Abstract
Byrsonima is the third largest genus (about 200 species) in the Malpighiaceae family, and one of the most common in Brazilian savannas. However, there is no molecular phylogeny available for the genus and taxonomic uncertainties at the generic and family level still remain. Herein, we sequenced the complete chloroplast genome of B. coccolobifolia and B. crassifolia, the first ones described for Malpighiaceae, and performed comparative analyses with sequences previously published for other families in the order Malpighiales. The chloroplast genomes assembled had a similar structure, gene content and organization, even when compared with species from other families. Chloroplast genomes ranged between 160,212 bp in B. crassifolia and 160,329 bp in B. coccolobifolia, both containing 115 genes (four ribosomal RNA genes, 28 tRNA genes and 83 protein-coding genes). We also identified sequences with high divergence that might be informative for phylogenetic inferences in the Malpighiales order, Malpighiaceae family and within the genus Byrsonima. The phylogenetic reconstruction of Malpighiales with these regions highlighted their utility for phylogenetic studies. The comparative analyses among species in Malpighiales provided insights into the chloroplast genome evolution in this order, including the presence/absence of three genes (infA, rpl32 and rps16) and two pseudogenes (ycf1 and rps19).
Introduction
The chloroplast is an organelle that belongs to the family of plastids, playing an essential part in plant growth and development. Its main role is the photosynthesis, but it is also responsible for synthesis of amino acids, fatty acids, lipid components of their membranes and pigments, besides participating in the assimilation of nitrogen1. This organelle possesses its own genetic material, a circular and double-stranded DNA molecule, comprising about 120 genes (encoding ribosomal RNA, transfer RNA and proteins), and ranging in size between 107–218 kb2. Chloroplast genomes commonly present a highly conserved quadripartite structure formed by two inverted repeats (IRa and IRb), one large and another small single copy region (LSC and SSC, respectively)3. Nevertheless, some structural rearrangements may be observed, such as inversions, translocations, variation in copy number of tandem repeats and indels4. Chloroplast genome sequencing has contributed to solve phylogenetic and taxonomic problems in several groups5–7, to identify species by providing barcodes8,9 and to help in the conservation of endangered species10.
Malpighiales is a large order of Angiosperms11 and, partly because of its size, many of the phylogenetic relationships between its members are still not resolved12. The family Malpighiaceae Juss. is the third largest of the order5 and due to its high ecological and morphological diversity the family presents some taxonomic difficulties13. Morphological13–15 and molecular16,17 data support the monophyly of Malpighiaceae, although they are not sufficient to resolve relationships among groups within the family17. Davis and Anderson17 suggested the use of a large number of slow evolving genes to help solving phylogenetic relationships within the family. Such type of markers could be chloroplast genes, due its generally slow evolutionary rates. However, to date, no chloroplast genome of the family Malpighiaceae has been published.
Byrsonima Rich. ex Kunth (popularly known as “murici” in Brazil) is one of the largest genera within the family Malpighiaceae18, including about 200 species. Native to the American continent, the genus has 97 species occurring in Brazil19, seven of which are endangered20. Up to now there are only two studies addressing the taxonomy and phylogeny of the genus18,21. In 1897, Niendzu22 proposed to split the genus into two subgenera, based on stamen morphology. More recently, Elias18 proposed to characterize two subgenera according to their flower color: one group (Byrsonima subg. Macrozeugma) with flowers displaying five, white or pink petals and the other (Byrsonima subg. Byrsonima) with all the petals, or just the posterior ones, yellow. Representing the first subgenus mentioned above, with pink flowers, there is Byrsonima coccolobifolia Kunth, popularly known as “murici-rosa”. On the other hand, Byrsonima crassifolia (L.) Kunth, commonly called “murici-amarelo”, is a typical representative of the subgenus Byrsonima. These two species have economic importance due to the use of their wood and fruits by both the food industry and popular trade23,24. Both species are common in Brazilian savannas (cerrado), including the disjunct savanna areas in the Amazon, where they are among the most common tree species25. Outside Brazil, there are occurrence records of B. coccolobifolia in Bolivia, Venezuela and Guyana26. Byrsonima crassifolia has a broader distribution, and is found from Mexico to Paraguay27. Despite its ecological and economic importance there is no molecular study addressing the taxonomy and phylogeny of the genus Byrsonima. The complete chloroplast genome of two Byrsonima subgenera representatives may be used to detect regions of high sequence divergence that could help resolve taxonomic uncertainties in the genus and in the Malpighiaceae family in general.
In the present study, we sequenced and performed a comparative analysis of the complete chloroplast genome of two species of the genus Byrsonima, B. coccolobifolia and B. crassifolia. We assessed regions of high sequence divergence between the two Byrsonima species to provide markers for phylogenetic and genetic studies. Furthermore, we compared the chloroplast genomes of Malpighiaceae family with those available for other families belonging to the Malpighiales in order to increase the knowledge about chloroplast genome evolution in this order and provide markers for further phylogenetic studies.
Results
Genome content and organization of the chloroplast genome in Byrsonima species
Sequencing of genomic libraries generated about 5GB (20 million reads) and 7GB (32 million reads) of raw data for B. coccolobifolia and B. crassifolia, respectively. The data was used to assemble both chloroplast genomes with a high mean coverage, 1074X for B. coccolobifolia and 805X for B. crassifolia.
The chloroplast genomes of B. crassifolia and B. coccolobifolia exhibited similar structure and organization (Table 1, Fig. 1). The length of B. coccolobifolia chloroplast genome was 160,329 bp divided in four different regions, a pair of inverted repeated regions (IRa and IRb, 26,986 bp each) separated by two single copy regions, one large (LSC, 88,524 bp) and one small (SSC, 17,833 bp). The B. crassifolia chloroplast genome followed the same quadripartite structure, slightly shorter: IR was 26,975 bp each, LSC 88,448 bp and SSC 17,814 bp, for a total of 160,212 bp for the whole genome. The overall GC content was similar for the two species, 36.76% for B. coccolobifolia and 36.77% for B. crassifolia. Among the LSC, SSC and IR regions, the highest GC content was found in the IR regions (42.4% for both species). The GC content for the rRNA (55.42%) and tRNA (53.11%) genes was the highest among all coding regions, compatible to what has been observed in other studies28–30. This relatively higher GC content in rRNA and tRNA genes explains the higher GC content of IR regions, since they contain a great number of these genes.
Table 1.
