Summary
Protein synthesis relies on several translational GTPases (trGTPases), related proteins that couple the hydrolysis of GTP to specific molecular events on the ribosome. Most bacterial trGTPases, including IF2, EF-Tu, EF-G, and RF3, play well-known roles in translation. The cellular functions of LepA (also termed EF4) and BipA (also termed TypA), on the other hand, have remained enigmatic. Recent studies provide compelling in vivo evidence that LepA and BipA function in biogenesis of the 30S and 50S subunit, respectively. These findings have important implications for ribosome biogenesis in bacteria. Because the GTPase activity of each of these proteins depends on interactions with both ribosomal subunits, some portion of 30S and 50S assembly must occur in the context of the 70S ribosome. In this review, we introduce the trGTPases of bacteria, describe the new functional data on LepA and BipA, and discuss the how these findings shape our current view of ribosome biogenesis in bacteria.
Abbreviated summary (for Graphical Abstract)
LepA and BipA are paralogs of EF-G highly conserved among bacteria. Recent studies show that these GTPases act in biogenesis of the 30S and 50S subunit, respectively. An important implication of these findings is that, for each subunit, some portion of the assembly process occurs in the context of the 70S ribosome.
The P-loop GTPases represent a large family of proteins involved in various cellular processes including translation, signal transduction, cell motility, intracellular transport, protein trafficking, and chromosome partitioning (Leipe et al., 2002). There is an important subfamily of P-loop GTPases termed the translational GTPases (trGTPases). Characteristic of the trGTPases is a domain architecture in which the highly-conserved GTPase (G) domain is followed by a β barrel domain (domain II). In bacteria, there are nine trGTPases including initiation factor IF2; elongation factor EF-Tu and similar proteins SelB and CysN; and elongation factor EF-G and similar proteins RF3, TetM, BipA, and LepA (Margus et al., 2007). With the sole exception of CysN, these trGTPases bind the 70S ribosome and exhibit ribosome-dependent GTP hydrolysis activity (Burdett, 1996; Carlson et al., 2017; De Laurentiis and Wieden, 2015; deLivron and Robinson, 2008). CysN is a component of the enzyme ATP sulfurylase, which forms activated sulfate (adenosine 5′-phosphosulfate; APS) from ATP and sulfate, an obligate step in the metabolic assimilation of sulfur (Leyh and Suo, 1992). GTP hydrolysis by the CysN subunit regulates APS formation in many bacteria (Liu et al., 1998; Margus et al., 2007). Structures of IF2, EF-Tu, SelB, EF-G, RF3, TetM, BipA, and LepA on the ribosome have been determined by cryo-electron microscopy and/or X-ray crystallography (Fischer et al., 2016; Gagnon et al., 2014; Gao et al., 2009; Kumar et al., 2015; Li et al., 2013; Schmeing et al., 2009; Sprink et al., 2016; Zhou et al., 2012). These factors bind the ribosome in a similar manner with domains G and II making specific contacts to the 50S and 30S subunit, respectively, on the A-site side of the ribosome (Fig. 1). The G domain interacts with a conserved region of the 50S subunit including the sarcin-ricin loop (SRL) of the 23S rRNA, contacts critical for GTPase activation.
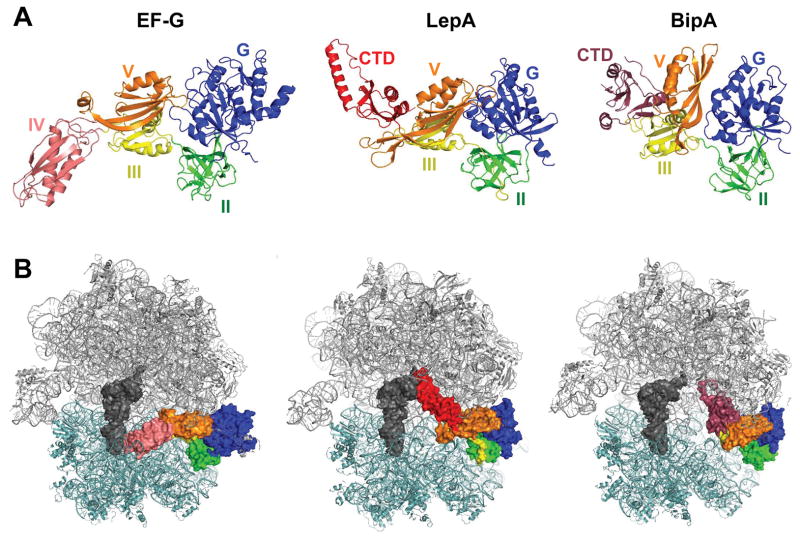
Figure 1. Homologous proteins EF-G, LepA, and BipA bind the ribosome similarly.
(A) Structures of EF-G, LepA, and BipA. Domains G (blue), II (green), III (yellow), and V (orange) are homologous among the factors. Unique domains include EF-G domain IV (pink), LepA C-terminal domain (red), and BipA C-terminal domain (raspberry). Images are based on PDB ID files 4V5F, 5J8B, and 4ZCI. (B) Structures of EF-G, LepA, and BipA bound to the 70S ribosome. 50S subunit, gray; 30S subunit, light teal; P-site tRNA; dark gray. Color coding of GTPase domains as in panel A. Images are based on PDB ID files 4V5F, 4W2E, and 5AA0.
