Abstract
The microbiota fulfils a key role in the training and function of the immune system, which contributes to the symbiosis between the host and complex microbial communities. In this study, we characterized the interplay between vaginal bacteria and local immune mediators during dysbiosis in selected women of reproductive age who were grouped according to Nugent’s criteria. The abundance of Gardnerella vaginalis and Bifidobacterium breve was increased in the intermediate dysbiotic status, while the presence of a plethora of non-resident bacteria characterized the group with overt vaginosis. In response to these increases, the anti-inflammatory IL1ra and pro-inflammatory IL2 increased, while the embryo trophic factors FGFβ and GMCSF decreased compared to the healthy milieu. A specific pattern, including IL1α, IL1β, IL8, MIG, MIP1α and RANTES, distinguished the intermediate group from the vaginosis group, while IL5 and IL13, which are secreted by Th2 cells, were significantly associated with the perturbation of the commensals Lactobacilli, Gardnerella and Ureaplasma. Summarizing, we postulate that although the dysbiotic condition triggers a pro-inflammatory process, the presence of a steady state level of Th2 may influence clinical manifestations. These results raise clinically relevant questions regarding the use of vaginal immunological markers as efficacious tools to monitor microbial alterations.
Introduction
The female reproductive tract harbours a symbiotic community that consists of host cells and bacterial communities1. Perturbation of the vaginal microbiota is associated with a large spectrum of urogenital diseases that affect many aspects of woman’s lives, including fertility and acquisition of sexually transmitted infections2–5. Indeed, the vaginal mucosa is susceptible to the entry of pathogens, where both local immune mediators and commensal microorganisms are the first line of host defence6–8.
Bacterial vaginosis (BV) is the most common vaginal dysmicrobism, diagnosed in 20–30% of women of reproductive age, with a prevalence of 50–60% in some high-risk sexual behaviour populations9. BV is caused by a reduction of the commensal Lactobacillus spp., elevated pH and a concomitant overgrowth of several strict or facultative anaerobic bacteria10. Thus, BV is a polymicrobial disease11–13 in which a single pathogen is not able to induce the disease. The predominance of Lactobacillus spp., as well as the presence of other lactic acid-producing bacteria, appears to be pivotal to counteract the growth of BV-associated bacteria and to reduce the susceptibility to other infections14–16.
Although approximately half of BV cases are clinically asymptomatic17, an increase of local pro-inflammatory effectors has been observed in women with BV18,19. Recent studies have reported the existence of biotic signals that are conducive to a vaginal protective function20. In this network, cytokines are interspecies communication molecules that coordinate the interactions of vaginal communities, including the growth of pathogenic microorganisms21,22. However, the clinical impact of the modulation of cytokines on vaginal dysbiosis remains to be elucidated.
We profiled the vaginal microbiome and the local immune fluctuation in selected women of reproductive age grouped according to the Nugent’s criteria with the goal of investigating the role of local immune mediators during the vaginal dysbiotic process.
In women with an intermediate Nugent score, we observed that the overgrowth of Gardnerella vaginalis was balanced by a high colonization of Bifidobacterium breve, which, overall, was able to guarantee a healthy vaginal equilibrium. Conversely, in women with BV, we observed a massive increase of non-resident vaginal species that paralleled a low presence of G. vaginalis.
The local immune response of the host was specifically stimulated by the vaginal dysbiosis, with a simultaneous increase of the anti-inflammatory IL1ra and the pro-inflammatory IL2 and a decrease of the embryo trophic factors FGFβ and GMCSF. Moreover, a specific pattern of mediators, including IL1α, IL1β, IL8, MIG, MIP1α and RANTES, distinguished the grade of vaginal dysbiosis observed in women with an intermediate Nugent score and in women with overt vaginosis.
Among the tested immune mediators, a significant association of the modulation of specific resident bacterial communities was highlighted for IL5 and IL13. The increase in the concentration of these two anti-inflammatory proteins was accompanied by a depletion of Lactobacilli ssp., G. vaginalis and Ureaplasma spp. The concomitant increase of the anti-inflammatory IL5 and IL13, secreted by Th2 cells, and the establishment of a vaginal dysmicrobism suggests a role for the Th2 activation in counteracting the Th1 response and in counteracting the presence of clinical symptoms.
Results
Characterization of women with vaginal dysbiosis
To characterize the vaginal dysbiotic process, we collected 62 cervical-vaginal samples from women who were diagnosed according to Nugent’s criteria, including 30 women with a Nugent score of 0–3 (Healthy), 15 women with Nugent score of 4–6 (Intermediate) and 17 women with Nugent score of 7–10 (Vaginosis). The sequencing of the vaginal microbial community yielded 3,496,234 high-quality reads, excluding samples with less than 10,000 reads, with an average of 53,788 reads/sample (range 10,531–190,162). We identified 15,769 operational taxonomic units (OTUs) across all the samples.
Bacterial diversity and comparisons of alpha diversity between cohorts
Out of the five distinct alpha diversity metrics (within-sample diversity), the Simpson, Shannon, observed species and PD_whole_tree metrics significantly characterized the dysbiotic vaginal microbiome (Table 1) from that of the healthy microbiome, while Chao1 did not exhibit significant variations. Overall, across the five metrics used, the intermediate and vaginosis groups showed higher alpha diversities than the healthy group.
