer to proximity imputation as described above. However, when implementing RFprx we use a slightly modified version that makes use of random splitting in order to increase computational speed. In random splitting, nsplit, a non-zero positive integer, is specified by the user. A maximum of nspit-split points are chosen randomly for each of the randomly selected mtrysplitting variables. This is in contrast to non-random (deterministic) splitting typically used by RF, where all possible split points for each of the potential mtrysplitting variables are considered. The splitting rule is applied to the nsplit randomly selected split points and the tree node is split on the variable with random split point yielding the best value, as measured by the splitting criterion. Random splitting evaluates the splitting rule over a much smaller number of split points and is therefore considerably faster than deterministic splitting.
The limiting case of random splitting is pure random splitting. The tree node is split by selecting a variable and the split-point completely at random—no splitting rule is applied; i.e. splitting is completely non-adaptive to the data. Pure random splitting is generally the fastest type of random splitting. We also apply RFprx using pure random splitting; this algorithm is denoted by RFprxR.
As an extension to the above methods, we implement iterated versions of RFprx and RFprxR. To distinguish between the different algorithms, we write and when they are iterated k ≥ 1 times. Thus, and indicates that the algorithms were iterated 5 times, while and indicates that the algorithms were not iterated. However, as this latter notation is somewhat cumbersome, for notational simplicity we will simply use RFprx to denote and RFprxR for .
2.3 On-the-fly-imputation (OTFI): RFotf and RFotfR
A disadvantage of the proximity approach is that OOB (out-of-bag) estimates for prediction error are biased [9]. Further, because prediction error is biased, so are other measures based on it, such as variable importance. The method is also awkward to implement on test data with missing values. The OTFI method [11] (strategy B) was devised to address these issues. Specific details of OTFI can be found in [11, 12], but for convenience we summarize the key aspects of OTFI below:
Only non-missing data is used to calculate the split-statistic for splitting a tree node.
When assigning left and right daughter node membership if the variable used to split the node has missing data, missing data for that variable is “imputed” by drawing a random value from the inbag non-missing data.
Following a node split, imputed data are reset to missing and the process is repeated until terminal nodes are reached. Note that after terminal node assignment, imputed data are reset back to missing, just as was done for all nodes.
Missing data in terminal nodes are then imputed using OOB non-missing terminal node data from all the trees. For integer valued variables, a maximal class rule is used; a mean rule is used for continuous variables.
It should be emphasized that the purpose of the “imputed data” in Step 2 is only to make it possible to assign cases to daughter nodes—imputed data is not used to calculate the split-statistic, and imputed data is only temporary and reset to missing after node assignment. Thus, at the completion of growing the forest, the resulting forest contains missing values in its terminal nodes and no imputation has been done up to this point. Step 4 is added as a means for imputing the data, but this step could be skipped if the goal is to use the forest in analysis situations. In particular, step 4 is not required if the goal is to use the forest for prediction. This applies even when test data used for prediction has missing values. In such a scenario, test data assignment works in the same way as in step 2. That is, for missing test values, values are imputed as in step 2 using the original grow distribution from the training forest, and the test case assigned to its daughter node. Following this, the missing test data is reset back to missing as in step 3, and the process repeated.
This method of assigning cases with missing data, which is well suited for forests, is in contrast to surrogate splitting utilized by CART [18]. To assign a case having a missing value for the variable used to split a node, CART uses the best surrogate split among those variables not missing for the case. This ensures every case can be classified optimally, whether the case has missing values or not. However, while surrogate splitting works well for CART, the method is not well suited for forests. Computational burden is one issue. Finding a surrogate split is computationally expensive even for one tree, let alone for a large number of trees. Another concern is that surrogate splitting works tangentially to random feature selection used by forests. In RF, candidate variables for splitting a node are selected randomly, and as such the candidate variables may be uncorrelated with one another. This can make it difficult to find a good surrogate split when missing values are encountered, if surrogate splitting is restricted to candidate variables. Another concern is that surrogate splitting alters the interpretation of a variable, which affects measures such as variable importance measures [11].
To denote the OTFI missing data algorithm, we will use the abbreviation RFotf. As in proximity imputation, to increase computational speed, RFotf is implemented using nsplitrandom splitting. We also consider OTFI under pure random splitting and denote this algorithm by RFotfR. Both algorithms are iterated in our studies. RFotf, RFotfR will be used to denote a single iteration, while denotes five iterations. Note that when OTFI algorithms are iterated, the terminal node imputation executed in step 4 uses inbag data and not OOB data after the first cycle. This is because after the first cycle of the algorithm, no coherent OOB sample exists.
Remark 1
As noted by one of our referees, missingness incorporated in attributes (MIA) is another tree splitting method which bypasses the need to impute data [19, 20]. Again, this only applies if the user is interested in a forest analysis. MIA accomplishes this by treating missing values as a category which is incorporated into the splitting rule. Let X be an ordered or numeric feature being used to split a node. The MIA splitting rule searches over all possible split values s of X of the following form:
Split A: {X ≤ s or X = missing} versus {X >s}.
Split B: {X ≤ s} versus {X >s or X = missing}.
Split C: {X = missing} versus {X = not missing}.
Thus, like OTF splitting, one can see that MIA results in a forest ensemble constructed without having to impute data.
2.4 Unsupervised imputation: RFunsv
RFunsv refers to OTFI using multivariate unsupervised splitting. However unlike the OTFI algorithm, the RFunsv algorithm is unsupervised and assumes there is no response (outcome) variable. Instead a multivariate unsupervised splitting rule [12] is implemented. As in the original RF algorithm, at each tree node t, a set of mtryvariables are selected as potential splitting variables. However, for each of these, as there is no outcome variable, a random set of ytryvariables is selected and defined to be the multivariate response (pseudooutcomes). A multivariate composite splitting rule of dimension ytry(see below) is applied to each of the mtrymultivariate regression problems and the node t split on the variable leading to the best split. Missing values in the response are excluded when computing the composite multivariate splitting rule: the split-rule is averaged over non-missing responses only [12]. We also consider an iterated RFunsv algorithm (e.g. implies five iterations, RFunsv implies no iterations).