Most ribosome-associated trGTPases play well-established roles in the cell. IF2 promotes the binding of initiator tRNA (fMet-tRNAfMet) to the small (30S) subunit of the ribosome and the subsequent docking of the large (50S) subunit during initiation (Antoun et al., 2003; Antoun et al., 2006; Goyal et al., 2015; Grigoriadou et al., 2007; Marshall et al., 2009; Pavlov et al., 2011). The GTP-bound form of IF2 is important for these functions, and the hydrolysis of GTP (which occurs upon subunit joining) facilitates subsequent IF2 release. EF-Tu catalyzes the binding of aminoacyl-tRNA (aa-tRNA) to the A site of the ribosome during elongation (Geggier et al., 2010; Gromadski and Rodnina, 2004; Johansson et al., 2012; Loveland et al., 2017; Ogle et al., 2002; Satpati et al., 2014; Voorhees et al., 2010). EF-Tu•GTP•aa-tRNA samples ribosomes carrying peptidyl-tRNA in the P site, and pairing between the anticodon of aa-tRNA and the codon in the 30S A site triggers GTP hydrolysis by EF-Tu. This causes release of the acceptor end of aa-tRNA from EF-Tu, allowing the aa-tRNA to move fully into the ribosome (A/A site) and participate in peptide bond formation. The GTP hydrolysis event is functionally irreversible and hence provides two independent opportunities for rejection of incorrect aa-tRNA substrates, increasing the overall accuracy of aa-tRNA selection. SelB resembles EF-Tu in structure and function but specifically facilitates incorporation of selenocysteinyl-tRNA at appropriate UGA codons (Fischer et al., 2016; Kotini et al., 2015; Zinoni et al., 1990). Such UGA codons are marked by a stem-loop structure (SECIS element), which lies just downstream in the mRNA. SelB binds selenocysteinyl-tRNASec exclusively and has a unique domain that interacts with the SECIS element, thereby directing site-specific incorporation of selenocysteine. EF-G catalyzes translocation, the movement of tRNAs (and paired codons) to their adjacent sites in the ribosome (Belardinelli et al., 2016; Chen et al., 2016; Cunha et al., 2013; Savelsbergh et al., 2003; Wasserman et al., 2016; Zhang et al., 2016; Zhou et al., 2014). Binding of EF-G•GTP to the pretranslocation complex (carrying deacylated tRNA in the P site and peptidyl-tRNA in the A site) results in rapid GTP hydrolysis, which is followed by a conformational change that “unlocks” the ribosome and promotes tRNA-mRNA movement. Hydrolysis of GTP speeds tRNA-mRNA movement and is critical for subsequent release of EF-G from the posttranslocation complex. TetM is structurally similar to EF-G, and confers resistance to the antibiotic tetracycline (Burdett, 1991; Burdett, 1996; Li et al., 2013). TetM•GTP binds the ribosome and catalyzes release of tetracycline from the 30S A site. This presumably occurs via conformational changes induced by TetM, although the detailed mechanism has yet to be elucidated. RF3 catalyzes the dissociation of release factor RF1 or RF2 (RF1/2) during termination (Freistroffer et al., 1997; Jin et al., 2011; Koutmou et al., 2014; Peske et al., 2014; Shi and Joseph, 2016; Zhou et al., 2012). RF3•GTP binds the posttermination ribosome (containing deacylated tRNA in the P site and RF1/2 in the A site) and promotes intersubunit rotation, which destabilizes RF1/2. Hydrolysis of GTP by RF3 allows dissociation of both termination factors from the ribosome. LepA and BipA are structurally similar to EF-G, with the three proteins sharing four homologous domains (Fig. 1) (Evans et al., 2008; Fan et al., 2015). The biological roles of LepA and BipA have been difficult to pin down.
A phylogenetic analysis of trGTPases in >200 representative bacterial genomes revealed that IF2, EF-Tu, EF-G, and LepA are the most ubiquitous (Margus et al., 2007). Genes encoding IF2, EF-Tu, and EF-G were found in all bacteria, while the lepA gene was found in all but one particular strain of Streptococcus pyogenes. BipA was found in most (86%) of the bacteria analyzed, often being absent in those lineages with minimal genomes. By comparison, RF3, a well-known participant in translation termination, was found in 62% of the bacteria analyzed. These observations imply that LepA and BipA play important roles in the bacterial cell.
LepA functions in 30S subunit biogenesis
In 1985, the lepA gene was originally identified as the gene upstream from the leader peptidase (lep) gene in Escherichia coli (March and Inouye, 1985). Dibb and Wolfe (1986) disrupted the lepA gene, and the null mutant conferred no obvious defects in growth or protein secretion (Dibb and Wolfe, 1986). Since then, other investigators have further analyzed the ΔlepA mutant and found only subtle phenotypes, such as increased sensitivity to tellurite (Shoji et al., 2010) and compromised fitness under conditions of low pH, low temperature, and high Mg2+ (the latter phenotypes being revealed only in growth competition assays) (Pech et al., 2011). Loss of LepA confers more obvious phenotypes in other organisms. In Helicobacter pylori, LepA was found to be essential under acidic conditions (Bijlsma et al., 2000). In Streptomyces coelicolor, loss of lepA leads to overproduction of an antibiotic (Badu-Nkansah and Sello, 2010). Eukaryotes also have LepA in the form of mitochondrial and chloroplast homologs. In Saccharomyces cerevisae, loss of LepA (Guf1) causes heat and cold sensitivity and reduced levels of cytochrome oxidase (Bauerschmitt et al., 2008). In mice, a mitochondrial lepA knockout leads to decreased spermatogenesis and male stertility (Gao et al., 2016). In Arabidopsis thaliana, deletion of cpLEPA impairs chloroplast development and increases sensitivity to light (Ji et al., 2012). These disparate phenotypes across various lineages have been puzzling and provided little clarity regarding the physiological role of LepA.
