Abstract
Purpose
Next generation sequencing (NGS) often identifies multiple rare predicted-deleterious variants (RDVs) in different genes associated with a recessive disorder in a given patient. Such variants have been proposed to contribute to digenicity/oligogenicity or “tri-allelism”, or to act as genetic modifiers.
Methods
Using the recessive ciliopathy Joubert syndrome (JBTS) as a model, we investigated these possibilities systematically, relying on NGS of known JBTS genes in a large JBTS and two control cohorts.
Results
65% of affected individuals had a recessive genetic cause, while 4.9% were candidates for di-/oligogenicity, harboring heterozygous RDVs in ≥2 genes, compared to 4.2–8% in controls (p = 0.66–0.21). Based on ExAC allele frequencies, the probability of cumulating RDVs in any two JBTS genes is 9.3%. We found no support for “tri-allelism” as no unaffected siblings carried the same bi-allelic RDVs as their affected relative. 60% of individuals sharing identical causal RDVs displayed phenotypic discordance. While 38% of affected individuals harbored RDVs in addition to the causal mutations, their presence did not correlate with phenotypic severity.
Conclusion
Our data offer little support for “tri-allelism” or digenicity/oligogenicity as clinically-relevant inheritance modes in JBTS. While phenotypic discordance supports the existence of genetic modifiers, identifying clinically-relevant modifiers remains challenging.
Keywords: oligogenicity, genetic modifiers, ciliopathies, Joubert syndrome
INTRODUCTION
Next-generation sequencing (NGS) has revolutionized the identification of genes underlying rare diseases, leading to the recognition that many Mendelian conditions are genetically heterogeneous1,2. The genes associated with a given disorder often function together in a biological pathway or network, leading to the reclassification of some seemingly disparate disorders into disease-groups unified by common pathophysiological mechanisms. Among the most striking examples are the ciliopathies, where individual disorders share causal genes and key phenotypic features (retinal dystrophy, fibro-cystic kidney disease, hepatic fibrosis, polydactyly)3.
Joubert syndrome (JBTS, MIM PS213300) is a prototypical ciliopathy, characterized by a pathognomonic hindbrain imaging finding called the Molar Tooth Sign (MTS)4,5. In addition to this core clinical feature, JBTS exhibits substantial phenotypic variability with 60% of individuals displaying additional features that span the entire spectrum of ciliopathy phenotypes6,7. JBTS follows a recessive inheritance pattern (or X-linked in the case of OFD1) with prominent genetic heterogeneity, since mutations in ~30 genes have been shown to cause this disorder6,8. Mutations in several of these genes also cause the more severe Meckel syndrome (MKS, MIM PS249000), which typically results in fetal or neonatal demise9.
Given the marked genetic heterogeneity of JBTS/MKS, identifying the genetic diagnosis is now typically accomplished using NGS, which often leads to the identification of rare, predicted-deleterious variants (RDVs) in ≥2 known JBTS genes in a given individual. Such findings have been suggested to lead to “oligogenicity,”10–21 a term which has been used to describe a number of distinct situations in the context of ciliopathies. Most commonly, oligogenicity refers to an inheritance mode in which variants in different genes contribute to disease causality. Digenic inheritance, in which single heterozygous RDVs in two different genes cause disease, has been proposed for a small number of families with nephronophthisis, a ciliopathy characterized by fibro-cystic renal disease (NPHP, MIM 256100) and in JBTS10–14. In addition, the combination of bi-allelic RDVs in one gene with a third RDV in a second gene has been proposed as a novel inheritance mode (called “tri-allelism”) in the related ciliopathy Bardet-Biedl syndrome (BBS, MIM 209900)11,15,16. Finally, given the prominent phenotypic variability observed in ciliopathies, the term oligogenicity has also been used to describe the idea that variants in other genes, present in addition to bi-allelic RDVs in one causal gene, may affect the phenotypic outcome8, a situation more specifically designated by the term “genetic modifiers”17–21.
While each of these situations has been proposed in a handful of families with diverse ciliopathies, it remains unclear how widespread such findings are, and how the occurrence of multiple variants in genes associated with the same recessive disorder should be interpreted and/or reported. To answer these questions, we took a systematic approach: Using NGS of 25 known JBTS genes in a large JBTS cohort and comparing our results with data from a similarly sequenced control cohort, the UK1958 birth cohort, and population data from ExAC, we investigated the evidence for (1) digenic/oligogenic inheritance, (2) “tri-allelism” and (3) genetic modifiers.
MATERIALS AND METHODS
Subject ascertainment and phenotypic data
Enrollment criteria for the University of Washington (UW) Joubert Syndrome Research Program and methodology for collection of clinical data were previously described6. All participants were enrolled with written informed consent for the study approved by the UW Institutional Review Board. Neurologically Normal Caucasian Control Panels (NINDS; Coriell panels NDPT020 and NDPT090 - http://ccr.coriell.org) were sequenced as controls.
Mutation identification
Using Molecular Inversion Probes (MIPs)22,23, we sequenced 25 genes associated with JBTS (Table S1) as previously described6. We included only samples with ≥25X coverage for >80% of the targeted base pairs and considered initially only high quality variants (Depth ≥25, Quality by Depth >5). We used the Combined Annotation Dependent Depletion (CADD) algorithm to estimate the predicted deleteriousness of variants (version 1.3; http://cadd.gs.washington.edu/score). Only rare deleterious variants (RDVs) were retained for analysis, as defined by a minor allele frequency (MAF) <0.2% in the Exome Variant Server (EVS, http://evs.gs.washington.edu/EVS/), and a CADD score ≥15. The CADD score cut-off was determined by maximizing the number of affected individuals “solved,” while minimizing false-positives (Figure S1), as previously described6. When DNA from unaffected siblings was available, the presence or absence of the causal RDVs identified in the proband was determined using targeted Sanger sequencing. We identified copy number variations (CNVs) using the relative read depth of individual MIPs across sequenced samples as previously described23.
