Abstract
Nine (9) different date palm (Phoenix dactylifera L.) cultivars from UAE, which differ in their flower timings were selected to determine the polymorphism and genetic relationship between these cultivars. Hereditary differences and interrelationships were assessed utilizing inter-simple sequence repeat (ISSR) and directed amplification of minisatellite DNA region (DAMD) primers. Analysis on eight DAMD and five ISSR markers produced total of 113 amplicon including 99 polymorphic and 14 monomorphic alleles with a polymorphic percentage of 85.45. The average polymorphic information content for the two-marker system was almost similar (DAMD, 0.445 and ISSR, 0.459). UPGMA based clustering of DAMD and ISSR revealed that mid-season cultivars, Mkh (Khlas) and MB (Barhee) grouped together to form a subcluster in both the marker systems. The genetic similarity analysis followed by clustering of the cumulative data from the DAMD and ISSR resulted in two major clusters with two early-season cultivars (ENg and Ekn), two mid-season cultivars (MKh and MB) and one late-season cultivar (Lkhs) in cluster 1, cluster 2 includes two late-season cultivars, one early-season cultivar and one mid-season cultivar. The cluster analysis of both DAMD and ISSR marker revealed that, the patterns of variation between some of the tested cultivars were similar in both DNA marker systems. Hence, the present study signifies the applicability of DAMD and ISSR marker system in detecting genetic diversity of date palm cultivars flowering at different seasons. This may facilitate the conservation and improvement of date palm cultivars in the future.
Keywords: Date palm (UAE), ISSR, DAMD, Genetic diversity
Introduction
Date palm (Phoenix dactylifera L.) is a monocotyledonous and dioecious species belonging to Arecaceae family. It has been economically, spiritually and ornamentally connected with the eastern Mediterranean life for a period of last 6000 years (El Hadrami and Al-Khayri 2012). Date palms need a long hot growing season and it is widely cultivated in arid regions of Middle East (Elmeer et al. 2011). Dates are proved to be antioxidant and anti-mutagenic assets and considered as a very good source of natural antioxidants (Mansouri et al. 2005). Date palm is one of the ultimate valuable fruiting tree in several arid and semiarid regions of the world and considered as a vital subsistence crop (Chao and Krueger 2007; Elsadig et al. 2017). The flowering habit and time of fruit set in date palm are important factors determining the commercial production of date fruits. Date palms were grouped into three cultivars: early, mid and late seasons. Due to its early fruiting nature, the early-season cultivar has more market value, because it reaches earlier to the market. Recent report showed the differences in hormone production and antioxidant metabolism in these three flowering type of date palms (Cheruth et al. 2015). In this context, the study is more demanding in determining the basic physiological mechanisms and molecular profiling underlying the flowering and fruiting behaviour of date varieties. It will be crucial to determine the molecular basis of flowering to improve the yield of early-bearing varieties.
DNA fingerprinting is mainly exploited for detecting the genetic diversity of plant species and for the identification of markers linked with specific traits (Kurup et al. 2009; Khanam et al. 2012). Inter-simple sequence repeats (ISSR) are highly discriminative, simple, fast, cost-effective, firm and reliable in identifying markers. It requires only a small quantity of the DNA sample and does not need any prior sequence information to design the primer. The PCR-based direct amplification of minisatellite DNA regions (DAMD) (Heath et al. 1993) and ISSR (Williams et al. 1990; Gupta et al. 1994) were effective tools for the hereditary diversity studies in plants. Both DNA markers display an exhaustive depiction of the nature and degree of diversity (Bhattacharya et al. 2005; Ranade et al. 2009). ISSR amplification is a comparatively recent technique which can distinguish closely related genotypes. Molecular marker studies have made great assistance to our knowledge on genetic diversity and the understanding on various crops (Zietkiewicz et al. 1994; Hamwieh et al. 2009).
