Abstract
Outcrossing and self-fertilization are fundamental strategies of sexual reproduction, each with different evolutionary costs and benefits. Self-fertilization is thought to be an evolutionary “dead-end” strategy, beneficial in the short term but costly in the long term, resulting in self-fertilizing species that occupy only the tips of phylogenetic trees. Here, we use volvocine green algae to investigate the evolution of self-fertilization. We use ancestral-state reconstructions to show that self-fertilization has repeatedly evolved from outcrossing ancestors and that multiple reversals from selfing to outcrossing have occurred. We use three phylogenetic metrics to show that self-fertilization is not restricted to the tips of the phylogenetic tree, a finding inconsistent with the view of self-fertilization as a dead-end strategy. We also find no evidence for higher extinction rates or lower speciation rates in selfing lineages. We find that self-fertilizing species have significantly larger colonies than outcrossing species, suggesting the benefits of selfing may counteract the costs of increased size. We speculate that our macroevolutionary results on self-fertilization (i.e. non-tippy distribution, no decreased diversification rates) may be explained by the haploid-dominant life cycle that occurs in volvocine algae, which may alter the costs and benefits of selfing.
Keywords: Haploid, phylogenetics, self-fertilization, sexual reproduction, sex, volvocine green algae
Outcrossing and self-fertilization are fundamental strategies of sexual reproduction, each with different evolutionary costs and benefits. Repeated origins of selfing have occurred in flowering plants (Stebbins 1974; Barrett 2002; Wright et al. 2013), bryophyte mosses (McDaniel et al. 2013), most animal phyla (Jarne and Auld 2006), brown algae (Luthringer et al. 2014), and in fungi (Whitehouse 1949; Yun et al. 1999; Billiard et al. 2011). The origin and distribution of selfing among species within a clade is usually explained by the “dead end” hypothesis, which posits short-term evolutionary advantages to selfing but long-term evolutionary costs (Stebbins 1957). Selfing may be favored in the short term through a 50% increase in transmission of genes to offspring (Williams 1975; Nagylaki 1976) and reproductive assurance when a species is locally rare or colonizing a new habitat (Darwin 1877; Baker 1955; Schoen et al. 1996; Barrett 2010). However, the evolution of selfing also imposes many costs, including inbreeding depression due to homozygosity in diploids and polyploids (Barrett 2002; Charlesworth 2006) and a reduced effective population size leading to reduced genetic variation and adaptive potential (Stebbins 1957; Pollak 1987; Wright et al. 2013). This reduced ability to adapt to changing environments may ensure that selfing lineages evolutionarily fail over longer timescales (Takebayashi and Morrell 2001). Furthermore, selfing is hypothesized to be evolutionarily irreversible, because once selfing has evolved the benefits of outcrossing may be insufficient to overcome the two-fold increase in genetic transmission (Lande and Schemske 1985). Thus, selfing is thought to have two important consequences: (1) negative net diversification rates (or at least reduced diversification rates relative to outcrossing); and (2) irreversibility to outcrossing (Takebayashi and Morrell 2001; Igic and Busch 2013). Taken together, these two consequences may make selfing an evolutionary dead-end strategy. Thus, selfing is predicted to generate a “tippy” phylogenetic distribution among living taxa, in which selfing evolves due to short-term benefits but does not persist evolutionarily due to reduced or negative diversification rates. Therefore, selfing is predicted to occupy only the tips of phylogenetic trees (Schoen et al. 1997; Takebayashi and Morrell 2001; Igic et al. 2008; Wright et al. 2013). Nevertheless, it is also possible that some origins of selfing may generate numerous descendant lineages (Stebbins 1957; Escobar et al. 2010; Goldberg et al. 2010; de Vos et al. 2014). This may occur because of higher diversification rates in selfing species (Hamrick and Godt 1996; Goldberg et al. 2010) or because insufficient time has passed for the macroevolutionary consequences of self-fertilization to take effect (Escobar et al. 2010).
Mating systems (including self-fertilization and outcrossing) may be linked to the evolution of body size, which may have consequences for these macroevolutionary patterns. The evolution of selfing versus outcrossing in angiosperms is thought to be linked to organism size, with larger species evolving dioecy (individuals only produce one type of sexual gamete, resulting in obligate outcrossing). Specifically, trees (larger size) are more likely to be dioecious than shrubs (medium size), and shrubs are more likely to be dioecious than herbs (smaller size), likely to avoid inbreeding depression (Darwin 1876; Bawa 1980; Lloyd 1982; Vamosi et al. 2003). It is assumed that a larger plant has a higher spatial density of reproductive cells (Maynard Smith 1978). This increased density of reproductive cells increases the probability of self-fertilization, thus increasing the selection for selfing avoidance, which can be achieved through dioecy (Maynard Smith 1978). Thus mating system evolution, including selfing and dioecy, may be linked to the evolution of body size.
The evolution of selfing may have different evolutionary consequences in haploid taxa relative to diploid taxa, given the potential for different inbreeding costs. Inbreeding depression is thought to be an important cost of self-fertilization, and inbreeding depression due to increased homozygosity is likely greater in diploid and polyploid taxa (Barrett 2002; Charlesworth 2006). Given this, in a haploid clade, we might expect to see reduced negative consequences of selfing over macroevolutionary timescales (e.g., tippy distributions of selfing taxa, negative or reduced diversification rates). However, most previous studies of the evolution of selfing have focused on diploid taxa.
Here, we analyze the macroevolution of mating systems in the haploid volvocine green algae. Specifically, we reconstruct the evolution of mating systems across the tree, test for a “tippy” distribution of self-fertilization among species on the tree, compare rates of diversification in selfing and outcrossing species, and test for a correlation between mating system and colony size.
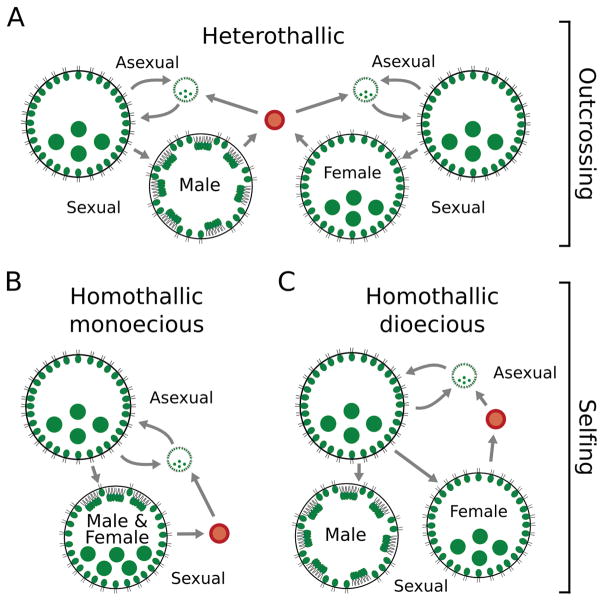
The volvocine green algae are a tractable model system for studying the evolution of mating systems. Previous studies showed that this group evolved relatively recently, approximately ~230 Myr ago (Herron et al. 2009), considerably younger than groups like animals (>900 Myr old stem age), plants (>400 Myr old), or fungi (>1,000 Myr old; Parfrey et al. 2011). Furthermore, several volvocine genomes have been sequenced (Merchant et al. 2007; Prochnik et al. 2010; Hanschen et al. 2016). Volvocine algae are haploid during the vegetative phase of their life cycle (Fig. 1), and many rounds of asexual reproduction typically occur between occasional rounds of sexual reproduction (Coleman 1979; Kirk 1998). Sexual reproduction generates the only diploid phase, a single-celled zygospore that germinates meiotically to re-enter the vegetative phase (Fig. 1). Importantly, extant species exhibit substantial diversity in mating systems. Three mating systems are found in the volvocine algae (Fig. 1). In heterothallic, obligately outcrossing species (Fig. 1A), distinct genotypes produce a single mating type or sex. Two variations of homothallic self-fertilization exist. In homothallic dioecious species (Fig. 1C), a single genotype is capable of sexually differentiating to produce both mating types or sexes, though in separate colonies. While dioecy necessarily implies outcrossing in obligately sexual organisms, in facultatively sexual volvocine algae, homothallic dioecious species can self-fertilize. In homothallic monoecious species (Fig. 1B), a single genetic strain is capable of sexually differentiating to produce hermaphroditic sexual colonies that produce both gamete types (Fig. 1). Little is known about selfing rates in homothallic species, though homothallic species are protandrous (sperm packets develop and liberate before egg development; Smith 1944), which is consistent with a relatively lower rate of selfing because sperm may fertilize another colony (Starr et al. 1980). Lastly, androdioecy (both hermaphrodite and male colonies; not shown) has been described in certain strains of Volvox africanus (Starr 1971; Nozaki et al. 2015a). Intra- and inter-colony selfing in monoecious and inter-colony fertilization of the same genetic strain in dioecious homothallic species have been observed (Darden Jr 1966; Starr et al. 1980; Nozaki et al. 2015b). Self-incompatibility has never been described in homothallic volvocine species.