Analysis of population data from ExAC and the UK1958 birth cohort
Per sample-exome sequencing data from 907 individuals in the UK1958 birth cohort (www.cls.ioe.ac.uk/ncds) was filtered for RDVs in the 25 JBTS genes using the same criteria as for our in house sequenced controls and JBTS cohort (MAF<0.2%, CADD≥15).
Only samples of European Ancestry in ExAC were considered to match the Caucasian NINDS control panel sequenced. NPHP1 was excluded for this analysis given that only deletions in this gene have been associated with JBTS. For each of the 24 genes analyzed, we calculated the joint RDV allele frequency by adding the frequencies of individual alleles that had a CADD score ≥15 and MAF<0.2% in European samples. Since each individual has two alleles, carrier rates for RDVs in each gene were estimated to be twice this joint allele frequency. To calculate the likelihood of harboring combinations of heterozygous RDVs in any two of 24 JBTS genes, we multiplied the carrier rates for each gene (independent probabilities), for all possible pairs of genes (1+2, 1+3, etc) and summed the results (Table S2).
Statistical Methods
Comparisons between groups were performed using Chi-square or exact binomial tests as appropriate. Significance of correlations between number of RDVs and number of phenotypic features was tested using Pearson’s correlation coefficient.
RESULTS
Solve rate of 65% in a classical recessive inheritance model
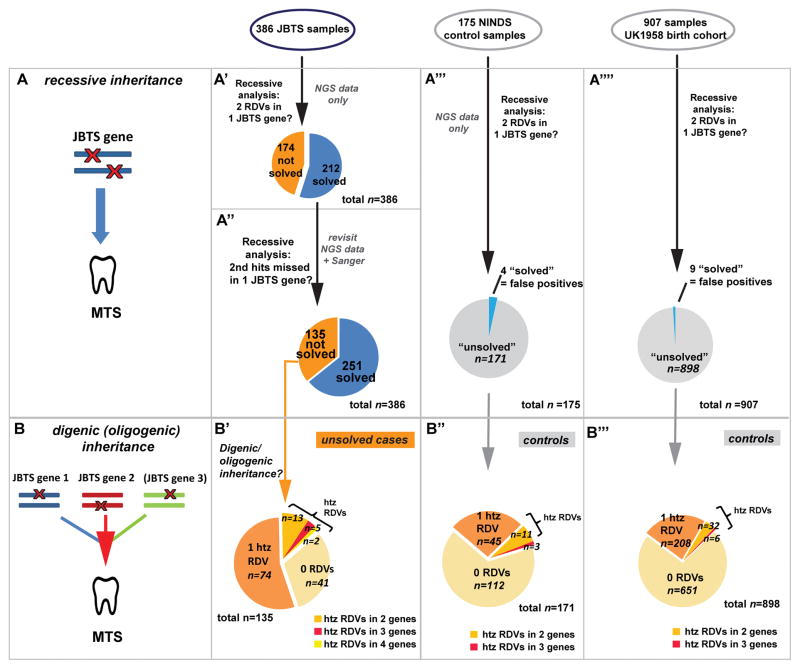
To estimate whether digenic/oligogenic inheritance modes may occur in JBTS, we first determined the proportion of individuals solved using a classical recessive model, i.e. bi-allelic mutations in one known JBTS gene (Figure 1A). We sequenced 25 known JBTS genes in 386 individuals from 333 families and 175 individuals without neurological disease from NINDS (Table S1). On average, 93% of targeted base pairs in affected samples and 95% in controls had ≥25X coverage. Rare deleterious variants (RDVs) were defined by a minor allele frequency (MAF) <0.2% in the Exome Variant Server and a CADD score ≥15. Using these methods, we were able to identify the presumed genetic cause in 212/386 affected individuals (55%; Figure 1A′), with a false positive rate of 2.3% (4/175 controls harbored two RDVs in one JBTS gene; Figure 1A‴). Among the remaining 174 “unsolved” individuals with JBTS, 48 had no RDVs in any of the 25 genes screened and 126 had single RDVs in at least one gene.
Figure 1. Recessive versus digenic/oligogenic inheritance models tested.
(A) Classical recessive inheritance requires bi-allelic (homozygous or compound heterozygous) rare deleterious variants (RDVs) in one disease gene to cause the disorder (represented by the molar tooth sign). (A′-A‴′) Recessive analysis of sequencing data for 25 Joubert syndrome (JBTS) genes in 386 affected individuals with JBTS (A′), 175 NINDS controls (A‴) and 907 UK1958 controls (A‴′) identified two RDVs in the same gene in 212 affected individuals (“solved samples”, blue in A′), in 4 NINDS and 9 UK1958 controls (“false positives”, blue in A‴ and A‴′). (A″) Reassessment of NGS and additional Sanger sequencing data increased the number of “genetically solved” affected individuals to 251 in a recessive model (65% solve rate).
(B) Digenic/oligogenic inheritance as considered here is based on presence of combined single heterozygous RDVs in ≥2 JBTS genes. (B′) Focusing on 135 individuals with JBTS without a recognized recessive genetic cause, we identified 20 individuals harbouring RDVs in two or more JBTS genes. (B″) 14/171 NINDS and (B‴) 38/898 UK1958 controls harboured RDVs in two or more JBTS genes.
NGS Next Generation Sequencing, MTS Molar Tooth Sign, htz heterozygous, RDVs Rare Deleterious Variants, JBTS Joubert Syndrome.
The large proportion of affected individuals carrying single heterozygous RDVs in JBTS genes suggests a second RDV in the same gene might have been missed. To identify second RDVs and search for additional recessive causes, we evaluated lower quality RDVs that had been excluded and previously generated Sanger sequencing data, searched for insertions/deletions in the 25 analyzed genes and sequenced novel JBTS genes (KIAA0586 and C2CD3). Including these results, we were able to identify recessive causes in 39 additional individuals, increasing the total solve rate to 65% (251/386; Figure 1A″).
