Abstract
Introduction: Hippocampal neuroanatomy is affected by genetic variations in dopaminergic candidate genes and environmental insults, such as early onset of chronic cannabis exposure. Here, we examine how hippocampal total and subregional volumes are affected by cannabis use and functional polymorphisms of dopamine-relevant genes, including the catechol-O-methyltransferase (COMT), dopamine transporter (DAT1), and the brain-derived neurotrophic factor (BDNF) genes.
Material and Methods: We manually traced total hippocampal volumes and automatically segmented hippocampal subregions using high-resolution MRI images, and performed COMT, DAT1, and BDNF genotyping in 59 male Caucasian young adults aged 18–30 years. These included 30 chronic cannabis users with early-onset (regular use at <16 years) and 29 age-, education-, and intelligence-matched controls.
Results: Cannabis use and dopaminergic gene polymorphism had both distinct and interactive effects on the hippocampus. We found emerging alterations of hippocampal total and specific subregional volumes in cannabis users relative to controls (i.e., CA1, CA2/3, and CA4), and associations between cannabis use levels and total and specific subregional volumes. Furthermore, total hippocampal volume and the fissure subregion were affected by cannabis×DAT1 polymorphism (i.e., 9/9R and in 10/10R alleles), reflecting high and low levels of dopamine availability.
Conclusion: These findings suggest that cannabis exposure alters the normal relationship between DAT1 polymorphism and the anatomy of total and subregional hippocampal volumes, and that specific hippocampal subregions may be particularly affected.
Keywords: : brain-derived neurotrophic factor, cannabis, catechol-O-methyltransferase gene, dopamine transporter gene, hippocampal subfields, hippocampus
Introduction
Cannabis is regularly used by ∼23 million individuals globally.1 Regular cannabis use may be detrimental to the brain,2 being associated with neuropsychological, emotional, and motivational impairments.3,4 The hippocampus in particular may be especially sensitive to the impact of heavy cannabis use, given its high concentration of cannabinoid receptors type 1 (CB1).5–7 This is evidenced by neuroimaging results of dose-dependent hippocampal volume reductions in regular cannabis users.8–11 However, hippocampal alteration has not been consistently observed across studies.2,12–18 Although it remains unclear why hippocampus volumetric reduction has not been consistently observed in cannabis users, some evidence suggests that noncannabis-specific vulnerability factors including dopamine gene polymorphism may have played a role in this heterogeneity.19,20 Importantly, dopamine gene polymorphism may also contribute to adverse mental health outcomes in cannabis users.21–24 Given the rising prevalence of cannabis use, treatment seekers, and the global trends toward decriminalization of cannabis products, it is vital to address the heterogeneity in cannabis use-related effects across users, by examining the contribution of dopamine gene polymorphism and cannabis use on hippocampal morphology.1
The neuroanatomy of the hippocampus and hippocampal subregions—subiculum, presubiculum, cornu ammonis (CA) subfields CA1-4, dentate gyrus (DG), and fimbria25—is affected by variation in the expression of genes implicated in dopamine regulation.26 These genes include the catechol-O-methyltransferase (COMT) gene, the dopamine transporter (DAT1) gene, and the brain-derived neurotrophic factor (BDNF) gene: all of which are highly expressed within the hippocampus.27–31 The COMT gene codes for the enzyme that metabolizes dopamine and may present as one of three variants (i.e., a single nucleotide polymorphism (SNP) at Val158Met, rs4680): val/val, val/met, and met/met, which are associated with low, medium, and high dopamine availability, respectively.32 The DAT1 gene, which is involved in dopamine transport and reuptake, consists of a polymorphic 40-base pair (bp) variable number of tandem repeats (VNTR) in an untranslated region, and has common variants including the 9-repeat (9/9R) and the 10-repeat allele (10/10R) associated with high and low dopamine availability, respectively.33,34 Lastly, the BDNF gene is not only involved in neuronal plasticity, but also in dopamine receptor expression. It may present with a SNP (Val66Met) (rs6265), which can lead to a significant reduction in BDNF trafficking to secretory granules and, as a result, to reduced BDNF production in met carriers.35–37
Human neuroimaging studies show that COMT, DAT1, and BDNF polymorphisms affect the neuroanatomy of the hippocampus27,28,38–43 and its subregions.44 These gene variants may further interact with cannabis use to affect cognitive functions ascribed to the hippocampus (e.g., verbal and visuospatial learning).45–47 Despite the relevance of dopamine gene polymorphism in hippocampal neuroanatomy and cannabis use, only one study to date has examined and revealed the influence of cannabis and COMT polymorphism on brain anatomy in regular cannabis users.19 It remains unclear whether polymorphisms in genes other than COMT, which are linked to dopamine regulation—such as DAT1 and BDNF—affect the volume of the hippocampus and its subregions in cannabis users.
We aim to address this gap and extend on our previous work19 by examining the combined effects of cannabis and genetic polymorphism (i.e., DAT1, COMT, and BDNF) on the volume of the hippocampus (quantified using a validated, highly reliable hand-tracing method)48,49 and its subregions. We hypothesized that reduced hippocampal (total and subregional) volumes would be apparent in cannabis users versus controls,2 and in those cannabis users carrying variants linked to high relative to low dopamine level (i.e., COMT met vs. val/val, DAT1 9/9R vs.10/10R, and BDNF val/val vs. met allele carriers),41,50 as elevated extracellular dopamine levels may mediate neurotoxic effects. We further explored whether hippocampal volumes would be affected by cannabis use×COMT–DAT1 cross-products.
Materials and Methods
Participants
We recruited 59 male participants from the general community, including 30 cannabis users and 29 controls matched by age, intelligence quotient (IQ), education, symptoms of depression, anxiety, and psychosis (Table 1).
Table 1.