In 2006, it was proposed that LepA acts as a “back-translocase,” catalyzing reverse tRNA-mRNA movement in the ribosome (Qin et al., 2006). This idea was attractive, due to the structural similarity between LepA and EF-G (Fig. 1). However, multiple laboratories have since been unable to confirm this biochemical activity (Balakrishnan et al., 2014; Ermolenko; Liu et al., 2010; Rodnina). Using toeprinting, which monitors the position of mRNA in the ribosome, Balakrishnan et al. (2014) found no evidence that LepA could catalyze reverse translocation in various ribosomal complexes, although the protein exhibited robust ribosome-dependent GTPase activity (Balakrishnan et al., 2014). Cooperman and co-workers used puromycin and fluorescent probes on mRNA and tRNA to interrogate the effects of LepA on translocation (Liu et al., 2010). Addition of LepA to ribosomes in the posttranslocation state promoted some movement of tRNA with respect to the 50S subunit, reducing puromycin reactivity of bound peptidyl-tRNA to an intermediate level. Codon-anticodon movement followed at a very slow rate (0.0009 s-1), virtually identical to that seen in the absence of LepA (Liu et al., 2010). Thus, while LepA can interact with the ribosome and compete with elongation factors (Liu et al., 2010; Liu et al., 2011), LepA does not substantially accelerate reverse codon-anticodon movement (i.e., back translocation) as initially proposed (Qin et al., 2006).
Important clues about the role of LepA came from Balakrishnan et al. (2014). Using ribosome profiling, they found that loss of LepA alters the average ribosome density (ARD) on hundreds of mRNAs in E. coli (Balakrishnan et al., 2014). The effects on ARD depend on the sequence of the translation initiation region, with a tendency for mRNAs with “strong” Shine-Dalgarno sequences to exhibit reduced translation efficiency in the absence of LepA. While clearly influencing ARD, LepA had virtually no effect on ribosome distribution along mRNA, arguing against a role in elongation. Indeed, direct measurements of elongation rates showed no difference in wild-type versus ΔlepA cells (Balakrishnan et al., 2014). Furthermore, Shoji et al. (2010) showed that ΔlepA does not alter the frequency of miscoding or frameshifting (-1 or +1, spontaneous or programmed) (Shoji et al., 2010). Together these data suggested that LepA primarily influences initiation, with little to no impact on elongation (Balakrishnan et al., 2014).
Balakrishnan et al. (2014) also screened for synthetic phenotypes by moving ΔlepA mutation into every strain of the Keio collection, a set of strains in which each non-essential gene is deleted (Baba et al., 2006). The results showed that ΔlepA confers a synthetic growth defect in strains compromised for gene regulation (dksA, molR), transport (tatB, tonB, tolR), respiration (ubiF, ubiG, ubiH) and ribosome assembly (rsgA) (Balakrishnan et al., 2014). Of these genes identified, rsgA is only one clearly linked to the ribosome and hence potentially most pertinent to LepA function. RsgA (also termed YjeQ) is a GTPase known to be important in 30S subunit biogenesis (Campbell and Brown, 2008; Goto et al., 2011; Leong et al., 2013), raising the possibility that LepA also contributes to this process. Balakrishnan et al. (2014) hypothesized that loss of LepA causes a defect in ribosome assembly that indirectly alters translation initiation in the cell (Balakrishnan et al., 2014).
To directly test the role of LepA in ribosome assembly, Gibbs et al. (2017) used SILAC (stable isotope labeling of amino acids in culture) (Ong et al., 2002) and mass spectrometry to determine r-protein composition of ribosomal particles in the presence and absence of LepA (Gibbs et al., 2017). They found that four r proteins (S3, S10, S14, and S21) are disproportionately underrepresented in 30S particles in the ΔlepA strain. These proteins assemble at a late stage of 30S subunit biogenesis and are involved in the folding of the 3′ major and minor domains of the 16S rRNA. In addition, 30S particles in the ΔlepA mutant contain elevated levels of precursor (17S) rRNA. This defect in 30S assembly did not appear to be an indirect consequence of altered protein production rates. Based on ribo-seq data, loss of LepA caused no appreciable decrease in small subunit protein or assembly factor production rates. Collectively, these data indicate that LepA functions in 30S subunit biogenesis. The genetic link between lepA and rsgA was further investigated by performing a similar SILAC analysis in ΔrsgA cells. Ribosomal protein composition of fractions from ΔrsgA cells showed accumulation of 30S particles lacking S2, S3, S10, and/or S21 (Gibbs et al., 2017). These particles, which presumably represent stalled pre-30S intermediates, resemble those seen in the absence of LepA. Thus, LepA and RsgA appear to play partially redundant roles in 30S subunit biogenesis (Gibbs et al., 2017).