Variant types and distribution differ between controls and individuals with JBTS
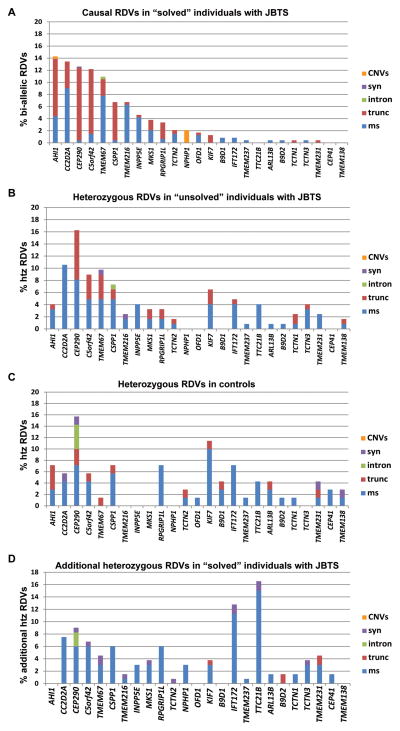
We next investigated whether the predicted functional effects of variants differed between affected individuals and controls by comparing the distribution and functional categories (truncating, missense, intronic, synonymous, indels) of RDVs identified in controls with the presumed causal RDVs in “solved” affected individuals. Truncating variants (stop-gain, canonical splice site and frameshift) comprised 54% of all causal variants in “solved” individuals, ranging between 9% (INPP5E) and 95% (CEP290 or CSPP1) (Figure 2A). In contrast, the overall proportion of truncating RDVs in controls was much lower (13/76=17% vs. 256/474=54%, p<0.0001, Chi-square). This difference was most striking for CEP290 RDVs (and to a lesser extent KIF7 RDVs), which were highly over-represented in controls and mostly non-truncating, while causal RDVs in these genes are almost exclusively truncating (Figure 2A and C).
Figure 2. Distribution and functional categories of RDVs found in affected individuals with Joubert syndrome and controls.
Distribution and RDV types by gene: (A) causal alleles in “solved” individuals with JBTS (two RDVs in the same JBTS gene), (B) “unsolved” individuals with JBTS, (C) NINDS controls (excluding the 4 individuals with two RDVs in one JBTS gene), and (D) heterozygous RDVs in “solved” affected individuals present in addition to the presumed causal alleles. Note the overrepresentation of CEP290 and KIF7 RDVs in controls (C), with a predominance of non-truncating RDVs; the mixed pattern of RDVs in unsolved samples (B), combining features from causal alleles (A) and from controls (C); the overrepresentation of RDVs in intra-flagellar transport-related genes (IFT172 and TTC21B) and the quasi absence of truncating additional RDVs as additional RDVs in “solved” affected individuals (D). RDV Rare Deleterious Variant, CNV Copy Number Variants, syn synonymous, intron intronic, trunc truncating, ms missense, htz heterozygous, JBTS Joubert syndrome.
We next compared the RDVs identified in “unsolved” affected individuals to the presumed-causal RDVs in “solved” affected individuals and to those identified in controls. Interestingly, the variant types in “unsolved” individuals appeared to display an intermediate pattern (Figure 2B). For instance, CEP290 and KIF7 RDVs were also over-represented in “unsolved” affected individuals like in controls, but truncating RDVs across all genes were significantly more common in “unsolved” individuals than in controls (52/162=32% in unsolved affected individuals vs. 13/76=17% in controls, p=0.02, Chi-square). This finding suggests that among the RDVs identified in “unsolved” affected individuals, some are contributing to disease, while others likely represent benign variants. It does not differentiate whether their putative contribution to disease follows a classical recessive model, where the second RDV in the same gene has been missed, versus these RDVs contributing to digenic/oligogenic inheritance.
Minimal evidence for oligogenicity in JBTS based on coding RDVs in JBTS genes
Evidence for digenic/oligogenic inheritance requires identifying heterozygous RDVs in ≥2 JBTS genes in individuals who do not have two RDVs in a single gene (Figure 1B). In our cohort, 20 affected individuals harbored single heterozygous RDVs in ≥2 JBTS genes (two genes in 13 individuals, three genes in five, and four genes in two; Figure 1B′ and Table S3). Of these, one individual had an affected sibling who did not carry the same two RDVs, excluding these variants as the cause. Therefore, a digenic/oligogenic inheritance mode could explain at most 4.9% (19/386) of JBTS. For comparison, 14/175 (8%) controls harbored single heterozygous RDVs in ≥2 genes (two genes in 11 individuals, and three genes in three; Figure 1B″; 19/386 vs 14/175: p=0.21, Chi-square).
To further assess the frequency of control individuals harboring RDVs in ≥2 JBTS genes in a larger control population, we analyzed per-sample exome sequencing data from 907 individuals in the UK1958 birth cohort for RDVs (MAF<0.2%, CADD≥15) in the 25 JBTS genes. Similar to the NINDS controls, we identified 9 individuals (1%) harboring two RDVs in one JBTS gene (likely “false positives”), 208 (23%) carrying a single heterozygous RDV in one JBTS gene and 38 carrying single heterozygous RDVs in two (32) or three (6) JBTS genes (38/907=4.2%; Figure 1A‴′ and B‴ and Table 1). This proportion does not statistically differ from that observed in our JBTS cohort (19/386=4.9% vs 38/907=4.2%, p=0.66, Chi-square).
Table 1.