Sample Sociodemographic, Substance Use Characteristics and Total and Subregional Hippocampus Volumes
| Cannabis users | Controls | t/χ2, p | |
|---|---|---|---|
| Demographics | |||
| Age (years) | 21.0 (2.3) | 22.4 (3.3) | 1.84, 0.07 |
| Males (n [%]) | 30 (100) | 29 (100) | — |
| Cannabis use | |||
| Duration (years) | 5.7 (2.4) | — | — |
| Age onset (years) | |||
| First try | 15.0 (1.1) | 16.7 (2.0) | 2.96, <0.05 |
| Regular use | 18.1 (2.0) | — | — |
| Joints | |||
| Daily | 2.5 (1.5) | — | — |
| Past month | 86.03 (56.85) | — | — |
| Past year | 987.52 (679.97) | — | — |
| Lifetime (cumulative) | 5203 (4192) | 5.1 (11.3) | 6.68, <0.01 |
| Alcohol use | |||
| Age of onset (years) | 15.0 (1.1) | 15.7 (1.5) | 2.20, <0.05 |
| Duration (years) | 5.7 (2.3) | 6.2 (3.1) | ns |
| Alcohol (units/week) | 5.3 (3.8) | 3.2 (2.6) | 2.35, <0.05 |
| Tobacco use | |||
| Current smokers (n) | 27 (90.0) | 9 (31.0) | 21.6, <0.01 |
| Age of onset (years) | 16.3 (1.5) | 16.3 (2.2) | ns |
| Duration of use (years) | 4.5 (2.7) | 4.9 (3.3) | ns |
| Cigarettes smoked daily (n) | 6.0 (5.0) | 2.4 (5.9) | 1.79, 0.08 |
| Hippocampus | |||
| Total volume | 4817.6 (482.0) | 4919.7 (465.6) | a |
| Subiculum | 10700.4 (879.8) | 10341.6 (1076.2) | a |
| Presubiculum | 7284.6 (549.0) | 7246.0 (753.8) | a |
| CA1 | 5566.9 (569.5) | 5243.3 (629.8) | a |
| CA2/3 | 16852.4 (1653.7) | 15595.6 (1779.7) | a |
| CA4/DG | 9385.2 (922.8) | 8821.4 (1024.0) | a |
| Fimbria | 993.2 (219.2) | 1123.6 (262.7) | a |
| Fissure | 552.5 (193.34) | 537.8 (164.0) | a |
| Genetic polymorphism | |||
| BDNF (n) | |||
| Met | 10 | 9 | ns |
| Val/val | 16 | 18 | |
| COMT (n) | |||
| Met | 23 | 22 | ns |
| Val/val | 7 | 7 | |
| DAT (n) | |||
| 9/9R | 15 | 13 | ns |
| 10/10R | 13 | 16 | |
| COMT×DAT (n) | |||
| Val/val + 10/10R | 4 | 3 | ns |
| Met + 10/10R and Val/val +9/9R | 10 | 17 | |
| Met + 9/9R | 14 | 9 | |
| Met + 10/10R and Val/val +9/9R | 10 | 17 | |
| Met + 9/9R | 14 | 9 | |
For all analyses, df=58. COMT=Val108/158; DAT1=DAT 3′ UTR VNTR; BDNF=Val66Met; Met=carriers of COMT or BDNF alleles val/met and met/met.
Results from multiple analyses are outlined in the article body for total hippocampal volume, and Supplementary Table S1 for hippocampal subregions.
CA, cornu ammonis; DG, dentate gyrus; BDNF, brain-derived neurotrophic factor; COMT, catechol-O-methyltransferase; ns, not significant; VNTR, variable number of tandem repeats; UTR, untranslated region.
All participants were recruited through web page and flyers, and underwent a comprehensive telephone interview screening on sociodemographic and substance use data. Participants' inclusion criteria were male gender, Caucasian ethnicity, aged 18–30 years, IQ >90, <5 lifetime use of psychoactive substances other than cannabis, nicotine, or alcohol. Cannabis users were included if they started smoking before age 16, if they smoked >14 weekly “joints” for >2 years, and if they tested positive for cannabinoids during the urine toxicology test. Controls were included if they smoked cannabis <15 times, did not smoke in the past month, and if they tested negative for any illicit drugs as ascertained by the urine toxicology test.
Exclusion criteria for all participants were (i) lifetime Axis I, DSM-IV disorder apart from nicotine use disorder, or cannabis use disorder in cannabis users; (ii) positive urine toxicology return for opiates, cocaine, amphetamines, and benzodiazepines; (iii) current psychoactive medications; (iv) history of chronic medical illness or neurological conditions; (v) head trauma with loss of consciousness >2 min; (vi) left-handedness; and (vii) uncorrected visual impairment, color blindness, and hearing impairment.
Procedure
Participants underwent a detailed medical history check, physical/neurological examinations, and urine and hair toxicology tests through immunometric assay kits (Instant-View, ASD, Inc., Poway, CA). The latter corroborated self-reported substance use through the Psychiatric Research Interview for Substance and Mental Disorders (PRISM).51 Verbal intelligence was assessed with the WAIS-III vocabulary subscale.52
All participants provided written informed consent after receiving a complete description of the study, and were financially compensated for any incurred cost. The study was approved by the Ethical and Clinical Research Committee where the study was run (CEIC-Parc de Salut Mar).
Genotyping
The COMT val108/158met (rs4680) and BDNF val66met (rs6265) SNP allelic variants were determined using the 59 exonuclease TaqMan assay with ABI 7900HT Sequence Detection System (real time PCR) supplied by Applied Biosystems. Primers and fluorescent probes were obtained from Applied Biosystems (TaqMan SNP genotyping assays: assay ID C_2255335_10 and C_11592758_10 for rs4680 and rs6265, respectively). Reaction conditions were those described in the ABI PRISM 7900HT user's guide. Endpoint fluorescent signals were detected on the ABI 7900, and the data were analyzed using SDS software, version 2.3 (Applied Biosystems).
DAT1 VNTR genotyping was performed using polymerase chain reaction (PCR).33 Primers used were Forward 5′-FAM-TGTGGTGTAGGGAACGGCCTGAG and reverse 5′-CTTCCTGGAGGTCACGGCTCAAGG. Each reaction mixture contained (i) one PCR amplification buffer and 0.3 PCR enhancer solution (Invitrogen, Carlsbad, CA), (ii) 3 mM MgSO4, 200 mM dNTPs, 0.2 mM of each primer, (iii) 1 U of Taq DNA polymerase (Invitrogen), and (iv) 50 ng of genomic DNA as template. Amplification conditions included an initial denaturation step of 5 min at 95°C, followed by 35 cycles of 30 sec at 95°C, 40 sec at 58°C, 45 sec at 72°C, and 5 min at 72°C. We used a total reaction volume of 10 μL. After PCR, we detected the products of allelic-specific amplifications (allele 9/9R, 450 bp, and allele 10/10R, 480 bp) through an automatic ABI 3730XL capillary sequencer. Data were analyzed through GeneMapper Software v3.5 (Applied Biosystems). Sequences for genotyping BDNF are described in detail elsewhere.53
After genotype determination, the sample was divided into subgroups based on (i) COMT polymorphism (i.e., val/val homozygote and met-allele carriers including val/met and met homozygous); (ii) DAT1 polymorphism (i.e., 9/9R and 10/10R allele carriers); (iii) BDNF polymorphism (i.e., val/val homozygote and met-allele carriers including val/met and met homozygous); and (iv) combinations of DAT1–COMT variants associated with different dopamine availability levels (Table 1).54–56 The distribution of COMT, DAT1, BDNF, and COMT–DAT1 genetic polymorphism was in Hardy–Weinberg equilibrium.
