Abstract
Ingestion of probiotics appears to have modest effects on the incidence of viral respiratory infection. The mechanism of these effects is not clear; however, there is evidence from animal models that the probiotic may have an effect on innate immune responses to pathogens. The purpose of this randomised, placebo-controlled study was to determine the effect of administration of Bifidobacterium animalis subspecies lactis Bl-04 on innate and adaptive host responses to experimental rhinovirus challenge. The effect on the response of chemokine (C-X-C motif) ligand 8 (CXCL8) to rhinovirus infection was defined as the primary endpoint for the study. 152 seronegative volunteers who had been supplemented for 28 days, 73 with probiotic and 79 with placebo, were challenged with RV-A39. Supplement or placebo administration was then continued for five days during collection of specimens for assessment of host response, infection, and symptoms. 58 probiotic and 57 placebo-supplemented volunteers met protocol-defined criteria for analysis. Probiotic resulted in higher nasal lavage CXCL8 on day 0 prior to virus challenge (90 vs 58 pg/ml, respectively, P=0.04, ANCOVA). The CXCL8 response to rhinovirus infection in nasal lavage was significantly reduced in the probiotic treated group (P=0.03, ANCOVA). Probiotic was also associated with a reduction in nasal lavage virus titre and the proportion of subjects shedding virus in nasal secretions (76% in the probiotic group, 91% in the placebo group, P=0.04, Fisher Exact test). The administration of probiotic did not influence lower respiratory inflammation (assessed by exhaled nitric oxide), subjective symptom scores, or infection rate. This study demonstrates that ingestion of Bl-04 may have an effect on the baseline state of innate immunity in the nose and on the subsequent response of the human host to rhinovirus infection. Clinicaltrials.gov registry number: NCT01669603.
Keywords: upper respiratory infection, innate immunity, common cold
1. Introduction
Probiotics are non-pathogenic living organisms that are ingested with the intent of providing a health benefit. The use of probiotics for treatment or prevention of illnesses related to intestinal infection or inflammation has been the primary focus of the use of these agents but recent studies have reported that ingestion of probiotics may be beneficial for prevention of upper respiratory illnesses (Hao et al., 2015). Systematic reviews of the use of probiotics for upper respiratory illnesses note that, while there appear to be modest effects on the incidence of illness, information about the role of probiotic bacterial strain and dose, efficacy in healthy adult populations, and mechanism of action against specific viral pathogens is incomplete (Esposito et al., 2014; Lehtoranta et al., 2014).
While potential mechanisms for an effect of probiotics on respiratory infection have not been defined, the interactions between the human microbiome and innate and adaptive immunity appear to be robust and complex (Winkler et al., 2007). The pathogenesis of common cold symptoms associated with rhinovirus infection is related to the innate inflammatory response to the virus. Multiple studies have demonstrated that nasal lavage concentrations of inflammatory cytokines, especially chemokine (C-X-C motif) ligand 8 (CXCL8), and the inflammatory response in the nose correlate with symptom severity (Barrett et al., 2006; Proud, 2008). Thus, interaction between the gut microbiome and the innate immune system is a potential mechanism by which a probiotic might affect viral upper respiratory disease. Studies in animal models suggest that the gut microbiome provides a low-level stimulation to the innate immune system that modulates the susceptibility of the host to viral infection (Pang and Iwasaki, 2012). The concept that innate ‘tone’ can be altered by non-pathogenic organisms to modify the host response to the viral infection was examined in this study of the effect of probiotics on host responses to rhinovirus infection.
Bifidobacterium animalis subsp. lactis Bl-04 (Bl-04) has been previously reported to decrease the risk of upper respiratory illness in the natural setting (West et al., 2014b). However, an extensive analysis of cytokines and immune cell profiles in plasma revealed no differences attributable to the probiotic in healthy individuals (West et al., 2014a). The purpose of this study was to conduct a detailed assessment of the effect of ingestion of Bl-04 on the host response to rhinovirus infection in the upper respiratory tract of human volunteers using the experimental rhinovirus challenge model.