Figure 1.
Diversity of mating systems in the volvocine green algae and their respective life cycles. A. In outcrossing (heterothallic) species, distinct genotypes (male on left and female on right) sexually differentiate producing either eggs or sperm. A diploid zygospore (red) is produced after fertilization. Sexual offspring hatch and enter the haploid, asexual phase of the life cycle. B. In selfing (homothallic) monoecious species, a single genotype is capable of producing both gamete types. Upon sexual differentiation, each sexual colony produces both sperm and eggs. C. In selfing (homothallic) dioecious species, a single genotype sexually differentiates, producing either eggs or sperm, but not both within the same colony. Cartoons in panels A–C are shown with anisogamous, Volvox-like morphology for illustrative purposes only.
In this study, we analyze the evolution of mating systems in the volvocine green algae. We first estimate a new phylogeny for the group. We next use ancestral state reconstruction to reconstruct the evolutionary history of mating systems within this group. We find numerous independent origins of homothallic self-fertilization and two reversions to heterothallic outcrossing. Then, using three quantitative metrics, we demonstrate that selfing does not exhibit a tippy phylogenetic pattern. We also find no evidence for lowered (or negative) diversification rates in selfing species. This unexpectedly non-tippy distribution may be due to a haploid-dominant life cycle in volvocine algae, which may alter the costs and benefits of selfing.
Material and Methods
IDENTIFICATION OF CHARACTER STATES
Trait data for each species and strain were compiled from published reports (Dataset S1, available on Dryad: doi:), including sexual traits (heterothallic/homothallic and dioecious/monoecious) and two colony size metrics (number of cells and cell/colony length). For consistency, we used the maximum reported values of these metrics. To avoid possible incorrect assignment of character-state data due to erroneous species identification or intraspecific variation, care was taken to ensure that data were based on particular strains within a species, rather than using data from individuals identified only to species or genus.
TREE ESTIMATION
We estimated a new volvocine phylogeny in order to include novel strains and species not included in previous analyses (including Gonium pectorale Russia, Volvox ferrisii, Pleodorina starrii, Pleodorina thompsonii, Volvox perglobator). A total of 19 of the 97 ingroup terminal taxa in this study were not included in previous analyses. The ingroup was defined as the smallest monophyletic clade containing Chlamydomonas reinhardtii and Volvox carteri, a group commonly referred to as the “volvocine green algae”. We generated a concatenated phylogeny using Bayesian Markov chain Monte Carlo implemented in MrBayes version 3.2.2 (Ronquist et al. 2012) using default parameters except as described below. The data matrix included sequences for 97 volvocine terminal taxa and seven outgroup taxa. The outgroup taxa represented different groups of non-volvocine algae, including two taxa from the immediate sister group and one taxa from each of five other major groups (Herron and Michod 2008). The sequence data consisted of five chloroplast genes (ATP synthase beta-subunit, atpB; P700 chlorophyll a-apoprotein A1, psaA; P700 chlorophyll a-apoprotein A2, psaB; photosystem II CP43 apoprotein, psbC; and the large subunit of Rubisco, rbcL). We did not perform multi-locus species-tree analyses since the chloroplast genes effectively belong to the same locus. The best fitting combination of partitioning scheme and nucleotide substitution models was determined using PartitionFinder version 2.1.1 (Lanfear et al. 2016) using AICc and a greedy search algorithm with branch lengths linked across partitions. A total of 15 possible partitions were initially defined (3 codon positions for 5 protein-coding chloroplast genes) and the best-fitting strategy included 11 data blocks (Table S1). Four independent Bayesian runs of four chains each (three heated chains and one cold chain) were run for 2×107 generations with a burn-in of 5×106 generations. Trees were sampled every 100 generations. We considered the runs to have adequately sampled the solution space when the standard deviation of split frequencies was below 5×10−3. Post burn-in trees were combined and assembled to construct a majority-rule consensus phylogram. Posterior probabilities for nodes were calculated using the pooled set of all post burn-in trees from the four runs (Fig. S1). An ultrametric tree, necessary for maximum-likelihood ancestral-state reconstruction using the R package diversitree (FitzJohn 2012), was calculated using a penalized likelihood function in the ape package (Paradis et al. 2004). A correlated model was used without age constraints, making the units for the chronogram, as well as subsequent rate estimates, arbitrary (The data matrix and trees are available on Dryad: doi:).
The topology and branch lengths of our new estimated phylogenetic tree are consistent with previously published chloroplast phylogenies (Nozaki et al. 2002; Herron and Michod 2008; Herron et al. 2009), and with a phylogenomic analysis of eleven green algae including Chlamydomonas reinhardtii, Gonium pectorale, and Volvox carteri (Hanschen et al. 2016). Our tree was also consistent with a phylogeny based on the nuclear internal transcribed spacer (ITS) for Volvox section Volvox (Fig. 4 of Isaka et al. 2012).
Sequence data for all five chloroplast genes were not available for all taxa (16.5% of the cells in the data matrix were missing). Previous simulation and empirical analyses suggest that this low level of missing data should be inconsequential (Wiens and Morrill 2011). Nevertheless, we constructed an additional Bayesian concatenated tree including only taxa for which sequence data for all five chloroplast genes are available (72 of the 97 ingroup OTUs remained). PartitionFinder was independently run on this dataset resulting in the same best-fitting partition scheme. This tree had no strongly supported topological differences from the tree generated from the full dataset (Fig. S1).