MRI data
MRI images were acquired with a 1.5T Signa Excite system (General Electric, Milwaukee, WI) equipped with an eight-channel phased-array head coil. High-resolution T1-weighted anatomical images were obtained by using a three-dimensional fast spoiled gradient inversion-recovery prepared sequence with 130 contiguous slices (TR, 11.8 msec; TE, 4.2 msec; flip angle, 15°; field of view, 30 cm; pixel matrix, 256×256; and slice thickness, 1.2 mm).
Images were transferred to a Linux workstation for preprocessing including reorientation, resizing to 1×1×1 mm resolution, and alignment to the Montréal Neurological Institute standard template (www.fmrib.ox.ac.uk/fsl). The same investigator (A.B.) manually delineated the hippocampus using Analyze software (version 9.0; Mayo Clinic, Rochester, MN), while being blind to group membership. Tracing was performed on coronally displayed MRI slices, from the caudal to the rostral portion of the brain, based on a previously validated protocol.48,57 Hippocampal boundaries were set, medially, by the cerebral spinal fluid (CSF) of the uncal cistern; laterally, by the CSF in the temporal horn of the lateral ventricle; and inferiorly by the parahippocampal white matter running medially from the temporal horn of the lateral ventricle. We used the protocol developed by Watson et al. to separate the hippocampus from the amygdala.58
Intraclass correlation coefficients (absolute agreement) were computed to assess tracing reliability, based on 10 randomly selected images. Intrarater reliability was 0.96 and 0.95, whereas inter-rater reliability against an experienced hippocampus tracer (V.L.) was 0.94 and 0.90, for the right and left hemisphere, respectively. We obtained intracranial volumes (ICVs) through the Voxel Brain Morphometry module (http://dbm.neuro.uni-jena.de/vbm/)59 of the Statistical Parametric Mapping software (www.fil.ion.ucl.ac.uk/spm/).
To assess hippocampal subregional effect, the hippocampus was also segmented using the FreeSurfer image analysis environment (http://surfer.nmr.mgh.harvard.edu/) version 5.3.0, into several subregions (i.e., fimbria, presubiculum, subiculum, CA1, CA2/3, CA4/DG fields, and hippocampus fissure).25
Statistical analyses
We ran a series of four repeated-measures analysis of covariances, with total and subregional hippocampal volumes as dependent variables, hemisphere (i.e., left and right) as repeated measure, and ICV as covariate. Factors in the first three models included COMT genetic polymorphisms (i.e., val/val vs. met), DAT1 genetic polymorphisms (9/9R vs. 10/10R), and BDNF genetic polymorphisms (val/val vs. met carriers including mostly val/met carriers and three met/met carriers), respectively, as well as cannabis use (i.e., user vs. nonuser). Factors in the exploratory model were cannabis use×cross-product of COMT–DAT1 genotypes associated with varying levels of dopamine availability54: (i) low dopamine, including carriers of alleles val/val and 10/10R; (ii) high dopamine, including carriers of met and 9/9R; (iii) medium dopamine, including carriers of alleles (a) val/val and 9/9R and (b) met and 10/10R and one cannabis using carrier of the val/val and 9/9R. In addition, we explored the impact of cannabis use as the only factor (without genetic polymorphism as a factor) on hippocampal volumes, using the same model already outlined.
Nicotine use (monthly cigarettes) and alcohol use were not included as covariate in the analyses. Monthly cigarettes were not associated with hippocampal volumes in cannabis users. Nine controls were tobacco smokers, from whom only two smoked >10 cigarettes per day. Alcohol use did not affect any analyses, at either a significant or trend level, when used as a covariate.
Finally, we ran partial correlations to explore the association between hippocampal total and subregional volumes and cannabis use levels (lifetime, past year, and past month cumulative number of joints) and monthly cigarettes, controlling for ICV. No outlier was observed in hippocampal volume distribution. All analyses were performed using SPSS version 19.0. For all the analyses, the significance threshold was p<0.05.
Discussion
Cannabis use and dopaminergic gene polymorphism were found to have both distinct and interactive effects on total and subregional hippocampal volumes. The left hippocampus volume was nonsignificantly reduced in cannabis users relative to controls (F52=3.70, p=0.06). This trend emerged when cannabis use was the only factor in the model, and dissipated when genetic polymorphism was added as an additional factor (COMT, DAT1, BDNF, and COMT×DAT1 cross-products, see Supplementary Table S1). Interestingly, cannabis-using samples from past studies, with consumption levels similar to those of this study (i.e., mean 15 years age of onset, smoking 22 days/month, 3 years duration), also show nonsignificant hippocampal differences relative to nonusing controls.12 In contrast, samples of heavier users with longer periods of use (i.e., daily use for >10 years)10,60 show hippocampal reductions relative to nonusers. Duration of heavy use may have played a role in the discrepancy of these findings, with longer duration of heavy use driving reduced hippocampi.2,10,12,60,61 This is unsurprising given the direct association between cannabinoid exposure and receptor organization on the hippocampus.62 A longer duration of heavy cannabis use may be required, before significant hippocampal reduction is observed. Accordingly, in this sample we found an association between smaller total hippocampal volumes and higher cumulative cannabis joints smoked during users' lifetime (r=−0.41, p=0.046), past year (r=−0.35, p=0.08), and past month (r=0.34, p=0.08), suggesting a dose-dependent relationship between cannabis use and hippocampal volume.
We also found significant effects of cannabis on specific subregions, with a larger volume in CA1 (i.e., in two of the four analyses, which used COMT and DAT1 as factors), CA2/3, and CA4 (i.e., in one analysis using COMT and cannabis as factors) in cannabis users relative to nonusers, when controlling for COMT gene polymorphism and ICV (Supplementary Table S1); this is alongside dose-dependent relationships between the volume of various subregions and cannabis use levels—including larger right CA1 and greater monthly/yearly joints (r53=0.29, p<0.05 and r53=0.25, p=0.07), larger right CA2/3 and greater monthly/yearly/lifetime joints (r53=40, p<0.005, r53=0.36, p<0.01 and r53=0.33, p<0.05), and larger right CA4 and greater monthly, yearly, and lifetime joints (r53=0.30, p<0.05, r53=0.26, p=0.06 and r53=0.23, p=0.09).
Total hippocampal volume and the fissure subregion were affected by cannabis×DAT1 polymorphism (i.e., 9/9R and in 10/10R alleles), reflecting high and low levels of dopamine availability (F1,52=4.93, p=0.031, see Fig. 1).
FIG. 1.