Table 1.
Comparison of bacterial diversity between cohorts.
| Healthy | Intermediate | Vaginosis | p values | |
|---|---|---|---|---|
| Chao1 | 400.86 ± 110.87 | 498.94 ± 147.60 | 431.37 ± 128.93 | 0.051; 0.54; 1 |
| Simpson | 1.54 ± 0.56 | 3.55 ± 1.83 | 3.63 ± 2.270 | 0.003; 0.003; 1 |
| Shannon | 1.25 ± 0.6 | 2.52 ± 0.86 | 2.51 ± 0.87 | 0.003; 0.003; 1 |
| Observed species | 154.54 ± 43.08 | 214.06 ± 49.49 | 203.17 ± 53.14 | 0.003; 0.009; 1 |
| PD whole tree | 8.36 ± 1.31 | 10.03 ± 1.56 | 9.75 ± 1.99 | 0.003; 0.03; 1 |
Bacterial diversity values are given as the mean ± standard deviation at a rarefaction depth of 10,000 sequences per sample. Alpha diversity was compared between groups by means of a non-parametric t-test using the compare_alpha_diversity.py script of QIIME. The p values are shown in the last column in the following order: Healthy vs Intermediate, Healthy vs Vaginosis, and Intermediate vs Vaginosis.
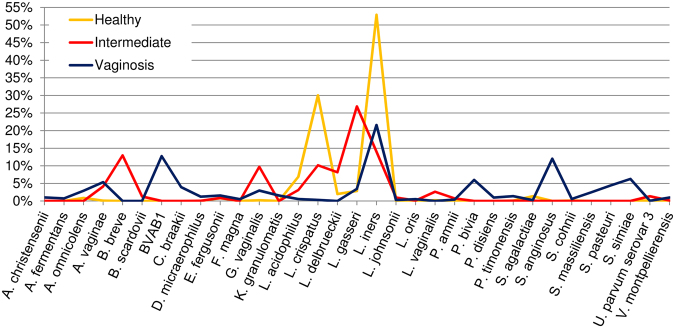
When accounting for the bacterial communities in the cohorts (Fig. 1), the intermediate and vaginosis groups showed more heterogeneity among Lactobacilli species when compared to the healthy group.
Figure 1.
The vaginal bacterial communities from patients with eubiotic and dysbiotic microbiota. The output of plot_taxa_summary.py of QIIME showing the relative abundance of the 33 predominant bacterial taxonomic groups, in alphabetical order, in the studied cohorts: Healthy (Nugent score 0–3), Intermediate (Nugent score 4–6) and Vaginosis (Nugent score 7–10).
Healthy women exhibited three predominant Lactobacilli species, namely L. acidophilus, L. crispatus and L. iners (Firmicutes), which were underrepresented in the intermediate and vaginosis groups. L. gasseri was the most abundant Lactobacillus in the intermediate group, while L. iners was the dominant Lactobacillus species in the vaginosis group.
Among the other microorganisms detected, the intermediate group showed an increase of Gardnerella vaginalis (Actinobacteria) and Ureaplasma parvum (Tenericutes) compared to the vaginosis and healthy groups. Bifidobacterium breve was only detected in the intermediate group.
The vaginosis cohort exhibited a vast change in specific species, which are shown in Table 2.
Table 2.
BV associated-bacteria.
| BV associated-bacteria |
|---|
| A. christensenii (Firmicutes) |
| A. fermentans (Firmicutes) |
| A. omnicolens (Fusobacteria) |
| A. vaginae (Actinobacteria) |
| BVAB1 |
| C. braakii (Gammaproteobacteria) |
| D. micraerophilus (Firmicutes) |
| E. fergusonii (Gammaproteobacteria) |
| F. magna (Firmicutes) |
| K. granulomatis (Gammaproteobacteria) |
| P. bivia (Bacteroidia) |
| P. disiens (Bacteroidia) |
| P. timonensis (Bacteroidia) |
| S. anginosus (Firmicutes) |
| S. cohnii (Firmicutes) |
| S. massiliensis (Firmicutes) |
| S. pasteuri (Firmicutes) |
| S. simiae (Firmicutes) |
| V. montpellierensis (Firmicutes) |
Bacterial identities that were uniquely identified in the Vaginosis cohort.
We assessed which OTUs were discriminative between cohorts, and significant differences of bacterial species were not observed according to the FDR p value.
We next compared the overall microbial diversity between groups using the unweighted and weighted UniFrac distance matrices. The results of the β-diversity (between sample diversity comparison) were visualized by a Principal Coordinates Analysis (PCoA). As expected, the bacterial composition of these vaginal communities clustered according to the clinical grouping. Indeed, the analysis of the β-diversity highlights three distinct microbial clusters in the unweighted UniFrac PCoA (Fig. 2). The same clustering is partially confirmed by the weighted UniFrac PCoA (Fig. 3), although it is graphically less evident.
Figure 2.
