Abstract
Biomarkers that guide therapy selection are gaining unprecedented importance as targeted therapy options increase in scope and complexity. In conjunction with high-throughput molecular techniques, therapy-guiding biomarker assays based upon immunohistochemistry (IHC) have a critical role in cancer care in that they inform about the presence of a protein target. Here, we describe the validation procedures for four clinically available IHC biomarker assays – PTEN, RB, MLH1 and MSH2 – for use as integral biomarkers in the nationwide NCI-MATCH (Molecular Analysis for Therapy Choice) EAY131 clinical trial. Validation procedures were developed through an iterative process based on collective experience and adaptation of broad guidelines from the United State Food and Drug Administration (FDA). The steps included primary antibody selection, assay optimization, development of assay interpretation criteria incorporating biological considerations and expected staining patterns, including indeterminate results, orthogonal validation, and tissue validation. Following assay lockdown, patient samples and cell lines were used for analytical and clinical validation. The assays were then approved as laboratory developed tests and used for clinical trial decisions for treatment selection. Calculations of sensitivity and specificity were undertaken using various definitions of gold standard references, and external validation was required for the PTEN IHC assay. In conclusion, validation of IHC biomarker assays critical for guiding therapy in clinical trials is feasible using comprehensive pre-analytical, analytical and post-analytical steps. Implementation of standardized guidelines provides a useful framework for validating IHC biomarker assays that allow for reproducibility across institutions for routine clinical use.
Keywords: immunohistochemistry, biomarker, validation, clinical trials, companion diagnostics
INTRODUCTION
Biomarker assessment is a critical component of cancer patient management. Towards that end, immunohistochemistry (IHC) plays a robust clinical role in tissue-based assessment of protein expression, particularly in solid tumors. Examples of this role include HER2 expression in breast cancer and gastroesophageal adenocarcinoma, estrogen receptor expression in breast cancer, and expression of mismatch repair (MMR) proteins in patients with colorectal adenocarcinoma or endometrial carcinoma.(1–4) Nonetheless, the availability of predictive biomarkers to support therapy selection remains limited in a variety of cancer types, and this area of unmet need is expected to evolve as use of targeted therapies, including antibodies as well as small molecules, continues to expand. As such needs expand, procedures to ensure standardized optimization and performance of such assays gain increasing importance.
Automation and technical advances in reagent chemistry, coupled with a broadening of the scope and quality of primary antibodies that recognize specific target epitopes in formalin-fixed paraffin-embedded (FFPE) tissue, have improved the quality and reliability of IHC in routine clinical practice. As with any complex in vitro diagnostic (IVD) assay, however, IHC is susceptible to variations that impact its performance, making ongoing quality monitoring a prerequisite to the clinical utility of any biomarker assessed by IHC. Nowhere is this more important than for IHC assays performed to guide therapy selection – henceforth referred to as therapy-guiding assays.
The Centers for Disease Control and Prevention Office of Population Genomics has provided the ACCE (analytic validity, clinical validity, clinical utility and ethical, legal and social implications) model(5) that can be applied to IHC (the scope of this manuscript applies to analytic validity). The FDA categorizes IHC assays as Class II or III IVD medical devices and has provided guidance to industry.(6,7) Such devices are subject to FDA clearance or approval, albeit the latter is not a requirement for clinical use under the Clinical Laboratory Improvement Amendments of 1988 (CLIA) or the Center for Medicare and Medicare Services (CMS) requirements.(8,9) Although broad guidelines for pre-analytical and analytical validation of IHC assays have been proposed (10–13) and IHC laboratory manuals are available(14), standardization and in-depth characterization of the steps required for ensuring reliable assay performance for therapy-guiding IHC biomarker assays remain underdeveloped. IHC assays performed in clinical trials to guide therapy are regulated by the FDA under the Investigational Device Exemptions (IDE) regulations.(15)
Herein, we describe the validation principles and processes that were followed for four IHC biomarker assays (phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase (PTEN), retinoblastoma-asociated protein (RB), DNA mismatch repair protein Mlh1 (MLH1), and DNA mismatch repair protein Msh2 (MSH2)) intended for use within the context of a nationwide clinical trial in the NCI National Clinical Trial Network, the NCI Molecular Analysis for Therapy Choice (NCI-MATCH) EAY131 phase II precision oncology trial (ClinicalTrials.gov ID#: NCT02465060).(16,17) Validation procedures for each of these IHC assays went beyond typical validations required for routine diagnostic IHC assays and are presented in detail as applied examples of analytical validation aimed at informing future standardization guidelines for IHC biomarker assays. All four assays described in this manuscript were successfully validated for use in the NCI-MATCH trial.
PRIMARY ANTIBODY SELECTION
Primary antibodies are produced most commonly by immunization of a mammal, commonly mouse or rabbit, with a synthetically produced peptide portion of the target protein. Commercial products are favored over “homegrown” and customized antibodies to increase the likelihood of stable supply. Antibodies may be monoclonal or polyclonal, and the vast majority is IgG1 or IgG2 subtype. No single primary antibody attribute predetermines its performance characteristics in an IHC assay, and it is not uncommon for multiple commercially available options to be equally suitable for a given purpose.
Selection of the primary antibody starts with a thorough understanding of the target protein, its encoding gene, and location of the epitope(s) within the broader protein structure. Ideally, the epitope is within a segment of the protein that is not (or only infrequently) impacted by mutations or deletions, unless the intended use is to detect a mutated gene product. Accordingly, where choices between primary antibodies that recognize various domains of a given protein exist, selection should be guided by the highest sensitivity potential for the intended application.(18,19) Selection of a primary antibody may be based further on data in published peer-reviewed scientific studies in which sensitivity and specificity are assessed in a research setting using orthogonal protein detection methods.
Vendor attributes are important to ensure adherence to supply continuity and minimal lot-to-lot variations or interruptions. In the United States, suppliers of Class II/III IVD products are required to provide data on performance characteristics and abide by Good Manufacturing Practices.(20) The antibody sources and details are summarized in Table 1.
Table 1.
Summary of primary antibodies.
| Clone | Host | Isotype | Vendor | Immunogen | Dilution | Selected References† | |
|---|---|---|---|---|---|---|---|
| PTEN | 6H2.1 | Mouse | IgG2 | Dako | Full-length protein | 1:100 | (21) |
| RB | LM95.1 | Mouse | IgG1 | EMD Millipore/Calbiochem | C-terminal fragment | 1:30 | n/a |
| MLH1 | G168-728 | Mouse | IgG2 | Millipore Sigma/Cell Marque | Not specified | 1:300 | (51–53) |
| MSH2 | FE11 | Mouse | IgG1 | EMD Millipore/Calbiochem | C-terminal fragment | 1:100 | (51–53) |
Peer-reviewed studies using the corresponding primary antibody.
Abbreviations: n/a=not available.