ISSR marker displayed its suitability to generate genetic diversity qualities examination. In addition, this marker exceedingly reproducible, cost-effective, and requires no earlier data of the sequence (Bornet et al. 2002). These confirmations suggest that ISSR could be an unprejudiced instrument to assess the progression of differing qualities in agronomically significant crops (Brantestam et al. 2004). The use of ISSR marker for detecting the genetic diversity of date palm was reported earlier by many researchers (Zehdi et al. 2004; Karim et al. 2010). The application of DAMD marker for detecting the polymorphism was also reported in different plant species (Hu et al. 2011; Singh et al. 2014). Currently, there is no information available for the use of DAMD markers in genetic diversity study of date palm cultivars. Therefore, the objectives of the present study were to determine the potential use of ISSR and DAMD markers technique to detect polymorphism and genetic relationship between date palm cultivars flowering at different seasons, which will enable the identification and seasonal production of good quality date palm cultivars of UAE.
Materials and methods
Experimental sites and date palm varieties
The date palm experimental trees were located and marked separately from Al-Foah Research Station of College of Food and Agriculture (270N and 220S latitude and 510W and 570E longitude), UAE University in Al Ain city in 160 km Eastern Abu Dhabi, United Arab Emirates. The index leaves of date palm were collected from the various stages like pre-flowering, flowering and post-flowering date palm cultivars. Three (3) cultivars from each of the three categories were selected. In each individual cultivar, three plants located and marked for analysis. Standard normal date palm cultivation practices and agriculture procedures was applied in this study. The varieties used in this study are given below:
(i) Early season—Nagal (ENg), Shaham (ESh), and Khanezi (EKn); (ii) Mid-season –Barhee (MB), Khlas (MKh), and Nabthasaif (MNb) and (iii) Late season—Lulu (LL), Khasab (LKhs), and Fardh (LF).
Plant DNA Isolation using CTAB method
The fresh leaf sample (3 g) of Phoenix dactylifera was grounded in liquid nitrogen and DNA extraction was performed using a CTAB method (Arif et al. 2010). Isolated DNA was resuspended in 50 µL of TE buffer. The purity and quantity of isolated DNA were determined using Nanodrop 2000 spectrophotometer (Thermo fisher, India).
PCR amplification using DAMD and ISSR primers
For DAMD marker analysis, eight out of ten markers showed clear amplification pattern for all the tested cultivars. The annealing temperature of primers varied from 42 to 61.5 °C for each primer (Table 1). In ISSR analysis, a set of ten primers were pre-screened, in which five primers resulted in clear amplification and the annealing temperature was optimized for each primer. PCR amplifications were carried out in a total volume of 20 μL containing 10 μL master mix, 2 μL primer (0.2 µM), 2 μL of 25 ng template DNA and 6 μL of DNase free water. The PCR conditions for DAMD marker was carried out based on Ranade et al. (2009) using Master cycler (Nexus gradient-Eppendorf). Similarly, ISSR was conducted with similar conditions based on Rout et al. (2009). The amplified products were separated using agarose gel electrophoresis on 1.5% agarose gel in 1X TBE buffer. The electrophoresis was done at 100 V for 3 h and the bands were visualized under UV light using a gel documentation system.
Table 1.
List of DAMD and ISSR Primers and their annealing temperatures
| Primer | Sequence | Annealing temperature (°C) |
|---|---|---|
| DAMD Markers | ||
| HBV5 | GGTGTAGAGAGGGGT (15) | 50.6 |
| HVR | CCTCCTCCCTCCT(13) | 44.0 |
| INS | ACAGGGGTGGGG(12) | 42.0 |
| URP9F | ATGTGTGCGATCAGTTGCTG (20) | 57.3 |
| URP2R | CCCAGCAACTGATCGCACAC (20) | 61.4 |
| URP25F | GATGTGTTCTTGGAGCCTGT (20) | 57.3 |
| M13 | GAGGGTGGCGGCTCT (15) | 56.0 |
| HBV3 | GGTGAAGCACAGGTG (15) | 50.6 |
| ISSR markers | ||
| N7 | GAGGAGGAGGC (11) | 38.0 |
| N8 | CACACACACACAGT (14) | 42.0 |
| N9 | ACACACACACACACACAG(18) | 53.7 |
| N10 | ACACACACACACACACAA (18) | 51.4 |
| N12 | ACACACACACACACACGA (18) | 53.7 |
Data scoring and analysis
Only clear and well-separated amplicons were determined. These bands were scored independently as either present (1) or absent (0). The PIC value was calculated using the formula PICi = 2fi (1–fi), where fi is the frequency of the amplified fragments and 1–fi is the frequency of non-amplified fragments (Roldan-Ruiz et al. 2000). The data were scored individually; a dendrogram was generated by cluster analysis using the UPGMA method based on Jaccard’s coefficient. The correlation coefficient (r) based on Mantel test was conducted using the NTSYS-pc software version 2.02e 9 (Rohlf 1998).