Comparison of population data from ExAC with observations from the JBTS cohort and the NINDs and UK1958 control cohorts for varying CADD score cut-offs
| CADD≥15 | CADD≥20 | CADD≥25 | truncating onlyb | |
|---|---|---|---|---|
| Joint allele frequency of all CC2D2A alleles in ExAC a (%) | 0.89 | 0.71 | 0.37 | 0.19 |
| CEP290 | 2.6 | 2.2 | 1.5 | 0.36 |
| AHI1 | 0.72 | 0.45 | 0.27 | 0.07 |
| TMEM67 | 0.6 | 0.55 | 0.32 | 0.15 |
| C5ORF42 | 1.55 | 1.17 | 0.6 | 0.16 |
| RPGRIP1L | 1.25 | 1 | 0.54 | 0.08 |
| INPP5E | 0.59 | 0.47 | 0.23 | 0.01 |
| CSPP1 | 1.11 | 0.89 | 0.49 | 0.10 |
| TMEM216 | 0.2 | 0.07 | 0.04 | 0.01 |
| TCTN1 | 0.6 | 0.53 | 0.33 | 0.06 |
| TCTN2 | 0.49 | 0.3 | 0.12 | 0.03 |
| TTC21B | 1.51 | 1.2 | 0.76 | 0.10 |
| KIF7 | 2.3 | 2 | 1.2 | 0.06 |
| TCTN3 | 0.36 | 0.24 | 0.11 | 0.03 |
| B9D1 | 0.86 | 0.51 | 0.15 | 0.01 |
| B9D2 | 0.25 | 0.2 | 0.09 | 0.00 |
| ARL13B | 0.79 | 0.69 | 0.17 | 0.01 |
| OFD1 | 0.42 | 0.3 | 0.17 | 0.03 |
| CEP41 | 0.29 | 0.23 | 0.16 | 0.01 |
| MKS1 | 1.08 | 0.7 | 0.49 | 0.01 |
| IFT172 | 2.35 | 1.5 | 0.96 | 0.03 |
| TMEM138 | 0.15 | 0.06 | 0.03 | 0.06 |
| TMEM231 | 0.87 | 0.75 | 0.39 | 0.06 |
| TMEM237 | 0.52 | 0.47 | 0.19 | 0.00 |
|
| ||||
| Solve rate (recessive model) in JBTS cohort | 251/386 | 243/386 | 170/386 | 111/386 |
|
| ||||
| False positive rate in NINDS controls | 4/175 | 3/175 | 3/175 | 1/175 |
|
| ||||
| False positive rate in UK1958 controls | 8/907 | 5/907 | 2/907 | 0/907 |
|
| ||||
| Observed rate of heterozygous RDVs in ≥2 JBTS genes in individuals with JBTS (% of total) | 4.9% (19/386) | 4.4% (17/386) | 3.1% (12/386) | 0.2% (1/386) |
|
| ||||
| Observed rate of heterozygous RDVs in ≥2 JBTS genes in NINDS control dataset | 8% (14/175) | 5.7% (10/175) | 2.3%(4/175) | 0 |
|
| ||||
| Observed rate of heterozygous RDVs in ≥2 JBTS genes in UK1958 birth cohort control dataset | 4.2% (39/907) | 2.9% (27/907) | 1.1% (10/907) | 0.2% (2/907) |
|
| ||||
| Calculated probability of finding heterozygous RDVs in any 2 of 24* JBTS genes based on ExAC joint allele frequencies | 9.3% | 3% | 1% | 0.04% |
Only individual alleles with a MAF <0.2% in ExAC were considered. Details for the calculations of disease prevalence and likelihood of RDVs in 2 JBTS genes are displayed in Supplementary Table S2.
Truncating variants are frameshift, stop-gain and canonical splice site variants.
NPHP1 was not considered for this calculation given that for this gene only deletions have been associated with JBTS. CADD Combined Annotation Dependent Depletion; RDV rare deleterious variant, JBTS Joubert Syndrome, ExAC Exome Aggregation Consortium, NINDS National Institute of Neurological Disease and Stroke.
While our findings indicate that combinations of RDVs in several JBTS genes do not cause disease in the majority of cases, they do not rule out disease causality in a minority of specific situations. Therefore, we further scrutinized the types of combined RDVs in the 19 affected individuals, 14 NINDS and 38 UK1958 controls. We found no significant difference in functional categories between affected individuals and either set of controls (Supplementary Figure S2A), and average CADD scores were comparable between cases and controls (Supplementary Figure S2B). The distribution of genes harboring RDVs was also not strikingly different, and no recurrent gene-specific association pattern was observed in affected individuals compared to controls (Table S3 and Supplementary Figure S2C).
High number of rare predicted-deleterious variants in JBTS genes in population data
We further compared the RDV frequencies observed in our NINDS controls to allele frequencies found in European samples in ExAC (Exome Aggregation Consortium, http://exac.broadinstitute.org)24. Using the same criteria for retaining ExAC variants as in our NINDS controls (MAF<0.2%, CADD≥15 or predicted truncating), and adding up the frequencies for individual alleles in each gene, the observed joint allele frequency by gene in European populations ranged from 0.15% for TMEM138 to 2.6% for CEP290, similar to the frequencies identified in NINDS controls (Table S4). Based on the allele frequencies from ExAC, the probability that a given individual carries heterozygous RDVs in any two of 24 JBTS genes (NPHP1 excluded) is 9.3% (Tables 1 and S2), similar to the 6.2% (11/175, Figure 1B″) observed in NINDS controls. This high prevalence of RDVs in two JBTS genes is not compatible with disease causality in the majority of cases, given the estimated joint prevalence of 1/25,000–1/30,000 for JBTS and MKS4,9. This observation remains true even when excluding RDVs with CADD scores <20 or <25 (Tables 1 and S2). In conclusion, combined RDVs in ≥2 JBTS genes occur commonly in unaffected individuals.