General information and comparison of chloroplast genomes of Byrsonima coccolobifolia and B. crassifolia.
| Characteristics | B. coccolobifolia | B. crassifolia | |
|---|---|---|---|
| Size (base pair; bp) | 160329 | 160212 | |
| LSC length (bp) | 88524 | 88448 | |
| SSC length (bp) | 17833 | 17814 | |
| IR length (bp) | 26986 | 26975 | |
| Number of genes | 139 | 139 | |
| Protein-coding genes | 94 | 94 | |
| tRNA genes | 37 | 37 | |
| rRNA genes | 8 | 8 | |
| Genes with intron(s) | 18 | 18 | |
| GC content | Total (%) | 36.76 | 36.77 |
| LSC (%) | 34.53 | 34.52 | |
| SSC (%) | 30.66 | 30.76 | |
| IR (%) | 42.4 | 42.4 | |
| CDS (%) | 37.74 | 37.72 | |
| rRNA (%) | 55.42 | 55.42 | |
| tRNA (%) | 53.11 | 53.01 | |
| Coding protein genes (%bp) | 50.2 | 50.2 | |
| Noncoding regions (%bp) | 49.8 | 49.8 | |
Figure 1.

Chloroplast genome circular map of Byrsonima coccolobifolia Kunth and B. crassifolia (L.) Kunth (Malpighiaceae) with annotated genes. Genes inside the circle are transcribed clockwise, genes outside are transcribed counter-clockwise. Genes are color coded according to functional groups. Boundaries of the small (SSC) and large (LSC) single copy regions and inverted repeat (IRa and IRb) regions are noted in the inner circle for each species. Picture of B. crassiflora was taken by Dr. Daniel L. Nickrent (source: http://www.phytoimages.siu.edu). Picture of B. coccolobifolia was provided by Maurício Mercadante.
Both species displayed the same gene content and order (Table 2, Fig. 1), with 118 genes (four ribosomal RNA genes, 30 tRNA genes and 84 protein-coding genes), 21 of which are duplicated in the IR region. These species showed a bias towards using thymine (T) and adenine (A) in the third position of the codon; among the 20 amino acids, 11 of them used mostly codons ending with T and 7 used codons ending with A (Supplementary Table S1). This event is probably a result of an A + T rich genome, also observed in other chloroplast genomes studied30–32. The two genomes have 19 genes containing introns (Table 1), 15 with one intron and four with two or more introns. The rpl32 gene (large ribosomal protein 32) contains three introns in B. coccolobifolia and four introns in B. crassifolia. In both species, the rps12 gene (small ribosomal protein 12) is trans-spliced, that is, this gene has one intron, and the 5′ end exon is located in the LSC region while the second exon (3′ end exon) is located in the IRb (and therefore is duplicated in the IRa). We also detected 10 genes that partially overlap their sequences: psbD/psbC, atpE/atpB, ycf1/ndhF, trnN-GUU/trnR-ACG and orf42/trnA-UGC. For both species the ycf1 gene (5,745 bp) has its start in SSC region, but its sequence goes forward through SSC/IRa boundary, causing a duplication of the 3′ end portion of the ycf1 gene in IRb and, therefore, producing a 1,389 bp ycf1 pseudogene.
Table 2.
Chloroplast genome gene content and functional classification in Byrsonima coccolobifolia Kunth and B. crassifolia (L.) Kunth.
| Gene group | Gene name | |||
|---|---|---|---|---|
| Ribosomal RNA genes | rrn4.5 | rrn5 | rrn16 | rrn23 |
| Transfer RNA genes | trnA-TGC * | trnC-CGA | trnD-GTC | trnE-TTC |
| trnF-GAA | trnfM-CAT | trnG-GCC* | trnG-TCC | |
| trnH-GTG | trnI-CAT * | trnK-UUU* | trnL-CAA | |
| trnL-TAA* | trnL-TAG | trnM-CAT | trnN-GTT | |
| trnP-GGG | trnP-TGG | trnQ-TTG | trnR-ACG | |
| trnR-TCT | trnS-GCT | trnS-GGA | trnS-TGA | |
| trnT-GGT | trnT-TGT | trnV-GAC | trnV-TAC* | |
| trnW-CCA | trnY-GTA | |||
| Small subunit of ribosome | rps2 | rps3 | rps4 | rps7 |
| rps8 | rps11 | rps12 * | rps14 | |
| rps15 | rps16* | rps18 | rps19 | |
| rps12_3end | ||||
| Large subunit of ribosome | rpl2 * | rpl14 | rpl16 | rpl20 |
| rpl22 | rpl23 | rpl32* | rpl33 | |
| rpl36 | ||||
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 |
| NADH dehydrogenase | ndhA* | ndhB * | ndhC | ndhD |
| ndhE | ndhF | ndhG | ndhH | |
| ndhI | ndhJ | ndhK | ||
| Photosystem I | psaA | psaB | psaC | psaI |
| psaJ | ycf3* | |||
| Photosystem II | psbA | psbB | psbC | psbD |
| psbE | psbF | psbH | psbI | |
| psbJ | psbK | psbL | psbM | |
| psbN | psbT | psbZ | ||
| Cytochrome b/f complex | petA | petB | petD | petG |
| petL | petN | |||
| ATP synthase | atpA | atpB | atpE | atpF* |
| atpH | atpI | |||
| Large subunit of rubisco | rbcL | |||
| Maturase | matK | |||
| Protease | clpP* | |||
| Envelope membrane protein | cemA | |||
| Subunit of acetyl-CoA-carboxylase | accD | |||
| c-type cytochrome synthesis | ccsA | |||
| Component of TIC complex | ycf1 | ycf1 Ψ | ||
| Hypothetical chloroplast reading frames | ycf2 | |||
| ORFs | orf42 | orf56 * | ycf 4 | ycf15 * |
| ycf68 * | ||||
*Genes containing introns; ΨPseudogene; genes in bold are located within the IR and therefore are duplicated.