Results and discussion
DAMD and ISSR analysis
In the present study, two dominant two PCR-based DNA markers, DAMD and ISSR were utilized for the screening of genetic polymorphism between different cultivars of date palm, which flowers in different seasons. A total of ten (10) DAMD and ISSR primers were tested in which eight (8) DAMD and five (5) ISSR showed reproducible banding pattern. In DAMD analysis, a total of 76 bands were amplified in which 69 polymorphic and 7 monomorphic bands were observed. Among the tested DAMD primers, the average number polymorphic bands developed were eight bands per primer. In each DAMD primers, the number of amplicon developed was ranged from a minimum of 3 and maximum of 16 bands. The percentage of polymorphic band (PPB) ranged from 71.42% for HBV3 to 100% for HVR, INS, URP9F, URP2R, and URP25F.
The overall percentage of polymorphic bands (PPB) was 90.76%. The polymorphic information content (PIC) varied from 0.383 for primer URP25F to 0.493 for primer URP2R. The average PIC value for the DAMD marker was 0.445 which confirms that these markers are highly polymorphic (Table 2). In ISSR markers, the number of amplified products ranged from six for primer N8 to a maximum of ten for N9 primer. The banding pattern revealed a total of 37 amplified products, in which 30 polymorphic and 7 monomorphic bands were observed. On an average, 80.14% of polymorphic bands were observed ranging from 50% for N10 primer to a maximum of 100% for N8 primer. The average PIC value (0.473) was found to be slightly high when compared to the DAMD markers ranging from 0.444 (N8) to 0.496 (N7). A similar range of PIC value for both DAMD (0.445) and ISSR (0.473) denotes these primers have similar levels of probability in determining the polymorphism among the selected genotypes.
Table 2.
DAMD and ISSR primer used for amplification
| S.no | Primers | M B | P B | T B | PPB | PIC |
|---|---|---|---|---|---|---|
| DAMD | ||||||
| 1 | HBV5 | 2 | 7 | 9 | 77.77 | 0.456 |
| 2 | HVR | 0 | 16 | 16 | 100 | 0.422 |
| 3 | INS | 0 | 6 | 6 | 100 | 0.465 |
| 4 | URP9F | 0 | 9 | 9 | 100 | 0.451 |
| 5 | URP2R | 0 | 13 | 13 | 100 | 0.493 |
| 6 | URP25F | 0 | 3 | 3 | 100 | 0.383 |
| 7 | M13 | 3 | 10 | 13 | 76.9 | 0.483 |
| 8 | HBV3 | 2 | 5 | 7 | 71.42 | 0.408 |
| Total | 7 | 69 | 76 | |||
| Average | 0.875 | 8.62 | 9.5 | 90.76 | 0.445 | |
| ISSR | ||||||
| 1 | N7 | 1 | 6 | 7 | 85.71 | 0.496 |
| 2 | N8 | 0 | 6 | 6 | 100 | 0.444 |
| 3 | N9 | 1 | 9 | 10 | 90 | 0.48 |
| 4 | N10 | 3 | 3 | 6 | 50 | 0.452 |
| 5 | N12 | 2 | 6 | 8 | 75 | 0.493 |
| Total | 7 | 30 | 37 | |||
| Average | 1.4 | 6 | 7.4 | 80.14 | 0.473 | |
| DAMD + ISSR | Grand total | 14 | 99 | 113 | 85.45 | 0.459 |
MB monomorphic band, PB polymorphic band, TB total band, PPB percentage of polymorphic band, PIC polymorphic information content