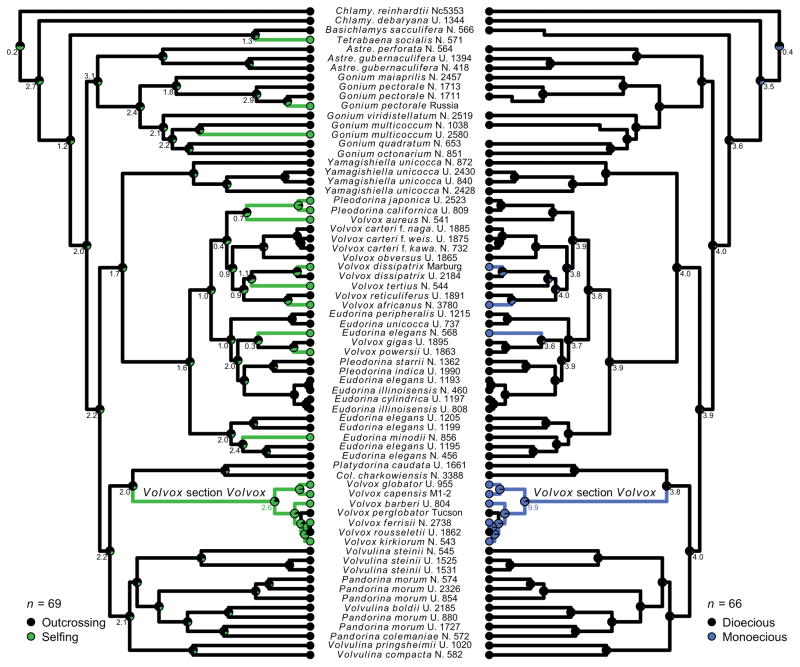
Our phylogenetic tree includes all described species from which sequence data were available (69 species). However, there are described species that were not included, either due to missing sequence data or unobserved mating system (10 species). In Volvox section Volvox (Fig. 2), there is one heterothallic, outcrossing species (V. prolificus, Iyengar 1933) and two homothallic, selfing species (V. merrilii and V. amboensis, Shaw 1922; Rich and Pocock 1933) that could not be included in our tree or in the tree-based analyses. There are also three other species which have been morphologically characterized but were lost from culture collections before genetic sequencing (Smith 1944). These species are the homothallic Pandorina morum (Coleman 1959), the homothallic dioecious Volvox pocockiae (Starr 1970), and the homothallic dioecious Volvox spermatosphaera (Powers 1908). Lastly, there are several species in which sexual reproduction has never been observed (Vitreochlamys, Chlamydomonas cribrum, Pleodorina thompsonii, and Volvox ovalis). Our ancestral-state reconstructions therefore do not include these 10 species (69 included species of 79 total species).
Figure 2.
Ancestral state reconstruction of selfing (left) and monoecy (right). Left, the evolution of outcrossing (black) and selfing (green). Right, the evolution of dioecy (black, for this analysis, outcrossing heterothallic species were treated as dioecious) and monoecy (blue). Three selfing species (Tetrabaena socialis, Gonium pectorale Russia, Gonium multicoccum UTEX 2580) break apart into unicells during sexual differentiation preventing assignment to monoecy or dioecy. Branch color refers to the most likely state inferred by maximum likelihood (ML) reconstruction. Pie charts at nodes represent scaled marginal likelihoods from ML reconstruction. Numbers at select nodes indicate Bayes factors (support for that character state against the next most likely state), which explicitly take phylogenetic uncertainty into account, colored by which state is most supported. Interpretation of Bayes factors (Kass and Raftery 1995): 0 to 2 barely worth mentioning, 2 to 6 positive, 6 to 10 strong, >10 very strong. Chlamy, Chlamydomonas; Astre, Astrephomene; Col., Colemanosphaera; N., NIES; U., UTEX.
SPECIES DELIMITATION
We ensured that our estimates of “tippiness” and diversification rates of selfing and outcrossing species were not biased by treating multiple genetically unique individuals of the same species as if they were distinct species. Doing so might artificially inflate the estimated speciation rates and impact our tests of tippiness. Therefore, we removed individuals of the same species, so that each species was represented by one terminal taxon in the tree. However, we cannot simply use taxonomic species since these species were often described without taking into account major differences in mating system (e.g., Gonium pectorale includes both selfing and outcrossing strains) and strong evolutionary divergence based on sequence data (e.g., Pandorina morum contains numerous highly diverged lineages). Thus, many named species may contain two or more distinct evolutionary species. Individuals of the same species were computationally identified using the single rate Poisson Tree Processes (PTP) method (Fujisawa and Barraclough 2013). A maximum likelihood approach with a default p-value of 0.001 was used. This approach identified 21 species as having multiple individuals present in the tree. Nine of these 21 hypothesized species from PTP were rejected because they represented morphologically and taxonomically distinct species (e.g., closely related homothallic V. ferrisii and heterothallic V. rousseletti were hypothesized to be the same species; Table S2). The remaining 12 species delineations, which assigned 31 individuals to 12 named species, were accepted (Table S2). There were 69 species in the final tree. Conspecific individuals were pruned from the tree using the R package ape (Paradis et al. 2004).
MEASURING PHYLOGENETIC SIGNAL
We tested the level of phylogenetic signal in traits to assess their lability across the volvocine tree. For homothallism/heterothallism and monoecy/dioecy, the D value (Fritz and Purvis 2010) was measured using the R package caper version 0.2 (Orme et al. 2012). For continuous traits related to colony size, Blomberg’s (2003) K and Pagel’s (1999) λ were measured using the R package phytools version 0.5–64 (Revell 2012; R Core Team 2013). All traits had statistically significant phylogenetic signal (Table S3, available on Dryad: doi:).
ANCESTRAL-STATE RECONSTRUCTION
Ancestral states were reconstructed using maximum likelihood and Bayesian methods. Heterothallism (outcrossing)/homothallism (selfing) and dioecy/monoecy were treated as two separate binary characters. We did this for two reasons. First, there were three homothallic species that break apart into unicells during sexual reproduction, preventing their assignment to either dioecy or monoecy. Second, coding the evolution of homothallism and monoecy together in a three-state model would result in some states (i.e., monoecious homothallic and dioecious homothallic) that would be present in fewer than ~10% of the species in the tree. Such rare states are known to lead to unreliable reconstructions using the type of likelihood methods used here (Davis et al. 2013).
The maximum likelihood analysis was performed using the R package diversitree version 0.9–9 (FitzJohn 2012). Several models of character evolution were evaluated for each character, including equal rates of change for all transitions between states (ER) and all rates different for all transitions between states (ARD). Model fit was compared using the AICc (Akaike 1974). The AICc is the Akaike information criterion with a small sample size correction (Burnham and Anderson 2002). The AICc should reveal the best-fitting model without including unnecessary parameters (Table S3). Alternative root state models were evaluated, by comparing their likelihoods while holding the best-fitting transition model constant. These models included an equal probability for each state, probabilities based on the frequency of each state among species on the tree, and fixing the root to each of the alternative states. We found that the ΔAICc values for alternative root-state models were indistinguishable (ΔAICc < 8.2×10−4 for all root-state models). We selected the root prior that was weighted based on the observed frequency of each state among taxa across the tree. Based on preliminary analyses, the use of alternative root-state priors did not substantially affect the resulting ancestral-state reconstructions. The state with the highest probability was considered the most likely for a given node. However, a state was considered to be significantly supported at a given node only if it was at least 7.39 times (if the natural logarithm of the ratio of two likelihoods is greater than 2) more likely than the alternative state (Pagel 1999). The likelihood ratio test was used to test for evolutionary irreversibility of selfing homothallism by comparing alternative maximum likelihood models.
Statistical support for estimated character states at internal nodes was further evaluated using Bayesian hypothesis testing implemented in BayesTraits version 2 (Pagel et al. 2004). We used this Bayesian approach to explicitly incorporate phylogenetic uncertainty, by analyzing a sample of trees. Every 1,000th post-burnin tree from the four runs was included, for a total of 600 trees. Outgroups and taxa without available character data were trimmed from these trees (e.g., monoecy vs. dioecy has never been reported for homothallic Tetrabaena socialis, Gonium pectorale Russia, and Gonium multicoccum UTEX 2580). Each tree was then ultrametricized using a penalized likelihood function (Sanderson 2002) with a correlated model without age constraints.