ASSAY OPTIMIZATION
Next steps include determination of optimal antibody dilution, antigen retrieval conditions, and incubation time. Other parameters, such as incubation temperature, are typically preset on most automated immunostainers designed for use in the clinical environment. The use of automated platforms is strongly recommended, particularly for therapy-guiding IHC biomarker assays. The starting points for assay optimization are the antibody manufacturer’s recommendations and/or conditions used in peer-reviewed publications. The ideal antibody dilution may be defined as the lowest antibody concentration that yields an optimal balance between sensitivity and specificity. Commercial antigen retrieval solutions offer a pH-based choice to modulate an epitope’s three-dimensional configuration with the aim of optimizing primary antibody binding. The ideal antigen retrieval solution minimizes exposure of tissues to stringent acidic or alkaline conditions and thus limits alteration of tissue integrity for morphologic evaluation. Incubation time should be adequate to allow antibody molecules to bind to epitope sites, but short enough to minimize non-specific binding. The use of “blocking” solutions that quench endogenous peroxidase enzymes is useful for most biomarkers. The combination of staining conditions that produces an optimal signal-to-noise ratio is then adopted for the remainder of the validation process. It should be noted that FDA-approved IHC biomarker assays have preset staining parameters with minimal leeway for variations by the end-user.(13)
ASSAY INTERPRETATION CRITERIA
The distribution of target in various tissue components determines IHC staining patterns. A priori knowledge of the biology of the protein provides a basis for assessing expected staining patterns (nuclear, cytoplasmic, membranous topography) and interpretation guidelines. Interpretation guidelines, including definition and handling of indeterminate results, should be defined during the validation process to ensure applicability to patient samples when an IHC assay is deployed. (Table 2) Image-assisted interpretation is not under consideration for the validations illustrated in this paper due to the characteristics and intended uses of the assays, but is likely to have a greater role in this process over time due to improved reproducibility.
Table 2.
Interpretation guidelines.
| Staining Pattern | Positive Therapy-guiding Alteration | Lack of Therapy-guiding Alteration | Indeterminate Results | |
|---|---|---|---|---|
| PTEN | Cytoplasmic | Loss of expression (Absent) |
Retained expression (Retained) |
|
| RB | Nuclear | |||
| MLH1 | ||||
| MSH2 |
Staining should be repeated in such a situation before indeterminate category is assigned.
For each therapy-guiding IHC assay, a standardized reporting template is required. The template should stipulate standardized reporting terminology. For instance, the use of terms such as “positive” or “negative” can be problematic in the context of biomarkers whose loss constitutes an actionable finding. In the assessment of mismatch repair (MMR) protein expression to evaluate microsatellite instability (MSI) status, for example, negative staining for MLH1 or MSH2 is a positive result that indicates the presence of high levels of MSI (MSI-H), so the use of such terms is ambiguous and can lead to untoward clinical consequences. Accordingly, the terminology should be made unambiguous and describe the results of the actual assay rather than the clinical implication of the result. In the reporting templates for MLH1, MSH2, PTEN, and RB – whose loss is what constitutes an actionable finding – results are best reported as “loss of expression” and “retained expression”.
PTEN
The phosphatase and tensin homolog (PTEN) is a tumor-suppressor gene located on chromosome 10q23.3 that encodes a dual-specificity phosphatase that acts as a dominant negative regulator of the PI3K/AKT signaling axis. Loss of PTEN expression results in constitutive AKT activation and promotes tumor growth and altered cancer cell metabolism, mainly via upregulation of the mammalian target of rapamycin (mTOR). Abnormalities in the PTEN/PI3K/AKT pathway have been detected in many human tumors, including endometrial carcinomas.(21–23) PTEN knock-out mice develop proliferative endometrial lesions, and germline PTEN mutations in human beings lead to Cowden syndrome.(24) Germline polymorphisms involving the PTEN gene have been identified in 60–80% of patients with PTEN hamartoma tumor-related syndromes (PHTS), a group of disorders that includes Cowden syndrome (CS), Bannayan-Riley-Ruvalcaba syndrome (BRRS), PTEN-related Proteus syndrome and Proteus-like syndrome. All of these syndromes are characterized by tissue overgrowth and benign tumors, but only Cowden syndrome and BRRS have a predisposition for cancer development.
In tumors with PTEN loss, the activated PI3K/AKT/mTOR pathway constitutes an attractive target of therapy. Loss of PTEN expression detected by IHC is regarded as the most accurate reflection of the loss of PTEN function and, as a result, serves to determine eligibility for therapies that target critical downstream nodes within the PI3K/AKT/mTOR axis in the NCI-MATCH trial.
PTEN is a cytosolic protein that is expressed ubiquitously in human tissues. Neoplasms with retained PTEN expression show a cytoplasmic and occasionally nuclear pattern of expression, whereas those with PTEN loss are composed of neoplastic cells that distinctly lack expression. Since PTEN loss is an acquired somatic event that may result from biallelic loss or mutations in PTEN (25,26), non-neoplastic tissue elements, including endothelial and stromal cells, retain expression and serve as an adjoining internal positive control, a feature that is particularly useful in cases with PTEN loss. (Figure 1) Tumor sampling is an issue, as heterogeneity in expression levels is common.
Figure 1.
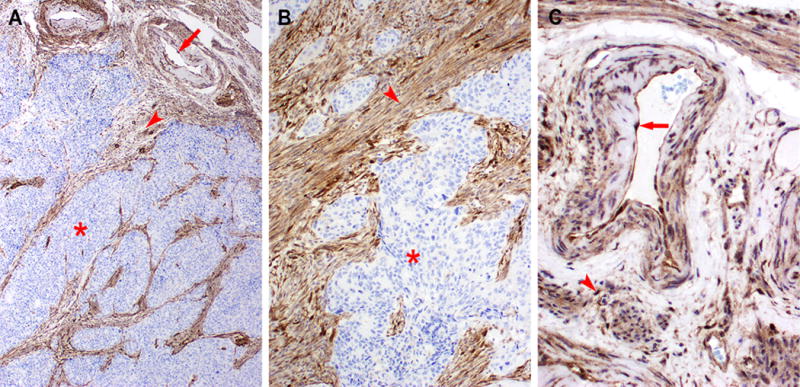
Example of PTEN loss in endometrioid endometrial adenocarcinoma. Low-power (A, 40×) and higher magnification (B, 100×) showing lack of labeling in tumor cells (*). Endothelial cells (red arrow) and stromal cells (red arrowhead) serve as a positive internal control.