BipA functions in 50S subunit biogenesis
BipA (BPI-inducible protein A) was first identified in Salmonella typhimurium as a protein produced when cells are treated with bactericidal/permeability increasing protein (BPI), an antimicrobial secreted by human neutrophils (Qi et al., 1995). BipA has since been implicated in the virulence of several human pathogens. In enteropathogenic E. coli (EPEC), loss of the factor leads to decreased cytoskeletal rearrangements in host cells, sensitivity to host defense peptides, and hypermotility (Farris et al., 1998; Grant et al., 2003). BipA influences thermoregulation of the E. coli capsule, leading to increased transcription of K-antigens at 37°C and reduced transcription at 20°C (Rowe et al., 2000). In E. coli K12, ΔbipA leads to reduced growth rate at 20°C (Pfennig and Flower, 2001), suggesting that BipA activity depends on temperature. The ΔbipA mutant also exhibits hypersensitivity to antibiotics that target the ribosome, such as chloramphenicol and tobramycin (Duo et al., 2008). In Pseudomonas aeruginosa, BipA is important for virulence, antimicrobial resistance, and biofilm formation (Neidig et al., 2013).
Biochemical studies showed that BipA, like other trGTPases, exhibits 70S-dependent GTP hydrolysis activity (deLivron and Robinson, 2008; Kumar et al., 2015). The structure of BipA is similar to LepA and EF-G except for a unique C-terminal domain (CTD, Fig. 1), which is crucial but not sufficient for ribosome binding (deLivron et al., 2009). The truncated factor BipAΔCTD exhibits elevated and ribosome-independent GTP hydrolysis activity, suggesting the CTD is necessary for GTPase control (deLivron et al., 2009). BipA can also bind ppGpp, the bacterial stress alarmone, and BipA•ppGpp appears to preferentially interact with the 30S subunit (deLivron and Robinson, 2008; Fan et al., 2015). Structural studies have revealed that, off the ribosome, BipA adopts virtually the same structure regardless of the nucleotide bound (GDPCP, GDP, ppGpp, or no nucleotide) (Fan et al., 2015; Kumar et al., 2015). On the ribosome, BipA•GDPCP exhibits a different, more compact structure (Kumar et al., 2015). Although structures of ribosome-bound BipA•ppGpp have yet to be reported, modeling of ppGpp in place of GDPCP predicts a steric clash between the 3′ diphosphate and the SRL of the 50S subunit (Kumar et al., 2015). This may explain why BipA•ppGpp associates with 30S rather than 70S particles. It was postulated that BipA acts as a stress factor, with ppGpp allosterically controlling ribosome binding specificity to regulate synthesis of virulence and stress factors (deLivron and Robinson, 2008). However, to our knowledge, no additional evidence has since materialized to bolster this hypothesis.
Compelling in vivo evidence suggests instead that BipA functions in biogenesis of the 50S subunit of the ribosome. Flower and coworkers screened for Tn5 insertion mutations that suppress the cold sensitive phenotype of ΔbipA in E. coli (Krishnan and Flower, 2008). They found several independent insertions, all of which mapped to rluC, which encodes a pseudouridine synthase that modifies U955, U2504, and U2580 of the 23S rRNA. It was further shown that loss of RluC or a triple substitution of its three target nucleotides (U955, U2504, U2580) could suppress the cold-sensitive and capsule synthesis phenotypes of the ΔbipA mutant (Krishnan and Flower, 2008). As ribosome modification and assembly are coordinated processes, this raised the possibility that BipA plays a role in 50S biogenesis. Indeed, further work by Flower and coworkers showed that loss of bipA leads to accumulation of pre-50S particles and elevated levels of precursor 23S rRNA in cells grown at 20°C (Choudhury and Flower, 2015). Additionally, they found that deletion of deaD, encoding a DEAD-box helicase that acts on the 23S rRNA, exacerbates the cold-sensitivity and 50S assembly phenotypes of the ΔbipA mutant (Choudhury and Flower, 2015). Together, these data provide strong evidence that BipA participates in ribosome assembly. A defect in 50S biogenesis could be the basis of the pleiotropic phenotypes of ΔbipA reported previously (Duo et al., 2008; Farris et al., 1998; Grant et al., 2003; Neidig et al., 2013; Pfennig and Flower, 2001; Qi et al., 1995; Rowe et al., 2000).