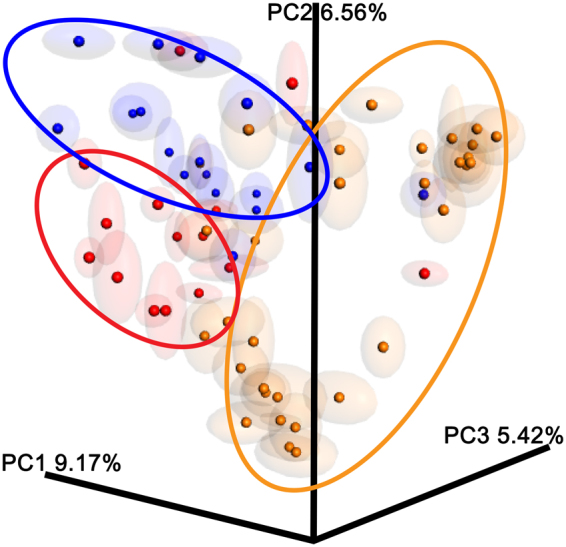
Unweighted UniFrac-based Principal Coordinates Analysis (PCoA). Unweighted UniFrac-based Principal Coordinates Analysis (PCoA) showing the clustering of bacterial communities according to the clinical grouping: Healthy (orange), Intermediate (red) and Vaginosis (blue). Each dot represents a sample. The Emperor PCoA plots were generated from the jackknifed_beta_diversity.py script of QIIME.
Figure 3.
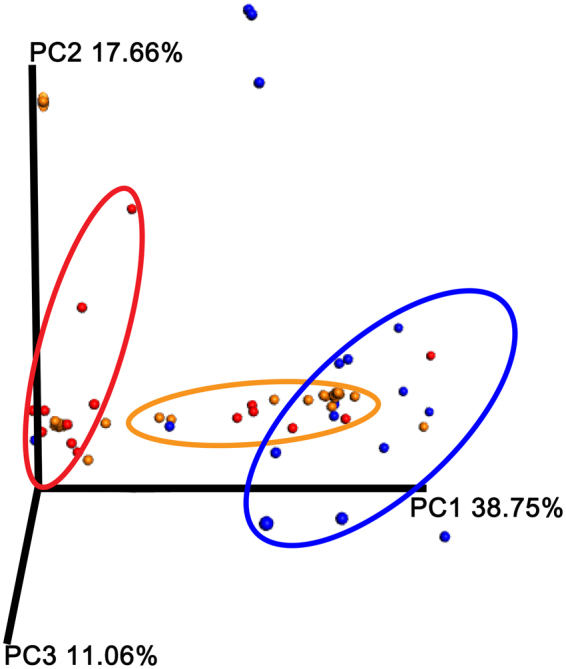
Weighted UniFrac-based Principal Coordinates Analysis (PCoA). Weighted UniFrac-based Principal Coordinates Analysis (PCoA) showing the clustering of bacterial communities according to the clinical grouping: Healthy (orange), Intermediate (red) and Vaginosis (blue). Each dot represents a sample. The Emperor PCoA plots were generated from the jackknifed_beta_diversity.py script of QIIME.
A one-way Analysis of Similarity (ANOSIM) statistical test was applied to the UniFrac distance matrices to test for significant differences according to the clinical grouping. ANOSIM attributed a significant difference to the grouping, shown by the weighted UniFrac (p = 0.001) and unweighted UniFrac (p = 0.001) measures, although the ability of the clinical grouping to explain the differences between cohorts is small (R = 0.31 and 0.21, respectively).
Correlations between microbiome and immune soluble factors
Forty-eight soluble immune mediators, including cytokines, chemokines and growth factors, were measured in the vaginal samples to address the local immune response to microbial changes, the results of which are shown in Table 3. We observed that an increase of the anti-inflammatory IL1ra and the pro-inflammatory IL2, and a decrease of the embryo trophic factors FGFβ and GMCSF, distinguished the dysbiotic from the eubiotic microbiomes.
Table 3.
Significant variations in soluble immune factors.