RB
The RB transcriptional corepressor 1 (RB1 gene is a tumor suppressor gene that encodes the RB protein, a negative cell cycle regulator.(27) Mutations and deletions of RB1 are common in many cancers, and inherited allelic loss of RB1 confers increased cancer susceptibility. The RB protein and its two family members, p107 and p130, regulate cell proliferation through transcriptional repression of genes involved in cell cycle transition from the G1 to S phase.(27) Loss of RB function allows unregulated cell cycle progression and promotes tumor growth. Specifically, cell cycle progression requires the dissociation of the RB/E2F complex, which is tightly regulated physiologically via RB phosphorylation. In tumors, constitutive disruption of the RB/E2F complex results from loss of RB expression through deletions or mutations, or from increased RB phosphorylation. With abrogation of RB-dependent cell cycle inhibition, cell cycle transition and commitment to cell division are coordinated by cyclin-dependent protein kinases (CDKs) which are emerging targets for therapy.(28) Palbociclib is a potent selective inhibitor of CDK4 and CDK6 with significant activity in breast cancer models.(29,30) Of note, palbociclib shows no activity in RB-deficient cells.(28)
RB is a nuclear protein that is expressed ubiquitously in human tissues. Neoplasms with retained RB expression show nuclear expression by IHC, while those with RB loss have neoplastic cells that distinctly lack nuclear reactivity. (Figure 2) Aside from the nuclear staining pattern, interpretation guidelines are similar to those detailed above for PTEN.
Figure 2.
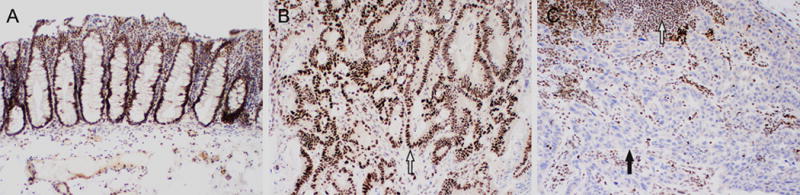
Examples of positive and negative RB expression by immunohistochemistry. (A and B, 100×) Positive RB expression. The nuclei of non-neoplastic colonic mucosa and submucosa (A) and colonic adenocarcinoma (B, white arrow) exhibit diffuse strong nuclear labeling. (C, 100×) Loss of RB expression in lung adenocarcinoma. Loss of nuclear staining in neoplastic cells (blue counterstain nuclei, black arrow), while adjacent non-neoplastic stromal cells and lymphocytes have retained nuclear expression (dark brown nuclei, white arrow).
MLH1 and MSH2
The genes mutL homolog 1(MLH1) and mutS homolog 2 (MSH2) are located on chromosome 3p21.3 and chromosome 2p21, respectively. Both belong to a family of genes known as MMR genes. MLH1 encodes a protein which heterodimerizes predominantly with PMS2, a MutL homolog, to form the MutL complex. MSH2 encodes a protein which heterodimerizes predominantly with MSH6, a MutS homolog, to form the MutS complex. Both complexes are essential for the detection and initiation of repair of DNA strand misalignment and basepair matching errors that occur during DNA replication. Loss-of-function of either MLH1 and its binding partner PMS2 or of MSH2 and its binding partner MSH6 results in error-prone DNA replication. This abnormality leads to, among other effects, alterations in the length of tandem DNA sequence repeats called microsatellites, a condition known as MSI or deficient mismatch repair (dMMR). Uncorrected mutations occur throughout the genome, termed hypermutation or tumor mutation burden. The extent of microsatellite alterations is assessed semi-quantitatively as MSI-low or MSI-high in DNA-based assays, with the latter having a higher degree of correlation with bona fide MMR genomic defects.
In most (~95%) cases, loss of MMR results from sporadic methylation-induced MLH1 promoter inactivation of both copies of the gene that can occur in a wide variety of tumors, especially colonic and endometrial adenocarcinomas (28). Somatic mutations occur occasionally. Germline mutations in MMR genes, most of which involve MLH1 or MSH2, result in the autosomal dominant Lynch syndrome, formerly hereditary non-polyposis colorectal cancer syndrome (HNPCC). These patients have a significantly increased risk for colorectal, gastric, small intestinal, liver, gallbladder, urothelial, brain, and skin tumors. Women also have an increased risk of ovarian and uterine endometrial carcinomas. Muir-Torre syndrome is a subtype of Lynch syndrome in which patients have skin neoplasms (sebaceous tumors and keratoacanthomas) in addition to an increased risk of developing visceral malignancies. Tumors with MLH1 or MSH2 loss, or other MMR defects, have distinctive clinical features compared to those without MMR defects. For example, colonic adenocarcinomas with MMR defects (e.g. loss of MLH1, MSH2, etc.) have a better stage-specific prognosis, and these patients do not benefit from 5-fluorouracil monotherapy.(31,32)
In view of the prognostic, therapeutic, and genetic implications of MMR aberrations in cancer, guidelines have been in place for many years to screen for and delineate their nature. In accordance with the commonly used guidelines from several professional organizations, screening is typically carried out by surveying the expression of MLH1, PMS2, MSH2 and MSH6 in tumor tissue using IHC. Tumors that exhibit loss of expression of one or more MMR genes are often reflexed for MSI evaluation using molecular techniques. Genetic counseling and genomic evaluation then may be pursued in line with the findings in the tumor and family history to identify family members with Lynch syndrome.(4)
Tumors with retained MLH1 or MSH2 expression as well as non-neoplastic cells have diffuse moderate-to-strong nuclear staining by IHC, whereas tumors that have lost MLH1 or MSH2 expression have a distinct absence of nuclear expression. (Figure 3) These tests were the best way to determine if patients had MMR, which had been shown to respond to PD-1 checkpoint inhibitors, a treatment in NCI MATCH.(33,34) Occasional cases have a speckled nuclear pattern on immunohistochemistry for MLH1 with complete loss of expression of the binding partner PMS2 and the presence of MSI-H/dMMR. Aside from this rare staining pattern, interpretation guidelines are similar to those detailed above for PTEN.
Figure 3.
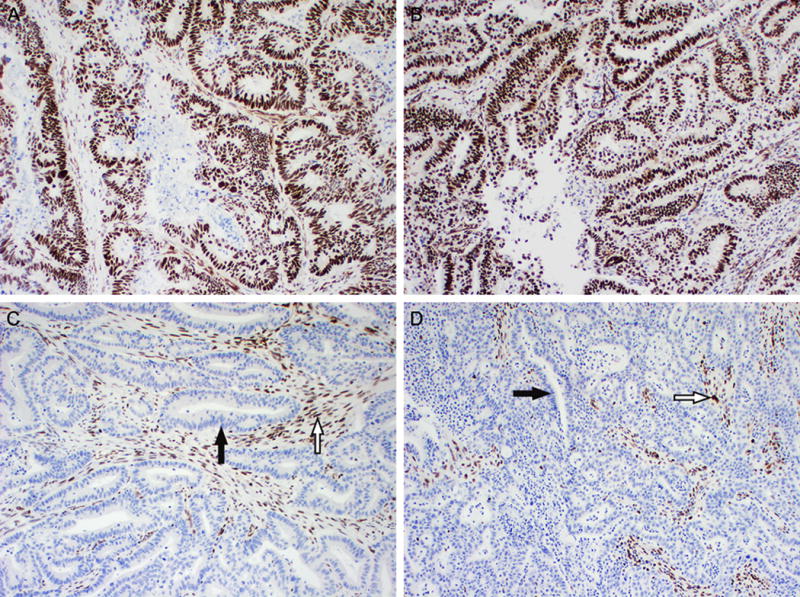
Examples of positive and negative MLH1 expression by immunohistochemistry. (A and B, 100×) Positive MLH1 expression. The nuclei of colonic adenocarcinoma (A) and endometrial endometrioid adenocarcinoma (B) exhibit diffuse strong staining. (C and D, 100×) Loss of MLH1 expression in colonic adenocarcinoma (C) and endometrial endometrioid adenocarcinoma (D). Loss of nuclear staining in neoplastic cells (black arrow), while adjacent normal stromal cells and lymphocytes have retained nuclear expression (white arrow). Similar staining patterns were seen for MSH2 (not shown).