Here is the description of the multivariate composite splitting rule. We begin by considering univariate regression. For notational simplicity, let us suppose the node t we are splitting is the root node based on the full sample size n. Let X be the feature used to split t, where for simplicity we assume X is ordered or numeric. Let s be a proposed split for X that splits t into left and right daughter nodes tL:= tL(s) and tR:= tR(s), where tL = {Xi ≤ s} and tR = {Xi > s}. Let nL = #tL and nR = #tR denote the sample sizes for the two daughter nodes. If Yi denotes the outcome, the squared-error split-statistic for the proposed split is
where ȲtL and ȲtR are the sample means for tL and tR respectively. The best split for X is the split-point s minimizing D(s, t). To extend the squared-error splitting rule to the multivariate case q > 1, we apply univariate splitting to each response coordinate separately. Let Yi = (Yi,1, …, Yi,q)T denote the q ≥ 1 dimensional outcome. For multivariate regression analysis, an averaged standardized variance splitting rule is used. The goal is to minimize
where ȲtLj and ȲtRj are the sample means of the j-th response coordinate in the left and right daughter nodes. Notice that such a splitting rule can only be effective if each of the coordinates of the outcome are measured on the same scale, otherwise we could have a coordinate j, with say very large values, and its contribution would dominate Dq(s, t). We therefore calibrate Dq(s, t) by assuming that each coordinate has been standardized according to
The standardization is applied prior to splitting a node. To make this standardization clear, we denote the standardized responses by . With some elementary manipulations, it can be verified that minimizing Dq(s, t) is equivalent to maximizing
| (1) |
For multivariate classification, an averaged standardized Gini splitting rule is used. First consider the univariate case (i.e., the multiclass problem) where the outcome Yi is a class label from the set {1, …, K} where K ≥ 2. The best split s for X is obtained by maximizing
where Zi(k) = 1{Yi=k}. Now consider the multivariate classification scenario r > 1, where each outcome coordinate Yi,j for 1 ≤ j ≤ r is a class label from {1, …, Kj} for Kj ≥ 2. We apply Gini splitting to each coordinate yielding the extended Gini splitting rule
| (2) |
where Zi(k),j = 1{Yi,j=k}. Note that the normalization 1/Kj employed for a coordinate j is required to standardize the contribution of the Gini split from that coordinate.
Observe that (1) and (2) are equivalent optimization problems, with optimization over for regression and Zi(k),j for classification. As shown in [21] this leads to similar theoretical splitting properties in regression and classification settings. Given this equivalence, we can combine the two splitting rules to form a composite splitting rule. The mixed outcome splitting rule Θ(s, t) is a composite standardized split rule of mean-squared error (1) and Gini index splitting (2); i.e.,
where p = q + r. The best split for X is the value of s maximizing Θ(s, t).
Remark 2
As discussed in [22], multivariate regression splitting rules patterned after the Mahalanobis distance can be used to incorporate correlation between response coordinates. Let ȲL and ȲR be the multivariate means for Y in the left and right daughter nodes, respectively. The following Mahalanobis distance splitting rule was discussed in [22]
where V̂L and V̂R are the estimated covariance matrices for the left and right daughter nodes. While this is a reasonable approach in low dimensional problems, recall that we are applying Dq(s, t) to ytryof the feature variables which could be large if the feature space dimension p is large. Also, because of missing data in the features, it may be difficult to derive estimators for V̂L and V̂R, which is further complicated if their dimensions are high. This problem becomes worse as the tree is grown because the number of observations decreases rapidly making estimation unstable. For these reasons, we use the splitting rule Dq(s, t) rather than Mq(s, t) when implementing imputation.
2.5 mForest imputation: mRFα and mRF
The missForest algorithm recasts the missing data problem as a prediction problem. Data is imputed by regressing each variable in turn against all other variables and then predicting missing data for the dependent variable using the fitted forest. With p variables, this means that p forests must be fit for each iteration, which can be slow in certain problems. Therefore, we introduce a computationally faster version of missForest, which we call mForest. The new algorithm is described as follows. Do a quick strawman imputation of the data. The p variables in the data set are randomly assigned into mutually exclusive groups of approximate size αp where 0 < α < 1. Each group in turn acts as the multivariate response to be regressed on the remaining variables (of approximate size (1−α)p). Over the multivariate responses, set imputed values back to missing. Grow a forest using composite multivariate splitting. As in RFunsv, missing values in the response are excluded when using multivariate splitting: the split-rule is averaged over non-missing responses only. Upon completion of the forest, the missing response values are imputed using prediction. Cycle over all of the ≈ 1/α multivariate regressions in turn; thus completing one iteration. Check if the imputation accuracy of the current imputed data relative to the previously imputed data has increased beyond an ε-tolerance value (see Section 3.3 for measuring imputation accuracy). Stop if it has, otherwise repeat until convergence.
To distinguish between mForest under different α values we use the notation mRFα to indicate mForest fit under the specified α. Note that the limiting case α = 1/p corresponds to missForest. Although technically the two algorithms missForest and mRFα for α = 1/p are slightly different, we did not find significant differences between them during informal experimentation. Therefore for computational speed, missForest was taken to be mRFα for α = 1/p and denoted simply as mRF.
3 Methods
3.1 Experimental design and data
Table 1 lists the nine experiments that were carried out. In each experiment, a pre-specified target percentage of missing values was induced using one of three different missing mechanisms [23]:
Table 1.
Experimental design used for large scale study of RF missing data algorithms.
| Missing Mechanism | Percent Missing | |
|---|---|---|
| EXPT-A | MCAR | 25 |
| EXPT-B | MCAR | 50 |
| EXPT-C | MCAR | 75 |
| EXPT-D | MAR | 25 |
| EXPT-E | MAR | 50 |
| EXPT-F | MAR | 75 |
| EXPT-G | NMAR | 25 |
| EXPT-H | NMAR | 50 |
| EXPT-I | NMAR | 75 |
Missing completely at random (MCAR). This means the probability that an observation is missing does not depend on the observed value or the missing ones.
Missing at random (MAR). This means that the probability of missingness may depend upon observed values, but not missing ones.
Not missing at random (NMAR). This means the probability of missingness depends on both observed and missing values.
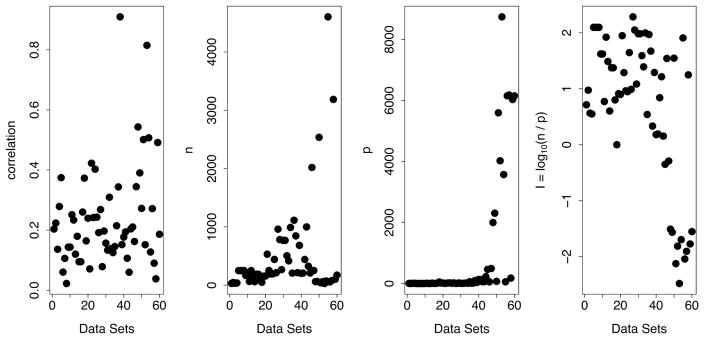
Sixty data sets were used, including both real and synthetic data. Figure 1 illustrates the diversity of the data. Displayed are data sets in terms of correlation (ρ), sample size (n), number of variables (p), and the amount of information contained in the data (I = log10(n/p)). The correlation, ρ, was defined as the L2-norm of the correlation matrix. If R = (ρi,j) denotes the p × p correlation matrix for a data set, ρ was defined to equal
Figure 1.