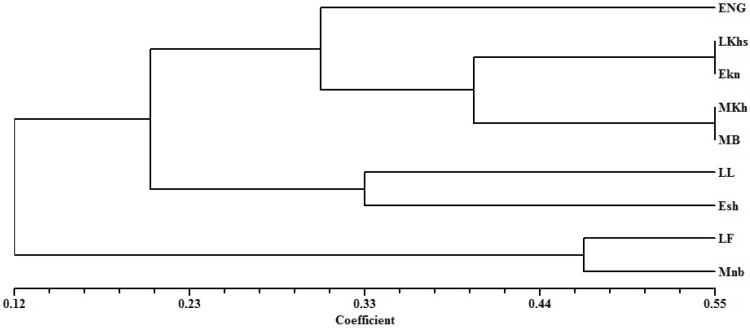
The similarity matrix was calculated based on the Jaccard coefficient. The similarity coefficient for the DAMD marker ranged from a minimum of 0.18 between the cultivar Mnb and Lkhs to a maximum of 0.66 between the cultivars MB and MKh (Table 3). The dendrogram of nine different varieties of date palm was constructed using the SAHN clustering based on UPGMA algorithm. The dendrogram revealed three major clusters for the tested cultivars (Fig. 1). In cluster 1, early cultivar ENg formed a cluster by joining with a subcluster formed among two mid-flowerings, one early-flowering and one late-flowering variety. The mid-flowering varieties MB and MKh showed a higher similarity of 0.66. In the same cluster, 0.6 similarities were noticed between late season cultivars LKhs and early-season cultivar EKn. In cluster 2, LL (Late lulu) produced 0.49 similarities with the early ESh. LF and MNb joined to form cluster 3 with 0.46 similarities between them.
Table 3.
Similarity matrix of date palm cultivars computed with Jaccard coefficient in DAMD primers
| ENG | LF | LKhs | MKh | MB | Ekn | Mnb | LL | Esh | |
|---|---|---|---|---|---|---|---|---|---|
| ENG | 1.0000000 | ||||||||
| LF | 0.3888889 | 1.0000000 | |||||||
| LKhs | 0.4905660 | 0.3600000 | 1.0000000 | ||||||
| MKh | 0.5636364 | 0.3888889 | 0.4629630 | 1.0000000 | |||||
| MB | 0.5660377 | 0.3584906 | 0.5510204 | 0.6600000 | 1.0000000 | ||||
| Ekn | 0.4230769 | 0.3404255 | 0.5952381 | 0.5416667 | 0.6136364 | 1.0000000 | |||
| Mnb | 0.2745098 | 0.4594595 | 0.1836735 | 0.3265306 | 0.3191489 | 0.2926829 | 1.0000000 | ||
| LL | 0.4736842 | 0.4313725 | 0.4528302 | 0.4482759 | 0.5283019 | 0.4417647 | 0.3404255 | 1.0000000 | |
| Esh | 0.3888889 | 0.2549020 | 0.3600000 | 0.3888889 | 0.4400000 | 0.4000000 | 0.2857143 | 0.4897959 | 1.0000000 |
Fig. 1.
UPGMA dendrogram of date palm cultivars generated for the DAMD marker
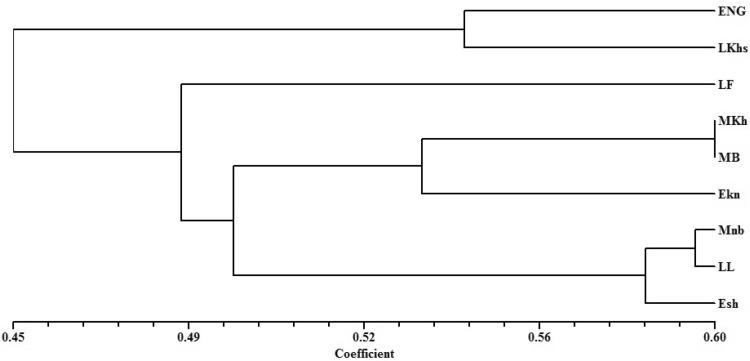
In ISSR analysis, the dendrogram resulted in two major clusters. The similarity index value between the tested cultivars varied from a minimum of 0.37 between MNb and LKhs to a maximum of 0.60 between MKh and MB (Table 4). In cluster 1, early-flowering Nagal showed 0.54 similarities with late-season LKhs. In cluster 2, late-flowering Fardh formed a cluster by joining with two subclusters (Fig. 2). In ISSR marker study, the highest similarity index value of 0.60 was revealed between MKh and MB, which confirms that both ISSR and DAMD showed similar range of polymorphism between two mid-flowering cultivars MKh and MB. Similar to DAMD cluster analysis, the MKh, MB and EKn was found to be grouped together in a subcluster for ISSR markers. This shows the similarity between the selected mid- and early-season genotypes in both marker systems.