Indeterminate results
In the interpretation of biomarker results, occasional cases will have difficult to interpret and/or uncertain results due to technical or nontechnical variances that may be attributable to pre-analytical, analytical, or post-analytical factors. Thus, validation documents and subsequent standard operating procedures need to address the processes by which indeterminate/equivocal results are handled. Absent or weak labeling of non-neoplastic elements (internal controls) usually indicates a pre-analytical issue (e.g. tissue fixation or processing, or inadequate sampling by biopsy) or an analytical problem in the IHC staining process. In such instances, the test may be repeated to exclude a possible one-off issue. If unresolved, the sample may be regarded as indeterminate. The indeterminate category should also include situations in which tissue is insufficient, for example due to extensive necrosis or inadequate sampling.
Criteria for handling cases with heterogeneous staining results also should be clarified. For example, in cases with heterogeneous staining for PTEN or RB, wherein only a subset of neoplastic cells shows loss of expression, the antigen may be regarded as retained since the cellular subsets with retention would not be responsive respectively to AKT- or CDK4/6-targeted therapies.
VALIDATION SAMPLES
Cell Lines
Cell lines with known mutation and expression profiles can offer a useful tool for orthogonal validation after the IHC assays are finalized and locked for use. Preparation of cell pellets that are fixed in 10% neutral buffered formalin and embedded in paraffin (FFPE) can be used to perform IHC assays, analogous to cytology cell blocks. As cell lines are a convenient source of well-characterized cells that can be used for a variety of DNA and RNA assays, the performance of IHC on FFPE cell line blocks allows direct comparison of an IHC assay to other protein-level or DNA/RNA-level assays. A list of cell lines used in our validations is in Table 3.
Table 3.
Summary of cell lines
| Cell Line | Derivation | Wild-Type | Mutated | IHC Staining | |
|---|---|---|---|---|---|
| PTEN | MDA-MB-468 | Breast Adenocarcinoma | X | Absent | |
| MDA-MB-231 | Breast Adenocarcinoma | X | Retained | ||
| RB | A2058 | Melanoma | X | Absent | |
| A549 | Lung carcinoma | X | Retained | ||
| BJ | Fibroblast | X | Retained | ||
| MLH1 | HCT116 | Colonic Adenocarcinoma | X | Absent | |
| SW480 | Colonic Adenocarcinoma | X | Retained | ||
| MSH2 | LoVo | Colonic Adenocarcinoma | X | Absent | |
| SW480 | Colonic Adenocarcinoma | X | Retained |
The PTEN-mutant (mut) and wild-type (wt) cell lines were obtained from the MD Anderson Cancer Center (MDACC) Characterized Cancer Cell Line Core Facility. IHC results for the PTENmut cell line (MDA-MB-468) demonstrated absence of protein staining, whereas the PTENwt cell line (MDA-MB-231) demonstrated retained PTEN expression. These results were identical to those previously obtained using reverse protein phase array expression studies (data not shown).
Similarly, the RB1mut cell line A2058 (derived from derived from melanoma) demonstrated complete absence of RB staining, whereas the cell lines A549 (lung carcinoma) and BJ (fibroblast) with RB1wt demonstrated positive staining. (Figure 4) These cell lines were purchased from the American Tissue Culture Collection (Manassas, Virginia).
Figure 4.
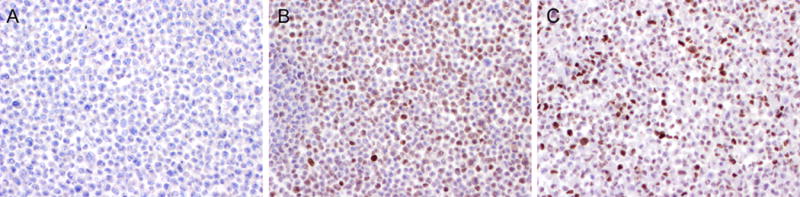
(A) Melanoma cancer cell line A2058 with RB1 mutation demonstrating complete loss of RB expression. (B) Lung cancer cell line A549with wild-type RB1 showing retained RB expression. (C) Normal human fibroblast cell line BJ with wild-type RB1 showing retained RB expression.
The MLH1mut cell line HCT116 demonstrated complete absence of MLH1 staining, whereas the cell line SW480 with MLH1wt had positive staining by IHC. Similarly, the MSH2mut cell line LoVo demonstrated complete absence of MSH2 staining, and the SW480 with MSH2wt had retained staining. These cell lines were purchased from the American Tissue Culture Collection.
Patient Samples
A major component of validating patient tumor testing entails the use of patient samples as the source of materials to demonstrate an appropriate spectrum of staining across a particular tumor type, or diverse tumor types. The validations we performed included a main cohort of tumors of specific histologic types commonly evaluated with a particular assay (e.g. colorectal adenocarcinoma for MLH1/MSH2) as well as a wider array of available tumor samples with molecular and/or FISH results that were used for reasons of convenience. In addition, when feasible, samples from selected patient cohorts (e.g. Cowden syndrome, Muir-Torre syndrome, etc.), wherein the performance of the assay is predictable, were also used to provide further confirmation of assay performance and reliability (see below).
STABILITY, REPRODUCIBILITY, REPEATABILITY
For each of the 4 biomarkers, assay stability and reproducibility were tested as follows: From each of 5 FFPE blocks of tumor tissue with retained expression, 10 sections were cut at the beginning of a consecutive 10-day validation period and stored at room temperature. Analysis was performed each day on a single unstained FFPE slide from each of those 5 blocks on a rotating basis by three different histotechnologists. In addition, for each of the biomarkers, assay repeatability was tested as follows: from each of 3 FFPE blocks of tumor tissue with retained expression, 3 sections were stained on three different racks of a Leica Bond autostainer. Each of the assays showed consistent performance across these validation experiments. Together, these data demonstrate operator-independent assay reproducibility, with stable performance over consecutive days of operation, regardless of rack position, without significant loss in epitope reactivity.