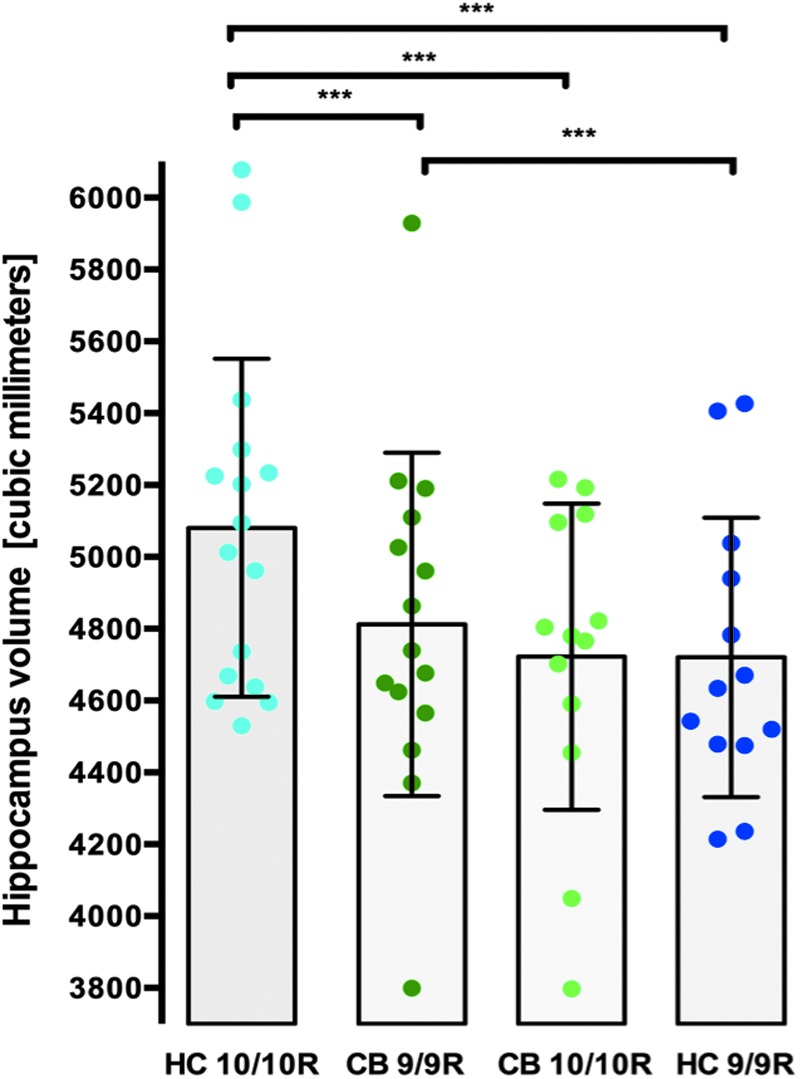
Hippocampal volumes by DAT1 polymorphism variants (10/10R and 9/9R) in controls (HC) and cannabis users (CB), with larger volumes in HC 10/10R carriers relative to CB 10/10R and CB 9/9R carriers; smaller volumes in HC 9/9R carriers versus CB 9/9R carriers and HC 10/10R carriers.
Across all groups, average hippocampal volume was largest in control carriers of the 10/10R allele, followed by cannabis-using carriers of the 9/9R and the 10/10R allele, and finally by control carriers of the 9/9R allele. 10/10R-carrying cannabis users had significantly smaller hippocampi than 10/10R-carrying controls in the left hemisphere (t27=7.81, p<0.001, Cohen'd r=−0.8, large effect size) and right hemisphere (t26=4.07, p<0.0001; Cohen'd r=−0.7, large effect size). However, the inverse pattern was observed in 9/9R carriers, with 9/9R-carrying cannabis users having larger left hippocampal volumes than 9/9R-carrying controls (t26=4.07, p<0.0001, Cohen'd r=−0.6, large effect size). This may be related to the DAT1 genotype affecting total hippocampal volumes in controls (i.e., 9/9R and 10/10R carriers showed the smallest and largest volumes, respectively—left: t27=2.69, p=0.012 and right t27=1.28, p=0.21), but not in cannabis users (i.e., 9/9R and 10/10R carriers had similar hippocampal volumes—left, t26 = −0.68, p=0.50 and right, t26 = −0.26, p=0.79). Notably, the cannabis×DAT1 effect on total hippocampal volumes may be driven by specific subregion (i.e., hippocampal fissure, Supplementary Table S1) where this effect was also observed. The interaction between dopamine genetic polymorphism (DAT1) and cannabis may partly explain the mixed evidence to date on smaller hippocampal volumes in cannabis users than in controls. This notion remains unexplored, as genetic polymorphism remains underinvestigated in neuroimaging studies of cannabis users.2
Our findings suggest that carrying the 9/9R allele of DAT1 predates smaller hippocampi, and cannabis use may reverse this process. Although the underlying mechanism remains unclear, we speculate that cannabis exposure may selectively interact with DAT1 gene variants to alter the normal effect of dopaminergic transmission on neuroanatomy.63–65 Chronic cannabis use may alter “optimal” extracellular dopamine levels (e.g., lower synthesis/release and/or contributing to hypersensitivity of postsynaptic receptors)66 and compromise neuronal growth.67 The DAT1 gene influences neuronal growth, survival, and differentiation, and is furthermore highly expressed within the hippocampus.67,68 Importantly, emerging evidence shows that dopaminergic genes (e.g., DAT1)63,69 mediate the effects of specific cannabinoid compounds on the brain. It remains unclear whether these effects change for distinct cannabinoid compounds that have very different effects on neuroanatomy,2,70 neural activity,71 and emotion processing72—such as the neuroprotective cannabidiol73,74 and the psychotogenic tetrahydrocannabinol.75
Apart from the interactive effect of cannabis×DAT1 on hippocampal fissure volume, the total and subregional volumes of the hippocampus were not affected by either COMT, DAT1, or BDNF polymorphism alone, nor by cannabis×COMT or cannabis×BDNF interaction. This is consistent with some38,76,77 but not other findings to date that demonstrated an effect of COMT (i.e., volume reduction in val vs. met allele carriers)28,50 and BDNF (i.e., volume reduction in met vs. val allele carriers).41 We may have failed to detect an effect of COMT, DAT1, or BDNF polymorphism due to the small sample size, or the relatively lower duration of heavy cannabis use in our user sample. Yet, our findings suggest that neuroanatomy may be affected by a multitude of genetic and environmental (e.g., substance use) factors.