2. Methods
Subjects
Healthy adult volunteers were recruited from the University of Virginia (UVa) community. This study was reviewed and approved by the by the Institutional Review Board for Health Sciences Research at the University of Virginia. Written informed consent was obtained prior to study participation and the study was conducted in compliance with Good Clinical Practices and in accordance with the Declaration of Helsinki. Subjects were compensated for participation. Volunteers susceptible to rhinovirus type 39, as evidenced by a serum neutralising antibody titre of ≤1:4, were invited to participate. Volunteers who had significant underlying respiratory or gastrointestinal disease, an acute illness, or any condition that would potentially compromise assessment of the study endpoints were excluded. The complete inclusion/exclusion criteria are provided in the Supplementary material.
Study material
The probiotic supplement was Bl-04 (Danisco US, Madison, WI, USA). The daily dose of probiotic was provided in a sachet containing a minimum of 2×109 cfu of Bl-04 mixed with 1 g of sucrose as a carrier. The placebo was provided as a sachet containing 1 g of sucrose that was identical to the study product in appearance, smell, and taste. Subjects were advised to mix the study product into drink that was not hot or alcoholic. The probiotic and placebo were randomised in a 1:1 allocation ratio by the sponsor using commercially available software (nQuery Advisor 7.0, Statistical Solutions, Boston, MA, USA) in blocks of six and numbered sequentially. All study personnel were blinded to the randomisation scheme until the study database was locked. Volunteers were assigned a subject number as they enrolled in the study and then provided numbered study product corresponding to their subject number.
Conduct of the study
The study was conducted as a randomised, double-blind, placebo controlled trial with two parallel arms. Three cohorts of volunteers, with 55, 61 and 74 subjects randomised to treatment per cohort, were studied between January and December, 2013. On day -28 of the study, all eligible volunteers had a baseline nasal lavage for CXCL8 concentration and analysis of exhaled nitric oxide (eNO) and then began the supplement. Volunteers returned to the study site on days -21, -14, and -7 to monitor compliance with the study protocol. On study day 0, after collection of eNO, blood for antibody to RV-A39, nasal lavage for CXCL8 and viral PCR, and a symptom assessment, all volunteers were challenged with the study virus. Study supplement was continued for the next five days during which the volunteers returned to the study site daily for assessment of symptoms, eNO, and collection of nasal lavage for CXCL8 and quantitative viral culture for RV-A39. Volunteers returned a final time at day 28 for collection of convalescent serum for antibody to RV-A39. During the supplementation period, days -28 to 5, the volunteers were instructed to avoid ingestion of other probiotics or probiotic containing foods and were prohibited from taking antibiotics or anti-inflammatory agents.
Assessment of compliance and blinding
Compliance with study supplements was assessed by counting used sachets of the study products and by qPCR for Bl-04 on stool specimens collected at day 0. For the PCR, faecal samples were collected from the subjects on study day 0 and stored at −80 °C. Samples were processed to extract DNA and analysed using MagMAX™ Total Nucleic Acid Isolation Kit (Applied Biosystems, Bridgewater, NJ, USA) followed by PCR inhibitor removal with OneStep-96™ PCR Inhibitor Removal Kit (Zymo Research, Irvine, CA, USA). Bl-04 was quantified from the samples with strain specific primers (forward primer 5′-CTTCCCAGAAGGCCGGGT-3′ and reverse primer 5′-CGAGGCCACGGTGCTCATATAGA-3′) using Fast SYBR Green Master mix (Applied Biosystems) and the ABI-PRISM FAST 7500 sequencing detection system (Applied Biosystems) with melt-curve analysis. The detection limit was approximately 475 genomes per reaction. The adequacy of study blinding was assessed on day 0, after 28 days of treatment but prior to virus challenge.
Challenge virus
The challenge virus used for this study was rhinovirus-A39. This challenge pool was produced under GMP conditions and was used for this study under FDA IND #15241. All subjects were inoculated with approximately 100 tissue culture infectious dose 50 (TCID50) of virus by intranasal drops.