| Healthy | Intermediate | Vaginosis | Healthy vs Intermediate | Healthy vs Vaginosis | Intermediate vs Vaginosis | |
|---|---|---|---|---|---|---|
| cytokines | ||||||
| IL1α | 62 ± 16 | 35 ± 6 | 295 ± 118 | 0.999 | 0.08 | 0.03 |
| IL1β | 43 ± 28 | 16 ± 7.5 | 215 ± 126 | 0.999 | 0.064 | 0.017 |
| IL1ra | 1.3E5 ± 2.5E4 | 3E5 ± 4.6E4 | 1.5E9 ± 1E9 | 0.014 | 0.000 | 0.605 |
| IL2 | 1.4 ± 0.44 | 4.3 ± 0.6 | 4 ± 0.9 | 0.000 | 0.020 | 0.999 |
| IL3 | 121 ± 9 | 87 ± 10 | 97 ± 10 | 0.028 | 0.277 | 0.999 |
| IL17 | 12 ± 1.9 | 5 ± 0.5 | 7 ± 1 | 0.013 | 0.209 | 0.999 |
| IL18 | 108 ± 21 | 216 ± 135 | 1373 ± 593 | 0.999 | 0.000 | 0.001 |
| MIF | 219 ± 113 | 780 ± 606 | 2353 ± 890 | 0.999 | 0.019 | 0.237 |
| LIF | 22 ± 1.3 | 12 ± 1.5 | 23 ± 2.7 | 0.000 | 0.999 | 0.002 |
| SCF | 23 ± 3.3 | 12 ± 2.8 | 18 ± 2.9 | 0.021 | 0.999 | 0.207 |
| TNFα | 14 ± 1.7 | 14 ± 2.3 | 31 ± 6 | 0.999 | 0.002 | 0.025 |
| TNFβ | 14 ± 1.8 | 9 ± 0.3 | 10 ± 0.6 | 0.013 | 0.999 | 0.271 |
| chemokynes | ||||||
| IL8 | 157 ± 26 | 127 ± 45 | 351 ± 95 | 0.646 | 0.430 | 0.047 |
| MCP1 | 9 ± 1.6 | 4.6 ± 0.5 | 5 ± 0.5 | 0.003 | 0.061 | 0.999 |
| MIG | 309 ± 119 | 188 ± 122 | 488 ± 178 | 0.08 | 0.999 | 0.016 |
| MIP1α | 1.7 ± 0.3 | 1.4 ± 0.4 | 2 ± 0.4 | 0.561 | 0.180 | 0.012 |
| RANTES | 11 ± 3.7 | 17 ± 12 | 24 ± 6 | 0.406 | 0.137 | 0.005 |
| growth factors | ||||||
| FGFβ | 32 ± 3 | 17 ± 1.2 | 18 ± 1.6 | 0.008 | 0.019 | 0.999 |
| GMCSF | 109 ± 4.5 | 81 ± 6.4 | 75 ± 4.6 | 0.015 | 0.000 | 0.593 |
Data are shown as the mean value (pg/mL) ± standard error of mean. Comparisons between groups were performed using a non-parametric Kruskal-Wallis test. The p values are adjusted for multiple comparisons. Significant p values are highlighted in bold.
A specific pattern of soluble immune mediators distinguished the grade of vaginal dysmicrobism. Specifically, a significant decrease of the proteins IL3, LIF, SCF, TNFβ, MCP1 and IL17 was associated with the intermediate group compared to the healthy group (Table 3). A significant increase of IL18, MIF and TNFα characterized the vaginosis group compared to the healthy group (Table 3). In addition, a panel of cytokines, including IL1α, IL1β, IL8, MIG, MIP1α and RANTES, exclusively differentiated the vaginal inflammatory process of the women who were diagnosed with an intermediate dysmicrobism from those with vaginosis (Table 3).
To evaluate the possible relationship between the local immune response and the bacterial composition, the concentrations of each immune factors were included as variables in the BIO-ENV rank-correlation procedure, where single variables or combinations of variables are selected based on how they best explain differences among samples.
Based on the weighted UniFrac distance matrix, several immune factors showed a high correlation with the different microbial patterns observed among samples. In the dysbiotic microbiota, many of the significantly upregulated cytokines, such as IL18, IL2, IL1ra, MIF, RANTES, TNFα, MIP1α and IL8, were confirmed to correlate with the alteration of the vaginal composition. Other immune factors, such as IL5, IL13, IL6, IL15, IL9, GROα, MIP1β and IFNγ, exhibited a correlation with the altered vaginal milieu, although their amount did not significantly change between cohorts.
Based on the unweighted UniFrac distance matrix, the soluble factors that correlated with the microbiome composition were MIF, IL18, MIP1β, TNFα and IL1ra (Table 4).
Table 4.
Output of the BIOENV rank-correlation procedure.
| ρ (weighted UniFrac DM) | |||
|---|---|---|---|
| IL18 | 0.273 | IL6 | 0.17 |
| IL2 | 0.264 | MIP1α | 0.166 |
| IL1ra | 0.234 | IL15 | 0.15 |
| MIF | 0.234 | IL8 | 0.127 |
| IL5 | 0.219 | IL9 | 0.127 |
| IL13 | 0.184 | GROα | 0.113 |
| RANTES | 0.175 | MIP1β | 0.112 |
| TNFα | 0.175 | IFNγ | 0.109 |
| ρ (unweighted UniFrac DM) | |||
| MIF | 0.243 | TNFα | 0.102 |
| IL18 | 0.157 | IL1ra | 0.101 |
| MIP1β | 0.137 | — | — |
Correlations between the 48 immune mediators and both weighted and unweighted UniFrac distance matrices are shown. The table shows the results for ρ ≥ 0.1. The results were generated from the beta_diversity.py output using the compare_categories.py script (metric BIOENV) of QIIME. Abbreviations: ρ: Spearman’s rank correlation coefficient; DM: distance matrix.
Next, we explored whether any individual bacterium correlated with an increase or decrease of specific immune factors. The associations were performed after rarefying the otu_table_biom (depth 10,000 sequences) and using the observation_metadata_correlation.py script with the Fisher_z_transform.
Interleukin IL5 showed a significant correlation (FDR p < 0.01) with G. vaginalis, L. delbrueckii, L. acidophilus, L. johnsonii, L. gasseri, L. crispatus and L. iners, while IL13 was significantly (FDR p < 0.01) associated with G. vaginalis, L. delbrueckii, L. acidophilus, L. johnsonii, L. gasseri, L. crispatus, L. iners and U. parvum serovar 3. The association of other bacteria with the remaining analysed immune factors was not statistically significant when the p value was corrected for the false discovery rate (FDR).