Our third exploratory analysis suggests that the volume of the hippocampus is affected by COMT–DAT1 combinations associated with different levels of dopamine concentration. Hippocampal volumes were (at a trend level) affected by cannabis×COMT–DAT1 cross-products (F2,50=2.62, p=0.08) and significantly affected by the cross-product of hemisphere×cannabis×COMT–DAT1 (F2=3.62, p=0.034). Interestingly, the analysis on COMT–DAT1 products linked to different levels of dopamine availability revealed that the volume of the left hippocampus was the largest within controls, but not cannabis users, who carried alleles linked to high versus low and medium dopamine level. Similarly, other reports show that regional brain volumes (i.e., within the caudate)78 and activity45,54,55,79 are affected by the epistasis between dopamine-related genes. We warrant the conduct of studies to examine the role of gene×gene interaction on dopamine availability and neuroanatomy in large normative samples, including cannabis users.55 Finally, in this model, hippocampal volumes were not affected by cannabis alone (F1,50=0.82, p=0.37) and COMT–DAT1 cross-products (F2,50=0.2.29, p=0.11).
Interestingly, we found emerging effects for larger subregional volumes in cannabis users. These effects are surprising given the significant evidence for dose-dependent total hippocampal volume reduction, and subregional volume reduction in cannabis users.2,10,80,81 This discrepancy may be due to the different method of delineating the total hippocampus (i.e., manual tracing) and its subregions (i.e., automated segmentation). Automated segmentation methods may be more susceptible to magnetic resonance image quality factors.82 Our magnetic resonance images presented with artifacts along the temporal lobe, which may have reduced the accuracy of automated segmentations. Alternatively, cannabis may have differential effects in distinct subregions of the hippocampus, given the different distribution of cannabinoid receptors across subregions.83
In our study, total hippocampal reduction in cannabis users relative to controls is presented in the left hemisphere, whereas subregional volume enlargement in cannabis users relative to controls is presented in the right hemisphere, which may point to a hemisphere-specific effect as well. Importantly, the finding of larger subregions in cannabis users was inconsistently found across the series of four statistical analyses examining the effect of cannabis and four distinct genetic polymorphisms (COMT, DAT1, BDFN, and COMT×DAT1 cross-products) on the hippocampus. Of the four analyses, larger CA2/3 subregions were detected in only two analyses, and larger CA2 and CA4 were apparent in only one analysis (Supplementary Table S1). The inconsistent findings on larger hippocampal subregions must be interpreted with caution and warrant replication in a separate sample of regular cannabis users.
This study has several limitations. Our small sample size of 59 participants limited the power to detect complex effects of COMT, DAT1, and cannabis. Still, large effect sizes in the cannabis×DAT1 effects on hippocampal volumes (i.e., r=0.6 and r=0.8) warrant replication in large normative cohorts, where the impact of additional genetic polymorphism relevant to neuroanatomy can be measured with adequate power—including cannabis receptor-1 (CNR1)43 and P2 promoter region (rs2097603).28 Such work is warranted to unravel the complex gene–environmental interplay that affects neuroanatomical alterations in cannabis users. Using Bonferroni correction for multiple comparison, we used a stringent significance threshold p<0.01 (to control for the four independent variables used to predict hippocampal volumes—COMT, DAT1, BDNF, and COMT–DAT1) and found that only sparse results survived (i.e., cannabis×COMT–DAT1, and cannabis×hemisphere on the fissure).
Furthermore, although we have only considered the effect of cannabis use and genetic polymorphism on hippocampus volume, genetic predisposition may similarly interact with cannabis use to affect the neuroanatomy of other subcortical structures (e.g., ventral striatum),84 and should be considered in future studies. Finally, we could not discern the impact of nicotine from that of cannabis on hippocampal volumes.85 Although we found no significant correlation between total hippocampal volumes and monthly cigarettes smoked, we cannot conclusively rule out the effect of nicotine on hippocampal volumes, given the low level of cigarette use in our participant sample (average of <7 cigarettes daily).
Conclusions
Our findings suggest that although cannabis alone may subtly affect hippocampal neuroanatomy, given the trend-level and dose-dependent reduction observed in cannabis users, the interaction between cannabis and DAT1 polymorphism was a stronger predictor of hippocampal volume differences between cannabis users and controls. Hippocampal neuroanatomical differences were observable between DAT1 9/9R and 10/10R-carrying controls but not in cannabis users in our sample, suggesting that cannabinoid exposure may alter the normal relationship between DAT1 polymorphism and hippocampal neuroanatomy. The increasing potency of cannabis, rates of treatment-seeking users, and availability of cannabis products due to recent decriminalization policies warrant the identification of vulnerability factors within cannabis users for adverse neurobiological outcomes, such that targeted prevention and intervention strategies may be developed. We warrant the examination of large-scale adult and adolescent samples from existing consortia collating genetic, neural, and behavioral data (ENIGMA Addiction http://enigma.ini.usc.edu/ongoing/enigma-addiction-working-group/86 and IMAGEN http://imagen-europe.com/en/consortium.php)87 to study the role of additional genetic polymorphisms (e.g., CNR1) and nongenetic factors on neuroanatomy (e.g., mental health, socioeconomic status, comorbid substance use, and cannabinoid compounds), and psychosocial outcomes in cannabis users, and to ultimately minimize cannabis-related harms.
Supplementary Material
Abbreviations Used
- 10/10R
10-repeat allele
- BDNF
brain-derived neurotrophic factor
- bp
base pair
- CA
cornu ammonis
- CB1
cannabinoid receptors type 1
- CNR1
cannabis receptor-1
- COMT
catechol-O-methyltransferase
- CSF
cerebral spinal fluid
- DAT1
dopamine transporter
- DG
dentate gyrus
- ICV
intracranial volume
- IQ
intelligence quotient
- MRI
magnetic resonance imaging
- PCR
polymerase chain reaction
- PRISM
Psychiatric Research Interview for Substance and Mental Disorders
- SNP
single nucleotide polymorphism
- UTR
untranslated region
- VNTR
variable number of tandem repeats
Acknowledgments
This study has been done in part with Spanish grants: Plan Nacional Sobre Drogas, Ministerio de Sanidad y Consumo: PNSD PI101/2006, and PNSD PI041731/2011 (RMS); and with the support of the Generalitat de Catalunya/Support a les activitats del Grups de Recerca: SGR 2009/1435, and SGR 2014/1411 (RMS). CSM is supported by a Miguel Servet contract from the Carlos III Health Institute (CPII16/00048). SB has been funded by the National Insitute for Health Research (NIHR), UK through a Clinician Scientist award (NIHR-CS-11-001) and also received support from the Medical Research Council (MRC), UK (MR/J012149/1; MC_PC_14105 v.2). MT is supported by Instituto de Salud Carlos III-FEDER (RD16/0017/0010). JAC is recipient of Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq, Brazil) productivity fellowship (1A). The present study was supported by a CNPq grant (CNPq/MS/SCTIE/DECIT No 26/2014 – Pesquisas sobre Distúrbios Neuropsiquiátricos; 466805/2014-4). JAC has a grant from University Global Partnership Network (UGPN) – Global priorities in cannabinoid research excellence.