Two models of character evolution (ER, ARD) were analyzed in the Bayesian analyses. Bayes factors (BF) were used to find the best-fitting model. The BF was estimated based on twice the difference between the highest harmonic mean log likelihood for each model, calculated from nine independent MCMC runs of 7,500,000 generations (with a burn-in period of 500,000 generations, Table S3). The best-fitting model of evolution was used to statistically test which state was most likely to be present at specific nodes of interest (those near evolutionary transitions as predicted by the likelihood analysis). When no model of character evolution was strongly preferred (BF < 2), nodes were also tested under the alternative model. For each node of interest, the ancestral character state was tested by estimating a BF from five independent MCMC runs of 5,500,000 generations (each with a burn-in of 500,000 generations) in which the node in question was constrained to one state or the other. A state was considered strongly supported at a given node when the BF was >2 relative to the alternative state. Uniform priors and gamma-distributed hyperpriors seeded from a uniform distribution were used to seed all rate parameters.
TEST OF PHYLOGENETIC TIPPINESS
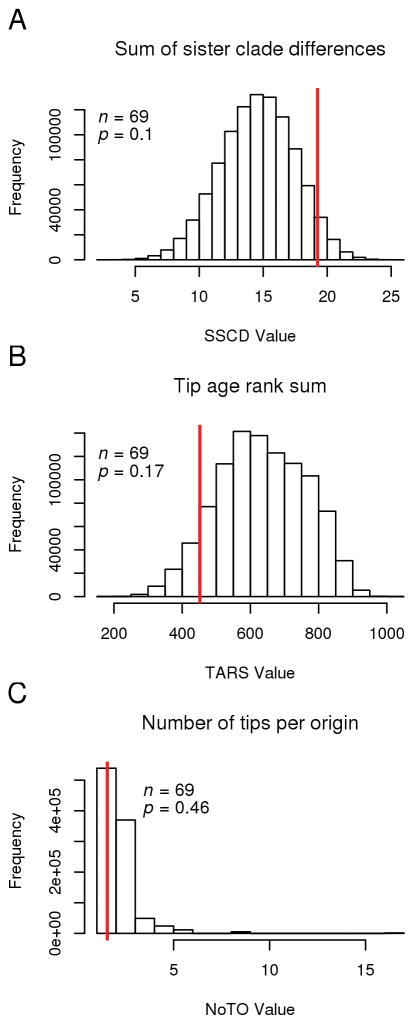
If selfing tends to lead to extinction, we predicted that homothallic self-fertilization may be restricted to recent tips of a tree (“tippiness”). To test for a tippy distribution, three tests of tippiness were performed (Bromham et al. 2016). First, we estimated the sum of sister-clade differences. This index measures trait clustering by assigning tips the value 0 or 1, assigning each node the absolute difference of the two daughter tip/nodes then summing across all nodes. Second, we estimated tip age rank sum, which compares the summed lengths of tips for each state, Third, we estimated the number of tips (i.e., species) per origin, which compares the observed number of tips per origin of trait to a null model with the same frequency of states. These metrics were estimated using the R package phylometrics version 0.0.1 (Hua et al. 2016). This package calculates two sided p-values by using a threshold Brownian motion model to simulate 1,000,000 binary traits with the same frequency of states as the observed data and compares the observed value to the simulated distribution (Bromham et al. 2016). Significant deviations from the null, Brownian motion model indicate that the observed trait shows a tippy distribution. Each analysis was repeated using both a phylometric tree (branch lengths indicate evolutionary change) and an ultrametric tree (branch lengths indicate time). Both trees yielded qualitatively identical statistical results.
We also tested whether our sampling of taxa allowed sufficient power to detect a significant tippy distribution in the selfing species. To do this, we repeated these analyses after re-assigning selected selfing species to be outcrossing (Table S4). The two manipulations were designed to artificially decrease the number of selfing species per origin of selfing and the average tip length of selfing taxa. The two replicates differed in which species were altered. These analyses resulted in significantly tippy distributions for all three metrics in at least one manipulation (Table S4). Thus, these analyses show that the number of taxa sampled was sufficient to obtain significant results for all three metrics, and so negative results need not be caused by insufficient statistical power.
ESTIMATING DIVERSIFICATION RATES
To complement our analyses of phylogenetic tippiness, we used BiSSE models (Binary State Speciation and Extinction; Maddison et al. 2007) to estimate state-dependent speciation and extinction rates. Specifically, we tested whether selfing (homothallic) species had lower diversification rates than outcrossing (heterothallic) species, as predicted given their lower expected macroevolutionary success. The diversification rate is the rate of speciation minus the rate of extinction. We also tested whether selfing lineages had the predicted negative diversification rates (extinction rate > speciation rate).
BiSSE analyses were performed using the R package diversitree version 0.9–9 (FitzJohn 2012). Twelve models were evaluated (Table S5), including (1) unconstrained (selfing and outcrossing states are associated with different rates of speciation and extinction, with different rates of transition between the states), (2) same as (1), but constraining speciation rates of selfing and outcrossing species to be equal, (3) same as (1), but constraining extinction rates to be equal, (4) constraining transition rates between states to be equal, but allowing speciation and extinction rates to differ, (5) constraining both speciation and extinction rates to be equal, but allowing transition rates to differ, (6) constraining both speciation and transition rates to be equal (but allowing extinction rates to vary), (7) constraining both extinction and transition rates to be equal (but allowing speciation rates to vary), and (8) constraining speciation, extinction, and transition rates to be equal. The remaining models tested the hypothesized irreversibility of self-fertilization, including (9) constraining transitions from selfing to outcrossing to be zero, (10), constraining the speciation rate to be equal between states and the reversal rate (selfing to outcrossing) to be zero, (11) constraining the extinction rate to be equal and the reversal rate to be zero, and (12) constraining both the speciation and extinction rates to be equal and the reversal rate to be zero (Table S5). Model fit was compared using the AICc (Akaike 1974). The model constraining only the extinction rates to be equal had the best fit. The estimates from this model were used to evaluate whether diversification rates were lower in selfing lineages.
We also used the best-fitting BiSSE model to perform ancestral-state reconstructions. To do this, alternative root state models were first evaluated, by comparing their likelihoods while holding the best-fitting model of speciation, extinction, and transition rates constant. These root-state models included (1) an equal probability for each state, (2) relative probabilities for each state based on the frequency of each state among species on the tree, and fixing the root state to be (3) outcrossing or (4) selfing. We found that the ΔAICc values for alternative root state models were low (ΔAICc < 1.78 for all reconstructions), showing that they all had similar fit. Therefore, the root prior was weighted based on the observed frequency of each state among taxa across the tree (FitzJohn 2012). Based on preliminary analyses, the use of alternative root-state priors did not substantially affect the resulting ancestral-state reconstructions. We then performed ancestral-state reconstructions using the best-fitting model. Across the tree, the state with the highest probability was considered the most likely for a given node.
We acknowledge that BiSSE models may be somewhat problematic when applied to our data. First, our analyses include only 69 taxa, and BiSSE may perform poorly when relatively few species are included in the analysis. Specifically, BiSSE may infer rates with reduced accuracy and precision, and have little power to distinguish between alternative models (Davis et al. 2013; Gamisch 2016). However, we show that our data do have sufficient power to strongly distinguish between models (and select relatively complex ones). Second, our species sampling may be poor relative to the total number of species that likely exist (based on the current rate of species descriptions; Isaka et al. 2012; Nozaki et al. 2014, 2015a). Furthermore, there is no reliable estimate for how many species actually exist with each character state. Therefore, we did not include a correction for incomplete sampling of each state. Instead, we simply assumed that the species included in our tree represented a reasonable estimate of the true frequency of each state. Finally, some authors have noted potential problems in using BiSSE to infer differences in speciation and extinction rates associated with different states (e.g., Maddison and FitzJohn 2015; Rabosky and Goldberg 2015). Furthermore, a common alternative approach (BAMM; Rabosky 2014) does not directly associate rates with states (Meyer and Wiens 2017).