Table 4.
Similarity matrix date palm cultivars computed with Jaccard coefficient in ISSR primers
| ENG | LF | LKhs | MKh | MB | Ekn | Mnb | LL | Esh | |
|---|---|---|---|---|---|---|---|---|---|
| ENG | 1.0000000 | ||||||||
| LF | 0.5000000 | 1.0000000 | |||||||
| LKhs | 0.5454545 | 0.3947368 | 1.0000000 | ||||||
| MKh | 0.4000000 | 0.4871795 | 0.400000 | 1.0000000 | |||||
| MB | 0.5263158 | 0.5384615 | 0.5454545 | 0.6000000 | 1.0000000 | ||||
| Ekn | 0.5227273 | 0.4375000 | 0.4285714 | 0.4772727 | 0.5952381 | 1.0000000 | |||
| Mnb | 0.4583333 | 0.4400000 | 0.3695652 | 0.4468085 | 0.4893617 | 0.5800000 | 1.0000000 | ||
| LL | 0.4318182 | 0.4772727 | 0.4000000 | 0.4523810 | 0.5365854 | 0.5319149 | 0.5957447 | 1.0000000 | |
| Esh | 0.4761905 | 0.5238095 | 0.4102564 | 0.5000000 | 0.4418605 | 0.4791667 | 0.5744681 | 0.5952381 | 1.0000000 |
Fig. 2.
UPGMA dendrogram of date palm cultivars generated for the ISSR marker
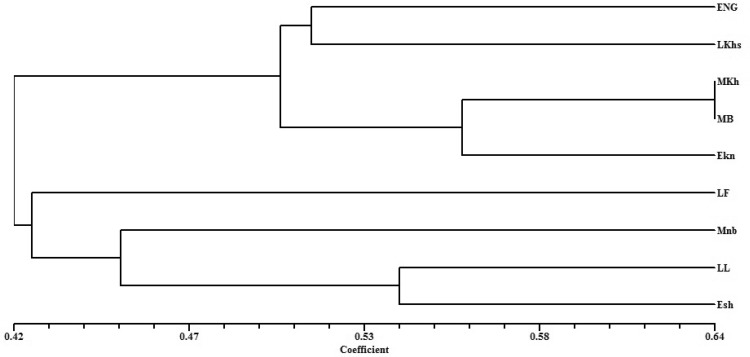
In the present study, a total of 13 primers, eight (8) DAMD and five (5) ISSR produced 113 bands of which 99 bands were found to be polymorphic with an 85.5% polymorphism. The average polymorphic information content for the 13-cumulative primer was found to be 0.459. The similarity index value for the cumulative primers varied from 0.273 to 0.635. The maximum similarity value was observed between Mkh and MB cultivars and the least (0.273) was between Lkhs and Mnb cultivars. UPGMA clustering-based dendrogram was constructed using the similarity value, which resulted in formation of two major clusters, cluster 1 and cluster 2 (Fig. 3). In cluster 1, two early-season cultivars (ENg and EKn), two mid-season cultivars (MKh and MB) and one late-season cultivar (LKhs) were grouped together, in which the mid-season cultivar formed a subcluster. Cluster 2 was formed by grouping of two late-season cultivars, one early-season cultivar and one mid-season cultivar. Mantel’s test was performed to find the correlation coefficient (r) between the tested primers (Mantel 1967). Three data sets viz, DAMD, ISSR and cumulative data were analysed with MxComp in NtSys software. A significant correlation coefficient was observed between DAMD and cumulative data with a maximum value of 0.86 followed by ISSR vs cumulative and DAMD vs ISSR (Table 5). These results showed a positive correlation between DAMD and cumulative data as reported by Singh et al. (2014). The lower correlation between the ISSR and DAMD markers may be due to specificity of each marker to different target regions in the genome (see Figs. 4, 5).
Fig. 3.
UPGMA dendrogram of date palm cultivars generated for the cumulative data of DAMD and ISSR markers
Table 5.