Informed Consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, and the applicable revisions at the time of the investigation. Informed consent was obtained from all participants for being included in the study.
Author Disclosure Statement
JAC is co-inventor (Mehoulam R, JC, Guimaraes FS, AZ, JH, Breuer A) of the patent “Fluorinated CBD compounds, compositions and uses thereof. Pub. No.: WO/2014/108899. International Application No.: PCT/IL2014/050023” Def. US no. Reg. 62193296; 29/07/2015; INPI on 19/08/2015 (BR1120150164927). The University of São Paulo has licensed the patent to phytecs Pharm (USP Resolution No. 15.1.130002.1.1). The University of São Paulo has an agreement with Prati-Donaduzzi (Toledo, Brazil) to “develop a pharmaceutical product containing synthetic cannabidiol and prove its safety and therapeutic efficacy in the treatment of epilepsy, schizophrenia, Parkinson's disease, and anxiety disorders”. JAC has received travel support from and is medical advisor of BSPG-Pharm. For all other authors, no competing financial interests exist.
References
- 1.UNODC. World Drug Report 2015: United Nations Office on Drugs and Crime, 2015
- 2.Lorenzetti V, Solowij N, Yücel M. The role of cannabinoids in neuroanatomic alterations in cannabis users. Biol Psychiatry. 2016;79:e17–e31 [DOI] [PubMed] [Google Scholar]
- 3.Meier MH, Caspi A, Ambler A, et al. Persistent cannabis users show neuropsychological decline from childhood to midlife. Proc Natl Acad Sci U S A. 2012;109:E2657–E2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkow ND, Swanson JM, Evins AE, et al. Effects of cannabis use on human behavior, including cognition, motivation, and psychosis: a review. JAMA Psychiatry. 2016;73:292–297 [DOI] [PubMed] [Google Scholar]
- 5.Burns HD, Van LK, Sanabria-Bohorquez S, et al. [18F]MK-9470, a positron emission tomography (PET) tracer for in vivo human PET brain imaging of the cannabinoid-1 receptor. Proc Natl Acad Sci U S A. 2007;104:9800–9805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass M, Dragunow M, Faull RL. Cannabinoid receptors in the human brain: a detailed anatomical and quantitative autoradiographic study in the fetal, neonatal and adult human brain. Neuroscience. 1997;77:299–318 [DOI] [PubMed] [Google Scholar]
- 7.Herkenham M, Lynn AB, Johnson MR, et al. Characterization and localization of cannabinoid receptors in rat brain: a quantitative in vitro autoradiographic study. J Neurosci. 1991;11:563–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzetti V, Solowij N, Fornito A, et al. The association between regular cannabis exposure and alterations of human brain morphology: an updated review of the literature. Curr Pharm Des. 2014;20:2138–2167 [DOI] [PubMed] [Google Scholar]
- 9.Batalla A, Bhattacharyya S, Yucel M, et al. Structural and functional imaging studies in chronic cannabis users: a systematic review of adolescent and adult findings. PLoS One. 2013;8:e5582–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocchetti M, Crescini A, Borgwardt S, et al. Is cannabis neurotoxic for the healthy brain? A meta-analytical review of structural brain alterations in non-psychotic users. Psychiatry Clin Neurosci. 2013;67:483–492 [DOI] [PubMed] [Google Scholar]
- 11.Martin-Santos R, Fagundo AB, Crippa JA, et al. Neuroimaging in cannabis use: a systematic review of the literature. PsycholMed. 2010;40:383–398 [DOI] [PubMed] [Google Scholar]
- 12.Cousijn J, Wiers RW, Ridderinkhof KR, et al. Grey matter alterations associated with cannabis use: results of a VBM study in heavy cannabis users and healthy controls. NeuroImage. 2012;59:3845–3851 [DOI] [PubMed] [Google Scholar]
- 13.Medina KL, Nagel BJ, Park A, et al. Depressive symptoms in adolescents: associations with white matter volume and marijuana use. J Child Psychol Psychiatry. 2007;48:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina KL, Schweinsburg AD, Cohen-Zion M, et al. Effects of alcohol and combined marijuana and alcohol use hippocampal volume and asymmetry. Neurotoxicol Teratol. 2007;29:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzilos GK, Cintron CB, Wood JBR, et al. Lack of hippocampal volume change in long-term heavy cannabis users. Am J Addict. 2005;14:64–72 [DOI] [PubMed] [Google Scholar]
- 16.Block RI, O'Leary DS, Ehrhardt JC, et al. Effects of frequent marijuana use on brain tissue volume and composition. Neuroreport. 2000;11:491–496 [DOI] [PubMed] [Google Scholar]
- 17.Wilson W, Mathew R, Turkington T, et al. Brain morphological changes and early marijuana use. A MRI and PET study. J Addict Dis. 2000;19:1–22 [DOI] [PubMed] [Google Scholar]
- 18.Palamar JJ, Fenstermaker M, Kamboukos D, et al. Adverse psychosocial outcomes associated with drug use among US high school seniors: a comparison of alcohol and marijuana. Am J Drug Alcohol Abuse. 2014;40:438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batalla A, Soriano-Mas C, Lopez-Sola M, et al. Modulation of brain structure by catechol-O-methyltransferase Val(158) Met polymorphism in chronic cannabis users. Addict Biol. 2014;19:722–732 [DOI] [PubMed] [Google Scholar]
- 20.Behan AT, Hryniewiecka M, O'Tuathaigh CM, et al. Chronic adolescent exposure to delta-9-tetrahydrocannabinol in COMT mutant mice: impact on indices of dopaminergic, endocannabinoid and GABAergic pathways. Neuropsychopharmacology. 2012;37:1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henquet C, Di Forti M, Morrison P, et al. Gene-environment interplay between cannabis and psychosis. Schizophr Bull. 2008;34:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction. 2009;104:518–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volkow ND, Wang G-J, Telang F, et al. Decreased dopamine brain reactivity in marijuana abusers is associated with negative emotionality and addiction severity. Proc Natl Acad Sci. 2014;111:E3149–E3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Winkel R. Family-based analysis of genetic variation underlying psychosis-inducing effects of cannabis: sibling analysis and proband follow-up. Arch Gen Psychiatry. 2011;68:148–157 [DOI] [PubMed] [Google Scholar]
- 25.Van Leemput K, Bakkour A, Benner T, et al. Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus. 2009;19:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto M, Weickert CS, Akil M, et al. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience. 2003;116:127–137 [DOI] [PubMed] [Google Scholar]