Virus isolation and serology
Nasal lavage specimens collected on day 0, before virus challenge, were tested by a multiplex PCR for the detection of unsuspected viral respiratory infections. Nasal lavage collected on study days 1–5 was cultured for rhinovirus by standard methods (Turner et al., 1998). Sera were tested for neutralising antibody to RV-A39 by a standard microtiter assay (Gwaltney et al., 1989). Volunteers who had RV-A39 isolated from at least one post-inoculation specimen or had at least a 4-fold increase in serum neutralising antibody to RV-A39 were considered infected with the study virus. Viral titres were determined in the original nasal wash specimens stored at −80 °C by culturing serial 10-fold dilutions in microtiter plates of MRC-5 cells as previously described (Turner et al., 1999).
Symptom assessment
Symptom scoring was done daily on each of the five days after virus challenge using the standardised Wisconsin Upper Respiratory Symptom Survey WURSS-21 questionnaire (Barrett et al., 2005, 2006). This questionnaire assesses both symptom severity and functional status. The total symptom score reported in this study includes only the symptom severity evaluations.
Assessment of airway inflammation
CXCL8 concentration was measured in nasal lavage using a commercially available ELISA assay (R&D Systems, Minneapolis, MN, USA) as previously described (Turner et al., 1998). A panel of cytokines and chemokines (transforming growth factor (TGF)-β1, granulocyte-colony stimulating factor (GCSF), granulocyte-macrophage colony-stimulating factor (GMCSF), interferon (IFN)-γ, interleukin (IL)-1α, IL-12p70, IL-15, chemokine (C-C motif) ligand (CCL)20, IL-1β, IL-6, IL-10, chemokine (C-X-C motif) ligand 10 (CXCL10), CCL2, CCL3, CCL5, tumour necrosis factor (TNF)-α) were measured in nasal lavage fluid using a commercially available multiplex assay (Aushon BioSystems, Inc., Billerica, MA, USA). The eNO was measured with commercially available instruments and reagents (NIOX MINO, Aerocrine, Inc., New Providence, NJ, USA). This assay has a functional range of 5–300 ppb. The measurements were done according to the published guidelines of the American Thoracic Society (Anonymous, 1999).
Statistical analysis
The primary outcome variable for this study was defined by the study protocol as comparison of the CXCL8 response to rhinovirus infection in nasal lavage in the probiotic and placebo groups. The planned analysis cohort was those volunteers who were susceptible to RV-A39 by neutralising antibody titre, had no virus detected in the nasal lavage on day 0, and who were infected and completed the study. Based on previous studies (Barrett et al., 2006; Turner et al., 1998, 1999) a sample size of 60 subjects/treatment arm was sufficient to detect approximately a 50% reduction in the change in nasal lavage concentration of CXCL8 in response to rhinovirus infection after probiotic treatment with pα=0.05 (two-sided) and pβ=0.2 (one-sided). We planned to randomise up to 80 subjects per arm to assure at least 60 subjects per treatment arm in the analysis cohort. For the secondary comparisons, a Bonferonni two-sided P≤0.05 decision rule was established a priori as the null hypothesis rejection criterion. A complete description of the statistical methods for the study is provided as Supplementary material.
3. Results
Subjects
789 subjects signed consent for participation in the trial. The study was conducted in three cohorts, two in the spring of 2013 and one in the fall of 2013. The flow of subjects through the study is shown in Figure 1. 190 subjects were randomised to treatment and, after withdrawals and per protocol exclusions, 115 (58 probiotic, 57 placebo), were available for analysis. The pivotal analysis for the study was done on those subjects, 51 probiotic and 55 placebo, who were infected with rhinovirus. The study population was drawn primarily from the UVa student population and the demographics of the study reflect that population. Age, gender, race and ethnicity were comparable between the two study groups (Table 1).
Figure 1.