We did find some unusual ancestral reconstructions from BiSSE. Specifically, this analysis inferred 22 reversions from self-fertilization to outcrossing (Fig. S2). These results seemed particularly problematic in that numerous large clades of mostly or all outcrossing species were strongly supported as having selfing ancestors (Fig. S2; including the most recent common ancestor (MRCA) of Astrephomene and Gonium, three ancestors within the Gonium genus, three ancestors within the polyphyletic Pandorina and Volvulina genera, the MRCA of Eudorina elegans UTEX 1205 and Eudorina elegans NIES 456, and three ancestors in the smallest clade containing Eudorina peripheralis UTEX 1215 and Eudorina illinoisensis UTEX 808). While this result does not overturn our main conclusion (that outcrossing can evolve from selfing), it nevertheless seems unlikely. Therefore, we used BiSSE primarily to estimate speciation and extinction rates associated with selfing and outcrossing, rather than for ancestral-state reconstructions.
TEST OF REPRODUCTION AND SIZE
To test for a correlation between self-fertilization and colony size, as observed in angiosperms (Darwin 1876; Bawa 1980; Lloyd 1982; Vamosi et al. 2003), we used phylogenetic t-tests, implemented in the R package phytools version 0.5–64 (Revell 2012). Phylogenetic t-tests used two metrics of size: (1) natural logarithm of colony length, and (2) rounds of cell divisions (i.e., log2[cell number]). P-values were adjusted for multiple comparisons following Benjamini and Hochberg (1995).
Results
EVOLUTION OF MATING SYSTEMS
Self-fertilization and mating systems in the volvocine algae have a complex evolutionary history. Taking phylogenetic uncertainty into account in the Bayesian ancestral state reconstructions, heterothallic outcrossing is the ancestral state in this group (Fig. 2). Homothallism, the genetic capacity to self fertilize, has evolved 11 times (Fig. 2). Monoecy, in which both gamete types are produced within a colony (Fig. 1B), was modeled as a separate binary trait from homothallism. Monoecy has evolved at least four times (Fig. 2). Furthermore, when comparing the reconstructions of selfing and monoecy (Fig. 2), there are no transitions between dioecious homothallism and monoecious homothallism, only transitions between heterothallic outcrossing and homothallic selfing (whether monoecious or dioecious, Fig. 3).
Figure 3.
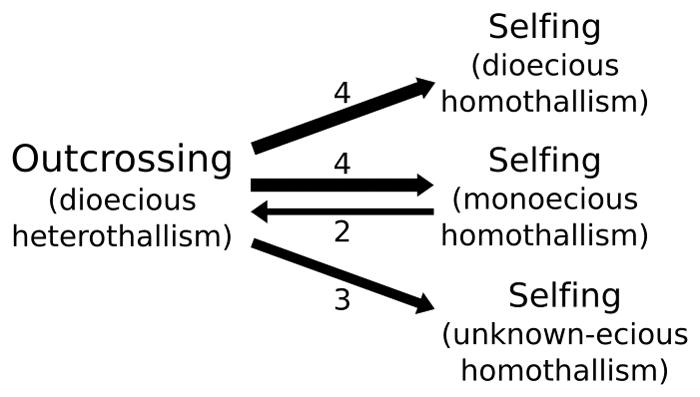
Summary of the number of observed transitions between mating systems. This minimum number of transitions was counted after assigning each ancestor to the most likely state. We treated the origin of selfing in Pleodorina japonica, Pleodorina californica, and Volvox aureus as one transition, although the ancestral state of the ancestor to these three species is ambiguous. Three homothallic species break apart into unicells during sexual differentiation preventing assignment to monoecy or dioecy (unknown-ecious homothallism).
Two independent reversions from selfing to outcrossing were inferred, in Volvox perglobator and V. rousseletii (Fig. 2). However, the rate of origin of selfing was approximately four times greater than the rate of reversion back to outcrossing (Table S3). These reversions to outcrossing were accompanied (on the same branches) by reversions from monoecy to dioecy (Fig. 2). The rate of reversal (monoecy to dioecy) was approximately 12 times greater than the rate of evolution of monoecy (Table S3).
To further test for evolutionary reversals from selfing back to outcrossing, three models were compared, one constraining the rate of loss of selfing to zero, one constraining the rate of loss of selfing to be equal to the rate of gain, and a third that did not constrain the rate of loss of selfing. Using the likelihood ratio test, the unconstrained model allowing selfing to reverse is strongly supported (χ2 = 15.70, p = 7.4×10−5). Similarly, when comparing BiSSE models, the model allowing selfing to reverse is strongly supported (χ2 = 29.36, p = 6.0×10−8, Table S5).
Given the repeated, relatively recent origins of selfing (Fig. 2), the “tippiness” of this trait was tested (Bromham et al. 2016). Using three metrics of trait distribution, homothallic self-fertilization is not significantly tippy in the volvocine algae (p > 0.096, Figs. 4, S3, Table S4). Selfing species do not have shorter branches than outcrossing species, demonstrating their long-term evolutionary persistence (Fig. 4B). Selfing species do not have fewer species per origin of selfing than a null distribution (Fig. 4C). Finally, selfing species are not sparsely distributed across the volvocine tree (Fig. 4A), demonstrating that considerable speciation of selfing species has occurred (Fig. 4A, C). We used manipulated datasets to demonstrate that there was sufficient power to detect tippy distributions, given the number of taxa in the tree (Table S4). Therefore, these results are not due to insufficient statistical power.
Figure 4.
Quantification of “tippiness” of homothallic self-fertilization across the volvocine phylometric tree. A. Sum of sister clade differences, which measures trait clustering by assigning tips (species) the value 0 or 1, assigning each node the absolute difference of the two daughter tip/nodes, then summing across all nodes. B. Tip age rank sum, which compares the branch length associated with tips (species) for each state. C. Number of tips per origin, which compares the observed number of tips per origin of the trait to a Brownian motion null model with the same trait frequency among species as in the observed data. For all panels, the histogram represents a null distribution calculated from 1,000,000 simulations of a threshold Brownian motion model with the same trait frequencies as in the observed data (red line). This analysis was repeated with an ultrametric tree (Fig. S3), which gave very similar results.
We also directly estimated speciation and extinction rates of selfing and outcrossing species (Table S5). We found that the best-fitting BiSSE model had unequal speciation and transition rates between the selfing and outcrossing states, but equal (and very low) extinction rates. The next-best model (ΔAIC=1.97) had all rates different between states, including extinction rates. All other models had substantially poorer fit (ΔAIC>10). Contrary to the predictions of the dead-end hypothesis (i.e. lower and possibly negative diversification rates in selfing lineages), the best-fitting model showed that selfing species have a positive net diversification rate (13.45 species per time unit), which is three times higher than the speciation and diversification rates of outcrossing species (4.19 species/time unit). Consistent with the initial ancestral-state reconstructions that ignored state-dependent speciation and extinction, this analysis inferred that the transition rate from selfing to outcrossing is non-zero (11.30), and much higher than the outcrossing to selfing transition rate (1.37). However, this extreme difference in rates may be an artifact (see Methods). Models in which the transition rate from selfing to outcrossing was set to zero were strongly rejected (all ΔAIC>25 relative to the best-fitting model).
The evolution of mating systems was significantly correlated with increased colony size (Fig. S1). We found that selfing species have significantly larger colonies than outcrossing species (phylogenetic t-test, adj. p < 0.011; Table S6, available on Dryad: doi:).