QUALITY CONTROL
Ongoing quality control
For all therapy-guiding IHC assays, on-slide positive controls are applied on one edge of the slide containing the patient sample. Positive controls are sections of tissue known to express the target antigen detectable using the same epitope retrieval and staining protocols as the patient tissue. Conversely, negative controls are defined as sections of tissue known to be negative for the target antigen under similar staining conditions. The use of a negative control wherein the primary antibody is omitted or an irrelevant primary antibody is used is no longer required in clinical IHC.
Lot-to-Lot Consistency
In accordance with guidelines of the College of American Pathologists (CAP), the performance of a new lot of antibody is compared with that of the antibody lot that is about to be depleted.(10) This procedure is applicable for all clinical IHC assays, including all therapy-guiding IHC assays.
VALIDATION RESULTS
ACCURACY
Sensitivity and Specificity
The diagnostic accuracy of a laboratory assay is defined as the extent of its agreement with a reference standard, with a reference standard being “the best available method to establishing the presence or absence of the target condition”.(35–37) Selection of a reference standard for a particular assay is predicated on the biology of the analyte being measured and the inherent characteristics of the tool used to measure it. Estimates of sensitivity and specificity are among the most commonly employed measures of diagnostic accuracy. On the basis of definitions adopted by the FDA(35) from the Clinical and Laboratory Standards Institute Harmonized Terminology Database(36) and the STARD (Standards for Reporting of Diagnostic Accuracy) Initiative(37), sensitivity of a test is the “proportion of subjects with the target condition in whom the test is positive”, while specificity is the “proportion of subjects without the target condition in whom the test is negative”.(35)
Cross-validation of PTEN
Analytical performance of the PTEN IHC assay was determined through cross-validation by measuring positive and negative concordances with an IHC PTEN assay previously validated at another institution, the Memorial Sloan Kettering Cancer Center (MSKCC). As such, the MSKCC assay was considered the benchmark (comparator) against which the performance of the MDACC PTEN IHC assay was compared for the purposes of analytic validation. The selection of this benchmark was based on the fact that genomic alterations that lead to loss of PTEN expression include biallelic loss or mutations in the PTEN gene whose detection requires FISH and NGS, respectively, neither of which has been shown to be superior to IHC in identifying loss of PTEN function.(25,26) The PTEN IHC assay at MSKCC is performed on identical autostainers (Leica Biosystems, Buffalo Grove, Illinois) using the same antibody clone and titer (Dako, clone 6H2.1, 1:100). However, notable differences between the MSKCC and MDACC staining protocols included the duration of antigen retrieval time (30 vs. 20 minutes, respectively) and primary antibody incubation time (30 vs. 15 minutes, respectively), which produced a signal of stronger intensity. To validate the MDACC PTEN IHC assay, 31 sections from FFPE blocks containing tumor tissue were stained at MSKCC and interpreted by pathologists at both institutions without knowledge of the results at the other institution. Results were concordant in 29/31 (94%) cases (95% CI: 0.771–0.989), with an analytical sensitivity of 92% (95% CI lower limit: 0.715) and specificity of 100% (95% CI lower limit: 0.560). (Supplementary Table S1). Analysis of discordant results in two false-negative cases relative to comparator was included in the validation report. These two cases had moderate to weak labeling on the MSKCC IHC, which was either not noted on MDACC stains or attributed to non-specific staining at the tissue edge or within areas of necrosis. One of the cases assessed for mutations using a clinically-validated next-generation sequencing (NGS) mutation screening panel(38) was found to be negative for PTEN mutations.
Cross-validation of RB
For the RB IHC assay, RB1 mutation analysis using the aforementioned clinically-validated NGS panel was considered the benchmark comparator. Forty-eight human tumors and 6 normal human control tissues were selected from the pathology files at MDACC on the basis of available mutation results and adequate residual tissue specimens. The selection of this benchmark was based on the fact that cases with RB1 mutations were expected to lack RB protein expression.(29,39) Using these criteria, the RB IHC biomarker assay had a sensitivity and a specificity of 100% (95% CI lower limit: 0.828 and 0.858, respectively): all tumors with RB1 mutations had loss of RB expression using the RB IHC assay. (Supplementary Tables S2 and S3)
Cross-validation of MLH1 and MSH2
For the MLH1 IHC assay, MSI testing and MLH1 gene promoter methylation were considered as the benchmark comparators against which MLH1 expression by IHC was assessed. For the MSH2 IHC assay, MSI testing was considered as the benchmark comparator. The selection of these benchmarks for MLH1 and MSH2 was based on guidelines and recommendations based on specific patterns of inactivation of the genes encoding each of these MMR proteins.(40,41) Two cases had no MSI data, but patients were verified to harbor MSH2 germline mutations through germline sequencing after genetic counseling. It was our premise that a tumor that is microsatellite-stable would not be expected to have a deleterious mutation in an MMR gene, including MLH1 and MSH2. Forty tumors comprised of with known MLH1 (20 cases) or MSH2 (20 cases) loss were selected from the surgical pathology archives of MDACC for the validation studies. The validation set consisted of whole-tissue sections of various tumor types, including cases of colorectal, endometrioid, and esophageal adenocarcinoma; urothelial carcinoma, and tubulovillous adenomas.
Sensitivity was defined as the percentage of samples that had loss of MLH1 or MSH2 expression and classified as MSI-high, and that had respectively MLH1 promoter methylation or MSH2 mutation. Specificity was defined as the percentage of samples that had retained MLH1 or MSH2 expression and that were classified as MSI-stable. The sensitivity and specificity of the IHC MLH1 and MSH2 assays were 100% (95% CI lower limit: 0.799 for each). (Supplementary Tables S4, S5, S6)
Selected samples from specific relevant clinical context
Samples from patients with a specific clinical context (Cowden syndrome, retinoblastoma, hereditary osteosarcoma) were identified from the pathology files at MDACC. Electronic medical records were reviewed for pathologic and clinical features. Formalin-fixed paraffin-embedded tissue blocks were sectioned, and slides were stained per respective IHC protocols.
PTEN IHC assay performance in tumors from patients with Cowden syndrome
Cowden syndrome is an autosomal dominant genetic syndrome characterized by germline mutations in PTEN manifested by multiple benign skin tumors (including sclerotic fibromas and trichilemmomas), gastrointestinal hamartomatous polyps, and an increased risk for various cancers (including those arising in the thyroid, endometrium, breast, and kidney). Earlier studies suggested that tumors associated with Cowden syndrome have loss of PTEN expression assessed by IHC.(42)
Specimens from three patients tested previously to have PTEN germline mutations were determined to have Cowden syndrome by MDACC genetic counselors. Tumors tested include metastatic breast carcinoma, trichilemmoma, and sclerotic fibroma. As expected, all tumors (3/3, 100%) had complete loss of PTEN by IHC, with positive internal non-neoplastic tissue controls.