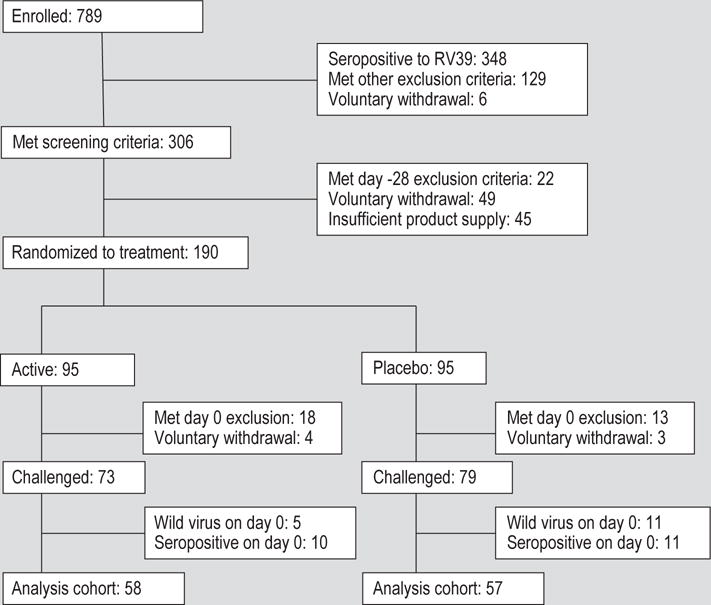
Enrolment and disposition of study subjects over the course of the study.
Table 1.
Demographics of the study participants included in the data analysis.
| Probiotic (n=58) | Placebo (n=57) | P-value | |
|---|---|---|---|
| Gender | 0.3391 | ||
| Male | 19 (33%) | 24 (42%) | |
| Female | 39 (67%) | 33 (57%) | |
| Race | 0.2702 | ||
| Asian | 2 (3%) | 8 (14%) | |
| Black | 3 (5%) | 2 (4%) | |
| White | 52 (90%) | 46 (81%) | |
| Other | 1 (2%) | 1 (2%) | |
| Ethnicity | 0.8572 | ||
| Hispanic | 3 (5%) | 4 (7%) | |
| Non-Hispanic | 54 (93%) | 52 (91%) | |
| Unknown | 1 (2%) | 1 (2%) | |
| Age (yrs, mean (SD)) | 22 (6) | 23 (7) | 0.9753 |
Fisher’s Exact test.
Pearson Exact test.
Wilcoxon Rank Sum test.
Compliance and blinding
Based on counting of used sachets, 5/58 (9%) of probiotic-treated and 10/57 (18%) of placebo-treated volunteers missed at least one dose of the study treatment. Two of the volunteers, both in the probiotic group, missed more than one dose of treatment. Forty (69%) of 58 probiotic treatment volunteers had Bl-04 detected in stool on day 0. The majority of volunteers in both treatment groups believed they were taking the placebo preparation but there was no evidence of unblinding (P=0.56, Likelihood Ratio Chi-square test).
Effect of probiotic on nasal inflammatory response to rhinovirus
Nasal lavage CXCL8 concentrations were comparable in the probiotic and placebo groups at day -28 prior to probiotic treatment (medians 130 and 106 pg/ml, respectively). On day 0 prior to challenge, the CXCL8 concentrations in the lavage from placebo-treated volunteers were significantly decreased from day -28 and were significantly lower than in the probiotic-treatment group (P=0.04, ANCOVA, Figure 2A). After rhinovirus infection, the concentration of CXCL8 in nasal lavage increased, compared to day 0, in both treatment groups, but these increases were statistically significant only for the placebo group on study days 3, 4 and 5. Comparison of the treatment groups revealed that the CXCL8 response to virus infection over the course of the 5 days after virus challenge was significantly reduced in the probiotic treatment group (Figure 2B, geometric mean ratio, probiotic:placebo=0.65, P=0.03, ANCOVA). The planned pivotal analysis, the change in CXCL8 from day 0 to day 3, was not different between the two study groups (P=0.14).
Figure 2.