Summary values for the 60 data sets used in the large scale RF missing data experiment. The last panel displays the log-information, I = log10(n/p), for each data set.
| (3) |
This is similar to the usual definition of the L2-norm for a matrix, but where we have modifed the definition to remove the diagonal elements of R which equal 1, as well as the contribution from the symmetric lower diagonal values.
Note that in the plot for p there are 10 data sets with p in the thousands—these are a collection of well known gene expression data sets. The right-most plot displays the log-information of a data set, I = log10(n/p). The range of values on the log-scale vary from −2 to 2; thus the information contained in a data set can differ by as much as ≈ 104.
3.2 Inducing MCAR, MAR and NMAR missingness
The following procedures were used to induce missigness in the data. Let the target missigness fraction be 0 < γNA < 1. For MCAR, data was set to missing randomly without imposing column or row constraints to the data matrix. Specifically, the data matrix was made into a long vector and nγNA of the entries selected at random and set to missing.
For MAR, missing values were assigned by column. Let Xj = (X1,j, …, Xn,j) be the n-dimensional vector containing the original values of the jth variable, 1 ≤ j ≤ p. Each coordinate of Xj was made missing according to the tail behavior of a randomly selected covariate Xk, where k ≠ j. The probability of selecting coordinate Xi,j was
where F(x) = (1 + exp(−3x))−1 and Bj were i.i.d. symmetric 0–1 Bernoulli random variables. With this method, about half of the variables will have higher missigness in those coordinates corresponding to the right tail of a randomly selected variable (the other half will have higher missigness depending on the left tail of a randomly selected variable). A total of nγNA coordinates were selected from Xj and set to missing. This induces MAR, as missing values for Xj depend only on observed values of another variable Xk.
For NMAR, each coordinate ofXj was made missing according to its own tail behavior. A total of nγNA values were selected according to
Notice that missingness in Xi,j depends on both observed and missing values. In particular, missing values occur with higher probability in the right and left tails of the empirical distribution. Therefore, this induces NMAR.
3.3 Measuring imputation accuracy
Accuracy of imputation was assessed using the following metric. As described above, values of Xj were made missing under various missing data assumptions. Let (11,j, …, 1n,j) be a vector of zeroes and ones indicating which values of Xj were artificially made missing. Define 1i,j = 1 if Xi,j is artificially missing; otherwise 1i,j = 0. Let be the number of artificially induced missing values for Xj.
Let 𝒩 and 𝒞 be the set of nominal (continuous) and categorical (factor) variables with more than one artificially induced missing value. That is,
Standardized root-mean-squared error (RMSE) was used to assess performance for nominal variables, and misclassification error for factors. Let be the n-dimensional vector of imputed values for Xj using procedure ℐ. Imputation error for ℐ was measured using
where . To be clear regarding the standardized RMSE, observe that the denominator in the first term is the variance of Xj over the artificially induced missing values, while the numerator is the MSE difference of Xj and over the induced missing values.
As a benchmark for assessing imputation accuracy we used strawman imputation described earlier, which we denote by 𝒮. Imputation error for a procedure ℐ was compared to 𝒮 using relative imputation error defined as
A value of less than 100 indicates a procedure ℐ performing better than the strawman.
3.4 Experimental settings for procedures
Randomized splitting was invoked with an nsplitvalue of 10. For random feature selection, mtrywas set to . For random outcome selection for RFunsv, we set ytryto equal . Algorithms RFotf, RFunsv and RFprx were iterated 5 times in addition to being run for a single iteration. For mForest, the percentage of variables used as responses was α = .05, .25. This implies that mRF0.05 used up to 20 regressions per cycle, while mRF0.25 used 4. Forests for all procedures were grown using a nodesizevalue of 1. Number of trees was set at ntree= 500. Each experimental setting (Table 1) was run 100 times independently and results averaged.
For comparison, k-nearest neighbors imputation (hereafter denoted as KNN) was applied using the impute.knnfunction from the R-package impute[24]. For each data point with missing values, the algorithm determines the k-nearest neighbors using a Euclidean metric, confined to the columns for which that data point is not missing. The missing elements for the data point are then imputed by averaging the non-missing elements of its neighbors. The number of neighbors k was set at the default value k = 10. In experimentation we found the method robust to the value of k and therefore opted to use the default setting. Much more important were the parameters rowmaxand colmaxwhich control the maximum percent missing data allowed in each row and column of the data matrix before a rough overall mean is used to impute the row/column. The default values of 0.5 and 0.8, respectively, were too low and led to poor performance in the heavy missing data experiments. Therefore, these values were set to their maximum of 1.0, which greatly improved performance. Our rationale for selecting KNN as a comparison procedure is due to its speed because of the large scale nature of experiments (total of 100×60×9 = 54, 000 runs for each method). Another reason was because of its close relationship to forests. This is because RF is also a type of nearest neighbor procedure—although it is an adaptive nearest neighbor. We comment later on how adaptivity may give RF an advantage over KNN.
4 Results
Section 4.1 presents the results of the performance of a procedure as measured by relative imputation accuracy, ℰR(ℐ). In Section 4.2 we discuss computational speed.