Discussion
The use of high-throughput sequencing techniques shed new light on the high variability and complexity of the vaginal microbiome. Our in vivo results agree with recent studies that have shown that Nugent score, based on the quantification of only three bacterial morphotypes, is not sufficiently informative to clearly define a vaginal dysbiotic status23.
Consistent with this issue is the observation of a high colonization by Bifidobacterium breve, neglected by Nugent Score, in the vaginal microbiome of women with an intermediate dysmicrobism. B. breve is able to counteract the suboptimal colonization by the lactate-producing Lactobacilli spp.24, guaranteeing a healthy vaginal equilibrium through the same mechanism exploited by Lactobacilli, that is, the production of lactic acid. It is noteworthy that, by neglecting the presence of all lactate producing bacteria and the identification of Lactobacilli species, the Nugent score criteria led to the overestimation of the dysbiotic status and to an unnecessary therapeutic intervention being suggested.
In addition, the Nugent score neglects the dynamic of interspecies communication between specific species of Lactobacilli and opportunistic pathogens. In the group of healthy women, L. crispatus25 and L. acidophilus26 inversely correlated with the presence of Gardnerella vaginalis, while in women with an intermediate Nugent Score, L. gasseri negatively correlated with Atopobium vaginae27, a frequently predominant microorganism in women with BV.
The novelty of our study consists of the in vivo demonstration that specific vaginal immune profiles significantly correlate with the microbial composition and clinical manifestation. This finding may help in the diagnosis of BV, predict the recurrence of vaginal dysbiosis and help follow the recovery after treatment. We observed that a subset of pro-inflammatory mediators, typically involved in the chronic inflammation process, synergized with a subset of cytokines that are involved in the switch toward the Th2 immune response. This network likely supports a dysbiotic condition, regardless of the host pro-inflammatory Th1 response, which is normally effective at restoring the eubiosis state.
More precisely, the women with an intermediate Nugent Score showed a significant decrease in several pro-inflammatory cytokines, including IL328 and IL1713. This event, together with the poor T cell-stimulating activity of G. vaginalis29, which was predominant in the intermediate group, may explain the absence or weakness of clinical symptoms11. Moreover, B. breve, which was uniquely identified in the intermediate group, can induce Th1 polarization of lymphocytes30, supporting their own growth by the production of IL231 and determining the resistance toward the infections. This mechanism may support a biotic balance assuring a healthy vaginal milieu.
In women with vaginosis, a massive pro-inflammatory response was observed, supported by a significant increase of IL1832, a constituent of the inflammasome complex32. The increase in concentration of other components of the inflammasome, such as IL1β, and the increase of several chemokines, such as IL8, MIG, MIP1α and RANTES, which are involved in the recruitment of leukocytes at the site of inflammation33, strongly differentiated women with vaginosis from women with an intermediate Nugent Score. This observation confirms the results of previous in vitro studies that showed an immunological shift towards a Th1-dominated profile during episodes of bacterial dysmicrobism34.
A new observation that emerged from our study is the significant association between specific commensal microorganisms and the modulation of local cytokines and chemokines, some of which have never been described before. Specifically, IL5 and IL13, which are secreted by Th2 cells35 and are known to inhibit both the cell-mediated immune response and several macrophage functions, were statistically associated with Lactobacilli, Gardnerella and Ureaplasma species.
Based on the slight increase of these two cytokines (Table 5) and the depletion of some commensal bacteria (Fig. 1) in women with dysmicrobism, we suggest that the Th2 response is maintained at a steady state level of activation alongside the host Th1 response. The increase of the two anti-inflammatory cytokines is expected to significantly rise in asymptomatic women with an altered microbiome, explaining the absence or the weakness of symptoms. Furthermore, the steady-state Th2 response, together with a blunted Th1 response, could lead to immunologic tolerance causing chronic recurrent vaginal dysbiosis36.
Table 5.
Soluble immune factors related to changes of the microbial composition.
| Healthy | Intermediate | Vaginosis | Healthy vs Intermediate | Healthy vs Vaginosis | Intermediate vs Vaginosis | |
|---|---|---|---|---|---|---|
| IL5 | 0.29 ± 0.02 | 0.29 ± 0.04 | 0.35 ± 0.08 | 0.686 | 0.999 | 0.999 |
| IL13 | 0.82 ± 0.06 | 0.75 ± 0.07 | 1 ± 0.19 | 0.592 | 0.999 | 0.580 |
Data are shown as the mean value (pg/mL) ± standard error of mean. Comparisons between groups were performed using a nonparametric Kruskal-Wallis test. The p values are adjusted for multiple comparisons.
Since IL5 and IL13 are strongly influenced by the alteration of eubiotic conditions, regardless of the specific pathogens causing the alteration of vaginal milieu, they could be further studied as indirect markers of vaginal disorder.