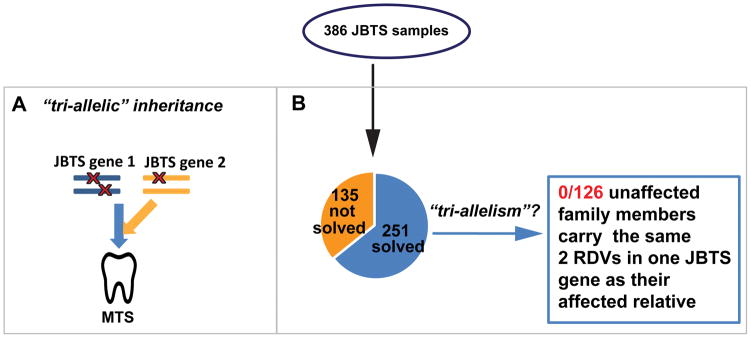
No evidence for “tri-allelism” in JBTS
The presence of two RDVs in one JBTS gene does not exclude the possibility that a third allele (in a different gene) might be required for disease occurrence, a mechanism previously described as “tri-allelism” (Figure 3A). If “tri-allelism” were the inheritance mode in a family, unaffected relatives should carry the same two alleles in one JBTS gene as the probands, presumably lacking the third allele. To address this possibility in JBTS, we sequenced 126 unaffected siblings of 87 probands with known genetic cause (Figure 3B). In our cohort, we did not identify any unaffected individuals who harbored the same two RDVs as their affected family member(s) (expected proportion of siblings segregating two RDVs in one JBTS gene regardless of phenotype = ¼ = 32/126 vs. observed 0/126 p<0.0001, exact.). While our findings do not rule out “tri-allelism” in all families, it is at most an exceptional occurrence in JBTS.
Figure 3. Investigating “tri-allelism” in Joubert syndrome (JBTS).
(A) Proposed “tri-allelic” inheritance: bi-allelic rare deleterious variants (RDVs) in a first gene AND one additional heterozygous RDV in a second gene are required to cause the disorder (represented by the characteristic Molar Tooth Sign (MTS) of JBTS). (B) In this JBTS cohort, no unaffected family members were identified carrying the same two presumed causal RDVs in one JBTS gene as their affected relative.
Phenotypic discrepancies between individuals sharing the same causal mutations support the existence of genetic modifiers
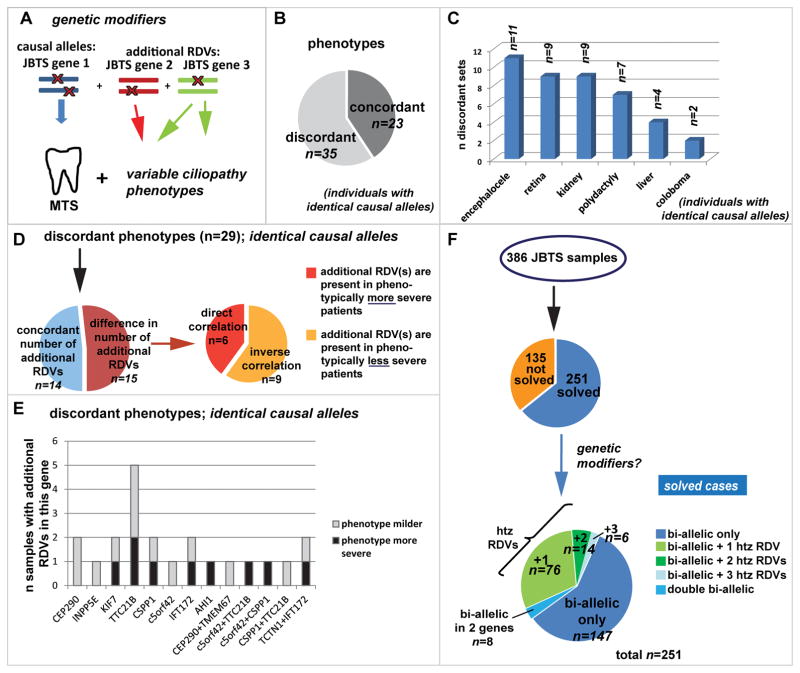
The highly variable phenotypic presentation in JBTS prompted the hypothesis that RDVs in JBTS genes, present in addition to the bi-allelic causal RDVs, act as genetic modifiers (Figure 4A)8. Since some genotype-phenotype correlations have been identified,6,25–28 substantial phenotypic variability could be explained by allele-specific consequences. To control for the influence of the causal mutations, we considered 58 sets of individuals sharing identical causal alleles, including families with multiple affected individuals, as well as unrelated individuals with the same causal mutations (Supplementary Figure S3A). A discordant phenotype, where other ciliopathy features were present in addition to the MTS in some but not other affected individuals, was observed in 35/58 sets (60%; Figure 4B). This included progressive features such as retinal or renal disease (discrepant in 9 situations each) and non-progressive, readily recognizable features such as encephalocele (n=11), polydactyly (n=7) or coloboma (n=2) (Figure 4C). These observations support the hypothesis that genetic modifiers affect the severity of JBTS.
Figure 4. Investigating genetic modifiers among JBTS genes in Joubert syndrome.
(A) Genetic modifiers: Mutations in JBTS gene 2 and 3 modify the phenotypic outcome of the disorder, which is caused by bi-allelic mutations in JBTS gene 1. (B) Pie-chart indicating the proportion of affected individuals sharing identical causal RDVs but for whom phenotypic features are discordant. (C) Bar graph showing the discordant phenotypic features present or not in addition to the Molar Tooth Sign in individuals with shared causal RDVs. (D) The left pie chart indicates for 29 sets of affected individuals with shared causal alleles but discordant phenotypes, the proportion with concordant (blue) or discordant (red) numbers of additional RDVs. Note that phenotypic discordance was observed despite concordant numbers of additional RDVs 50% of the time. The right pie chart shows for those individuals with discordant phenotypes AND discordant RDV numbers (n=15), the proportion of patients in whom the additional RDVs correlated with more severe and with less severe phenotypes. (E) For individuals with shared causal alleles: Bar graph indicating the number of instances where presence of additional RDVs in each of the displayed genes corresponded with a milder (grey) or with a more severe (black) phenotype than in individuals lacking these additional RDVs. (F) Considering the entire JBTS cohort (n=386 individuals), we identified additional heterozygous (htz) RDVs in 96/251(=38%) “genetically solved” individuals with JBTS. Eight affected individuals carried two RDVs in each of two different genes.