4.1 Imputation Accuracy
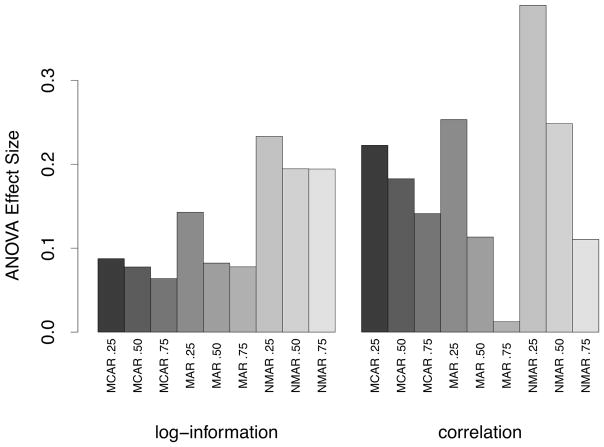
In reporting the values for imputation accuracy, we have stratified data sets into low, medium and high-correlation groups, where correlation, ρ, was defined as in (3). Low, medium and high-correlation groups were defined as groups whose ρ value fell into the [0, 50], [50, 75] and [75, 100] percentile for correlations. Results were stratified by ρ because we found it played a very heavy role in imputation performance and was much more informative than other quantities measuring information about a data set. Consider for example the log-information for a data set, I = log10(n/p), which reports the information of a data set by adjusting its sample size by the number of features. While this is a reasonable measure, Figure 2 shows that I is not nearly as effective as ρ in predicting imputation accuracy. The figure displays the ANOVA effect sizes for ρ and I from a linear regression in which log relative imputation error was used as the response. In addition to ρ and I, dependent variables in the regression also included the type of RF procedure. The effect size was defined as the estimated coefficients for the standardized values of ρ and I. The two dependent variables ρ and I were standardized to have a mean of zero and variance of one which makes it possible to directly compare their estimated coefficients. The figure shows that both values are important for understanding imputation accuracy and that both exhibit the same pattern. Within a specific type of missing data mechanism, say MCAR, importance of each variable decreases with missingness of data (MCAR 0.25, MCAR 0.5, MCAR 0.75). However, while the pattern of the two measures is similar, the effect size of ρ is generally much larger than I. The only exceptions being the MAR 0.75 and NMAR 0.75 experiments, but these two experiments are the least interesting. As will be discussed below, nearly all methods performed poorly here.
Figure 2.
ANOVA effect size for the log-information, I = log10(n/p), and correlation, ρ (defined as in (3)), from a linear regression using log relative imputation error, log10(ℰR(ℐ)), as the response. In addition to I and ρ, dependent variables in the regression included type of RF procedure used. ANOVA effect sizes are the estimated coefficients of the standardized variable (standardized to have mean zero and variance 1). This demonstrates the importance of correlation in assessing imputation performance.
4.1.1 Correlation
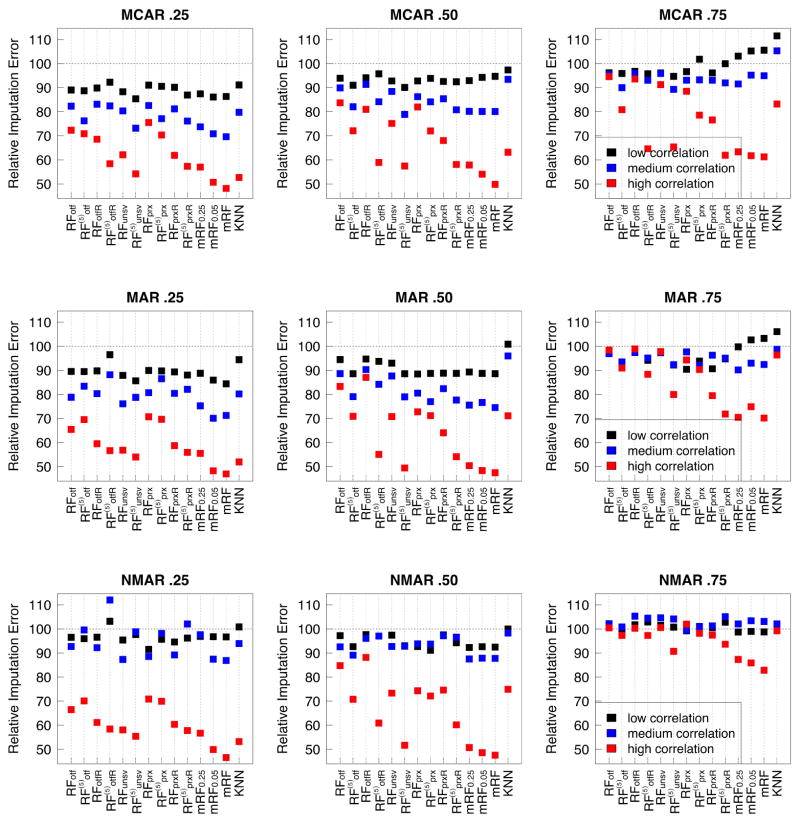
Figure 3 and Table 2, which have been stratified by correlation group, show the importance of correlation for RF imputation procedures. In general, imputation accuracy generally improves with correlation. Over the high correlation data, mForest algorithms were by far the best. In some cases, they achieved a relative imputation error of 50, which means their imputation error was half of the strawman’s value. Generally there are no noticeable differences between mRF (missForest) and mRF0.05. Performance of mRF0.25, which uses only 4 regressions per cycle (as opposed to p for mRF), is also very good. Other algorithms that performed well in high correlation settings were (proximity imputation with random splitting, iterated 5 times) and (unsupervised multivariate splitting, iterated 5 times). Of these, tended to perform slightly better in the medium and low correlation settings. We also note that while mForest also performed well over medium correlation settings, performance was not superior to other RF procedures in low correlation settings, and sometimes was worse than procedures like . Regarding the comparison procedure KNN, while its performance also improved with increasing correlation, performance in the medium and low correlation settings was generally much worse than RF methods.
Figure 3.
Relative imputation error, ℰR(ℐ), stratified and averaged by level of correlation of a data set. Procedures are: RFotf, (on the fly imputation with 1 and 5 iterations); RFotfR, (similar to RFotf and but using pure random splitting); RFunsv, (multivariate unsupervised splitting with 1 and 5 iterations); RFprx, (proximity imputation with 1 and 5 iterations); RFprxR, (same as RFprx and but using pure random splitting); mRF0.25, mRF0.05, mRF (mForest imputation, with 25%, 5% and 1 variable(s) used as the response); KNN (k-nearest neighbor imputation).
Table 2.