RB IHC assay performance in retinoblastomas and retinoblastoma-associated osteosarcoma
Retinoblastoma is rare cancer that arises from the retina and most often occurs in early childhood. Retinoblastoma may be hereditary (40%) or sporadic (60%), with hereditary tumors developing in infants and often bilaterally. These tumors characteristically harbor deleterious biallelic (or homozygous) mutations in RB1 (most common), or have deletions of both RB1 loci located on chromosome 13q14. Patients with germline RB1 mutations are susceptible for other tumors, including osteosarcoma and urothelial carcinomas.
Specimens from seven patients included retinoblastomas from six patients and a right tibia osteosarcoma from a patient with a history of early-onset bilateral retinoblastomas. As expected, all tumors (7/7, 100%) had complete loss of RB expression by IHC, with positive internal non-neoplastic tissue controls.
MLH1 and MSH2 IHC assay performance in tumors from patients with Lynch and Muir-Torre syndromes
Muir-Torre syndrome is an autosomal-dominant genetic variant of Lynch syndrome characterized by germline mutations in MMR genes, including MLH1 and MSH2. These patients have multiple benign skin tumors (sebaceous tumors and keratoacanthomas), and an increased risk for visceral cancers. Prior studies suggest that tumors in patients with Lynch and Muir-Torre syndrome with germline mutations in MLH1 or MSH2 have, respectively, loss of MLH1 or MSH2 expression.(43–46)
Four patients with germline MLH1 (n=2) or MSH2 (n=2) mutations were selected by MDACC genetic counselors. Sebaceous adenomas from three patients and an urothelial carcinoma from one patient were tested. As expected, all tumors (4/4, 100%) had complete loss of MLH1 or MSH2 as assessed by IHC, with positive internal non-neoplastic tissue controls.
PRECISION
Inter-pathologist Scoring Concordance
MLH1 and MSH2 IHC assays performed on the main patient set (n=40) were interpreted independently by two pathologists (JDK and WLW) according to interpretation guidelines summarized above in Table 2. Results were concordant (40/40, 100%) (95% CI: 0.943–1.000) for both assays, and the interpretation guidelines were applicable to all cases evaluated.
External Proficiency Testing
As part of ongoing quality assurance and improvement, the MDACC clinical IHC Laboratory participates in a nationwide external quality assessment survey, the DNA Mismatch Repair Proficiency Testing (2011- current). The CAP administers this proficiency test twice a year. Unstained FFPE slides from a single sample are provided to laboratories to perform MMR IHC studies, including MLH1 and MSH2. Laboratories are blinded to the MLH1 and MSH2 expression status and must render interpretation based on their in-house assay. The results are then compared with other participating and reference laboratories. In all five years, the results of MLH1 and MSH2 staining performed at MDACC were concordant with testing performed at all participating institutions with no unacceptable interpretations rendered. No commercial proficiency tests exist for PTEN and RB IHC assays.
DISCUSSION
In this paper, we provide details of validation procedures for four IHC assays currently used in conjunction with molecular tests to guide therapy for patients enrolled in the NCI-MATCH trial. (Figure 5) The aim of these procedures was to establish the clinical validity of the biomarker tests in question as adjunct tools for targeted therapy and ensure that their performance characteristics meet acceptable quality limits and standards. In addressing the various facets of therapy-guiding IHC biomarker assay development in a way that exceeds typical validation procedures for clinical diagnostic assays, the approach outlined in this manuscript strives to ensure that no details are omitted in the deployment of such assays in the clinical setting.
Figure 5.
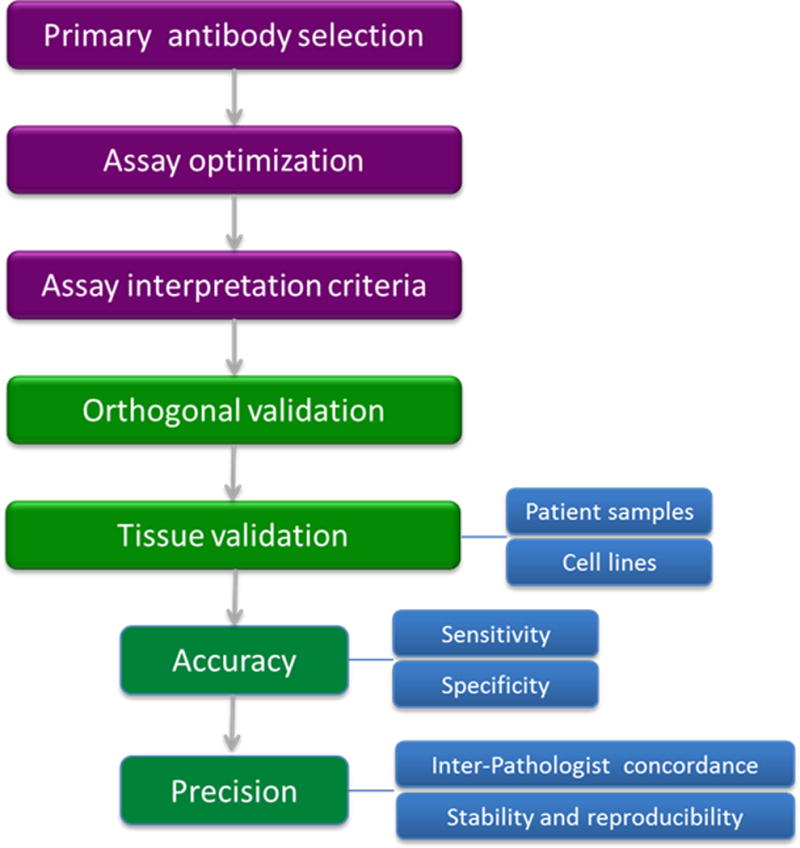
Outline of validation steps for therapy-guiding immunohistochemistry assays.
We have proposed the creation of specialized accreditation program to create certified advanced companion diagnostics facilities. (47–49) Such a program would require close collaborations between stakeholders in the medical laboratory and pathology field, oncology, and pharmaceutical industry. The integration of stringently validated therapy-guiding IHC biomarker assays into such a paradigm is can provide further refinement of personalized therapy selection in specific contexts, particularly where genomic or epigenetic lesions that cause loss of expression of a particular biomarker (e.g. MLH1 or PTEN) might not be readily detectable by mutation screening assays. (Figure 6) A similar recommendation to strengthen the oversight and accreditation of laboratories performing biomarker tests for targeted therapies was recently advocated by The Health and Medicine Division (formerly, Institute of Medicine), a division of the National Academies of Sciences, Engineering, and Medicine.(50)
Figure 6.