Comparative analysis of chloroplast genomes within Malpighiales
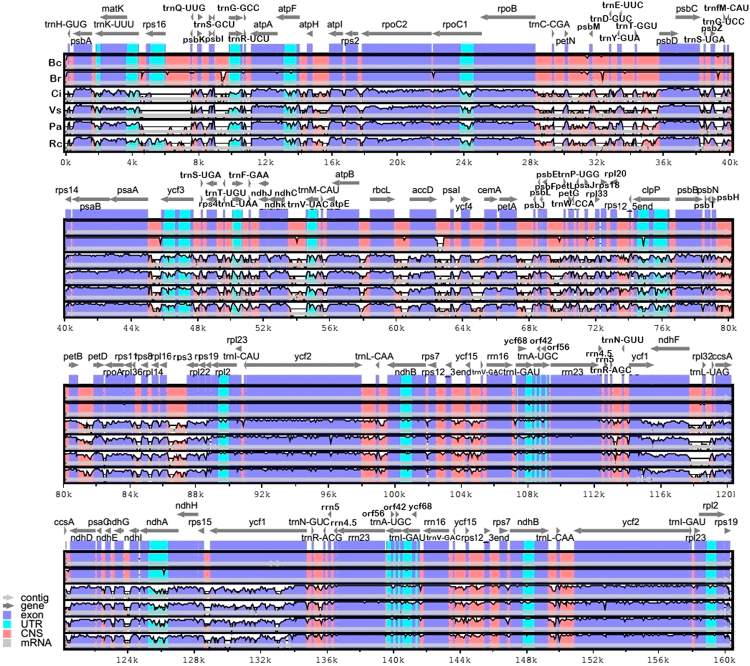
The analysis performed on the mVista software33 showed the level of similarity for the whole sequence of the chloroplast genome of the nine Malpighiales species analyzed (Supplementary Fig. S1). We observed highly conserved sequences within the families, thus to facilitate visualization we only include one member of each family and the two Byrsonima species in Fig. 2. Overall, the comparative genomic analyses showed low sequence divergence between the two Byrsonima species. The highest levels of divergence were found in intergenic regions, namely psbK-psbI, trnS-trnR, rpoC1-rpoC2, trnY-trnE, accD-psaI, psaJ-rpl33 and clpP intronII. Apart from these regions, when comparing the nine species from different families, we observed some coding sequences with low similarity levels (below 70%): accD, matK, rpoA, ycf2, ycf1 and rps7. Phylogenetic relationships within the order, reconstructed using these coding regions showed a concordant topology and boostrap values similar to the results obtained with complete chloroplasts, derived from all 1–1 orthologs (62 groups) (Fig. 3).
Figure 2.
Comparisons of percentage identity of chloroplast genomes for six species belonging to five different families within the order Malpighiales. Bc: Byrsonima coccolobifolia; Br: Byrsonima crassifolia (Malpighiaceae); Ci: Chrysobalanus icaco (Chrysobalanaceae); Vs: Viola seoulensis (Violaceae); Pa: Populus alba (Salicaceae), Rc: Ricinus communis (Euphorbiaceae). The percentage of identity is shown in the vertical axis, ranging from 50% to 100%, while the horizontal axis shows the position within the chloroplast genome. Each arrow displays the annotated genes and direction of their transcription in the reference genome (Byrsonima coccolobifolia). Genome regions are color coded as exon, untranslated region (UTR), conserved noncoding sequences (CNS) and mRNA.
Figure 3.
Maximum likelihood trees for the order Malpighiales inferred from complete chloroplast genomes of nine species of the order (using all putative 1–1 orthologs - right) and from five highly variable coding sequences identified in this study (accD, matK, rpoA, ycf2 and rps7 - left). Bootstrap values are indicated above branches.
Evolutionary rates varied widely among genes across the nine Malpighiales species analyzed (Supplementary Table S2). In general, the Ka/Ks values were lower than 0.5 for almost all genes (ca. 90%). Six genes related to photosynthesis (psbD, psbE, psbF, psbL, psbN and psbT) presented the lowest evolutionary rates (Ka/Ks = 0.0002 to 0.07), exhibiting a uniform rate across most of the species evaluated. Nineteen genes returned Ka/Ks rates higher than 0.5 and lower than 1 in at least one of the species. The genes rps14, psaI, cemA, rpl23, ycf2, ycf15, ycf68 and ycf1 showed Ka/Ks rate higher than 0.5 and lower than 1 for three or more species. The genes matK, clpP, infA and ccsA showed Ka/Ks values higher than 1 for one species and other five genes (atpE, ycf15, ycf68, orf42 and ycf1) presented these high rates for at least two species. The two Byrsonima species showed similar substitution rates and Ka/Ks ratio for most genes (ca. 77%), except for 25 genes that showed differences in Ka/Ks ratio higher than 5%. Fifteen of these genes (rps4, ndhJ, rbcL, accD, cemA, clpP, psbJ, petD, rps11, rpl22, rpl2, ycf68, orf56, ccsA and ndhI) were evolving faster in B. coccolobifolia than in B. crassifolia, on the other hand, ten genes (rps16, rpoC1, ndhK, atpE, rpoA, rps3, rps7, ndhF, rpl32 and ndhA) were evolving faster in B. crassifolia.
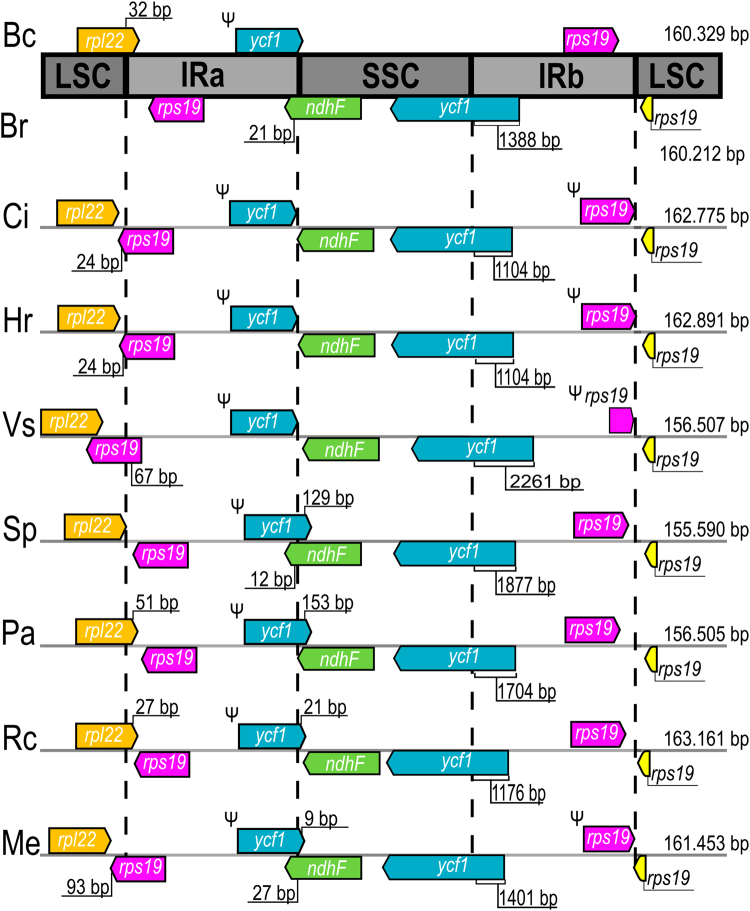
Figure 4 shows a comparison between boundary regions of the chloroplast genome of species in the order Malpighiales. The position of the SSC/IRb junction in all compared species is found within the ycf1 gene, therefore creating a pseudogene of the 5′ end of this gene (ycf1Ψ) in the IRa region. The ycf1Ψ size varies from 1,104 bp (in Chrysobalanus icaco L. and Hirtella racemosa Lam.) to 2,261 bp (Viola seoulensis Nakai). In both Byrsonima species the ycf1Ψ size was the same length, 1,388 bp. Regarding the LSC/IRa borders in B. coccolobifolia and B. crassifolia, they are located in the 3′ end of rpl22 gene, duplicating 32 nucleotides of this gene in the IRb. Populus alba L. and Ricinus communis L. showed the same pattern as the Byrsonima species. In C. icaco, H. racemosa and Manihot esculenta Crantz the location of LSC/IRa junction is in the 3′ end of the rps19 gene, thus creating an rps19 pseudogene in the IRb region. Salix purpurea L. presents the LSC/IRa boundary in the intergenic space between rpl22 and rps19. In V. seoulensis the limit between LSC and IR regions is located in the 5′ end of the rps19 gene, turning the gene copy in the IRb region into a pseudogene of 67 bp.