Taken together, our findings provide new in vivo insights into how different commensal bacterial species preserve the vaginal symbiotic equilibrium through their interplay with specific local immune mediators. These results raise clinically relevant questions regarding the role of vaginal immunological markers as crucial tool of surveillance of microbial alteration.
Methods
Patients and samples
Sixty-two immunocompetent women who fulfilled the inclusion eligibility criteria were included in this study. All women were Caucasian, of reproductive age (32–40 years old), were not pregnant, had no current use of hormonal or barrier contraceptive products, vaginal douching, tobacco or alcohol abuse, were not hospitalized or had systemic use of medication for chronic diseases or antibiotics/probiotics (oral or topical) within the 6 months prior to sample collection, and had no intercourse in the day prior to sampling. Microbiological criteria excluded concomitantly sexually transmitted viral infections, including HSV I-II and HPV.
We identified patients among women that attended the Gynaecology Service of the Institute for Mother and Child Health IRCCS Burlo Garofolo of Trieste, Italy, as outpatients.
According to Nugent’s criteria, 17 women were diagnosed with bacterial vaginosis (Vaginosis, score 7–10) and 15 with an intermediate dysbiotic status (Intermediate, score 4–6) while the remaining 30 women were diagnosed as healthy (score 0–3).
Vaginal samples were collected 7 days before the first day of the menstrual period. Under speculum examination, samples were collected using a 200 mm polyethylene Cervex brush device37 (Rovers Medical Devices B.V., The Netherlands) by a single gentle 360° rotation of the cytobrush at the cervical os and were suspended in 1.5 ml of TE buffer. Each sample was divided into 3 (500 µl) aliquots and stored at −80 °C.
Sample processing and Ion Torrent Sequencing
DNA extraction was carried out using the NucliSENS® easyMAG® system (BioMèrieux, Gorman, North Carolina, USA) with an elution volume of 50 µl. All DNA samples were stored at −80 °C prior to further processing.
A real time EvaGreen PCR (EvaGreen® dye, Fisher Molecular Biology, Waltham, USA) was performed with the degenerate primer 27FYM (5′-AGR GTT YGA TYM TGG CTC AG-3′) and the primer U534R, targeting the V1-V3 region (500 bp) to allow for the construction of rich libraries. A nested PCR, targeting the V3 region38, was performed with the primers B338F_P1-adaptor (B338F 5′-ACTCCTACGGGAGGCAGC-3′) and U534R_A_barcode (U534R 5′-ATTACCGCGGCTGCTGG-3′) in conjunction with the IonXpress Barcode Adapter to obtain the 200 bp V3 region template for sequencing analysis39. Negative controls, including a no template control, were processed with the clinical samples. The PCR reactions were performed using the Kapa 2 G HiFi Hotstart ready mix 2 × (Kapa Biosystems, Massachusetts, USA), which has robust amplification that is necessary for NGS sequencing40,41, and 400 ng/µL BSA, with the following temperature cycling conditions: 5 min at 95 °C, 30 sec at 95 °C, 30 sec at 59°/57 °C, 45 sec at 72 °C and a final elongation step at 72 °C for 10 min.
Quantification of dsDNA was assessed with a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, California, USA) and the pooled-library was diluted to a concentration of 100 pM.
Template preparation was performed using the Ion PGM Hi-Q View kit on the Ion OneTouch™ 2 System (Life Technologies, Gran Island, New York, USA) and sequenced using the Ion PGM Hi-Q View sequencing kit (Life Technologies, New York, USA) with the Ion PGM™ System technology.
Data Analysis
QIIME 1.8.0142 was used to process the sequence data. High quality (Q > 20) sequences were demultiplexed and filtered by quality using split_libraries_fastq.py with the default parameters, except for the length parameter (at least 150 bp). Operational taxonomic units (OTUs) were defined at 97% similarity and clustered against the Vaginal 16S rRNA gene Reference Database, which was constructed by Fettweis et al.43, using open-reference OTU picking44 with a uclust clustering tool45. Prior to further analysis, singleton OTUs and samples with low sequencing depth were removed (less than 10,000 reads). Chao1, PD whole tree, Shannon, Observed species and Simpson reciprocal metrics were used to assess alpha diversity (within-sample diversity), while beta diversity (between sample diversity comparison) was assessed with weighted and unweighted UniFrac distance matrices46,47 and presented with principal coordinates analysis (PCoA). The robustness of the identified clusters was investigated using jackknifing (randomly resampling sequences without replacement). Differences in community composition between cohorts were investigated using analysis of similarity (ANOSIM, 999 permutations) and Kruskal-Wallis test, implemented in QIIME.
Correlations between the immune mediators, for which normalized amounts were written in the mapping file, and both weighted and unweighted UniFrac distance matrices (generated from the beta_diversity.py script) were assessed by the compare_categories.py script (metric BIOENV) of QIIME, exploiting the Spearman rank-order coefficient. A Spearman coefficient close to 1 signifies a highly positive correlation between immune mediators and the distance matrices; a value close to 0 signifies no association; and a value approaching -1 signifies a highly negative correlation.
To survey the association between microbial identities and the increase or decrease of specific immune factors, the observation_metadata_correlation.py script (with the Fisher_z_transform p-value assignment) of QIIME was used. The otu_table_biom was rarefied to a depth of 10,000 sequences/sample and the normalized amounts of the immune markers were written in the mapping file.