Implications for ribosome assembly
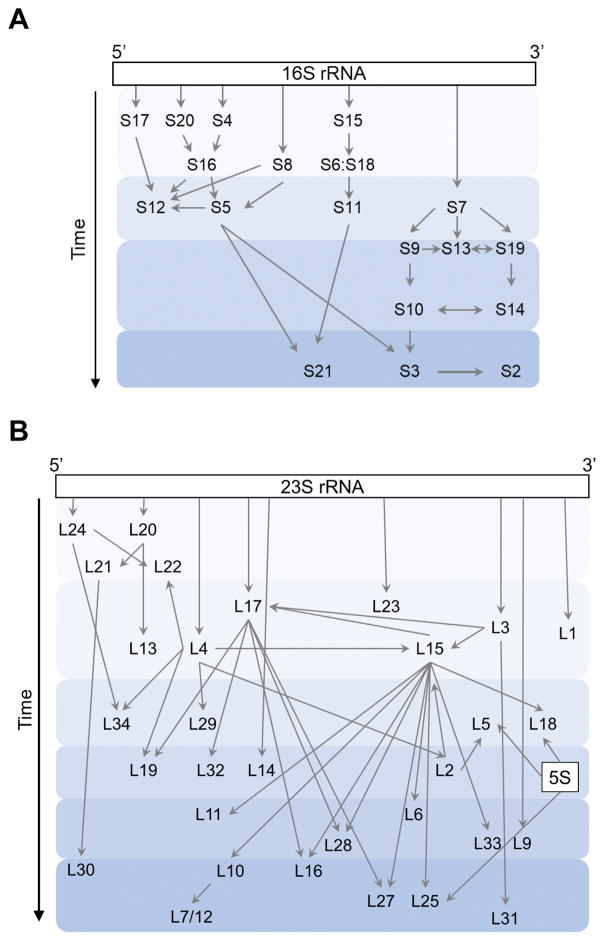
The bacterial 70S ribosome is a ~2.5 MDa enzyme composed of three large rRNAs (16S, 23S, 5S) and ~50 r proteins. Biogenesis of the ribosome is complicated and begins with transcription of the three rRNA molecules, which are usually co-transcribed in a single operon. The primary transcript is cleaved by endonucleases into precursor rRNAs, which are later trimmed to form mature 16S, 23S, and 5S rRNA. Assembly of each subunit involves the folding of rRNA and binding of r proteins in a generally hierarchical manner (Fig. 2). Transcription and cleavage, rRNA modification, r protein binding, and rRNA folding are coordinated, although there are many random-order events in the assembly process, resulting in multiple parallel pathways (Mulder et al., 2010; Talkington et al., 2005). Early studies showed that each ribosomal subunit can self-assemble in vitro using purified components (Nierhaus and Dohme, 1974; Traub and Nomura, 1968). However, self-assembly is slow and requires non-physiological conditions such as elevated temperature and Mg2+ concentration. In the cell, ribosome assembly is considerably faster and more efficient due to dozens of assembly factors (AFs), including ribonucleoprotein-binding (RNP-binding) proteins, modification enzymes, helicases, chaperones, and GTPases (Shajani et al., 2011; Wilson and Nierhaus, 2007) (Table 1).
Figure 2. Assembly maps.
Protein binding during assembly of the small (A) or large (B) subunit is depicted schematically. Arrows indicate hierarchical dependencies and shaded regions indicate temporal stages. Schemes are based on (Chen and Williamson, 2013; Davis et al., 2016).
Table 1.
A non-comprehensive list of E. coli proteins implicated in ribosome assembly
| Assembly factor1 | Type | Ribosomal subunit |
|---|---|---|
| RbfA | RNP binding | 30S |
| RimJ | RNP binding | 30S |
| RimM | RNP binding | 30S |
| RimP | RNP binding | 30S |
| YhbY | RNP binding | 50S |
| KsgA (RsmA) | Modification enzyme | 30S |
| RsmC | Modification enzyme | 30S |
| RlmA (RrmA) | Modification enzyme | 50S |
| RlmE (RrmJ) | Modification enzyme | 50S |
| RluB | Modification enzyme | 50S |
| RluC | Modification enzyme | 50S |
| RluD | Modification enzyme | 50S |
| DeaD (RhlD) | Helicase | 50S |
| DbpA (RhlC) | Helicase | 50S |
| SrmB (RhlA) | Helicase | 50S |
| RhlE | Helicase | 50S |
| DnaK/DnaJ/GrpE | Chaperone | 30S, 50S |
| GroES/GroEL | Chaperone | 50S |
| Era | GTPase | 30S |
| RsgA (YjeQ) | GTPase | 30S |
| LepA (EF4) | GTPase | 30S |
| Der (EngA) | GTPase | 50S |
| YihA (EngB) | GTPase | 50S |
| BipA (TypA) | GTPase | 50S |
Assignments based on (Keseler et al., 2017; Shajani et al., 2011; Wilson and Nierhaus, 2007) and references therein.
Assembly of the small and large subunits have generally been considered separately, as though they are independent processes. However, factors such as LepA and BipA, which are important for 30S and 50S biogenesis, respectively, each require the 70S ribosome for GTP hydrolysis. This implies that at least a portion of the assembly process occurs in the context of the 70S ribosome. It has been proposed for example, that LepA acts late in the assembly process by binding a precursor 70S particle and catalyzing a conformational change in the 30S subunit head domain that provides another opportunity for correct folding of the 3′ domain of 16S rRNA (Gibbs et al., 2017). This activity is in line with other trGTPases, such as EF-G, TetM, and RF3, which promote conformational changes in the 70S ribosome as part of their biological function.