Relative imputation error ℰR(ℐ).
| Low Correlation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MCAR | MAR | NMAR | ||||||||
| .25 | .50 | .75 | .25 | .50 | .75 | .25 | .50 | .75 | ||
| RFotf | 89.0 | 93.9 | 96.2 | 89.5 | 94.5 | 97.2 | 96.5 | 97.2 | 100.9 | |
|
|
88.7 | 91.0 | 95.9 | 89.5 | 88.6 | 93.5 | 96.0 | 92.6 | 98.8 | |
| RFotfR | 89.9 | 94.1 | 96.8 | 89.8 | 94.7 | 97.8 | 96.6 | 97.6 | 101.7 | |
|
|
92.3 | 95.8 | 95.8 | 96.5 | 93.7 | 94.2 | 103.2 | 97.0 | 102.9 | |
| RFunsv | 88.3 | 92.8 | 96.2 | 87.9 | 93.0 | 97.3 | 95.4 | 97.4 | 101.6 | |
|
|
85.4 | 90.1 | 94.7 | 85.7 | 88.6 | 92.2 | 97.7 | 93.0 | 100.8 | |
| RFprx | 91.1 | 92.8 | 96.7 | 89.9 | 88.5 | 90.4 | 91.5 | 92.7 | 99.2 | |
|
|
90.6 | 93.9 | 101.8 | 89.8 | 88.7 | 93.9 | 95.7 | 91.1 | 99.3 | |
| RFprxR | 90.2 | 92.6 | 96.2 | 89.4 | 88.8 | 90.7 | 94.6 | 97.5 | 100.5 | |
|
|
86.9 | 92.4 | 100.0 | 88.1 | 88.8 | 94.8 | 96.2 | 94.3 | 102.8 | |
| mRF0.25 | 87.4 | 92.9 | 103.1 | 88.8 | 89.3 | 99.8 | 96.9 | 92.3 | 98.7 | |
| mRF0.05 | 86.1 | 94.3 | 105.3 | 86.0 | 88.7 | 102.7 | 96.8 | 92.6 | 99.0 | |
| mRF | 86.3 | 94.7 | 105.6 | 84.4 | 88.6 | 103.3 | 96.7 | 92.5 | 98.8 | |
| KNN | 91.1 | 97.4 | 111.5 | 94.4 | 100.9 | 106.1 | 100.9 | 100.0 | 101.7 | |
| Medium Correlation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MCAR | MAR | NMAR | ||||||||
| .25 | .50 | .75 | .25 | .50 | .75 | .25 | .50 | .75 | ||
| RFotf | 82.3 | 89.9 | 95.6 | 78.8 | 88.6 | 97.0 | 92.7 | 92.6 | 102.2 | |
|
|
76.2 | 82.1 | 90.0 | 83.4 | 79.1 | 93.4 | 99.6 | 89.1 | 100.8 | |
| RFotfR | 83.1 | 91.4 | 96.0 | 80.3 | 90.3 | 97.4 | 92.2 | 96.1 | 105.3 | |
|
|
82.4 | 84.1 | 93.1 | 88.2 | 84.2 | 95.1 | 112.0 | 97.1 | 104.5 | |
| RFunsv | 80.4 | 88.4 | 95.9 | 76.1 | 87.7 | 97.5 | 87.3 | 92.7 | 104.7 | |
|
|
73.2 | 78.9 | 89.3 | 78.8 | 79.0 | 92.4 | 98.8 | 92.8 | 104.2 | |
| RFprx | 82.6 | 86.3 | 93.1 | 80.7 | 80.5 | 97.7 | 88.6 | 93.8 | 99.5 | |
|
|
77.1 | 84.1 | 93.3 | 86.5 | 77.0 | 92.1 | 98.1 | 93.7 | 101.0 | |
| RFprxR | 81.2 | 85.4 | 93.1 | 80.4 | 82.4 | 96.3 | 89.2 | 97.2 | 101.3 | |
|
|
76.1 | 80.8 | 92.0 | 82.1 | 77.7 | 95.1 | 102.1 | 96.6 | 105.1 | |
| mRF0.25 | 73.8 | 80.2 | 91.6 | 75.3 | 75.6 | 90.2 | 97.6 | 87.5 | 102.1 | |
| mRF0.05 | 70.9 | 80.1 | 95.2 | 70.1 | 76.6 | 93.0 | 87.4 | 87.9 | 103.4 | |
| mRF | 69.6 | 80.1 | 95.0 | 71.3 | 74.6 | 92.4 | 86.9 | 87.8 | 103.1 | |
| KNN | 79.8 | 93.5 | 105.3 | 80.2 | 96.0 | 98.7 | 93.9 | 98.3 | 102.1 | |
| High Correlation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MCAR | MAR | NMAR | ||||||||
| .25 | .50 | .75 | .25 | .50 | .75 | .25 | .50 | .75 | ||
| RFotf | 72.3 | 83.7 | 94.6 | 65.5 | 83.3 | 98.4 | 66.5 | 84.8 | 100.4 | |
|
|
70.9 | 72.1 | 80.9 | 69.5 | 70.9 | 91.0 | 70.1 | 70.8 | 97.3 | |
| RFotfR | 68.6 | 81.0 | 93.6 | 59.5 | 87.1 | 98.9 | 61.2 | 88.2 | 100.3 | |
|
|
58.4 | 58.9 | 64.6 | 56.7 | 55.1 | 88.4 | 58.4 | 60.9 | 97.3 | |
| RFunsv | 62.1 | 75.1 | 91.3 | 56.8 | 70.8 | 97.8 | 58.1 | 73.3 | 100.6 | |
|
|
54.2 | 57.5 | 65.4 | 54.0 | 49.4 | 80.0 | 55.4 | 51.7 | 90.7 | |
| RFprx | 75.5 | 82.0 | 88.5 | 70.7 | 72.8 | 94.3 | 70.9 | 74.3 | 102.0 | |
|
|
70.4 | 72.0 | 78.6 | 69.7 | 71.2 | 90.3 | 70.0 | 72.2 | 98.2 | |
| RFprxR | 61.9 | 68.1 | 76.6 | 58.7 | 64.1 | 79.5 | 60.4 | 74.6 | 97.5 | |
|
|
57.3 | 58.1 | 61.9 | 55.9 | 54.1 | 71.9 | 57.8 | 60.2 | 93.7 | |
| mRF0.25 | 57.0 | 57.9 | 63.3 | 55.5 | 50.4 | 70.5 | 56.7 | 50.7 | 87.3 | |
| mRF0.05 | 50.7 | 54.0 | 61.7 | 48.3 | 48.4 | 74.9 | 49.9 | 48.6 | 85.9 | |
| mRF | 48.2 | 49.8 | 61.3 | 47.0 | 47.5 | 70.2 | 46.6 | 47.6 | 82.9 | |
| KNN | 52.7 | 63.2 | 83.2 | 52.0 | 71.1 | 96.4 | 53.2 | 74.9 | 99.2 | |
4.1.2 Missing data mechanism
The missing data mechanism also plays an important role in accuracy of RF procedures. Accuracy decreased systematically when going from MCAR to MAR and NMAR. Except for heavy missingness (75%), all RF procedures under MCAR and MAR were more accurate than strawman imputation. Performance in NMAR was generally poor unless correlation was high.