Figure 4.
Details of boundary positions between inverted repeat regions (IR) and large and small single copy regions (LSC and SSC) among nine chloroplast genomes within the order Malpighiales. Bc: Byrsonima coccolobifolia; Br: B. crassifolia (Malpighiaceae); Ci: Chrysobalanus icaco; Hr: Hirtela racemosa (Chrysobalanaceae); Vs: Viola seoulensis (Violaceae); Sp: Salix purpurea, Pa: Populus alba (Salicaceae), Rc: Ricinus communis, Me: Manihot esculenta (Euphorbiaceae). Both Byrsonima species sequences are represented together at the top of the figure given that there are no differences between their boundaries. The direction of arrows shows the direction of transcription (right is forward and left is reverse). Ψ indicates a pseudogene. Length of arrows is illustrative. Number of base pairs (bp) indicates distance from the boundary to the end of the gene. Complete chloroplast genome sizes are noted on the right-hand side of the panel.
Repeated sequences analysis of Byrsonima species
The IMEx software34 found 427 small single repeats (SSR) in the B. coccolobifolia chloroplast genome and 414 in B. crassifolia (Supplementary Table S3). Most of the SSR discovered were mononucleotide repetitions (ca. 79%), varying from seven to 16 nucleotides long. About 57% of the SSR were mononucleotides sequences containing repetitions of adenine (A) or thymine (T). Repeats of di- and tri-nucleotides were also abundant, representing together 20% of the SSR found for both species. For dinucleotide SSR, the number of repeats ranged from four to seven, but for tri-, tetra- and penta-nucleotide SSRs, they had mostly three motif repetitions, except for two sequences with four repeats (Supplementary Table S3). The REPuter35 screening discovered three categories of dispersed repeats: forward (F), palindrome (P) and reverse (R) (Table 3). In the B. coccolobifolia chloroplast genome we found 15 repeats (F = 6; P = 8; R = 1) and 19 in B. crassifolia (F = 9; P = 9; R = 1), with motif length ranging from 30 bp to 57 bp. Most of the repeated sequences were located in the ycf2 gene (18 for each species) and intergenic spacers (IGS) (10 and 18, for B. coccolobifolia and B. crassifolia, respectively).
Table 3.
Distribution of repeated sequences in the chloroplast genome of Byrsonima coccolobifolia and B. crassifolia.
| Type | Location | Region | Repeated sequence | Size (bp) |
|---|---|---|---|---|
| F | ycf2 | IRa | ATATCGTCACTATCATCAATATCGTCACTATCATCAATATCGTCACTATCATCAATA | 57 |
| P | ycf2 | IRa/IRb | TATTGATGATAGTGACGATATTGATGATAGTGACGATATTGATGATAGTGACGATAT | 57 |
| P | ycf2 | IRa/IRb | TATTGATGATAGTGACGATATTGATGATAGTGACGATATTGATGATAGTGACGATAT | 57 |
| F | ycf2 | IRb | ATATCGTCACTATCATCAATATCGTCACTATCATCAATATCGTCACTATCATCAATA | 57 |
| P | trnQ-rps16 | LSC | AGAGATCTAATCCCATTGATTGAATTCAATCAATGGGATTAGATCTCT | 48 |
| F | trnS-trnQ* | LSC | TATACTATTAGATACTACTATATACTATTAGTATACTATTAGATACTA | 48 |
| P | petN-trnT* | LSC | AGATAGTATGGTAGAAAGAAATATATATATTTCTTTCTACCATACTAT | 48 |
| P | petA-petL | LSC | CTTTTCGATTTTATACGTATAAATTTATACGTATAAAATCGAAAAG | 46 |
| F | ycf2 | IRa | ATATCGTCACTATCATCAATATCGTCACTATCATCAATA | 39 |
| P | ycf2 | IRa/IRb | TATTGATGATAGTGACGATATTGATGATAGTGACGATAT | 39 |
| P | ycf2 | IRa/IRb | TATTGATGATAGTGACGATATTGATGATAGTGACGATAT | 39 |
| F | ycf2 | IRb | ATATCGTCACTATCATCAATATCGTCACTATCATCAATA | 39 |
| R | rbcL-accD | LSC | AGAATTAAGAGAATTAAAATTAAGAGAATTAAGA | 34 |
| F | psaB and psaA | LSC | ACCGATATTGCACACCATCATTTAGCTATTGCA | 33 |
| P | petN-psbM | LSC | TTTAATTTAAATTGAATTCAATTTAAATTAAA | 32 |
| P | trnR-trnS and ycf2 | LSC/IRa | ATATATGTTTGGAATAGATTCCATTTTGAGA | 31 |
| F | trnR-trnS and ycf2 | LSC/IRa | TCTCAAAATGGAATCTATTCCAAACATATAT | 31 |
| F | psbK-psbI* | LSC | ATACTATTAGATACTACTATATACTATTAG | 30 |
| F | psbK-psbI* | LSC | ATACTATTAGATACTACTATATACTATTAG | 30 |
*Repeats that appear only in B. crassifolia. Types of repeats are F (forward), P (palindrome) and R (reverse).