A growing body of evidence indicates that, in all organisms, ribosome assembly is checked through quality control mechanisms (Karbstein, 2013). In eukaryotes, a number of AFs mimic r proteins or translation factors and bind specific regions of the pre-40S, occluding the functional sites of the subunit until maturation is complete (Strunk et al., 2011). For example, Rio2, Tsr1, and Dim1 occupy binding sites for eIF1, eIF1A, and initiator tRNA, thereby preventing premature translation initiation (Strunk et al., 2011). Intriguingly, final assembly of the 40S subunit includes a functional “test drive” in which 80S complexes lacking mRNA and tRNA are formed and subsequently dissociated prior to canonical translation initiation (Strunk et al., 2012). The test drive begins with a pre-40S particle bound by several AFs (Enp1, Pno1, Dim1, Rio2, Tsr1, and Nob1). Initiation factor eIF5B promotes 60S subunit docking in a GTP-dependent manner, resulting in an 80S complex and release of Rio2. The ATPase Fap1 then promotes intersubunit rotation, causing dissociation of Dim1 and Tsr1 (Ghalei et al., 2017). Release factor homolog Dom34 and ATPase Rli1 then function to break apart the 80S complex, using the energy of ATP hydrolysis. The endonuclease Nob1 cleaves the ends of the precursor 18S rRNA either before or after subunit dissociation (Strunk et al., 2012). The mature 40S subunit is then ready to bind mRNA and start translation initiation. This test drive effectively checks the functional centers of the ribosome and licenses the mature 40S to enter the translation pool.
Similar quality control mechanisms are likely at play in bacteria, with late-stage assembly events occurring in the context of the 70S ribosome. Recent work from Varshney and colleagues suggests that ribosome assembly coincides with translation initiation in E. coli (Shetty and Varshney, 2016). Depletion of initiator tRNAfMet inhibited ribosome maturation, as indicated by cold sensitivity and elevated levels of unprocessed rRNA. Expression of mutant tRNAfMet with basepair substitutions in the anticodon stem also conferred defects in ribosome assembly, and 70S particles with precursor 17S rRNA accumulated in such strains. Based on these and additional data, the authors proposed that the final stages of ribosome assembly take place during the ribosome’s virgin round of initiation (Shetty and Varshney, 2016). This model can rationalize several earlier observations: (i) the presence of precursor rRNAs in polysomes (Mangiarotti et al., 1974; Srivastava and Schlessinger, 1988), (ii) specific inhibition of ribosome assembly by lamotrigine, which targets IF2 (Stokes et al., 2014), and (iii) genetic interactions between IF2 and assembly factors (Campbell and Brown, 2008).
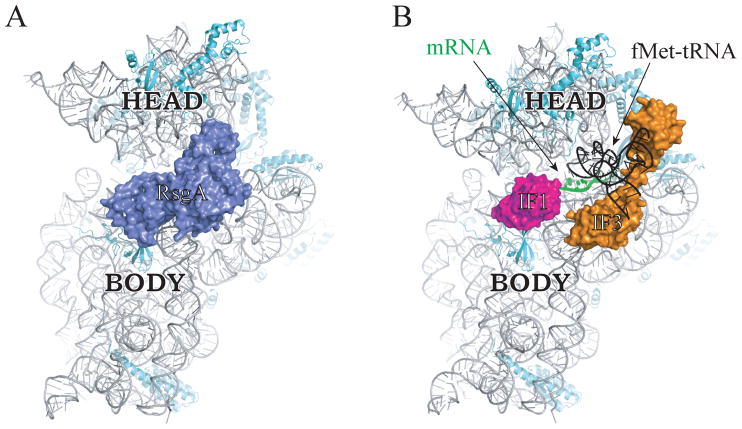
An alternative hypothesis is that assembly involves formation of a 70S complex that contains AFs in the binding sites of tRNA and initiation factors, more akin to the “test drive” described in eukaryotes. Recent cryo-EM studies show that RsgA binds the interface surface of the 30S subunit, with domains occupying the sites for IF1, IF3, and initiator tRNA (Lopez-Alonso et al., 2017) (Fig. 3), similar to how the eukaryotic AFs occlude the same sites of pre-40S subunits. A pre-30S particle bound by AFs including RsgA may participate in a translation-like cycle in which the 50S subunit docks and is subsequently released, events coordinated with late-stage 30S maturation. IF2 could mediate 50S docking in the absence of initiator tRNA, as eIF5B does in the eukaryotic case, rationalizing the functional links between IF2 and ribosome assembly (Stokes et al., 2014; Campbell and Brown, 2008). In this scenario, LepA would presumably act in later events, in the context of the 70S ribosome. Clearly, future work will be necessary to test these two hypotheses and gain further insight into ribosome assembly in bacteria.
Figure 3. RsgA binds the 30S subunit in a way that occludes IF1, IF3, and initiator tRNA.
Interface view of the 30S subunit bound by RsgA (A) or by IF1, IF3, fMet-tRNA, and mRNA (B), as determined by cryo-EM studies (Hussain et al., 2016; Lopez-Alonso et al., 2017). 16S rRNA, gray; r proteins, cyan; RsgA, blue; IF1, magenta; IF3, orange; mRNA, green; fMet-tRNA, black. Head and body regions of the subunit are labeled. Images are based on PDB ID files 5NO3 and 5LMV.
Acknowledgments
We thank B. Roy, L. Ying, and D. Watkins for comments on the article. The authors’ research on trGTPases is supported by a grant from the NIH (R01 GM072528 to K.F.). The authors declare no conflict of interest.