4.1.3 Heavy missingness
Accuracy degraded with increasing missingness. This was especially true when missingness was high (75%). For NMAR data with heavy missingness, procedures were not much better than strawman (and sometimes worse), regardless of correlation. However, even with missingness of up to 50%, if correlation was high, RF procedures could still reduce the strawman’s error by one-half.
4.1.4 Iterating RF algorithms
Iterating generally improved accuracy for RF algorithms, except in the case of NMAR data, where in low and medium correlation settings, performance sometimes degraded. We believe this is a real effect but have no explanation for this behavior except that it reflects the difficulty in dealing with NMAR.
4.2 Computational speed
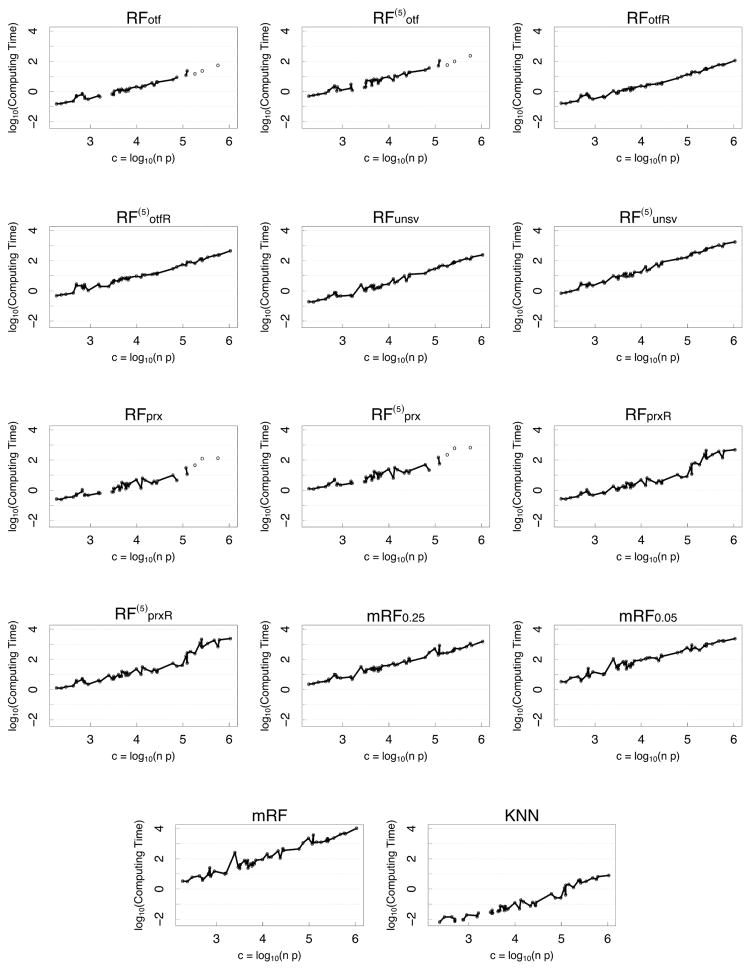
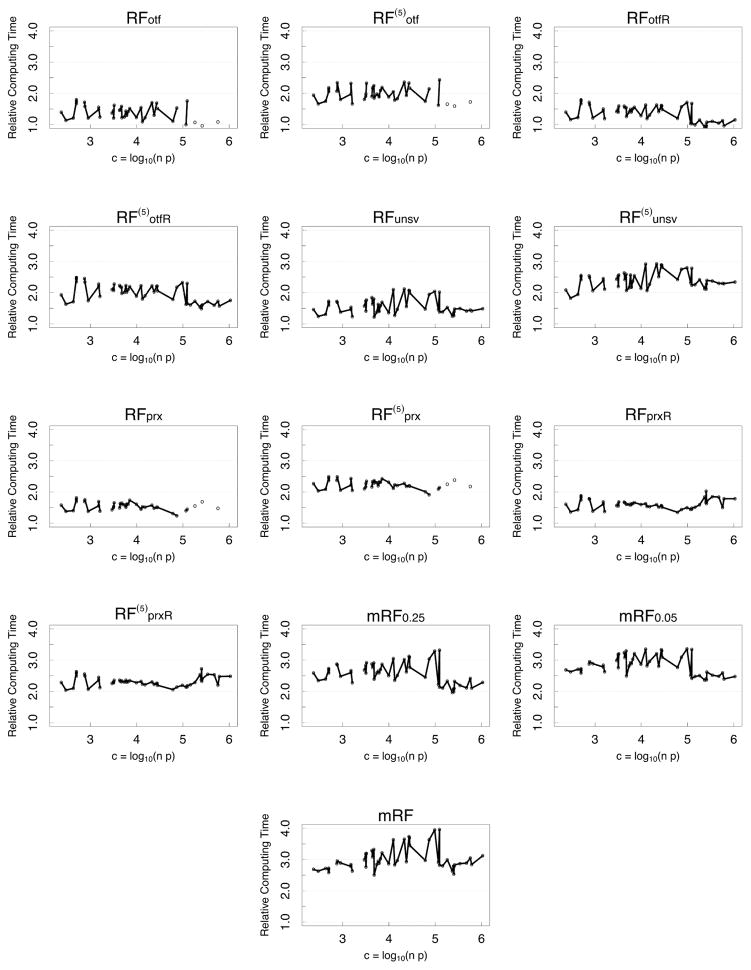
Figure 5 (see Appendix) displays the log of total elapsed time of a procedure averaged over all experimental conditions and runs, with results ordered by the log-computational complexity of a data set, c = log10(np). The fastest algorithm is KNN which is generally 3 times faster on the log-scale, or 1000 times faster, than the slowest algorithm, mRF (missForest). To improve clarity of these differences, Figure 6 (see Appendix) displays the relative computational time of procedure relative to KNN (obtained by subtracting the KNN log-time from each procedure’s log-time). This new figure shows that while mRF is 1000 times slower than KNN, the multivariate mForest algorithms, mRF0.05 and mRF0.25, improve speeds by about a factor of 10. After this, the next slowest procedures are the iterated algorithms. Following this are the non-iterated algorithms. Some of these latter algorithms, such as RFotf, are 100 times faster than missForest; or only 10 times slower than KNN. These kinds of differences can have a real effect when dealing with big data. We have experienced settings where OTF algorithms can take hours to run. This means that the same data would take missForest 100’s of hours to run, which makes it questionable to be used in such settings.
Figure 5.
Log of computing time for a procedure versus log-computational complexity of a data set, c = log10(np).