Highly divergent regions between Byrsonima species
The level of divergence between the two Byrsonima species was variable depending on the region of the chloroplast compared (Supplementary Fig. S2), with nucleotide diversity (π) ranging from 0.000345 (rpoB gene) to 0.065574 (atpA-atpF intergenic spacer). The IGS showed higher average π (0.002664) than the protein coding (0.000623), intronic (0.000895) and tRNA regions (which proved to be very conserved, π = 0). Among the 20 regions with the highest values of π (all > 0.005), 18 were IGS and only two were protein coding genes (Table 4). Some regions exhibited neighboring sequences with high π values (Supplementary Fig. S2). Thus, we calculated the divergence values for the combined tandem sequences. Among these tandems, one region of 625 bp between the genes rpoA and rpl36 exhibited a high π value (0.011475 – Table 4). The gene ycf1 showed no divergence between the two Byrsonima species (π = 0), whereas its ycf1Ψ pseudogene, located in the IRb, had a π of 0.002747, higher than the average for IGS.
Table 4.
Twenty most divergent regions of chloroplast genome based on a comparison between Byrsonima coccolobifolia Kunth and B. crassifolia (L.) Kunth.
| Region | Nucleotide diversity (π) | Total number of mutations (η) | Region length (bp) |
|---|---|---|---|
| atpA-atpF | 0.065574 | 4 | 61 |
| ccsA-ndhD | 0.040000 | 10 | 250 |
| rpoA-rps11 | 0.029851 | 2 | 80 |
| psbT-psbN | 0.015385 | 1 | 65 |
| trnH-GUG-psbA | 0.014337 | 4 | 279 |
| psbI-trnS-GCU | 0.011765 | 1 | 85 |
| trnG-UCC-trnfM-CAU | 0.011765 | 2 | 172 |
| rpoA-rps11-rpl36 | 0.011475 | 7 | 625 |
| psbZ-trnG-UCC | 0.011050 | 6 | 712 |
| rps11 | 0.009639 | 4 | 417 |
| psaI-ycf4 | 0.008869 | 4 | 453 |
| rpl32-trnL-UAG | 0.007874 | 3 | 381 |
| rps11-rpl36 | 0.007813 | 1 | 128 |
| rpl14-rpl16 exon II | 0.007246 | 1 | 139 |
| trnK-UUU-rps16 | 0.006289 | 3 | 518 |
| psaJ-rpl33 | 0.005859 | 3 | 555 |
| petD-rpoA | 0.005682 | 1 | 176 |
| matK-trnK-UUU | 0.005587 | 4 | 716 |
| rpl32 | 0.005556 | 1 | 184 |
| rps16-trnQ-UUG | 0.005178 | 8 | 1,575 |
Discussion
In this study, the whole chloroplast genomes of Byrsonima coccolobifolia and B. crassifolia were sequenced and analyzed. The comparative analysis of these genomes and other species of the order Malpighiales has brought insights about chloroplast genome evolution in this order. Moreover, this study identified sequences suitable for use in future evolutionary studies in the order Malpighiales, in the family Malpighiaceae and in the genus Byrsonima, in order to clarify phylogenetic relationships and resolve taxonomic uncertainties.
Although gene content and organization were generally similar in the species analyzed within the order Malphighiales, some striking differences were found among them. One remarkable variation among the species analyzed is the presence or absence of three protein coding genes. The rps16 and rpl32 genes were absent in the single Violaceae species analyzed (V. seoulensis) and also in the Salicaceae family (P. alba and S. purpurea). The gene infA was lacking in both species of Malpighiaceae, B. coccolobifolia, B. crassifolia, in V. seoulensis and in one of the two species of Euphorbiaceae, M. esculenta. Thus, the evolutionary change leading the absence/presence of infA gene in the chloroplast genome even within a family appears to have occurred several times within the order Malpighiales. The absence of some genes, including these three particular genes, has been described in other plant species36–40. Some studies have shown that infA41, rpl3238 and rps1642 genes that were missing in the chloroplast genome of certain species have been transferred to the nuclear genome. Further investigation will be needed to check if the three genes lacking in the chloroplast genome of these Malpighiales species analyzed were transferred to another genome compartment or were completely lost.
Another important characteristic of the chloroplast genome that is useful for evolutionary studies is the location of the boundaries among the four chloroplast regions. Evaluating their contraction and expansion can shed some light on the evolution of some taxa32. From our results we noticed that the length variation in the IR regions created some pseudogenes, like the ycf1Ψ or rps19Ψ. The ycf1Ψ pseudogene is present in all studied species, whereas the rps19Ψ pseudogene is only present in C. icaco, H. racemosa (Chrysobalanaceae), V. seoulensis (Violaceae) and M. esculenta (Euphorbiaceae); in the other Malpighiales species the rps19 gene is fully duplicated. Thus, the contraction/expansion of IR regions, creating pseudogenes, has occurred more than once in the order Malpighiales.
Even though, as expected, sequence divergence among families was higher than within a family, in general, the chloroplast genomes within Malpighiales are still conserved, as observed in other flowering plants2. High levels of divergence among families were found for the accD, matK, rpoA, ycf2, ycf1 and rps7 genes. Most of these sequences have already been used for phylogenetic studies43–46, and our analyses showed that these regions (excluding ycf1) were in fact very informative for inferring phylogenetic relationships within the order, with results comparable to those obtained from complete chloroplast genomes. Moreover, these topologies were concordant with the most complete phylogenetic study performed for the order so far5. These results highlight the utility of the highly divergent regions identified herein for phylogenetic inference in the Malpighiales order.
The slow evolutionary rates and the low Ka/Ks ratio detected in the Malpighiales species analyzed are expected for chloroplast genomes in general47. The genes with the lowest evolutionary rates were photosynthesis genes (psbD, psbE, psbF, psbL, psbN and psbT), an evolutionary pattern common in photosynthetic plants48. Among the genes with highest evolutionary rates ycf1, ycf15 and ycf68 do not have a known function and its high Ka/Ks ratio may show that they play a non-essential role in plant cells. These results, together with the differences found between the two Byrsonima species in Ka/Ks ratios for 25 genes, are evidence that evolutionary rates in the chloroplast genome in Malpighiales vary strongly among genes and lineages.
Repetitive sequences have been reported in the chloroplast genome of many plant lineages49,50. These types of markers are used for a wide range of evolutionary and population genetic studies51,52. Byrsonima coccolobifolia and B. crassifolia showed the same motifs of SSR markers, but in general the B. coccolobifolia chloroplast genome presented more SSR loci than B. crassifolia. In terms of dispersed repeats, both species shared most of the repeated sequences, but three repeats were found only in B. crassifolia. Interestingly, dispersed repeats were found mainly in protein coding sequences, and 18 (of the 30 repeats in B. coccolobifolia and 36 in B. crassifolia) were contained in the ycf2 genes, whereas other two were found in the psaA and psaB genes. This result does not follow the tendency of organelar genomes, since most repeated sequences in chloroplast genomes are located in intergenic sequences53–55. However, a greater amount of dispersed repeats was also found in coding sequences in five species of the genus Epimedium L. (Berberidaceae7).