Discussion
The reproductive strategy of self-fertilization has long been thought to be an evolutionary dead-end, where short-term benefits, such as reproductive assurance and transmission advantages, are countered by long-term costs, such as reduced or negative net diversification rates and irreversibility (Stebbins 1957; Takebayashi and Morrell 2001; Igic and Busch 2013; Wright et al. 2013). Thus, self-fertilization is expected to occupy the tips of phylogenetic trees. Alternatively, a self-fertilizing lineage may produce numerous selfing species (Goldberg et al. 2010; Johnson et al. 2011) and may not display the predicted lower net diversification rate (Johnson et al. 2011; Gamisch et al. 2015). These large groups of selfing species may be due to temporal variation in selection pressures, resulting in temporary macroevolutionary success of selfing species (Goldberg et al. 2010) or to insufficient time having passed for the macroevolutionary consequences of self-fertilization to take effect (Escobar et al. 2010). We reconstructed the evolutionary history of self-fertilization in the volvocine algae, and showed that heterothallic outcrossing is ancestral in the volvocine algae and that homothallic self-fertilization evolved eleven times (Fig. 2). However, contrary to expectations, we found that the volvocine algae underwent two reversals from selfing to outcrossing (Figs. 2, 3). Furthermore, we found that they lack the “tippy” phylogenetic distribution expected if they led to an evolutionary dead-end, based on three different metrics (Fig. 4). Lastly, we used BiSSE to estimate speciation and extinction rates of selfing and outcrossing species, and inferred that selfing species have positive diversification rates that are higher than those of outcrossing species. The BiSSE analysis also inferred transitions from selfing to outcrossing (Table S5).
These results are contingent on several important caveats. First, the phylogenetic tree used here was based upon five chloroplast genes rather than a multi-locus nuclear gene dataset (which is currently unavailable for the volvocine algae). There are several phenomena, including incomplete lineage sorting and chloroplast capture, which may cause incongruence between the true species tree and the chloroplast gene tree. However, results from the available nuclear data are congruent with the chloroplast phylogeny, including (1) a phylogenomic analysis including Chlamydomonas reinhardtii, Gonium pectorale, and Volvox carteri (Hanschen et al. 2016), and (2) a phylogenetic tree from the nuclear internal transcribed spacer (ITS) for Volvox section Volvox (Isaka et al. 2012). The congruence between nuclear and chloroplast trees suggests that the chloroplast tree accurately reconstructs the species tree, which supports our inferred reversals to outcrossing. This congruence also suggests that the chloroplast tree may not be heavily influenced by incomplete lineage sorting or chloroplast capture, at least in the particularly relevant Volvox section Volvox (Fig. 2). Furthermore, the chloroplast genome should be less influenced by incomplete lineage sorting given its reduced effective population size (Birky 1983). Second, many new species of volvocine algae continue to be described (Isaka et al. 2012; Nozaki et al. 2014, 2015a). This incomplete taxon sampling may effect both ancestral-state reconstructions as well as phylogenetic analyses of tippiness (i.e., results might change if more species were added). Our taxon sampling of described species is relatively complete (69 of 79 species, 87%). Third, it is possible that future studies of algae phylogeny may reveal that the volvocine algae had a selfing (homothallic) ancestor. Thus, we cannot exclude the possibility that evolution of selfing in volvocine algae represents reversion to ancestral selfing. However, even if this were true, it would not necessarily overturn our conclusions within volvocine algae regarding reversals to outcrossing within Volvox and the lack of both tippiness and reduced diversification rates associated with selfing.
Reversals from selfing to outcrossing are predicted to be rare (Lande and Schemske 1985; Takebayashi and Morrell 2001). However, we found that Volvox section Volvox (Fig. 2) had two independent reversions from homothallic selfing to heterothallic outcrossing (Volvox perglobator and Volvox rousseletii, Fig. 2). Furthermore, the evolutionary model allowing evolutionary reversal from selfing to outcrossing is strongly supported over one in which reversals are not allowed. While detailed studies of selfing and outcrossing rates in natural populations are necessary, this group of closely related species may be highly valuable for studying both reversals from selfing to outcrossing and the genetic mechanism underlying this transition.
The two inferred transitions from selfing to outcrossing occur in Volvox section Volvox, in which the ancestor is inferred to have been monoecious and selfing (Figs. 2, 3). It may be intuitive that dioecious selfing species would be more likely to revert to dioecious outcrossing than a monoecious selfing species. We speculate that the timing of sexual differentiation during development may complicate this intuition. If sexual differentiation in a self-fertilizing species occurs early in development (before cell lineages leading to reproductive cells have diverged), dioecy may result because all reproductive cell lineages in a colony have similarly differentiated. In contrast, if sexual differentiation in a selfing species occurs later in development (after cell lineages leading to reproductive cells have diverged), monoecy may result because reproductive cell lineages in a colony have independently differentiated. Therefore, Volvox section Volvox may revert to outcrossing more frequently only because there are more species in this group than among other origins of self-fertilization.
Despite the repeated evolution of selfing, homothallic species do not currently exhibit the “tippy” phylogenetic distribution (Fig. 4) predicted under the standard model that assumes short-term benefits but long-term costs of self-fertilization (Wright et al. 2013). Instead, we found that selfing is reversible (Fig. 2), that selfing species are able to persist and speciate (Fig. 4), and that selfing species have a higher net diversification rate than outcrossing species (Table S5). Selfing species may be able to persist and speciate if the benefits of selfing are higher than the costs. The benefits of selfing include (1) reproductive assurance, which may be a substantial benefit in patchy volvocine habitats such as lacustrine freshwater (Kirk 1998), and (2) increased contribution of genetic material to sexual offspring, compared with outcrossing individuals (Williams 1975; Nagylaki 1976). The costs of selfing include reduced genetic variation, reduced effective population size, and reduced rates of adaptation (Stebbins 1957; Pollak 1987; Wright et al. 2013). An additional cost of selfing is inbreeding depression, particularly in diploid and polyploid taxa (Barrett 2002; Charlesworth 2006). However, the volvocine algae have haploid-dominant life cycles (Coleman 2012), and costly effects of inbreeding depression are not expected in the haploid phase (Taylor et al. 2007). Inbreeding depression may exist in the dormant diploid zygospore, though little is known about the selection pressures upon these diploid zygospores. The costs of self-fertilization (i.e., inbreeding depression) are likely reduced in the haploid volvocine algae. This suggests that differences in the costs and benefits of selfing may explain why the volvocine algae do not display the tippy distribution of selfing species observed in diploid and polyploid taxa (Barrett 2002; Jarne and Auld 2006) and why selfing species do not have lower diversification rates than outcrossing species. Similarly, mosses have haploid-dominant life cycles (Shaw et al. 2011) and in mosses, hermaphroditic and dioecious species have similar diversification rates (McDaniel et al. 2013).
In this study, we found that the colonies of selfing volvocine species are significantly larger than those of outcrossing volvocine species (Table S6). Angiosperms show the opposite pattern, with larger species more likely to be outcrossing (Darwin 1876; Bawa 1980; Wright et al. 2013). We speculate two possible explanations for this pattern. First, increased colony size and selfing share the evolutionary cost of decreased effective population size (Lynch and Conery 2003; Smith et al. 2013) but have different evolutionary benefits: increased colony size decreases predation rates (Bell 1985; Boraas et al. 1998) while selfing leads to beneficial reproductive assurance (Baker 1955). If species with larger colonies are already paying the cost of decreased effective population size, evolving selfing may be highly beneficial, especially if fitness costs are nonlinear. In this way, a species with larger colonies experiences the benefits of both reduced predation and reproductive assurance, but experiences little additional cost due to reduced effective population size. Second, species with larger colonies may be rarer (Lynch and Conery 2003; Smith et al. 2013), thus making the beneficial reproductive assurance provided by self-fertilization even more important. Given the inherently fragmented nature of volvocine habitats across landscapes (freshwater lakes; Kirk 1998), the benefit of reproductive assurance may be substantial, especially when colonizing a novel habitat patch (Baker 1955; Pannell and Barrett 1998).