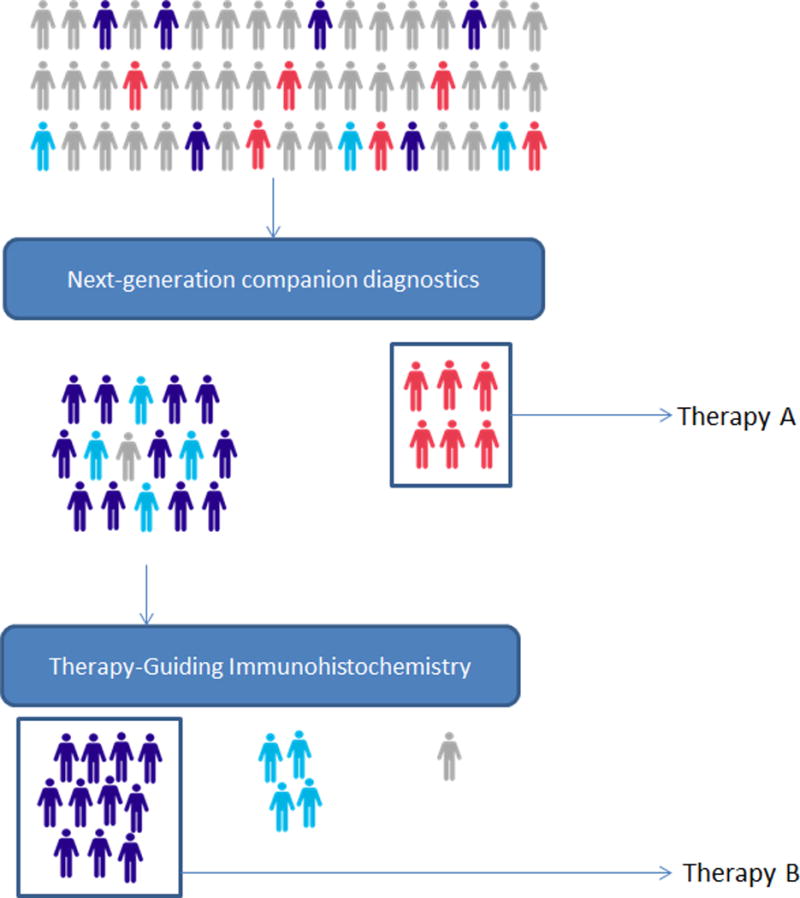
Screening-refinement model for the selection of targeted therapies. Next-generation companion diagnostics, which include next-generation sequencing and possibly large-scale proteomics, provide a screening platform for potential therapy selection. Precision therapy-guiding companion diagnostics, mostly including immunohistochemistry, may provide further refinement of inclusion/exclusion decisions for targeted therapies.
No specific guidelines exist for standardized validation of therapy-guiding biomarker assays. Components of the validation of the four IHC assays described here were collated from various sources and represent an iterative process that was developed in collaboration with an advisory team overseeing biomarker support for the NCI-MATCH trial. It is hoped that the validation steps outlined herein may provide a blueprint to inform development of standardized validation guidelines that will gain wide acceptance by regulatory agencies and payers. Ideally, the blueprint would be developed by consensus through collaborative efforts between key stakeholders, including the pathology and molecular diagnostics communities, oncologists, and the pharmaceutical industry.
Supplementary Material
Acknowledgments
Support: The authors thank the staff of the Clinical Immunohistochemistry Laboratory at the MD Anderson Cancer Center, especially Janet Quiñones and Victor Ortega; the NCI-MATCH support team at MD Anderson, especially Dr. Geeta Mantha, Dr. Mark Routbort, Dr. Ignacio Wistuba, Dr. Jaime Rodriguez-Canales and Barbara Mino for assistance with external validation studies for PTEN. The authors also thank the NCI-MATCH Manuscript Committee for editorial review of this manuscript. This study was supported by the National Institutes of Health/National Cancer Institute award U10CA180820 and Cancer Center Support Grant award P30 CA016672.
Footnotes
Conflict of interest disclosure: The authors declare no potential conflicts of interest.
References
- 1.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134(6):907–22. doi: 10.1043/1543-2165-134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–56. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartley AN, Washington MK, Ventura CB, Ismaila N, Colasacco C, Benson AB, 3rd, et al. HER2 Testing and Clinical Decision Making in Gastroesophageal Adenocarcinoma: Guideline From the College of American Pathologists, American Society for Clinical Pathology, and American Society of Clinical Oncology. Arch Pathol Lab Med. 2016;140(12):1345–63. doi: 10.5858/arpa.2016-0331-CP. [DOI] [PubMed] [Google Scholar]
- 4.Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017:JCO2016719807. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 5.Haddow JE, Palomaki GE. ACCE: A Model Process for Evaluating Data on Emerging Genetic Tests. In: Khoury M, Little J, Burke W, editors. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. Oxford University Press; 2003. pp. 217–33. [Google Scholar]
- 6.21CFR864.1860 - Code of Federal Regulations, Title 21, Volume 8 [current as of May 19, 2016].
- 7.United States Food and Drug Administration. In Vitro Companion Diagnostic Devices: Guidance for Industry and Food and Drug Administration Staff. 2011 < https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-meddev-gen/documents/document/ucm262327.pdf>.
- 8.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–51. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgibbons PL, Bradley LA, Fatheree LA, Alsabeh R, Fulton RS, Goldsmith JD, et al. Principles of analytic validation of immunohistochemical assays: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2014;138(11):1432–43. doi: 10.5858/arpa.2013-0610-CP. [DOI] [PubMed] [Google Scholar]
- 11.Goldsmith JD, Fitzgibbons PL, Swanson PE. Principles of Analytic Validation of Clinical Immunohistochemistry Assays. Adv Anat Pathol. 2015;22(6):384–7. doi: 10.1097/PAP.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein NS, Hewitt SM, Taylor CR, Yaziji H, Hicks DG, Members of Ad-Hoc Committee On Immunohistochemistry S Recommendations for improved standardization of immunohistochemistry. Appl Immunohistochem Mol Morphol. 2007;15(2):124–33. doi: 10.1097/PAI.0b013e31804c7283. [DOI] [PubMed] [Google Scholar]
- 13.United States Food and Drug Administration. Guidance for Submission of Immunohistochemistry Applications to the FDA. 2011 < https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM094015.pdf>.