Mantel correlation between the genetic similarities obtained from ISSR and DAMD and cumulative data among Phoenix dactylifera cultivars
| Marker system | Correlation coefficient (r) | P value |
|---|---|---|
| DAMD vs ISSR | 0.30 | 0.94 |
| DAMD vs Cumulative | 0.86 | 1 |
| ISSR vs Cumulative | 0.613 | 0.99 |
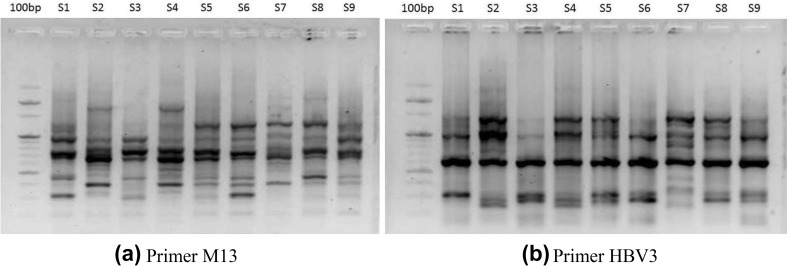
Fig. 4.
DAMD banding pattern of different cultivars of Phoenix dactylifera using primers, a M13 and b HBV3. Lane 1: 100 bp marker, lane 2–10: cultivars of Phoenix dactylifera
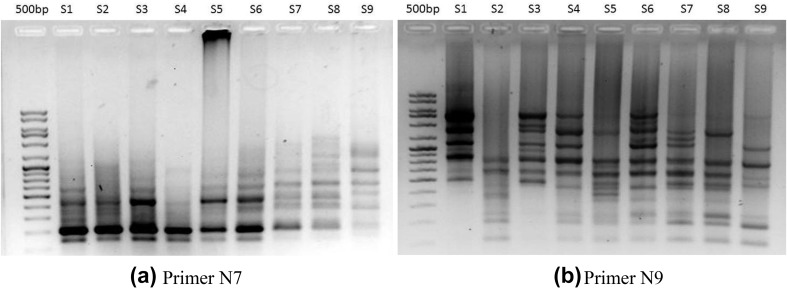
Fig. 5.
ISSR banding pattern of different cultivars of Phoenix dactylifera using primers, a N7 and b N9. Lane 1: 100 bp marker, lane 2–10: cultivars of Phoenix dactylifera
The genetic diversity study has great importance in crop improvement as well as for conservation and management of genetic resources. In the present study, the use of DAMD and ISSR marker revealed the genetic diversity among the different season varieties of date palm cultivated in UAE. In both DAMD and ISSR marker system, the similarity index value between the two mid-season varieties (MKh and MB) was maximum, which shows these cultivars have high similarity between them. An average PIC value of 0.459 denotes the potential role of these markers for the genetic diversity studies of date palm. The use DAMD markers for studying the genetic diversity, cultivar identification, and genetic associations was reported in many plants like Triticum aestivum (Babeli et al. 1997), Oryza sativa (Zhou et al. 1997), Punica granatum (Narzary et al. 2009), Morus spp. (Bhattacharya Ranade 2001) and Cucumis sativus (Hu et al. 2011). In date palm, the use of ISSR marker for revealing high-level polymorphism in Tunisian date palm germplasm was reported (Zehdi et al. 2004). Karim et al. (2010) reported the use of ISSR markers for the genetic diversity study of high -quality fruit date palm varieties. The current study confirms the importance of DAMD and ISSR marker system in detecting polymorphism of important date palm cultivars.
Conclusion
The present study confirms the potential use of DAMD and ISSR DNA markers for determination of genetic diversity and relationship among different date palm varieties. The similar range of PIN value and the clustering pattern for both DAMD and ISSR primer signifies that these primers have great potential for genetic diversity studies in date palm. The genetic diversity study date palm cultivars flowering at different seasons will be beneficial for the conservation and improvement of date palm cultivars.
Acknowledgements
The authors would like to thank the Department of Aridland Agriculture, College of food and Agriculture, United Arab Emirates University, Al Ain, United Arab Emirates, for the facilities. The farm supervisor and labourers of Al-Foah experimental station are gratefully acknowledged. This research is supported by the UAEU Program for Advanced Research-UPAR (Grant: UPAR # 31F045), received by the principal investigator Dr. Abdul J. Cheruth.