Conclusions
The reproductive strategy of self-fertilization has long been thought to be an evolutionary dead-end, which repeatedly evolves due to short-term benefits. In contrast, long-term costs are thought to reduce speciation and increase extinction of self-fertilizing lineages. The transmission costs of outcrossing are expected to prevent reversals to outcrossing once selfing has evolved. Therefore, self-fertilizing species are expected to occupy the tips of the phylogenetic tree. In this study, we test these predictions with phylogenetic analyses in volvocine green algae. We show that the volvocine green algae had repeated origins of self-fertilization. However, we also found the volvocine algae demonstrate repeated reversals from selfing to outcrossing. Contrary to predictions, we do not infer a reduced diversification rate in self-fertilizing volvocine species. Furthermore, selfing homothallic species currently lack the expected “tippy” phylogenetic distribution. We suggest that these results may be partially explained by the haploidy of volvocine green algae, which reduces the effects of inbreeding depression, ameliorating an important potential cost of selfing.
Supplementary Material
Acknowledgments
We would like to thank Patrick J. Ferris, Dinah R. Davison, Elizabeth C. Miller, and Daniel S. Moen for technical assistance, Deborah E. Shelton for collecting strains, Dinah R. Davison, Sarah P. Otto, Michael Barker, and anonymous reviewers for their helpful comments and discussion. We gratefully acknowledge the support of the National Aeronautics and Space Administration (NNX13AH41G, NNX15AR33G, Cooperative Agreement Notice 7), the National Institute of Health (GM084905), the National Science Foundation (MCB-1412395, DEB-1457701), and MEXT/JSPS KAKENHI Grants-in-Aid for Scientific Research (15K14590 and 16H02518).
Footnotes
Author contributions
E.R.H. conceived the project, collected species character data, described strains, performed statistical analyses, and wrote the manuscript; M.D.H. conceived the project and collected species character data; J.J.W. conceived experimental design; H.N. collected species character data; R.E.M. conceived the project; all authors contributed to preparation of the manuscript.
LITERATURE CITED
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Baker HG. Self-compatibility and establishment after “long-distance” dispersal. Evolution. 1955;9:347–349. [Google Scholar]
- Barrett SCH. The evolution of plant sexual diversity. Nat Rev Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. Understanding plant reproductive diversity. Philos Trans R Soc B Biol Sci. 2010;365:99–109. doi: 10.1098/rstb.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa KS. Evolution of dioecy in flowering plants. Annu Rev Ecol Syst. 1980;11:15–39. [Google Scholar]
- Bell G. The origin and evolution of germ cells as illustrated by the Volvocales. In: Halvorson HO, Monroy A, editors. The origin and evolution of sex. Alan R. Liss; New York: 1985. pp. 221–256. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev. 2011;86:421–442. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- Birky CW. Relaxed cellular controls and organelle heredity. Science. 1983;222:468–475. doi: 10.1126/science.6353578. [DOI] [PubMed] [Google Scholar]
- Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Boraas ME, Seale DB, Boxhorn JE. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol Ecol. 1998;12:153–164. [Google Scholar]
- Bromham L, Hua X, Cardillo M. Detecting macroevolutionary self-destruction from phylogenies. Syst Biol. 2016;65:109–127. doi: 10.1093/sysbio/syv062. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multi-model inference: a practical information-theoretic approach. Springer-Verlag; New York: 2002. [Google Scholar]
- Charlesworth D. Evolution of plant breeding systems. Curr Biol. 2006;16:726–735. doi: 10.1016/j.cub.2006.07.068. [DOI] [PubMed] [Google Scholar]
- Coleman A. Sexual isolation in Pandorina morum. J Protozool. 1959;6:249–264. [Google Scholar]
- Coleman AW. A comparative analysis of the Volvocaceae (Chlorophyta) J Phycol. 2012;48:491–513. doi: 10.1111/j.1529-8817.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- Coleman AW. Sexuality in colonial green flagellates. In: Levandowsky M, Hutner SH, editors. Biochemistry and Physiology of Protozoa. Academic Press; New York: 1979. pp. 307–340. [Google Scholar]
- Darden WH., Jr Sexual differentiation in Volvox aureus. J Protozool. 1966;13:239. doi: 10.1111/j.1550-7408.1966.tb01901.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. Different forms of flowers on plants of the same species. John Murray; London: 1877. [Google Scholar]
- Darwin C. The effects of cross and self fertilization in the vegetable kingdom. 2. John Murray; London: 1876. [Google Scholar]
- Davis MP, Midford PE, Maddison W. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol Biol. 2013;13:38. doi: 10.1186/1471-2148-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos JM, Wuest RO, Conti E. Small and ugly? Phylogenetic analyses of the “selfing syndrome” reveal complex evolutionary fates of monomorphic primrose flowers. Evolution. 2014;68:1042–1057. doi: 10.1111/evo.12331. [DOI] [PubMed] [Google Scholar]
- Escobar JS, Cenci A, Bolognini J, Haudry A, Laurent S, David J, Glémin S. An integrative test of the dead-end hypothesis of selfing evolution in Triticeae (Poaceae) Evolution. 2010;64:2855–2872. doi: 10.1111/j.1558-5646.2010.01045.x. [DOI] [PubMed] [Google Scholar]
- FitzJohn RG. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–1092. [Google Scholar]
- Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv Biol. 2010;24:1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Barraclough TG. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: A revised method and evaluation on simulated data sets. Syst Biol. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A. Notes on the statistical power of the binary state speciation and extinction (BiSSE) model. Evol Bioinforma. 2016;12:165–174. doi: 10.4137/EBO.S39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A, Fischer GA, Comes HP. Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum) in light of the hypothesis of selfing as an evolutionary dead end. BMC Evol Biol. 2015;15:192. doi: 10.1186/s12862-015-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igic B. Species selection maintains self-incompatibility. Science. 2010;330:493–495. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Hamrick J, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philos Trans Biol Sci. 1996;351:1291–1298. [Google Scholar]
- Hanschen ER, Marriage TN, Ferris PJ, Hamaji T, Toyoda A, Fujiyama A, Neme R, Noguchi H, Minakuchi Y, Suzuki M, Kawai-Toyooka H, Smith DR, Luria V, Karger A, Kirschner MW, Sparks H, Anderson J, Bakaric R, Durand PM, Michod RE, Nozaki H, Olson BJSC. The Gonium pectorale genome demostrates co-option of cell cycle regulation during the evolution of multicellularity. Nat Commun. 2016;7:11370. doi: 10.1038/ncomms11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci USA. 2009;106:3254–3258. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Michod RE. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution. 2008;62:436–451. doi: 10.1111/j.1558-5646.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Hua X, Bromham L, Freckleton R. Phylometrics: an R package for detecting macroevolutionary patterns, using phylogenetic metrics and backward tree simulation. Methods Ecol Evol. 2016;7:806–810. [Google Scholar]
- Igic B, Busch JW. Is self-fertilization an evolutionary dead end? New Phytol. 2013;198:386–397. doi: 10.1111/nph.12182. [DOI] [PubMed] [Google Scholar]
- Igic B, Lande R, Kohn JR. Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci. 2008;169:93–104. [Google Scholar]
- Isaka N, Kawai-Toyooka H, Matsuzaki R, Nakada T, Nozaki H. Description of two new monoecious species of Volvox sect. Volvox (Volvocaceae, Chlorophyceae), based on comparative morphology and molecular phylogeny of cultured material. J Phycol. 2012;48:759–767. doi: 10.1111/j.1529-8817.2012.01142.x. [DOI] [PubMed] [Google Scholar]
- Iyengar MOP. Contributions to our knowledge of the colonial Volvocales of south India. J Linn Soc London, Bot. 1933;49:323–373. [Google Scholar]