- 27.Durston S, Fossella JA, Casey BJ, et al. Differential effects of DRD4 and DAT1 genotype on fronto-striatal gray matter volumes in a sample of subjects with attention deficit hyperactivity disorder, their unaffected siblings, and controls. Mol Psychiatry. 2005;10:678–685 [DOI] [PubMed] [Google Scholar]
- 28.Honea R, Verchinski BA, Pezawas L, et al. Impact of interacting functional variants in COMT on regional gray matter volume in human brain. Neuroimage. 2009;45:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kambeitz JP, Bhattacharyya S, Kambeitz-Ilankovic LM, et al. Effect of BDNF val(66)met polymorphism on declarative memory and its neural substrate: a meta-analysis. Neurosci Biobehav Rev. 2012;36:2165–2177 [DOI] [PubMed] [Google Scholar]
- 30.Scheepers FE, de Wied CC, Hulshoff Pol HE, et al. The effect of clozapine on caudate nucleus volume in schizophrenic patients previously treated with typical antipsychotics. Neuropsychopharmacology. 2001;24:47–54 [DOI] [PubMed] [Google Scholar]
- 31.Rossato JI, Bevilaqua LR, Izquierdo I, et al. Dopamine controls persistence of long-term memory storage. Science. 2009;325:1017–1020 [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Lipska BK, Halim N, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandenbergh DJ, Persico AM, Hawkins AL, et al. Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics. 1992;14:1104–1106 [DOI] [PubMed] [Google Scholar]
- 34.Heinz A, Goldman D, Jones DW, et al. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology. 2000;22:133–139 [DOI] [PubMed] [Google Scholar]
- 35.Benjamin S, McQuoid DR, Potter GG, et al. The brain-derived neurotrophic factor Val66Met polymorphism, hippocampal volume, and cognitive function in geriatric depression. Am J Geriatr Psychiatry. 2010;18:323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269 [DOI] [PubMed] [Google Scholar]
- 37.Le Foll B, Gallo A, Le Strat Y, et al. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol. 2009;20:1–17 [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi T, Hashimoto R, Mori T, et al. The association between the Val158Met polymorphism of the catechol-O-methyl transferase gene and morphological abnormalities of the brain in chronic schizophrenia. Brain. 2006;129(Pt 2):399–410 [DOI] [PubMed] [Google Scholar]
- 39.Shook D, Brady C, Lee PS, et al. Effect of dopamine transporter genotype on caudate volume in childhood ADHD and controls. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McIntosh AM, Baig BJ, Hall J, et al. Relationship of catechol-O-methyltransferase variants to brain structure and function in a population at high risk of psychosis. Biol Psychiatry. 2007;61:1127–1134 [DOI] [PubMed] [Google Scholar]
- 41.Cao B, Bauer IE, Sharma AN, et al. Reduced hippocampus volume and memory performance in bipolar disorder patients carrying the BDNF val66met met allele. J Affect Disord. 2016;198:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molendijk ML, Bus BA, Spinhoven P, et al. A systematic review and meta‐analysis on the association between BDNF val66met and hippocampal volume—a genuine effect or a winners curse? Am J Med Genet B Neuropsychiatr Genet. 2012;159:731–740 [DOI] [PubMed] [Google Scholar]
- 43.Schacht JP, Hutchison KE, Filbey FM. Associations between cannabinoid receptor-1 (CNR1) variation and hippocampus and amygdala volumes in heavy cannabis users. Neuropsychopharmacology. 2012;37:236–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabl U, Meyer BM, Diers K, et al. Additive gene–environment effects on hippocampal structure in healthy humans. J Neurosci. 2014;34:9917–9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertolino A, Blasi G, Latorre V, et al. Additive effects of genetic variation in dopamine regulating genes on working memory cortical activity in human brain. J Neurosci. 2006;26:3918–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prata DP, Mechelli A, Fu CH, et al. Opposite effects of catechol-O-methyltransferase Val158Met on cortical function in healthy subjects and patients with schizophrenia. Biol Psychiatry. 2009;65:473–480 [DOI] [PubMed] [Google Scholar]
- 47.Solowij N, Battisti R. The chronic effects of cannabis on memory in humans: a review. Curr Drug Abuse Rev. 2008;1:81–98 [DOI] [PubMed] [Google Scholar]
- 48.Velakoulis D, Wood SJ, Wong MTH, et al. Hippocampal and amygdala volumes according to psychosis stage and diagnosis: a magnetic resonance imaging study of chronic schizophrenia, first-episode psychosis, and ultra-high-risk individuals. Arch Gen Psychiatry. 2006;63:139–149 [DOI] [PubMed] [Google Scholar]
- 49.Morey RA, Petty CM, Xu Y, et al. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehrlich S, Morrow EM, Roffman JL, et al. The COMT Val108/158Met polymorphism and medial temporal lobe volumetry in patients with schizophrenia and healthy adults. NeuroImage. 2010;53:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torrens M, Serrano D, Astals M, et al. Diagnosing comorbid psychiatric disorders in substance abusers: validity of the Spanish versions of the Psychiatric Research Interview for Substance and Mental Disorders and the Structured Clinical Interview for DSM-IV. Am J Psychiatry. 2004;161:1231–1237 [DOI] [PubMed] [Google Scholar]
- 52.Wechsler D. WAIS-III & WMS-III technical manual. The Psychological Corporation: San Antonio, 1997 [Google Scholar]
- 53.Cuyàs E, Verdejo-García A, Fagundo AB, et al. The influence of genetic and environmental factors among MDMA users in cognitive performance. PLoS One. 2011;6:e2720–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertolino A, Di GA, Blasi G, et al. Epistasis between dopamine regulating genes identifies a nonlinear response of the human hippocampus during memory tasks. Biol Psychiatry. 2008;64:226–234 [DOI] [PubMed] [Google Scholar]
- 55.Prata DP, Mechelli A, Fu CH, et al. Epistasis between the DAT 3′ UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proc Natl Acad Sci U S A. 2009;106:13600–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wetherill RR, Jagannathan K, Lohoff FW, et al. Neural correlates of attentional bias for smoking cues: modulation by variance in the dopamine transporter gene. Addict Biol. 2012;19:294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velakoulis D, Pantelis C, McGorry PD, et al. Hippocampal volume in first-episode psychoses and chronic schizophrenia: a high-resolution magnetic resonance imaging study. Arch Gen Psychiatry. 1999;56:133–141 [DOI] [PubMed] [Google Scholar]
- 58.Watson C, Andermann F, Gloor P, et al. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42:1743–1750 [DOI] [PubMed] [Google Scholar]