Figure 6.
Relative log-computing time (relative to KNN) versus log-computational complexity of a data set, c = log10(np).
5 Simulations
5.1 Sample size
We used simulations to study the performance of RF as the sample size n was varied. We wanted to investigate two questions: (1) Does the relative imputation error improve with sample size? (2) Do these values converge to the same or different values for the different RF imputation algorithms?
For our simulations, there were 10 variables X1, …, X10 where the true model was
where ε was simulated independently from a N(0, 0.5) distribution (here N(μ, v) denotes a normal distribution with mean μ and variance v). Variables X1 and X2 were correlated with a correlation coefficient of 0.96, and X5 and X6 were correlated with value 0.96. The remaining variables were not correlated. Variables X1,X2,X5,X6 were N(3, 3), variables X3,X10 were N(1, 1), variable X8 was N(3, 4), and variables X4,X7,X9 were exponentially distributed with mean 0.5.
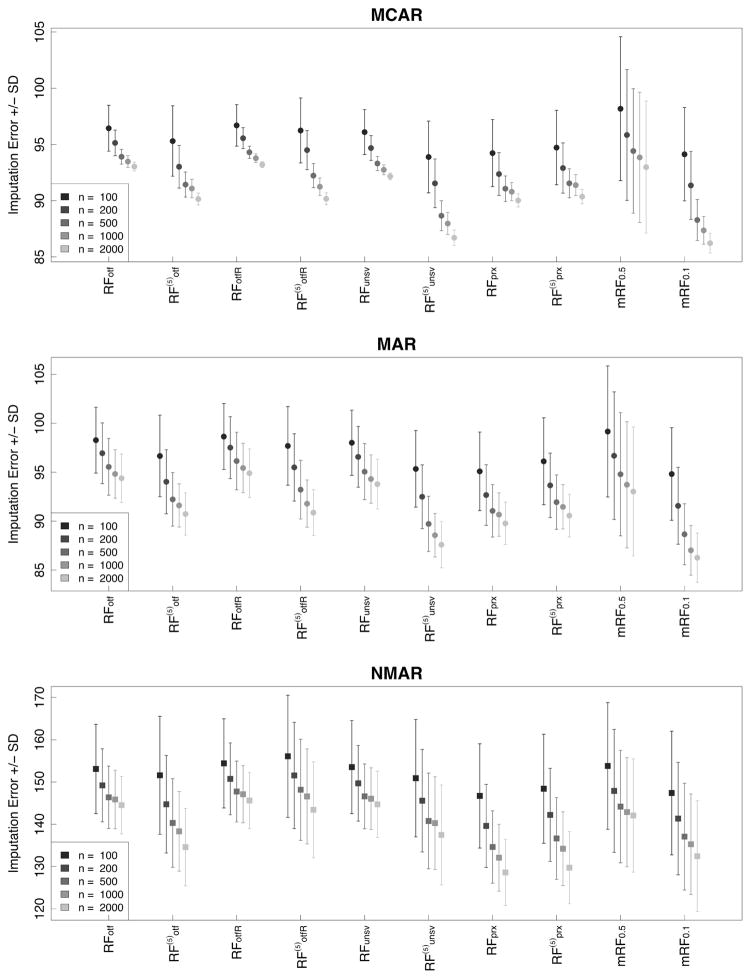
The sample size (n) was chosen to be 100, 200, 500, 1000, and 2000. Data was made missing using the MCAR, MAR, and NMAR missing data procedures described earlier. Percentage of missing data was set at 25%. All imputation parameters were set to the same values as used in our previous experiments as described in Section 3.4. Each experiment was repeated 500 times and the relative imputation error, ℰR(ℐ), recorded in each instance. Figure 4 displays the mean relative imputation error for a RF procedure and its standard deviation for each sample size setting. As can be seen, values improve with increasing n. It is also noticeable that performance depends upon the RF imputation method. In these simulations, the missForest algorithm mRF0.1 appears to be best (note that p = 10 so mRF0.1 corresponds to the limiting case missForest). Also, it should be noted that performance of RF procedures decrease systematically as the missing data mechanism becomes more complex. This mirrors our previous findings.
Figure 4.
Mean relative imputation error ± standard deviation from simulations under different sample size values n = 100, 200, 500, 1000, 2000.
5.2 Pattern mixture models
In pattern-mixture models [25, 26], the outcome is changed when an independent variable is missing. This is considered a challenging missing data scenario, therefore we sought to investigate performance of procedures in this setting. We used simulations to compare imputation performance of RF methods when the missing data mechanism was MCAR, MAR, and NMAR for the X features, and where the outcome Y was simulated with and without a pattern-mixture model (PMM). Independent variables X1, …, X10 were simulated as in Section 5.1. The PMM was defined as
where ε was simulated independently from a N(0, 0.5) distribution. Pattern-mixture missingness was induced by M1 which was set to M1 = 1 if X1 was missing and M1 = 0 if X1 was not missing. This results in the value of Y being affected by the missingness of X1. Missingness for X1, …, X10 was induced by MCAR, MAR, and NMAR as in Section 5.1. The sample size was chosen to be n = 2000. Simulations were repeated 500 times.
The results are displayed in Table 3. Relative imputation error for each procedure is given for each of the three X missing data mechanisms with and without a PMM for Y. Because methods such as mForest do not use Y in their imputation procedure, only the results from simulations without PMM are displayed for such procedures. As shown in Table 3, mRF0.1 generally had the best imputation accuracy (as before, note that mRF0.1 corresponds to the limiting case missForest because p = 10). For PMM simulations, OTF and proximity methods, which use Y in their model building, generally saw a degradation in imputation performance, especially when missingness forX was NMAR. This shows the potential danger of imputation procedures which include Y, especially when missingness in X has a complex relationship to Y. A more detailed study of this issue will be undertaken by the authors in a follow up paper.
Table 3.