- 14.Clinical and Laboratory Standards Institute. Quality Assurance for Design Control and Implementation of Immunohistochemistry Assays. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. (CLSI document I/LA28-A2). [Google Scholar]
- 15.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145(1):30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Abrams J, Conley B, Mooney M, Zwiebel J, Chen A, Welch JJ, et al. National Cancer Institute’s Precision Medicine Initiatives for the new National Clinical Trials Network. Am Soc Clin Oncol Educ Book. 2014:71–6. doi: 10.14694/EdBook_AM.2014.34.71. [DOI] [PubMed] [Google Scholar]
- 17.Brower V. NCI-MATCH pairs tumor mutations with matching drugs. Nature biotechnology. 2015;33(8):790–1. doi: 10.1038/nbt0815-790. [DOI] [PubMed] [Google Scholar]
- 18.Adam P, Baumann R, Schmidt J, Bettio S, Weisel K, Bonzheim I, et al. The BCL2 E17 and SP66 antibodies discriminate 2 immunophenotypically and genetically distinct subgroups of conventionally BCL2-“negative” grade 1/2 follicular lymphomas. Hum Pathol. 2013;44(9):1817–26. doi: 10.1016/j.humpath.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Salvatorelli L, Parenti R, Leone G, Musumeci G, Vasquez E, Magro G. Wilms tumor 1 (WT1) protein: Diagnostic utility in pediatric tumors. Acta Histochem. 2015;117(4–5):367–78. doi: 10.1016/j.acthis.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51(2):189–99. [PubMed] [Google Scholar]
- 21.Garg K, Broaddus RR, Soslow RA, Urbauer DL, Levine DA, Djordjevic B. Pathologic scoring of PTEN immunohistochemistry in endometrial carcinoma is highly reproducible. Int J Gynecol Pathol. 2012;31(1):48–56. doi: 10.1097/PGP.0b013e3182230d00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 23.Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57(21):4736–8. [PubMed] [Google Scholar]
- 24.Stambolic V, Tsao MS, Macpherson D, Suzuki A, Chapman WB, Mak TW. High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/- mice. Cancer Res. 2000;60(13):3605–11. [PubMed] [Google Scholar]
- 25.Lotan TL, Wei W, Ludkovski O, Morais CL, Guedes LB, Jamaspishvili T, et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol. 2016 doi: 10.1038/modpathol.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djordjevic B, Hennessy BT, Li J, Barkoh BA, Luthra R, Mills GB, et al. Clinical assessment of PTEN loss in endometrial carcinoma: immunohistochemistry outperforms gene sequencing. Mod Pathol. 2012;25(5):699–708. doi: 10.1038/modpathol.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14(5):297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadoo KA, Gucalp A, Traina TA. Palbociclib: an evidence-based review of its potential in the treatment of breast cancer. Breast Cancer (Dove Med Press) 2014;6:123–33. doi: 10.2147/BCTT.S46725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J, Thijssen B, McDermott U, Garnett M, Wessels LF, Bernards R. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35(37):4829–35. doi: 10.1038/onc.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–30. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 31.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. The New England journal of medicine. 2003;349(3):247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015;372(26):2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.United States Food and Drug Administration. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests: Guidance for Industry and Food and Drug Administration Staff. 2011 < https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm071287.pdf>.
- 36.Clinical and Laboratory Standards Institute. Harmonized Terminology Database. ( http://htd.clsi.org/listallterms.asp) (accessed: May 9 2017)
- 37.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Clin Chem. 2003;49(1):7–18. doi: 10.1373/49.1.7. [DOI] [PubMed] [Google Scholar]
- 38.Singh RR, Patel KP, Routbort MJ, Reddy NG, Barkoh BA, Handal B, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. The Journal of molecular diagnostics: JMD. 2013;15(5):607–22. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Valverde JR, Alonso J, Palacios I, Pestana A. RB1 gene mutation up-date, a meta-analysis based on 932 reported mutations available in a searchable database. BMC Genet. 2005;6:53. doi: 10.1186/1471-2156-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchanan DD, Tan YY, Walsh MD, Clendenning M, Metcalf AM, Ferguson K, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32(2):90–100. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoffel EM, Mangu PB, Gruber SB, Hamilton SR, Kalady MF, Lau MW, et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol. 2015;33(2):209–17. doi: 10.1200/JCO.2014.58.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Zaid T, Ditelberg JS, Prieto VG, Lev D, Luthra R, Davies MA, et al. Trichilemmomas show loss of PTEN in Cowden syndrome but only rarely in sporadic tumors. J Cutan Pathol. 2012;39(5):493–9. doi: 10.1111/j.1600-0560.2012.01888.x. [DOI] [PubMed] [Google Scholar]
- 43.Jessup CJ, Redston M, Tilton E, Reimann JD. Importance of universal mismatch repair protein immunohistochemistry in patients with sebaceous neoplasia as an initial screening tool for Muir-Torre syndrome. Hum Pathol. 2016;49:1–9. doi: 10.1016/j.humpath.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Roberts ME, Riegert-Johnson DL, Thomas BC, Thomas CS, Heckman MG, Krishna M, et al. Screening for Muir-Torre syndrome using mismatch repair protein immunohistochemistry of sebaceous neoplasms. J Genet Couns. 2013;22(3):393–405. doi: 10.1007/s10897-012-9552-4. [DOI] [PubMed] [Google Scholar]
- 45.Djordjevic B, Broaddus RR. Laboratory Assays in Evaluation of Lynch Syndrome in Patients with Endometrial Carcinoma. Surg Pathol Clin. 2016;9(2):289–99. doi: 10.1016/j.path.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodfellow PJ, Billingsley CC, Lankes HA, Ali S, Cohn DE, Broaddus RJ, et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J Clin Oncol. 2015;33(36):4301–8. doi: 10.1200/JCO.2015.63.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khoury JD, Catenacci DV. Next-generation companion diagnostics: promises, challenges, and solutions. Arch Pathol Lab Med. 2015;139(1):11–3. doi: 10.5858/arpa.2014-0063-ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruver AM, Schade AE. Advanced Companion Diagnostics Facilities: Opportunity Favors the Prepared Laboratory. Arch Pathol Lab Med. 2015;139(10):1201. doi: 10.5858/arpa.2015-0095-LE. [DOI] [PubMed] [Google Scholar]
- 49.Khoury JD. The evolving potential of companion diagnostics. Scand J Clin Lab Invest Suppl. 2016;245:S22–5. doi: 10.1080/00365513.2016.1206444. [DOI] [PubMed] [Google Scholar]
- 50.Graig LA, Phillips JK, Moses HL, editors. Biomarker Tests for Molecularly Targeted Therapies: Key to Unlocking Precision Medicine. Washington, DC: The National Academies Press; 2016. [PubMed] [Google Scholar]
- 51.Bartley AN, Luthra R, Saraiya DS, Urbauer DL, Broaddus RR. Identification of cancer patients with Lynch syndrome: clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev Res (Phila) 2012;5(2):320–7. doi: 10.1158/1940-6207.CAPR-11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RS, Grayson W, Redston M, Diwan AH, Warneke CL, McKee PH, et al. Site and tumor type predicts DNA mismatch repair status in cutaneous sebaceous neoplasia. Am J Surg Pathol. 2008;32(6):936–42. doi: 10.1097/pas.0b013e31815b0cc2. [DOI] [PubMed] [Google Scholar]
- 53.Djordjevic B, Barkoh BA, Luthra R, Broaddus RR. Relationship between PTEN, DNA mismatch repair, and tumor histotype in endometrial carcinoma: retained positive expression of PTEN preferentially identifies sporadic non-endometrioid carcinomas. Mod Pathol. 2013;26(10):1401–12. doi: 10.1038/modpathol.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