Lack of correlation between additional RDVs in JBTS genes and phenotypic severity
A correlation between presence of additional RDVs and phenotypic outcomes (Figure 4A) would further support the existence of genetic modifiers. Among 29 sets of affected individuals with shared causal alleles and discordant phenotypic features, for whom NGS data was available in all individuals, we found no difference in number of additional RDVs for 14 sets (Figure 4D). 15/29 sets with discordant phenotypes had differing numbers of additional RDVs, but these did not correlate with the presence of additional phenotypic features (Pearson’s correlation coefficient −0.06) (Figure 4D; Table S5). We further questioned whether additional RDVs in specific genes correlated with phenotypic severity (Figure 4E) or whether additional RDVs were more common with specific causal genes (Supplementary Figure S3C), but did not detect any obvious gene-specific trends.
To expand our analysis to the entire cohort, we analyzed all heterozygous RDVs present in addition to the two causal RDVs in 251 “solved” individuals and searched for a correlation with disease severity. While additional heterozygous RDVs in ≥1 JBTS gene were observed in 38% of individuals (96/251; Figure 4F), their presence/number did not correlate with presence/absence of additional phenotypic features (Pearson’s correlation coefficient −0.02; Table S5 and Supplementary Figure S3B). Interestingly, the distribution and functional categories of additional heterozygous RDVs observed in “solved” individuals with JBTS diverged from those in controls; almost no additional truncating alleles were present in “solved” individuals, while variants in TTC21B and IFT172 were overrepresented compared to controls (Figure 2D). The presence of additional RDVs in these two genes did however not correlate with disease severity (Figure 4E).
In eight affected individuals, we identified two RDVs in each of two different JBTS genes (Table S6). Remarkably, only one of these eight patients (UW050-6) had a more severe phenotype (encephalocele) compared to his affected relative carrying the same RDVs in only one of the genes. No major phenotypic differences were present in the other individuals, indicating that even the presence of two RDVs in a second JBTS gene does not necessarily enhance phenotypic severity.
DISCUSSION
Joubert syndrome is a recessive disorder
Our findings lend little support for coding RDVs in JBTS genes acting through non-Mendelian inheritance modes as clinically relevant disease mechanisms.
We found no support for “tri-allelism”, as previously reported in Bardet-Biedl syndrome15, based on the absence of unaffected siblings carrying the same bi-allelic causal RDVs as their affected relative. Other recent work has failed to support this particular inheritance mode in other ciliopathies 29–31.
Digenic inheritance, which is well-documented and occasionally supported by functional studies in other disorders11,32, has been proposed in ciliopathies based on isolated reports of individuals harboring heterozygous coding variants in two ciliopathy genes. Our systematic analysis found little evidence to support oligogenicity based on coding RDVs in 25 known JBTS genes: In our JBTS cohort, only a small number of individuals harbor RDVs in ≥2 JBTS genes, indicating that such digenic/oligogenic inheritance relying on combinations of RDVs in JBTS genes could at most account for 5% of JBTS. Moreover, the probability of combined occurrence of RDVs in any two JBTS genes is 9.3% based on ExAC allele frequencies, a probability that increases with the number of JBTS genes identified. Accordingly, we identified high rates of control individuals with RDVs in ≥2 JBTS genes in the NINDS and UK1958 control datasets, indicating that such combinations of RDVs do not cause disease in the majority of cases. While this does not exclude digenic/oligogenic inheritance in a minority of cases through specific combinations of alleles or genes, we did not identify such situations in our analysis.
The high overall proportion of heterozygous truncating RDVs in “unsolved” affected individuals suggests that some of these RDVs are contributing to disease. Based on our experience identifying additional second “hits” in individuals with single RDVs, these heterozygous RDVs most likely act in a recessive inheritance mode where second RDVs in the same gene were missed with NGS due to incomplete coverage, poor-quality sequence, difficulties detecting small indels and larger copy-number variants 33 or location of variants outside the targeted regions (non-coding variants).
Oligogenicity as a disease mechanism for JBTS cannot be fully excluded by our work, since we did not evaluate variants in genes not associated with JBTS, nor did we evaluate non-coding variants. In fact, oligogenicity could only be fully excluded if a bona fide recessive cause were identified in every family with JBTS. However, until stronger genetic or functional evidence for alternative inheritance modes exists, we feel that digenicity/oligogenicity and “tri-allelism” based on RDVs in JBTS should not be routinely included in clinical genetic counseling for JBTS.
Lessons learned from population data: importance of gene-specific variant patterns for clinical variant interpretation
Allele frequencies in controls and ExAC revealed much higher than expected “carrier” rates for RDVs in JBTS genes. In fact, 9 individuals from the UK1958 cohort and 4 individuals in the NINDS control cohort have two RDVs in one JBTS gene. If these RDVs actually caused JBTS, the prevalence would be 1–2%, drastically higher than the observed 1/50,000–80,0004. While segregation analysis could not be performed for the control cohorts (due to lack of parental samples), it is likely that many of these unaffected individuals carry the RDVs in trans. This indicates that a large proportion of rare alleles in controls are not disease-causing despite prediction algorithms. Consistent with this hypothesis, the RDVs were much more often non-truncating in controls compared to affected individuals. For genes such as CEP290, CSPP1 and C5ORF42, in which disease alleles are almost exclusively truncating6,34,35, this suggests that non-truncating alleles are unlikely to cause JBTS or other ciliopathies, regardless of their predicted deleteriousness. Conversely, for genes such as CC2D2A, TMEM67 or INPP5E, missense variants are more likely to be pathogenic, since they are commonly causal alleles in affected individuals.