Based on the comparison of nucleotide diversity among regions between the two Byrsonima species analyzed, we suggest a set of 20 regions with high divergence, most of them intergenic sequences, to be used as a starting point for investigating potential markers for phylogenetic and phylogeographic studies in the genus Byrsonima. Until now, there has been no phylogenetic study of this genus, and taxonomic uncertainties still remain18. To look for polymorphic sequences in the chloroplast of some species is usually very time-consuming when no previous chloroplast genome information is available. In fact, a recent study by our group56 observed only three polymorphic regions after testing 15 of the most commonly used chloroplast regions for a phylogeographic study in B. coccolobifolia populations. The lack of available sequences for these regions hindered us from testing their utility in a phylogenetic context, but we expect that the highly divergent sequences identified here by comparison of B. cocolobifolia and B. crassifolia chloroplast genomes will offer new tools for genetic and evolutionary studies in species of this genus and of the Malpighiaceae family.
Material and Methods
Sample material and sequencing
Samples used in the study were collected in Amazonian savanna enclaves: Byrsonima coccolobifolia (voucher BHCB 169523) from Boa Vista (60°49′45′′W, 2°39′40′′N) and B. crassifolia (voucher BHBC 169445) from Alto Alegre (61°09′04′′W, 3°09′45′′N). Voucher specimens were deposited in BHCB herbarium (Herbarium of Departamento de Botânica, Universidade Federal de Minas Gerais). Genomic DNA was extracted from silica-dried leaves, using Novaes et al.57 protocol. DNA quality was assessed in a spectrophotometer Nanodrop 2000 (Thermo Scientific) and integrity was evaluated using a 0.8% agarose gel. In addition, DNA was quantified through fluorometry using Qubit 2.0. (Life Technologies). DNA samples from each species were used to prepare two separate libraries with Nextera kit (Illumina Inc., San Diego, CA), following manufacturer’s protocol. Different barcodes were used to identify DNA fragments derived from each species. To guarantee the intended fragments size, aliquots of each library were ran in 1% agarose gel and quantified by quantitative PCR, using a Library Quantification Kit – Illumina/Universal (Kapa Biosystems Inc., Wilmington, MA). Short fragments of approximately 600 bp from both libraries were combined and submitted for paired-end sequencing using a single lane on a MiSeq sequencer (Illumina Inc.).
Genome assembly and annotation
Raw sequences were submitted to the Sequence Read Archive (SRA accession number SRP109225). Pair-end Illumina raw reads where cleaned from adaptors and barcodes and then quality filtered using Trimmomatic58. Reads were trimmed from both ends, and individual bases with Phred quality score < 20 were removed, as well as more than three consecutive uncalled bases. Entire reads with a median quality score lower than 21 or less than 40 bp in length after trimming were discarded. After quality filter, reads were mapped to the chloroplast genome of the closest species with a chloroplast genome available (Chrysobalanus icaco L. – Chrysobalanaceae Juss.), using Bowtie2 v.2.2.659 in order to exclude reads of nuclear and mitochondrial origins. All putative chloroplast reads mapped to the Chrysobalanus reference above were then used for de novo assembly to reconstruct Byrsonima chloroplast genomes using SPAdes 3.6.160 with iterative K-mer sizes of 55, 87 and 121. De novo assembled chloroplast contigs were concatenated into larger contigs using Sequencher 5.3.2 (Gene Codes Inc., Ann Arbor, MI, USA) based on at least 20 bp overlap and 98% similarity. A “genome walking” technique using the Unix “grep” function was able to find any remaining reads that could fill any gaps between contigs that did not assemble in the initial set of analyses. Read coverage analysis was then conducted to determine the inverted repeat (IR) region boundaries and any misassembled contigs using Jellyfish v.2.2.361 and pipeline developed by M. McKain (https://github.com/mrmckain/Chloroplast-Genome-Assembly/tree/master/Coverage_Analysis).
Automatic annotation of B. coccolobifolia and B. crassifolia chloroplast genomes were generated by CpGAVAS62 and a circular representation of both sequences was drawn using the online tool GenomeVX63. The draft annotations given by CpGAVAS were then manually corrected using the Artemis software64 and other plastid genomes for comparison. The complete chloroplast genomes of B. coccolobifolia and B. crassifolia were automatically annotated and aligned in Verdant65. Differences between results from CpGAVAS and Verdant were manually confirmed and investigated in GenBank when necessary. Open reading frames identified by these softwares were reported when sequences followed two criteria: (1) have been described previously in other chloroplast genomes32,66, (2) were homologous to known genes (using the BLAST tool from GenBank). The complete chloroplast genome sequence and final annotations for both species were submitted to GenBank under the following accession numbers: MF359247 (B. coccolobifolia) and MF359248 (B. crassifolia).
Comparative analyses and evaluating regions of high divergence
Aiming to perform a comparative genomic analysis within the order Malpighiales, we chose two species of each family in the order with chloroplast genomes available on NCBI database: Euphorbiaceae, Chrysobalanaceae, Salicaceae and Violaceae (which had only one genome currently published – supplementary Table S4). Then, we used the software mVISTA in Shuffle-LAGAN mode33, with default parameters for other options, to compare the chloroplast genomes from the five different plant families, using the newly sequenced B. coccolobifolia annotated genome as a reference. In order to detect expansion or contraction of the IR regions boundaries between the four main parts of the annotated chloroplast genomes (LSC, IRa, SSC and IRb) were visually inspected among the nine species in the order Malpighiales using the Artemis software64.
The protein coding regions of these same nine chloroplast genomes were used to evaluate evolutionary rate variation within Malpighiales. For that, we calculated the rates of non-synonymous (Ka) and synonymous substitutions (Ks), as well as their ratio (Ka/Ks) using Model Averaging in the KaKs_Calculator67. In this instance, the Malpighiales plant species Passiflora edulis Sims (NC_034285.1) was used as a reference, and alignments of the protein-coding sequences (without stop codons) from the nine species were performed using the MUSCLE68 program in Mega 769.
Further comparisons between Byrsonima species were performed with the repetitive elements found in their chloroplast sequences. To analyze the presence of perfect microsatellites we used the Imperfect Microsatellite Extractor (IMEx) interface34, with minimum thresholds of seven for mononucleotide repeats, four for dinucleotide repeats and three for tri-, tetra-, penta-, and hexanucleotide repeats. REPuter software35 was used to detect tandem repeats. We set the parameters to localize forward, reverse, complementary and palindromic sequences with a minimum distance of 30 bp and 90% minimum identity.