Abbreviations
- DAMD
Directly amplified minisatellite DNA
- ISSR
Inter-simple sequence repeat
- PCR
Polymerase chain reaction
- Date palm genotypes
Nagal (ENg), Shaham (ESh), Khanezi (EKn) Barhee (MB), Khlas (MKh), Nabthasaif (MNb) Lulu (LL), Khasab (LKhs), Fardh (LF)
Compliance with ethical standards
Conflict of Interest
The authors declared that there is no conflict of interest.
Footnotes
Sreeramanan Subramaniam: Temporarily corresponding author upon acceptance.
Contributor Information
Sreeramanan Subramaniam, Email: sreeramanan@gmail.com.
Abdul Jaleel Cheruth, Phone: 00971-3-7134576, Email: abdul.jaleel@uaeu.ac.ae.
References
- Arif A, Bakir MA, Khan HA, Ahamed A, Ahmad HA, Farhan A, Ali A, Homaidan MA, Sadoon AH, Bahkali M, Shobrak A. simple method for DNA extraction from mature date palm leaves: impact of sand grinding and composition of lysis buffer. Int J Mol Sci. 2010;11(9):3149–3157. doi: 10.3390/ijms11093149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babeli PJ, Zhou Z, Somer DJ, Gustafson JP. PCR primed with minisatellite core sequences yields DNA fingerprinting probes in wheat. Theor Appl Genet. 1997;95:276–283. doi: 10.1007/s001220050560. [DOI] [Google Scholar]
- Bhattacharya E, Ranade SA. Molecular distinction amongst varieties of mulberry using RAPD and DAMD profiles. BMC Plant Biol. 2001;1(3):1–8. doi: 10.1186/1471-2229-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya E, Dandin SB, Ranade SA. Single primer amplification reaction methods reveal exotic and indigenous mulberry varieties are similarly diverse. J Biosci. 2005;30:669–675. doi: 10.1007/BF02703567. [DOI] [PubMed] [Google Scholar]
- Bornet BC, Muller FP, Branchard M. Highly informative nature of inter simple sequence repeat (ISSR) sequences amplified using tri- and tetra-nucleotide primers from DNA of cauliflower (Brassica oleracea var. ‘botrytis’ L.) Genome. 2002;45:890–896. doi: 10.1139/g02-061. [DOI] [PubMed] [Google Scholar]
- Brantestam AK, Bothmer RV, Dayteg C, Rashal I, Tuvesson S, Weibull J. Inter simple sequence repeat analysis of genetic diversity and relationships in cultivated barley of Nordic and Baltic origin. Hereditas. 2004;141(2):186–187. doi: 10.1111/j.1601-5223.2004.01867.x. [DOI] [PubMed] [Google Scholar]
- Chao CT, Krueger RR. The date palm (Phoenix dactylifera L.): overview of biology, uses, and cultivation. HortScience. 2007;42:1046–1311. [Google Scholar]
- Cheruth AJ, Kurup SS, Subramaniam S (2015) Variations in hormones and antioxidant status in relation to flowering in early, mid, and late varieties of date palm (Phoenix dactylifera) of United Arab Emirates. The Sci World J 2015:846104. 10.1155/2015/846104 [DOI] [PMC free article] [PubMed]
- El Hadrami A, Al-Khayri JM. Socioeconomic and traditional importance of date palm. Emir J Food Agric. 2012;24:371–385. [Google Scholar]
- Elmeer K, Sarwath H, Malek J. New microsatellite markers for assessment of genetic diversity in date palm (Phoenix dactylifera L.). 3. Biotech. 2011;1(2):91–97. doi: 10.1007/s13205-011-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsadig E, Aljuburi H, Elamin A, Gafar M. Impact of organic manure and combination of N P K S, on yield, fruit quality and fruit mineral content of Khenazi date palm (Phoenix dactylifera L.) cultivar. J Sci Agric. 2017;1:335–346. [Google Scholar]
- Gupta M, Chyi YS, Severson JR, Owen JL. Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor Appl Genet. 1994;89:998–1006. doi: 10.1007/BF00224530. [DOI] [PubMed] [Google Scholar]