- Jarne P, Auld JR. Animals mix it up too: The distribution of self-fertilization among hermaphroditic animals. Evolution. 2006;60:1816–1824. doi: 10.1554/06-246.1. [DOI] [PubMed] [Google Scholar]
- Johnson MTJ, FitzJohn RG, Smith SD, Rausher MD, Otto SP. Loss of sexual recombination and segregation is associated with increased diversification in evening primroses. Evolution. 2011;65:3230–3240. doi: 10.1111/j.1558-5646.2011.01378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes Factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Kirk DL. Volvox: Molecular-genetic origins of multicellularity and cellular differentiation. Cambridge University Press; Cambridge: 1998. [Google Scholar]
- Lande R, Schemske D. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2016;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Lloyd DG. Selection of combined versus separate sexes in seed plants. Am Nat. 1982;120:571–585. [Google Scholar]
- Luthringer R, Cormier A, Ahmed S, Peters AF, Cock JM, Coelho SM. Sexual dimorphism in the brown algae. Perspect Phycol. 2014;1:11–25. [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Maddison WP, FitzJohn RG. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst Biol. 2015;64:127–136. doi: 10.1093/sysbio/syu070. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. The evolution of sex. Cambridge University Press; Cambridge: 1978. [Google Scholar]
- McDaniel SF, Atwood J, Burleigh JG. Recurrent evolution of dioecy in bryophytes. Evolution. 2013;67:567–572. doi: 10.1111/j.1558-5646.2012.01808.x. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu L-H, Nelson DR, Sanderfoot Aa, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen C-L, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral J-P, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen C-J, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–50. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer ALS, Wiens JJ. Estimating diversification rates for higher taxa: BAMM can give problematic estimate of rates and rate shifts. Evolution. 2017 doi: 10.1111/evo.13378. in press. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. A model for the evolution of self-fertilization and vegetative reproduction. J Theor Biol. 1976;58:55–58. doi: 10.1016/0022-5193(76)90138-7. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Matsuzaki R, Yamamoto K, Kawachi M, Takahashi F. Delineating a new heterothallic species of Volvox (Volvocaceae, Chlorophyceae) using new strains of “Volvox africanus”. PLoS One. 2015a;10:1–16. doi: 10.1371/journal.pone.0142632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, Takahara M, Nakazawa A, Kita Y, Yamada T, Takano H, Kawano S, Kato M. Evolution of rbcL group IA introns and intron open reading frames within the colonial Volvocales (Chlorophyceae) Mol Phylogenet Evol. 2002;23:326–338. doi: 10.1016/s1055-7903(02)00030-1. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Ueki N, Misumi O, Yamamoto K, Yamashita S, Herron MD, Rosenzweig F. Morphology and reproduction of Volvox capensis (Volvocales, Chlorophyceae) from Montana, USA. Phycologia. 2015b;54:316–320. [Google Scholar]
- Nozaki H, Yamada TK, Takahashi F, Matsuzaki R, Nakada T. New “missing link” genus of the colonial volvocine green algae gives insights into the evolution of oogamy. BMC Evol Biol. 2014;14:37–47. doi: 10.1186/1471-2148-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme CDL, Freckleton RP, Thomas GH, Petzoldt T, Fritz SA, Isaac NJB, Pearse W. The caper package: comparative analysis of phylogenetics and evolution in R 2012 [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol. 1999;48:612–622. [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Pannell JR, Barrett SCH. Baker’s law revisited: Reproductive assurance in a metapopulation. Evolution. 1998;52:657–668. doi: 10.1111/j.1558-5646.1998.tb03691.x. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parfrey L, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics. 1987;117:353–360. doi: 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J. Further studies in Volvox, with descriptions of three new species. Trans Am Microsc Soc. 1908;28:141–175. [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris PJ, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk DL, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–6. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One. 2014:9. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Goldberg EE. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst Biol. 2015;64:340–355. doi: 10.1093/sysbio/syu131. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna: 2013. [Google Scholar]
- Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- Rich F, Pocock MA. Observations on the genus Volvox in Africa. Ann South African Museum. 1933;16:427–471. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Johnston MO, L’Heureux A-M, Marsolais JV. Evolutionary history of the mating system in Amsinckia (Boraginaceae) Evolution. 1997;51:1090–1099. doi: 10.1111/j.1558-5646.1997.tb03956.x. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Morgan MT, Bataillon T. How does self-pollination evolve? Inferences from floral ecology and molecular genetic variation. Philos Trans R Soc B Biol Sci. 1996;351:1281–1290. [Google Scholar]
- Shaw AJ, Szövényi P, Shaw B. Bryophyte diversity and evolution: Windows into the early evolution of land plants. Am J Bot. 2011;98:352–369. doi: 10.3732/ajb.1000316. [DOI] [PubMed] [Google Scholar]
- Shaw WR. Janetosphaera, a new genus, and two new species of Volvox. Philipp J Sci. 1922;20:477–508. [Google Scholar]
- Smith DR, Hamaji T, Olson BJSC, Durand PM, Ferris P, Michod RE, Featherston J, Nozaki H, Keeling PJ. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol. 2013;30:793–797. doi: 10.1093/molbev/mst002. [DOI] [PubMed] [Google Scholar]
- Smith GM. A comparative study of the species of Volvox. Trans Am Microsc Soc. 1944;63:265–310. [Google Scholar]
- Starr RC. Sexual reproduction in Volvox africanus. In: Parker BC, Brown RMJ, editors. Contributions in Phycology. Allen Press; Lawrence, Kansas: 1971. pp. 59–66. [Google Scholar]
- Starr RC. Volvox pocockiae, a new species with dwarf males. J Phycol. 1970;6:234–239. [Google Scholar]
- Starr RC, O’Neil R, Miller CE., III L-Glutamic acid as a mediator of sexual morphogenesis in Volvox capensis. Proc Natl Acad Sci U S A. 1980;77:1025–1028. doi: 10.1073/pnas.77.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Flowering plants: evolution above the species level. Harvard University Press; Cambridge, MA: 1974. [Google Scholar]
- Stebbins GL. Self fertilization and population variability in the higher plants. Am Nat. 1957;91:337–354. [Google Scholar]
- Takebayashi N, Morrell PL. Is self-fertilization an evolutionary dead end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am J Bot. 2001;88:1143–1150. [PubMed] [Google Scholar]
- Taylor PJ, Eppley SM, Jesson LK. Sporophytic inbreeding depression in mosses occurs in a species with separate sexes but not in a species with combined sexes. Am J Bot. 2007;94:1853–1859. doi: 10.3732/ajb.94.11.1853. [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Otto SP, Barrett SCH. Phylogenetic analysis of the ecological correlates of dioecy in angiosperms. J Evol Biol. 2003;16:1006–1018. doi: 10.1046/j.1420-9101.2003.00559.x. [DOI] [PubMed] [Google Scholar]
- Whitehouse HLK. Heterothallism and sex in the fungi. Biol Rev. 1949;24:411–447. doi: 10.1111/j.1469-185x.1949.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Morrill MC. Missing data in phylogenetic analysis: Reconciling results from simulations and empirical data. Syst Biol. 2011;60:719–731. doi: 10.1093/sysbio/syr025. [DOI] [PubMed] [Google Scholar]
- Williams GC. Sex and evolution. Princeton University Press; Princeton, New Jersey: 1975. [Google Scholar]
- Wright SI, Kalisz S, Slotte T. Evolutionary consequences of self-fertilization in plants. Proc R Soc B Biol Sci. 2013;280:1–10. doi: 10.1098/rspb.2013.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S-H, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Evolution. 1999;96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.