Phenotypic discrepancies support the existence of genetic modifiers
Previously identified gene-phenotype correlations indicate that the causal mutations play an important role in determining the phenotypic severity. However, phenotypic discrepancies between family members with ciliopathies have been reported36. Recent work considering a large cohort with multiple different ciliopathies found a high concordance rate between causal alleles and disease category33. Our work focusing on a single ciliopathy with variable phenotypic features shows that phenotypic discordance of both progressive and non-progressive features of JBTS is very common, even among individuals with identical causal alleles. These findings strongly support the existence of additional genetic and/or environmental factors that influence phenotypic outcome within a given ciliopathy disorder.
Lack of correlation between additional RDVs in JBTS genes and disease severity
Despite this strong evidence for genetic modifiers, we were unable to identify a correlation between additional RDVs in JBTS genes and presence of phenotypic features. However, we made the intriguing observation that almost no truncating variants are found among the additional RDVs in affected individuals with established recessive causes. This could indicate that affected individuals are intolerant to additional highly deleterious variants in JBTS genes, which, if present, would lead to early lethality or phenotypes more severe than JBTS. This observation could be further evaluated by performing a similar analysis in a MKS cohort where affected individuals might harbor more heterozygous truncating variants in addition to their causal RDVs.
One explanation to the lack of correlation between RDV number and phenotypic severity in our analysis could be that additional RDVs may have opposite effects on the phenotype, some enhancing disease severity while others may play a protective role. In this case, the type of RDVs rather than the number should correlate with disease severity. Alternatively, JBTS genes may not act as genetic modifiers of each other, but rather variants in other ciliary (or non-ciliary) genes may affect the phenotype in JBTS more strongly, as suggested by recent work in animal models37. Sequencing non-JBTS associated ciliopathy genes could address this hypothesis. Finally, the phenotypic variability may be caused by multiple relatively common variants of modest effect that are not predicted to be highly deleterious, including non-coding variants affecting gene expression levels as recently described in C.elegans38. Given the rarity of JBTS and its prominent genetic heterogeneity, these possibilities will be challenging to test using human genetics alone, so functional studies may play a crucial role in recognizing genetic modifiers among more common variants.
Limitations
The varying quality of available phenotypic information on study subjects represents one limitation that could affect our analysis of genetic modifiers. Indeed, we relied on medical records of varying completeness as a result of the world-wide recruitment necessary given the rarity of the disorder. In addition, low power of statistical analyses is consequent to small numbers, a limitation inherent to the study of very rare disorders. The fact that only 25 of the ~30 JBTS-associated genes were analyzed likely has little impact on the conclusions of this work, since adding more genes would increase the number of individuals carrying RDVs in ≥2 genes in both affected individuals and in controls. Direct comparisons of the UK cohort to our in-house sequenced cohorts need to be taken with caution given the different sequencing methodologies. However, this limitation is mitigated by the exclusive inclusion of high-quality variants for the analyses.
Caution in interpreting the significance of variant burden in recessive disease genes
As more genes are associated with human disorders, genome-wide sequencing will increasingly identify RDVs in multiple genes associated with recessive disease in any given individual. Our systematic study using JBTS is a call for cautious interpretation of their clinical significance. Incorporating gene-specific variant information and patient-specific clinical considerations is critical, so a good flow of information between diagnostic laboratories, disease-specialized research groups and clinicians remains the cornerstone for accurate interpretation of sequencing findings. Going forward, variant characterization is being improved on multiple fronts, including guidelines for variant interpretation39 and large-scale variant sharing and curation efforts such as the Clinical Genome Resource (ClinGen; https://www.clinicalgenome.org/40).
Supplementary Material
Acknowledgments
We are grateful to all individuals with Joubert syndrome and their families for their participation. We also thank the Joubert Syndrome and Related Disorders Foundation and the innumerable clinicians who have referred participants to us over many years. We thank Elizabeth Blue and Michael Bamshad for comments on the manuscript, Pascal Joset and Anita Rauch for assistance with acquisition of the exome sequencing data from the 1958 Birth Cohort. This work made use of data and samples generated by the 1958 Birth Cohort (NCDS), which is managed by the Centre for Longitudinal Studies at the UCL Institute of Education.
Funding: R.B.-G. was supported by a Swiss NSF grant Ambizione-SCORE PZ00P3_142404/1. D.D. received support from the National Institutes of Health R01NS064077, a March of Dimes Basil O’Connor Starter Scholar Research Award, The Arc of Washington Trust Fund, and private donations from families of children with Joubert syndrome. The work was also supported by the University of Washington Intellectual and Developmental Disabilities Research Center Genetics Core (National Institutes of Health U54HD083091). The 1958 Birth Cohort (NCDS) is managed by the Centre for Longitudinal Studies at the UCL Institute of Education, funded by the Economic and Social Research Council (grant number ES/M001660/1). Access to these resources was enabled via the 58READIE Project funded by Wellcome Trust and Medical Research Council (grant numbers WT095219MA and G1001799). A full list of the financial, institutional and personal contributions to the development of the 1958 Birth Cohort Biomedical resource is available at www.cls.ioe.ac.uk/ncds.
Footnotes
Conflict of interest
The authors have no financial or commercial conflict of interest to declare.