In order to identify regions of high genetic divergence between Byrsonima species that could potentially be informative for future phylogenetic studies in the genus, we calculated the genetic divergence between B. coccolobifolia and B. crassifolia across the entire chloroplast genome. Genetic divergence was calculated using nucleotide diversity (π) and total number of mutations (η) for coding genes, intron sequences and intergenic spacers (IGS) aligned with Verdant, using DnaSP 5.070.
Phylogenetic analysis
Phylogenetic relationships within the Malpighiales order were reconstructed using the complete set of species sampled in our comparative analysis (seven species available in NCBI plus the two Byrsonima described in our study) and two species of different orders as outgoup, Averrhoa carambola and Arabidopisis thaliana (KU569488.1 and NC000932.1, respectively). In order to evaluate the usefulness of the highly variable chloroplast regions identified within Malpighiales by mVista (accD, matK, rpoA, ycf2, ycf1 and rps7), we compared phylogenies inferred from two matrices: one using five of these highly variable sequences and other using putative 1–1 orthologs genes within Malpighiales order. Because the highly variable sequence ycf1 showed some inversions that hindered the alignment, we excluded this region of the phylogenetic analysis.
The highly variable sequences were extracted separately for each species, aligned using MUSCLE68 and concatenated to generate a matrix for the first input file. To create the 1–1 orthologs genes file, we extracted coding sequences from complete chloroplast genomes of 11 species and translated them using in house Perl scripts (available from the authors upon request). The protein sequences were used as input to OrthoMCL2 to predict homology relationships71. The groups of homologs that are present in one copy in all predicted chloroplast proteomes were considered as putative 1–1 orthologs (62 groups) and were individually aligned using MUSCLE68. We used the aligned protein sequences for each group to generate codon alignments using PAL2NAL72. Finally, we took the aligned codon sequences for each genome and concatenate them to generate a gene matrix that was used to create the second input file. Both alignment were verified and edited manually. The program PartitionFinder73 was used to identify the best-fit partitioning schemes and suitable evolution model for phylogeny estimation of each matrix. Finally, the best trees were inferred from Maximum likelihood (ML) analyses, performed with RAxML 8.3.274 in CIPRES Science Gateway75, using GTR + G model and 1000 rapid bootstrap replications for each matrix.
Data availability
The complete chloroplast sequences generated and analysed during the current study are available in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession numbers are described in the text).
Electronic supplementary material
Acknowledgements
This work was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We would like to thank M. McKain for use of unpublished scripts for chloroplast genome assembly and coverage analysis. Computer analyses were performed using the Smithsonian OCIO Hydra-3 computer cluster.
Author Contributions
M.B.L. and L.C.R.-M. conceived and designed research. E.K. and M.B.L. provided financial resources to research. A.P.A.M., L.C.R.-M. and E.K. conducted experiments. A.P.A.M., L.C.R.-M., R.S.O.B., A.G.N., M.C. and F.P.L. did computational analyses. A.P.A.M., L.C.R.-M., R.S.O.B. and M.B.L. analysed data. A.P.A.M., L.C.R.-M., A.G.N., R.S.O.B. and M.B.L. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Alison P. A. Menezes, Luciana C. Resende-Moreira and Renata S. O. Buzatti contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20189-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cooper, G. M. Chloroplasts and other plastids in The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates. Available from: https://www.ncbi.nlm.nih.gov/books/NBK9905/ (2000).
- 2.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer, J. D. Plastid chromosomes: structure and evolution in cell culture and somatic cell genetics of plants (ed. Bogorad L. K., Vasil, I.). San Diego (CA): Elsevier (1991).
- 4.Jheng, et al. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 2012;190:62–73. doi: 10.1016/j.plantsci.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Xi, et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. 2012;109:17519–17524. doi: 10.1073/pnas.1205818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Yao X, Tan Y, Gan Y, Corlett RT. Complete chloroplast genome sequence of the avocado: gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can. J. For. Res. 2016;46:1293–1301. doi: 10.1139/cjfr-2016-0199. [DOI] [Google Scholar]
- 7.Zhang, et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, et al. Plant DNA barcoding: from gene to genome. Biol. Rev. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 9.Hollingsworth PM, Li D-Z, van der Bank M, Twyford AD. Telling plant species apart withDNA: from barcodes to genomes. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20150338. doi: 10.1098/rstb.2015.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Chen X, Feng X, Woeste KE, Zhao P. Characterization of the complete chloroplast genome of the endangered species Carya sinensis (Juglandaceae) Conserv. Genet. Resour. 2016;6:1–4. [Google Scholar]
- 11.APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121 (2009).
- 12.Wurdack KJ, Davis CC. Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 2009;96:1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 13.Araújo JS, Azevedo AA, Silva LC, Meira RMSA. Leaf anatomy as an additional taxonomy tool for 16 species of Malpighiaceae found in the Cerrado area (Brazil) Plant Syst. Evol. 2010;286:117–131. doi: 10.1007/s00606-010-0268-3. [DOI] [Google Scholar]
- 14.Anderson WR. Floral conservatism in Neotropical Malpighiaceae. Biotropica. 1979;11:219–223. doi: 10.2307/2388042. [DOI] [Google Scholar]
- 15.Anderson WR. The origin of the Malpighiaceae - The evidence from morphology. Mem. N. Y. Bot. Gard. 1990;64:210–224. [Google Scholar]
- 16.Davis CC, Anderson WR, Donoghue MJ. Phylogeny Malpighiaceae: Evidence from chloroplast ndhF and trnL-F nucleotide sequences. Am. J. Bot. 2001;88:1830–1846. doi: 10.2307/3558360. [DOI] [PubMed] [Google Scholar]
- 17.Davis CC, Anderson WR. A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Am. J. Bot. 2010;97:2031–2048. doi: 10.3732/ajb.1000146. [DOI] [PubMed] [Google Scholar]
- 18.Elias, S. Revisão Byrsonima subg. Macrozeugma Nied. (Malpighiaceae). PhD Thesis. University of São Paulo (2004).
- 19.Byrsonima in Flora do Brasil. Jardim Botânico do Rio de Janeiro. http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB8827 (2020 under construction).
- 20.Amorim, A. M., Kutschenko, D. C., Judice, D. M. & Barros, F. S. M. Malpighiaceae. In: G. Martinelli & M. A. Moraes (orgs), Livro vermelho da flora do Brasil. Vol. 1. Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, Rio de Janeiro, p. 648–654 (2013).