The dataset was deposited in the SRA database (PRJNA361297).
Immune soluble factor quantification
The quantification of soluble immune factors was performed using a recently described platform that is based on a magnetic bead multiplex immunoassay (Luminex, Bio-Plex, BIO-RAD Laboratories, Milano, Italy), which simultaneously detects 48 analytes, including cytokines, chemokines and growth factors48.
Briefly, 50 μL of biological samples and standards were added in duplicate into a 96 multiwell plate containing the analyte beads. After incubation for 30 minutes at room temperature and washing, the antibody-biotin reporter was added and incubated for 10 minutes with streptavidin-phycoerythrin. The concentrations of the cytokines were determined using the Bio-Plex array reader (Luminex, Austin, TX). The Bio-Plex Manager software optimized the standard curves automatically and returned the data as Median Fluorescence Intensity (MFI) and concentration (pg/mL). To normalize the results, the total protein concentrations of samples were determined using a Bradford assay (Sigma-Aldrich, St. Louis, MO). Next, all cytokine and chemokine concentrations were normalized to total protein in the sample and were expressed as pg of immune marker⁄mL of total protein.
Stata (v. 13.1) and GraphPad Prism (v. 5) were used for statistical data analysis. The Kruskal-Wallis one-way analysis of variance was used for comparisons between groups. When a significant p-value was observed (p < 0.05), a multiple comparison test was used to determine which groups were different.
Accession Codes
Sequences have been deposited at NCBI under the accessions PRJNA361297.
Ethics approval and consent to participate
The protocol used in this study was approved by the Ethics Committee of the IRCCS Burlo Garofolo Institute, Trieste (RC 26/13). All participants provided written informed consent for the experiments involving human participants and gave permission to access their medical records. All experiments were performed in accordance with the Declaration of Helsinki.
Acknowledgements
This work was financially supported by the grant RC 26/13, IRCCS Burlo Garofolo Institute.
Author Contributions
Conceptualization, M.C.; Methodology, G.C. and N.Z.; Software, G.C. and D.L.; Investigation, G.C. and N.Z. F.D.S; Resources, F.D.S.; Data Curation, G.C. and N.Z.; Writing - Original Draft, G.C. and M.C.; Writing – Review & Editing M.C.; Visualization, G.C.; Funding Acquisition, M.C.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Srinivasan S, Fredricks DN. The human vaginal bacterial biota and bacterial vaginosis. Interdisciplinary perspectives on infectious diseases. 2008;2008:750479. doi: 10.1155/2008/750479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brotman RM. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. The Journal of clinical investigation. 2011;121:4610–4617. doi: 10.1172/JCI57172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillet E, et al. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC infectious diseases. 2011;11:10. doi: 10.1186/1471-2334-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nardis C, Mosca L, Mastromarino P. Vaginal microbiota and viral sexually transmitted diseases. Annali di igiene: medicina preventiva e di comunita. 2013;25:443–456. doi: 10.7416/ai.2013.1946. [DOI] [PubMed] [Google Scholar]
- 5.Gosmann, C. et al. Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women. Immunity, 10.1016/j.immuni.2016.12.013 (2017). [DOI] [PMC free article] [PubMed]
- 6.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. The New England journal of medicine. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 7.Lamont RF, et al. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG: an international journal of obstetrics and gynaecology. 2011;118:533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedges SR, Barrientes F, Desmond RA, Schwebke JR. Local and systemic cytokine levels in relation to changes in vaginal flora. The Journal of infectious diseases. 2006;193:556–562. doi: 10.1086/499824. [DOI] [PubMed] [Google Scholar]
- 9.Livengood CH. Bacterial vaginosis: an overview for 2009. Reviews in obstetrics & gynecology. 2009;2:28–37. [PMC free article] [PubMed] [Google Scholar]
- 10.Eschenbach DA. Bacterial vaginosis and anaerobes in obstetric-gynecologic infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 1993;16(Suppl 4):S282–287. doi: 10.1093/clinids/16.Supplement_4.S282. [DOI] [PubMed] [Google Scholar]
- 11.Ling Z, et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC genomics. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan S, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PloS one. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner HL, Dukes CD. Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified non-specific vaginitis. American journal of obstetrics and gynecology. 1955;69:962–976. doi: 10.1016/0002-9378(55)90095-8. [DOI] [PubMed] [Google Scholar]
- 14.Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Archives of gynecology and obstetrics. 2014;289:479–489. doi: 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 15.Forney, L. J., Foster, J. A. & Ledger, W. The vaginal flora of healthy women is not always dominated by Lactobacillus species. The Journal of infectious diseases 194, 1468-1469; author reply 1469–1470, 10.1086/508497 (2006). [DOI] [PubMed]
- 16.Oh JE, et al. Dysbiosis-induced IL-33 contributes to impaired antiviral immunity in the genital mucosa. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E762–771. doi: 10.1073/pnas.1518589113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwebke JR. Asymptomatic bacterial vaginosis: response to therapy. American journal of obstetrics and gynecology. 2000;183:1434–1439. doi: 10.1067/mob.2000.107735. [DOI] [PubMed] [Google Scholar]