- 59.Good CD, Johnsrude IS, Ashburner J, et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1 Pt 1):21–36 [DOI] [PubMed] [Google Scholar]
- 60.Yucel M, Solowij N, Respondek C, et al. Regional brain abnormalities associated with long-term heavy cannabis use. Arch Gen Psychiatry. 2008;65:694–701 [DOI] [PubMed] [Google Scholar]
- 61.Ashtari M, Avants B, Cyckowski L, et al. Medial temporal structures and memory functions in adolescents with heavy cannabis use. J Psychiatr Res. 2011;45:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burston JJ, Wiley JL, Craig AA, et al. Regional enhancement of cannabinoid CB1 receptor desensitization in female adolescent rats following repeated Delta-tetrahydrocannabinol exposure. Br J Pharmacol. 2010;161:103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhattacharyya S, Atakan Z, Martin-Santos R, et al. Preliminary report of biological basis of sensitivity to the effects of cannabis on psychosis: AKT1 and DAT1 genotype modulates the effects of delta-9-tetrahydrocannabinol on midbrain and striatal function. Mol Psychiatry. 2012;17:1152–1155 [DOI] [PubMed] [Google Scholar]
- 64.Bhattacharyya S, Fusar-Poli P, Borgwardt S, et al. Modulation of mediotemporal and ventrostriatal function in humans by Delta9-tetrahydrocannabinol: a neural basis for the effects of Cannabis sativa on learning and psychosis. Arch Gen Psychiatry. 2009;66:442–451 [DOI] [PubMed] [Google Scholar]
- 65.Bloomfield MA, Morgan CJ, Egerton A, et al. Dopaminergic function in cannabis users and its relationship to cannabis-induced psychotic symptoms. Biol Psychiatry. 2014;75:470–478 [DOI] [PubMed] [Google Scholar]
- 66.Bloomfield MA, Ashok AH, Volkow N, et al. The effects of Δ9-tetrahydrocannabinol on the dopamine system. Nature. 2016;539:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santiago M, Matarredona ER, Granero L, et al. Neurotoxic relationship between dopamine and iron in the striatal dopaminergic nerve terminals. Brain Res. 2000;858:26–32 [DOI] [PubMed] [Google Scholar]
- 68.Zhou QY, Palmiter RD. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209 [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharyya S, Crippa JA, Martin-Santos R, et al. Imaging the neural effects of cannabinoids: current status and future opportunities for psychopharmacology. CurrPharmDes. 2009;15:2603–2614 [DOI] [PubMed] [Google Scholar]
- 70.Yücel M, Lorenzetti V, Suo C, et al. Hippocampal harms, protection and recovery following regular cannabis use. Transl Psychiatry. 2016;6:e71–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winton-Brown TT, Allen P, Bhattacharyya S, et al. Modulation of auditory and visual processing by delta-9-tetrahydrocannabinol and cannabidiol: an FMRI study. Neuropsychopharmacology. 2011;36:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hindocha C, Freeman TP, Schafer G, et al. Acute effects of delta-9-tetrahydrocannabinol, cannabidiol and their combination on facial emotion recognition: a randomised, double-blind, placebo-controlled study in cannabis users. Eur Neuropsychopharmacol. 2015;25:325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Izzo AA, Borrelli F, Capasso R, et al. Non-psychotropic plant cannabinoids: new therapeutic opportunities from an ancient herb. Trends Pharmacol Sci. 2009;30:515–527 [DOI] [PubMed] [Google Scholar]
- 74.Zuardi AW, Crippa JA, Hallak JE, et al. A critical review of the antipsychotic effects of Cannabidiol: 30 years of a translational investigation. Curr Pharm Des. 2012;18:5131–5140 [DOI] [PubMed] [Google Scholar]
- 75.Di Forti M, Marconi A, Carra E, et al. Proportion of patients in south London with first-episode psychosis attributable to use of high potency cannabis: a case-control study. Lancet Psychiatry. 2015;2:233–238 [DOI] [PubMed] [Google Scholar]
- 76.Harrisberger F, Smieskova R, Schmidt A, et al. BDNF Val66Met polymorphism and hippocampal volume in neuropsychiatric disorders: a systematic review and meta-analysis. Neurosci Biobehav Rev. 2015;55:107–118 [DOI] [PubMed] [Google Scholar]
- 77.Barnes A, Isohanni M, Barnett JH, et al. No association of COMT (Val158Met) genotype with brain structure differences between men and women. PLoS One. 2012;7:e3396–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertolino A, Fazio L, Di GA, et al. Genetically determined interaction between the dopamine transporter and the D2 receptor on prefronto-striatal activity and volume in humans. J Neurosci. 2009;29:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caldu X, Vendrell P, Bartres-Faz D, et al. Impact of the COMT Val108/158 Met and DAT genotypes on prefrontal function in healthy subjects. Neuroimage. 2007;37:1437–1444 [DOI] [PubMed] [Google Scholar]
- 80.Chye Y, Suo C, Yücel M, et al. Cannabis-related hippocampal volumetric abnormalities specific to subregions in dependent users. Psychopharmacology. 2017;234:2149–2157 [DOI] [PubMed] [Google Scholar]
- 81.Lorenzetti V, Solowij N, Whittle S, et al. Gross morphological brain changes with chronic, heavy cannabis use. Br J Psychiatry. 2015;206:77–78 [DOI] [PubMed] [Google Scholar]
- 82.Jovicich J, Czanner S, Han X, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46:177–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glass M, Faull RLM, Dragunow M. Cannabinoid receptors in the human brain: a detailed anatomical and quantitative autoradiographic study in the fetal, neonatal and adult human brain. Neuroscience. 1997;77:29–9. [DOI] [PubMed] [Google Scholar]
- 84.Pagliaccio D, Barch DM, Bogdan R, et al. Shared predisposition in the association between cannabis use and subcortical brain structure. JAMA Psychiatry. 2015;72:994–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durazzo TC, Meyerhoff DJ, Nixon SJ. Interactive effects of chronic cigarette smoking and age on hippocampal volumes. Drug Alcohol Depend. 2013;133:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mackey S, Kan K, Chaarani B, et al. Genetic imaging consortium: from neuroimaging to genes. Prog Brain Res. 2016;224:203–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schumann G, Loth E, Banaschewski T, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–1139 [DOI] [PubMed] [Google Scholar]
References
Cite this article as: Batalla A, Lorenzetti V, Chye Y, Yücel M, Soriano-Mas C, Bhattacharyya S, Torrens M, Crippa JAS, Martín-Santos R (2018) The influence of DAT1, COMT, and BDNF genetic polymorphisms on total and subregional hippocampal volumes in early onset heavy cannabis users, Cannabis and Cannabinoid Research 3:1, 1–10, DOI: 10.1089/can.2017.0021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