References
- 1.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 2.Chong JX, Buckingham KJ, Jhangiani SN, et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badano JL, Mitsuma N, Beales PL, Katsanis N. The Ciliopathies: An Emerging Class of Human Genetic Disorders. Annu Rev Genom Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 4.Parisi MA, Glass I. Joubert syndrome and related disorders. 2003 doi: 10.1038/sj.ejhg.5201648. http://www.ncbi.nlm.nih.gov/books/NBK1325/ [DOI] [PubMed]
- 5.Maria BL, Hoang KBN, Tusa RJ, et al. “Joubert Syndrome” Revisited: Key Ocular Motor Signs With Magnetic Resonance Imaging Correlation. J Child Neurol. 1997;12:423–430. doi: 10.1177/088307389701200703. [DOI] [PubMed] [Google Scholar]
- 6.Bachmann-Gagescu R, Dempsey JC, Phelps IG, et al. Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J Med Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty D. Joubert Syndrome: Insights Into Brain Development, Cilium Biology, and Complex Disease. Semin Pediatr Neurol. 2009;16(3):143–54. doi: 10.1016/j.spen.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romani M, Micalizzi A, Valente EM. Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol. 2013;12:894–905. doi: 10.1016/S1474-4422(13)70136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barisic I, Boban L, Loane M, et al. Meckel-Gruber Syndrome: a population-based study on prevalence, prenatal diagnosis, clinical features, and survival in Europe. Eur J Hum Genet. 2015;23:746–752. doi: 10.1038/ejhg.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stokman M, Lilien M, Knoers N. Nephronophthisis. GeneReviews® [Internet] 2016 Available from: http://www.ncbi.nlm.nih.gov/books/NBK368475/
- 11.Schaffer AA. Digenic inheritance in medical genetics. J Med Genet. 2013;50:641–652. doi: 10.1136/jmedgenet-2013-101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoefele J, Wolf MTF, O’Toole JF, et al. Evidence of Oligogenic Inheritance in Nephronophthisis. J Am Soc Nephrol. 2007;18:2789–2795. doi: 10.1681/ASN.2007020243. [DOI] [PubMed] [Google Scholar]
- 13.Tory K, Lacoste T, Burglen L, et al. High NPHP1 and NPHP6 Mutation Rate in Patients with Joubert Syndrome and Nephronophthisis: Potential Epistatic Effect of NPHP6 and AHI1 Mutations in Patients with NPHP1 Mutations. J Am Soc Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- 14.Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet. 2012;44:193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsanis N, Ansley SJ, Badano JL, et al. Triallelic Inheritance in Bardet-Biedl Syndrome, a Mendelian Recessive Disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 16.Katsanis N, Eichers ER, Ansley SJ, et al. BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet. 2002;71:22–29. doi: 10.1086/341031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppieters F, Casteels I, Meire F, et al. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat. 2010;31:E1709–66. doi: 10.1002/humu.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie CM, Caridi G, Lopes VS, et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010;42:175–180. doi: 10.1038/ng.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna H, Davis EE, Murga-Zamalloa CA, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis EE, Zhang Q, Liu Q, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maglic D, Stephen J, Malicdan MCV, et al. TMEM231 Gene Conversion Associated with Joubert and Meckel-Gruber Syndromes in the Same Family. Hum Mutat. 2016;37:1144–1148. doi: 10.1002/humu.23054. [DOI] [PubMed] [Google Scholar]
- 22.O’Roak BJ, Vives L, Fu W, et al. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2015;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mougou-Zerelli S, Thomas S, Szenker E, et al. CC2D2A mutations in Meckel and Joubert syndromes indicate a genotype–phenotype correlation. Hum Mutat. 2009;30:1574–1582. doi: 10.1002/humu.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann-Gagescu R, Ishak GE, Dempsey JC, et al. Genotype–phenotype correlation in CC2D2A-related Joubert syndrome reveals an association with ventriculomegaly and seizures. J Med Genet. 2012;49:126–137. doi: 10.1136/jmedgenet-2011-100552. [DOI] [PubMed] [Google Scholar]
- 27.Iannicelli M, Brancati F, Mougou-Zerelli S, et al. Novel TMEM67 mutations and genotype-phenotype correlates in meckelin-related ciliopathies. Hum Mutat. 2010;31:E1319–E1331. doi: 10.1002/humu.21239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doherty D, Parisi MA, Finn LS, et al. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis) J Med Genet. 2010;47:8–21. doi: 10.1136/jmg.2009.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hichri H, Stoetzel C, Laurier V, et al. Testing for triallelism: analysis of six BBS genes in a Bardet-Biedl syndrome family cohort. Eur J Hum Genet. 2005;13:607–616. doi: 10.1038/sj.ejhg.5201372. [DOI] [PubMed] [Google Scholar]
- 30.Abu-Safieh L, Al-Anazi S, Al-Abdi L, et al. In search of triallelism in Bardet-Biedl syndrome. Eur J Hum Genet. 2012;20:420–427. doi: 10.1038/ejhg.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smaoui N, Chaabouni M, Sergeev YV, et al. Screening of the eight BBS genes in Tunisian families: no evidence of triallelism. Invest Ophthal Vis Sci. 2006;47:3487–3495. doi: 10.1167/iovs.05-1334. [DOI] [PubMed] [Google Scholar]
- 32.Lemmers RJLF, Tawil R, Petek LM, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaheen R, Szymanska K, Basu B, et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016;17:242. doi: 10.1186/s13059-016-1099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coppieters F, Lefever S, Leroy BP, Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 35.Tuz K, Bachmann-Gagescu R, O’Day DR, et al. Mutations in CSPP1 Cause Primary Cilia Abnormalities and Joubert Syndrome with or without Jeune Asphyxiating Thoracic Dystrophy. Am J Hum Genet. 2014;94:62–72. doi: 10.1016/j.ajhg.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaki MS, Sattar S, Massoudi RA, Gleeson JG. Co-occurrence of distinct ciliopathy diseases in single families suggests genetic modifiers. Am J Med Genet. 2011;155:3042–3049. doi: 10.1002/ajmg.a.34173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yee LE, Garcia-Gonzalo FR, Bowie RV, et al. Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling. PLoS Genet. 2015;11:e1005627. doi: 10.1371/journal.pgen.1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu V, Verster AJ, Schertzberg M, et al. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell. 2015;162:391–402. doi: 10.1016/j.cell.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 39.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the Clinical Genome Resource. NEJM. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.