Reference List
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. Embo J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. Embo J. 2006;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:20060008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badu-Nkansah A, Sello JK. Deletion of the elongation factor 4 gene (lepA) in Streptomyces coelicolor enhances the production of the calcium-dependent antibiotic. FEMS Microbiol Lett. 2010;311:147–151. doi: 10.1111/j.1574-6968.2010.02083.x. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmitt H, Funes S, Herrmann JM. The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J Biol Chem. 2008;283:17139–17146. doi: 10.1074/jbc.M710037200. [DOI] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Peske F, Wintermeyer W, Rodnina MV. Translocation as continuous movement through the ribosome. RNA Biol. 2016;13:1197–1203. doi: 10.1080/15476286.2016.1240140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Lie-A-Ling M, Nootenboom IC, Vandenbroucke-Grauls CM, Kusters JG. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J Infect Dis. 2000;182:1566–1569. doi: 10.1086/315855. [DOI] [PubMed] [Google Scholar]
- Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol. 1996;178:3246–3251. doi: 10.1128/jb.178.11.3246-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991;266:2872–2877. [PubMed] [Google Scholar]
- Campbell TL, Brown ED. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J Bacteriol. 2008;190:2537–2545. doi: 10.1128/JB.01744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MA, Haddad BG, Weis AJ, Blackwood CS, Shelton CD, Wuerth ME, Walter JD, Spiegel PC., Jr Ribosomal protein L7/L12 is required for GTPase translation factors EF-G, RF3, and IF2 to bind in their GTP state to 70S ribosomes. Febs J. 2017;284:1631–1643. doi: 10.1111/febs.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cui X, Beausang JF, Zhang H, Farrell I, Cooperman BS, Goldman YE. Elongation factor G initiates translocation through a power stroke. Proc Natl Acad Sci U S A. 2016;113:7515–7520. doi: 10.1073/pnas.1602668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Williamson JR. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol. 2013;425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P, Flower AM. Efficient assembly of ribosomes is inhibited by deletion of bipA in Escherichia coli. J Bacteriol. 2015;197:1819–1827. doi: 10.1128/JB.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation (Austin) 2013;1:e24315. doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR. Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell. 2016;167:1610–1622. e15. doi: 10.1016/j.cell.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurentiis EI, Wieden HJ. Identification of two structural elements important for ribosome-dependent GTPase activity of elongation factor 4 (EF4/LepA) Sci Rep. 2015;5:8573. doi: 10.1038/srep08573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLivron MA, Makanji HS, Lane MC, Robinson VL. A novel domain in translational GTPase BipA mediates interaction with the 70S ribosome and influences GTP hydrolysis. Biochemistry. 2009;48:10533–10541. doi: 10.1021/bi901026z. [DOI] [PubMed] [Google Scholar]
- deLivron MA, Robinson VL. Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J Bacteriol. 2008;190:5944–5952. doi: 10.1128/JB.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ, Wolfe PB. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duo M, Hou S, Ren D. Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl Microbiol Biotechnol. 2008;81:731–741. doi: 10.1007/s00253-008-1709-6. [DOI] [PubMed] [Google Scholar]
- Ermolenko DN. Personal communication.
- Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci U S A. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Hahm J, Diggs S, Perry JJ, Blaha G. Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli. J Biol Chem. 2015;290:20856–20864. doi: 10.1074/jbc.M115.659136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris M, Grant A, Richardson TB, O’Connor CD. BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol Microbiol. 1998;28:265–279. doi: 10.1046/j.1365-2958.1998.00793.x. [DOI] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Bock LV, Maracci C, Wang Z, Paleskava A, Konevega AL, Schroder GF, Grubmuller H, Ficner R, Rodnina MV, Stark H. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016;540:80–85. doi: 10.1038/nature20560. [DOI] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. Embo J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Lin J, Bulkley D, Steitz TA. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science. 2014;345:684–687. doi: 10.1126/science.1253525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Bai X, Zhang D, Han C, Yuan J, Liu W, Cao X, Chen Z, Shangguan F, Zhu Z, Gao F, Qin Y. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat Struct Mol Biol. 2016;23:441–449. doi: 10.1038/nsmb.3206. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalei H, Trepreau J, Collins JC, Bhaskaran H, Strunk BS, Karbstein K. The ATPase Fap7 Tests the Ability to Carry Out Translocation-like Conformational Changes and Releases Dim1 during 40S Ribosome Maturation. Mol Cell. 2017;67:990–1000. e3. doi: 10.1016/j.molcel.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs MR, Moon KM, Chen M, Balakrishnan R, Foster LJ, Fredrick K. Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc Natl Acad Sci U S A. 2017;114:980–985. doi: 10.1073/pnas.1613665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Kato S, Kimura T, Muto A, Himeno H. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. Embo J. 2011;30:104–114. doi: 10.1038/emboj.2010.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Belardinelli R, Maracci C, Milon P, Rodnina MV. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 2015;43:10700–10712. doi: 10.1093/nar/gkv869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O’Connor CD. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC) Mol Microbiol. 2003;48:507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x. [DOI] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70 S translation initiation complex formation. J Mol Biol. 2007;373:562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Wimberly BT, Kieft JS, Ramakrishnan V. Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation. Cell. 2016;167:133–144. e13. doi: 10.1016/j.cell.2016.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji DL, Lin H, Chi W, Zhang LX. CpLEPA is critical for chloroplast protein synthesis under suboptimal conditions in Arabidopsis thaliana. PLoS One. 2012;7:e49746. doi: 10.1371/journal.pone.0049746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci U S A. 2012;109:131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23:242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martinez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini SB, Peske F, Rodnina MV. Partitioning between recoding and termination at a stop codon-selenocysteine insertion sequence. Nucleic Acids Res. 2015;43:6426–6438. doi: 10.1093/nar/gkv558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, McDonald ME, Brunelle JL, Green R. RF3:GTP promotes rapid dissociation of the class 1 termination factor. Rna. 2014;20:609–620. doi: 10.1261/rna.042523.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan K, Flower AM. Suppression of DeltabipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J Bacteriol. 2008;190:7675–7683. doi: 10.1128/JB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Chen Y, Ero R, Ahmed T, Tan J, Li Z, Wong AS, Bhushan S, Gao YG. Structure of BipA in GTP form bound to the ratcheted ribosome. Proc Natl Acad Sci U S A. 2015;112:10944–10949. doi: 10.1073/pnas.1513216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Leong V, Kent M, Jomaa A, Ortega J. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. Rna. 2013;19:789–802. doi: 10.1261/rna.037523.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyh TS, Suo Y. GTPase-mediated activation of ATP sulfurylase. J Biol Chem. 1992;267:542–545. [PubMed] [Google Scholar]
- Li W, Atkinson GC, Thakor NS, Allas U, Lu CC, Chan KY, Tenson T, Schulten K, Wilson KS, Hauryliuk V, Frank J. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O) Nat Commun. 2013;4:1477. doi: 10.1038/ncomms2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang R, Varlamova O, Leyh TS. Regulating energy transfer in the ATP sulfurylase-GTPase system. Biochemistry. 1998;37:3886–3892. doi: 10.1021/bi971989d. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen C, Zhang H, Kaur J, Goldman YE, Cooperman BS. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc Natl Acad Sci U S A. 2011;108:16223–16228. doi: 10.1073/pnas.1103820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J Mol Biol. 2010;396:1043–1052. doi: 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Alonso JP, Kaminishi T, Kikuchi T, Hirata Y, Iturrioz I, Dhimole N, Schedlbauer A, Hase Y, Goto S, Kurita D, et al. RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Demo G, Grigorieff N, Korostelev AA. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017;546:113–117. doi: 10.1038/nature22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarotti G, Turco E, Ponzetto A, Altruda F. Precursor 16S RNA in active 30S ribosomes. Nature. 1974;247:147–148. doi: 10.1038/247147a0. [DOI] [PubMed] [Google Scholar]
- March PE, Inouye M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]
- Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidig A, Yeung AT, Rosay T, Tettmann B, Strempel N, Rueger M, Lesouhaitier O, Overhage J. TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 2013;13:77-2180–13–77. doi: 10.1186/1471-2180-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Zorzet A, Andersson DI, Ehrenberg M. Activation of initiation factor 2 by ligands and mutations for rapid docking of ribosomal subunits. Embo J. 2011;30:289–301. doi: 10.1038/emboj.2010.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci U S A. 2011;108:3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res. 2014;42:1812–1820. doi: 10.1093/nar/gkt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig PL, Flower AM. BipA is required for growth of Escherichia coi K12 at low temperature. Mol Genet Genomics. 2001;266:313–317. doi: 10.1007/s004380100559. [DOI] [PubMed] [Google Scholar]
- Qi SY, Li Y, Szyroki A, Giles IG, Moir A, O’Connor CD. Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol Microbiol. 1995;17:523–531. doi: 10.1111/j.1365-2958.1995.mmi_17030523.x. [DOI] [PubMed] [Google Scholar]
- Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Rodnina MV. Personal communication.
- Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J Bacteriol. 2000;182:2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpati P, Sund J, Aqvist J. Structure-based energetics of mRNA decoding on the ribosome. Biochemistry. 2014;53:1714–1722. doi: 10.1021/bi5000355. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, 4th Weir, JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Shetty S, Varshney U. An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1609550113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Joseph S. Mechanism of Translation Termination: RF1 Dissociation Follows Dissociation of RF3 from the Ribosome. Biochemistry. 2016;55:6344–6354. doi: 10.1021/acs.biochem.6b00921. [DOI] [PubMed] [Google Scholar]
- Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010;92:157–163. doi: 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprink T, Ramrath DJ, Yamamoto H, Yamamoto K, Loerke J, Ismer J, Hildebrand PW, Scheerer P, Burger J, Mielke T, Spahn CM. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv. 2016;2:e1501502. doi: 10.1126/sciadv.1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci U S A. 1988;85:7144–7148. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes JM, Davis JH, Mangat CS, Williamson JR, Brown ED. Discovery of a small molecule that inhibits bacterial ribosome biogenesis. Elife. 2014;3:e03574. doi: 10.7554/eLife.03574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M. Structure and function of E. coli ribosomes. V Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ng MY, Chen Y, Cooperman BS. Kinetics of initiating polypeptide elongation in an IRES-dependent system. Elife. 2016;5 doi: 10.7554/eLife.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. Rna. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F, Heider J, Bock A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci U S A. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]