Relative imputation error ℰR(ℐ) with and without a pattern-mixture model (PMM) for Y. Some procedures, such as mForest, do not use Y outcome in imputations. Therefore, their imputation performance is the same with or without PMM. For clarity, we therefore only report values without PMM for such procedures.
| MCAR | MCAR (PMM) | MAR | MAR (PMM) | NMAR | NMAR (PMM) | ||
|---|---|---|---|---|---|---|---|
| RFotf | 91.8 | 92.5 | 91.1 | 91.9 | 94.5 | 95.6 | |
|
|
88.9 | 90.1 | 87.6 | 88.7 | 88.0 | 91.5 | |
| RFprx | 88.8 | 90.1 | 86.7 | 87.5 | 84.8 | 88.0 | |
|
|
89.1 | 90.1 | 87.4 | 88.1 | 84.8 | 89.1 | |
| RFunsv | 90.9 | – | 90.5 | – | 94.6 | – | |
|
|
85.1 | – | 84.5 | – | 90.0 | – | |
| RFprxR | 88.3 | – | 86.1 | – | 92.3 | – | |
|
|
88.4 | – | 86.4 | – | 93.3 | – | |
| mRF0.5 | 91.7 | – | 89.8 | – | 92.9 | – | |
| mRF0.1 | 85.1 | – | 83.3 | – | 86.6 | – |
6 Conclusions
Being able to effectively impute missing data is of great importance to scientists working with real world data today. A machine learning method such as RF, known for its excellent prediction performance and ability to handle all forms of data, represents a poentially attractive solution to this challenging problem. However, because no systematic comparative study of RF had been attempted in missing data settings, we undertook a large scale experimental study of different RF imputation procedures to determine which methods performed best, and under what types of settings.
We found that correlation played a very strong role in performance of RF procedures. Imputation performance generally improved with increasing correlation of features. This held even with heavy levels of missing data and for all but the most complex missing data scenarios. When there is high correlation we recommend using a method like missForest which performed the best in such settings. Although it might seem obvious that increasing feature correlation should improve imputation, we found that in low to medium correlation, RF algorithms did noticeably better than the popular KNN imputation method. This is interesting because KNN is related to RF. Both methods are a type of nearest neighbor method, although RF is more adaptive than KNN, and in fact can be more accurately described as an adaptive nearest neighbor method. This adaptivity of RF may play a special role in harnessing correlation in the data that may not necessarily be present in other methods, even methods that have similarity to RF. Thus, we feel it is worth emphasizing that correlation is extremely important to RF imputation methods.
In big data settings, computational speed will play a key role. Thus, practically speaking, users might not be able to implement the best method because computational times will simply be too long. This is the downside of a method like missForest, which was the slowest of all the procedures considered. As one solution, we proposed mForest (mRFα) which is a computationally more efficient implementation of missForest. Our results showed mForest could achieve up to a 10-fold reduction in compute time relative to missForest. We believe these computational times can be improved further by incorporating mForest directly into the native C-library of randomForestSRC(RF-SRC). Currently mForest is run as an external R-loop that makes repeated calls to the impute.rfsrc function in RF-SRC. Incorporating mForest into the native library, combined with the openMP parallel processing of RF-SRC, could make it much more attractive. However, even with all of this, we still recommend some of the more basic OTFI algorithms like unsupervised RF imputation procedures for big data. These algorithms perform solidly in terms of imputation and can be up to a 100 times faster than missForest.
Acknowledgments
This work was supported by the National Institutes of Health [R01CA163739 to H.I.].
APPENDIX
Figures 5 and 6 display computational speed for different RF algorithms as a function of complexity.
References
- 1.Enders CK. Applied Missing Data Analysis. Guilford Publications; New York: 2010. [Google Scholar]
- 2.Rubin DB. Multiple imputation after 18+ years. J Am Stat Assoc. 1996;91:473–489. [Google Scholar]
- 3.Loh PL, Wainwright MJ. Advances in Neural Information Processing Systems. 2011. High-dimensional regression with noisy and missing data: provable guarantees with non-convexity; pp. 2726–2734. [Google Scholar]
- 4.Aittokallio T. Dealing with missing values in large-scale studies: microarray data imputation and beyond. Brief Bioinform. 2009;2(2):253–264. doi: 10.1093/bib/bbp059. [DOI] [PubMed] [Google Scholar]
- 5.Liao SG, et al. Missing value imputation in high-dimensional phenomic data: imputable or not, and how? BMC Bioinformatics. 2014;15:346. doi: 10.1186/s12859-014-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doove LL, Van Buuren S, Dusseldorp E. Recursive partitioning for missing data imputation in the presence of interaction effects. Comput Stat Data An. 2014;72:92–104. [Google Scholar]
- 7.Bartlett JW, Seaman SR, White IR, Carpenter JR. Multiple imputation of covariates by fully conditional specification: accommodating the substantive model. Stat Methods Med Res. 2015;24(4):462–487. doi: 10.1177/0962280214521348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 9.Breiman L. Manual–setting up, using, and understanding random forests V4.0. 2003 Available at https://www.stat.berkeley.edu/~breiman.
- 10.Liaw A, Wiener M. Classification and regression by randomForest. Rnews. 2002;2(3):18–22. [Google Scholar]
- 11.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 12.Ishwaran H, Kogalur UB. randomForestSRC: Random Forests for Survival, Regression and Classification (RF-SRC) R package version 2.4.2. 2017 http://cran.r-project.org.
- 13.Stekhoven DJ, Buhlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28(1):112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 14.Waljee AK, et al. Comparison of imputation methods for missing laboratory data in medicine. BMJ Open. 2013;3(8):e002847. doi: 10.1136/bmjopen-2013-002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 16.Van Buuren S. Multiple imputation of discrete and continuous data by fully conditional specification. Stat Methods Med Res. 2007;16:219–242. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
- 17.Shah AD, et al. Comparison of random forest and parametric imputation models for imputing missing data using MICE: a CALIBER study. American Journal of Epidemiology. 2014;179(6):764–774. doi: 10.1093/aje/kwt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Belmont, California: 1984. [Google Scholar]
- 19.Twala B, Jones MC, Hand DJ. Good methods for coping with missing data in decision trees. Pattern Recognition Letters. 2008;29(7):950–956. [Google Scholar]
- 20.Twala B, Cartwright M. Ensemble missing data techniques for software effort prediction. Intelligent Data Analysis. 2010;14(3):299–331. [Google Scholar]
- 21.Ishwaran H. The effect of splitting on random forests. Machine Learning. 2015;99(1):75–118. doi: 10.1007/s10994-014-5451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal M, Xiao Y. Multivariate random forests. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. 2011;1(1):80–87. [Google Scholar]
- 23.Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–592. [Google Scholar]
- 24.Hastie T, Tibshirani R, Narasimhan B, Chu G. impute: Imputation for microarray data. R package version 1.34.0. 2015 http://bioconductor.org.
- 25.Little RJA. Pattern-mixture models for multivariate incomplete data. J American Statistical Association. 1993;88(421):125–134. [Google Scholar]
- 26.Little RJA, Rubin DB. Statistical Analysis with Missing Data. John Wiley & Sons; 2014. [Google Scholar]