- 21.Anderson, W. R. Malpighiaceae, in: The botany of the Guayana Highland, Part XI. pp. 21–305 (1981).
- 22.Niendenzu, F. Malpighiaceae, in: Engler, A. (Ed.), Die Natürlichen Pflanzenfamilien. Leipizig, pp. 41–74 (1897).
- 23.Lorenzi, H. Árvores brasileiras: manual de identificação e cultivo de plantas arbóreas nativas do Brasil, vol 2, Plantarum. Nova Odessa, SP (1998).
- 24.Lorenzi, H. Árvores brasileiras: manual de identificação e cultivo de plantas arbóreas nativas do Brasil, vol 3, Plantarum. Nova Odessa, SP (2009).
- 25.Ratter JA, Bridgewater S, Ribeiro JF. Analysis of the floristic composition of the Brazilian cerrado vegetation III: Comparison of the woody vegetation of 376 areas. Edinburgh J. Bot. 2003;60:57–109. doi: 10.1017/S0960428603000064. [DOI] [Google Scholar]
- 26.Missouri Botanical Garden. TROPICOS Specimen Data Base. http://mobot 1.mobot.org/website (2002).
- 27.Anderson, W. R. Byrsosinimoideae, a new subfamily of Malpighiaceae. Leandra (1977).
- 28.Gao L, Yi X, Yang Y-X, Su Y-J, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol. Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang, et al. The Complete Chloroplast Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. PLoS One. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, et al. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016;17:820. doi: 10.3390/ijms17060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen RK, Saski C, Lee S, Hansen AK, Daniell H. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers ofrpl22 to the nucleus. Mol. Biol. Evol. 2011;28:835–847. doi: 10.1093/molbev/msq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nazareno AG, Carlsen M, Lohmann LG. Complete Chloroplast Genome of Tanaecium tetragonolobum: The first Bignoniaceae plastome. PLoS One. 2015;10:e0129930. doi: 10.1371/journal.pone.0129930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudunuri SB, Nagarajaram HA. IMEx: Imperfect Microsatellite Extractor. Bioinformatics. 2007;23:1181. doi: 10.1093/bioinformatics/btm097. [DOI] [PubMed] [Google Scholar]
- 35.Kurtz, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JS, Kim JH. Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea) PLoS One. 2013;8:e68180. doi: 10.1371/journal.pone.0068180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei, et al. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 2016;6:21669. doi: 10.1038/srep21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S, Jansen RK, Park S. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of therpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 2015;15:40. doi: 10.1186/s12870-015-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson, et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat 1. Am. J. Bot. 2015;102:1115–1127. doi: 10.3732/ajb.1500184. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz et al. 2015. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 53, 458–468 (2015).
- 41.Millen, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda, et al. Substitution of the gene for chloroplastrps16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- 43.Huang JL, Sun GL, Zhang DM. Molecular evolution and phylogeny of the angiosperm ycf2 gene. J. Syst. Evol. 2010;48:240–248. doi: 10.1111/j.1759-6831.2010.00080.x. [DOI] [Google Scholar]
- 44.Domenech, et al. A phylogenetic analysis of palm subtribe Archontophoenicinae (Arecaceae) based on 14 DNA regions. Bot. J. Linn. Soc. 2014;175:469–481. doi: 10.1111/boj.12179. [DOI] [Google Scholar]
- 45.Luo, et al. Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS One. 2014;9:e99016. doi: 10.1371/journal.pone.0099016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bodin SS, Kim JS, Kim JH. Phylogenetic inferences and the evolution of plastid DNA in Campynemataceae and the Mycoheterotrophic Corsia dispar D.L. Jones & B. Gray (Corsiaceae) Plant Mol. Biol. Report. 2016;34:192–210. doi: 10.1007/s11105-015-0914-6. [DOI] [Google Scholar]
- 47.Redwan RM, Saidin A, Kumar SV. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015;15:196. doi: 10.1186/s12870-015-0587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edh K, Widén B, Ceplitis A. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae) Mol. Ecol. 2007;16:4972–4983. doi: 10.1111/j.1365-294X.2007.03585.x. [DOI] [PubMed] [Google Scholar]
- 50.Choi KS, Chung MG, Park S. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): Comparative analysis and highly divergent regions. Front. Plant Sci. 2016;7:355. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong Y-Q, Gong X. Pollen-mediated gene flow promotes low nuclear genetic differentiation among populations of Cycas debaoensis (Cycadaceae) Tree Genet. Genomes. 2016;12:93. doi: 10.1007/s11295-016-1051-6. [DOI] [Google Scholar]
- 52.Roy, et al. Nuclear and chloroplast DNA variation provides insights into population structure and multiple origins of native aromatic rices of Odisha, India. PLoS One. 2016;11:e0162268. doi: 10.1371/journal.pone.0162268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raubeson, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Yang S, Li H, Yang J, Li D. Comparative chloroplast genomes of Camellia species. PLoS One. 2013;8:e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wuyun T, Du H, Wang D, Cao D. Complete chloroplast genome sequences of Eucommia ulmoides: genome structure and evolution. Tree Genet. Genomes. 2016;12:12. doi: 10.1007/s11295-016-0970-6. [DOI] [Google Scholar]
- 56.Resende-Moreira, et al. East-west divergence in central Brazilian Cerrado revealed by cpDNA sequences of a bird-dispersed tree species. Biochem. Syst. Ecol. 2017;70:247–253. doi: 10.1016/j.bse.2016.12.007. [DOI] [Google Scholar]
- 57.Novaes RML, Rodrigues JG, Lovato MB. An efficient protocol for tissue sampling and DNA isolation from the stem bark of Leguminosae trees. Genet. Mol. Res. 2009;8:86–96. doi: 10.4238/vol8-1gmr542. [DOI] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu, et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conant GC, Wolfe KH. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24:861. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 64.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKain MR, Hartsock RH, Wohl MM, Kellogg EA. Verdant: automated annotation, alignment and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 2017;33:130–132. doi: 10.1093/bioinformatics/btw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Do HDK, Kim JS, Kim J-H. Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae) Gene. 2013;530:229–235. doi: 10.1016/j.gene.2013.07.100. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Geno. Prot. Bioinfo. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Librado P, Rozas J. DnaSPv5: software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Stoeckert CJ, Jr., Roos. DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 74.Stamatakis A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environment Workshop (GCE), 14 Nov. 2010, New Orleans, 1–8 (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast sequences generated and analysed during the current study are available in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession numbers are described in the text).