- 18.Swidsinski A, et al. Gardnerella biofilm involves females and males and is transmitted sexually. Gynecologic and obstetric investigation. 2010;70:256–263. doi: 10.1159/000314015. [DOI] [PubMed] [Google Scholar]
- 19.Swidsinski A, et al. Presence of a polymicrobial endometrial biofilm in patients with bacterial vaginosis. PloS one. 2013;8:e53997. doi: 10.1371/journal.pone.0053997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz J, Isaacs-Cosgrove N, Holzman C, Marsh TL. Temporal Shifts in Microbial Communities in Nonpregnant African-American Women with and without Bacterial Vaginosis. Interdisciplinary perspectives on infectious diseases. 2008;2008:181253. doi: 10.1155/2008/181253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasiela M, Krzeminski Z, Kalinka J, Brzezinska-Blaszczyk E. [Correlation between levels of selected cytokines in cervico-vaginal fluid of women with abnormal vaginal bacterial flora] Medycyna doswiadczalna i mikrobiologia. 2005;57:327–333. [PubMed] [Google Scholar]
- 22.Kremleva, E. A. & Cherkasov, S. V. [Production of cytokines by vaginal epitheliocytes in the process of interaction with dominant and associative microsymbionts]. Zhurnal mikrobiologii, epidemiologii, i immunobiologii, 114–118 (2012). [PubMed]
- 23.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Translational research: the journal of laboratory and clinical medicine. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayo B, van Sinderen D, Ventura M. Genome analysis of food grade lactic Acid-producing bacteria: from basics to applications. Current genomics. 2008;9:169–183. doi: 10.2174/138920208784340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breshears LM, Edwards VL, Ravel J, Peterson ML. Lactobacillus crispatus inhibits growth of Gardnerella vaginalis and Neisseria gonorrhoeae on a porcine vaginal mucosa model. BMC microbiology. 2015;15:276. doi: 10.1186/s12866-015-0608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aroutcheva AA, Simoes JA, Faro S. Antimicrobial protein produced by vaginal Lactobacillus acidophilus that inhibits Gardnerella vaginalis. Infectious diseases in obstetrics and gynecology. 2001;9:33–39. doi: 10.1155/S1064744901000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Backer E, et al. Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC microbiology. 2007;7:115. doi: 10.1186/1471-2180-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitali B, et al. Dynamics of vaginal bacterial communities in women developing bacterial vaginosis, candidiasis, or no infection, analyzed by PCR-denaturing gradient gel electrophoresis and real-time PCR. Applied and environmental microbiology. 2007;73:5731–5741. doi: 10.1128/AEM.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertran T, et al. Slight Pro-Inflammatory Immunomodulation Properties of Dendritic Cells by Gardnerella vaginalis: The “Invisible Man” of Bacterial Vaginosis? Journal of immunology research. 2016;2016:9747480. doi: 10.1155/2016/9747480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez B, et al. The effects of Bifidobacterium breve on immune mediators and proteome of HT29 cells monolayers. BioMed research international. 2015;2015:479140. doi: 10.1155/2015/479140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Current opinion in immunology. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nascimento FS, et al. Evaluation of library preparation methods for Illumina next generation sequencing of small amounts of DNA from foodborne parasites. Journal of microbiological methods. 2016;130:23–26. doi: 10.1016/j.mimet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelhardt E, et al. Chemokines IL-8, GROalpha, MCP-1, IP-10, and Mig are sequentially and differentially expressed during phase-specific infiltration of leukocyte subsets in human wound healing. The American journal of pathology. 1998;153:1849–1860. doi: 10.1016/S0002-9440(10)65699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libby EK, Pascal KE, Mordechai E, Adelson ME, Trama JP. Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes and infection. 2008;10:439–446. doi: 10.1016/j.micinf.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Tomasiak-Lozowska MM, Bodzenta-Lukaszyk A, Tomasiak M, Skiepko R, Zietkowski Z. The role of interleukin 13 and interleukin 5 in asthma. Postepy Hig Med Dosw (Online) 2010;64:146–155. [PubMed] [Google Scholar]
- 36.Miller RL, Tomai MA, Harrison CJ, Bernstein DI. Immunomodulation as a treatment strategy for genital herpes: review of the evidence. International immunopharmacology. 2002;2:443–451. doi: 10.1016/S1567-5769(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 37.Mitra A, et al. Comparison of vaginal microbiota sampling techniques: cytobrush versus swab. Scientific reports. 2017;7:9802. doi: 10.1038/s41598-017-09844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of microbiological methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundquist A, et al. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC microbiology. 2007;7:108. doi: 10.1186/1471-2180-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Experimental cell research. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Quail MA, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fettweis JM, et al. Species-level classification of the vaginal microbiome. BMC genomics. 2012;13(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rideout JR, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 46.Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanotta N, et al. Candidate Soluble Immune Mediators in Young Women with High-Risk Human Papillomavirus Infection: High Expression of Chemokines Promoting Angiogenesis and Cell Proliferation. PloS one. 2016;11:e0151851. doi: 10.1371/journal.pone.0151851. [DOI] [PMC free article] [PubMed] [Google Scholar]