- Hamwieh A, Upada SM, Sarker A, Jung C, Baum M. Development of new microsatellite markers and their application in the analysis of genetic diversity in lentil. Breed Sci. 2009;59:77–86. doi: 10.1270/jsbbs.59.77. [DOI] [Google Scholar]
- Heath DD, Iwama GK, Devlin RH. PCR primed with VNTR core sequences yield species specific patterns and hypervariable probes. Nucleic Acids Res. 1993;21:5782–5785. doi: 10.1093/nar/21.24.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JB, Li JW, Wang LJ, Liu LJ, Si SW. Utilization of a set of high-polymorphism DAMD markers for genetic analysis of a cucumber germplasm collection. Acta Physiol Plant. 2011;33:227–231. doi: 10.1007/s11738-010-0525-7. [DOI] [Google Scholar]
- Karim K, Chokri B, Amel S, Wafa H, Richid H, Nouredine D. Genetic diversity of Tunisian date palm germplasm using ISSR markers. Int J Bot. 2010;6:182–186. doi: 10.3923/ijb.2010.182.186. [DOI] [Google Scholar]
- Khanam S, Sham A, Bennetgen JL, Mohammed AMA. Analysis of molecular marker-based characterization and genetic variation in date palm (Phoenix dactylifera L.) Aust J. Crop Sci. 2012;6:1236–1244. [Google Scholar]
- Kurup SS, Hedar YS, Al Dhaheri MA, El-Heawiety AY, Aly MAM, Alhadrami G. Morpho-physiological evaluation and RAPD markers-assisted characterization of date palm (Phoenix dactylifera L.) varieties for salinity tolerance. J Food Agric Env. 2009;7(3&4):503–507. [Google Scholar]
- Mansouri A, Embarek G, Kokkalou E, Kefalas P. Phenolic profile and antioxidant activity of the Algerian ripe date palm fruit (Phoenix dactylifera) Food Chem. 2005;89:411–420. doi: 10.1016/j.foodchem.2004.02.051. [DOI] [Google Scholar]
- Mantel NA. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209–220. [PubMed] [Google Scholar]
- Narzary D, Mahar KS, Rana TS, Ranade SA. Analysis of genetic diversity among wild pomegranates in Western Himalayas, using PCR methods. Sci Hortic. 2009;121:237–242. doi: 10.1016/j.scienta.2009.01.035. [DOI] [Google Scholar]
- Ranade SA, Rana TS, Narzary D. SPAR profile and genetic diversity amongst pomegranate (Punica granatum L.) genotypes. Physiol Mol Biol Plants. 2009;15:61–70. doi: 10.1007/s12298-009-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf FJ (1998) NTSYS-PC: numerical taxonomy and multivariate analysis system. Version 2.02, Exeter software. Setauket, New York
- Roldan-Ruiz I, Dendauw J, Vanbockstaele E, Depicker A, De Loose M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.) Mol Breed. 2000;6:125–134. doi: 10.1023/A:1009680614564. [DOI] [Google Scholar]
- Rout GR, Senapati SK, Aparajita S, Palai SK. Studies on genetic identification and genetic fidelity of cultivars banana using ISSR marker. Plant Omics J. 2009;2:250–258. [Google Scholar]
- Singh N, Bajpai R, Mahar KS, Tiwari V, Upreti DK, Rana TS. ISSR and DAMD markers revealed high genetic variability within Flavoparmelia caperata in Western Himalaya (India) Physiol Mol Biol Plants. 2014;20(4):501–508. doi: 10.1007/s12298-014-0256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK, Rubelik AR, Livak KJ, Rafalski A, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehdi S, Trifi M, Ould MSA, Rhouma A, Marrakchi M. Molecular characterization of Tunisian date palm germplasm using ISSR markers. J Genet Breed. 2004;56:77–83. [Google Scholar]
- Zhou Z, Bebeli PJ, Somers DJ, Gustafson JP. Direct amplification of minisatellite-region DNA with VNTR core sequences in the genus Oryza. Theor Appl Genet. 1997;95:942–949. doi: 10.1007/s001220050645. [DOI] [Google Scholar]
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprint by simple sequence repeat (SSR)—anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]