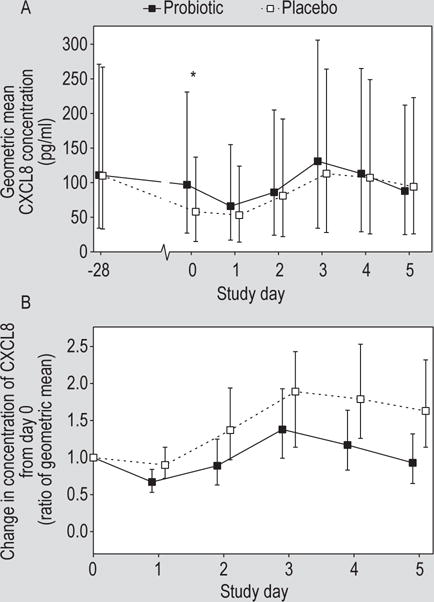
(A) Concentration of chemokine (C-X-C motif) ligand 8 (CXCL8) in the nasal lavage by study day (geometric mean ± 95% CI). The concentration on the day of rhinovirus challenge (day 0) was significantly lower in the placebo group (P=0.04, ANCOVA). (B) Change in CXCL8 concentration from day 0 prior to virus challenge in volunteers treated with probiotic or placebo. The overall change from baseline was significantly greater in the placebo than in the probiotic group geometric mean ratio, probiotic:placebo=0.65, P=0.03, ANCOVA.
Evaluation of CXCL10, IL-6, and GCSF in nasal lavage revealed a pattern similar to that seen for CXCL8 with a decrease in the nasal lavage concentration between days -28 and 0 in both treatment groups and a significant increase in the concentration of all three cytokines after virus challenge (Figure 3). There was no significant effect of the probiotic on any of these parameters. The concentration of IL-1β and CCL2 did not change significantly either prior to the virus challenge or in response to the viral infection. The other analytes tested were not consistently detectable in nasal lavage.
Figure 3.
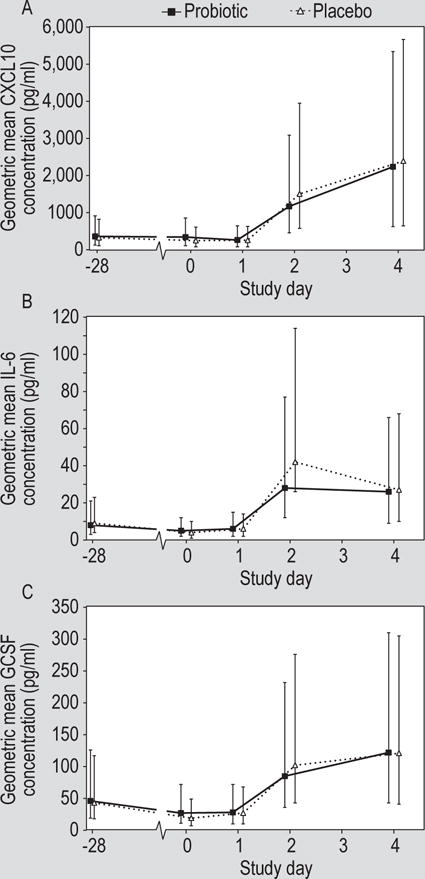
Concentration of (A) chemokine (C-X-C motif) ligand 10 (CXCL10), (B) interleukin 6 (IL-6) and (C) granulocyte-colony stimulating factor (GCSF) in the nasal lavage of volunteers treated with probiotic or placebo (geometric mean ± 95% CI). Rhinovirus challenge was on day 0.
Effect of probiotic treatment on rhinovirus infection and antibody response
Rhinovirus infection was documented in 51/58 (88%) of the volunteers challenged in the probiotic treatment group and 55/57 (96%) in the placebo group (P=0.3, Fisher Exact test). Nasal lavage viral titres over the five days post-challenge were significantly lower in the probiotic treated subjects (Figure 4A; P=0.03, ANOVA). Similarly, a post-hoc analysis found that the time to virus shedding was longer (P=0.02, logrank test) and the proportion of subjects with virus shedding was lower (76% in the probiotic group, 91% in the placebo group, Figure 4B; P=0.04, Fisher Exact test) in the probiotic treated group. There was no effect of probiotic on convalescent antibody titre. The median convalescent antibody titre in both groups was 1:8 (P=0.5, Mann Whitney test).
Figure 4.
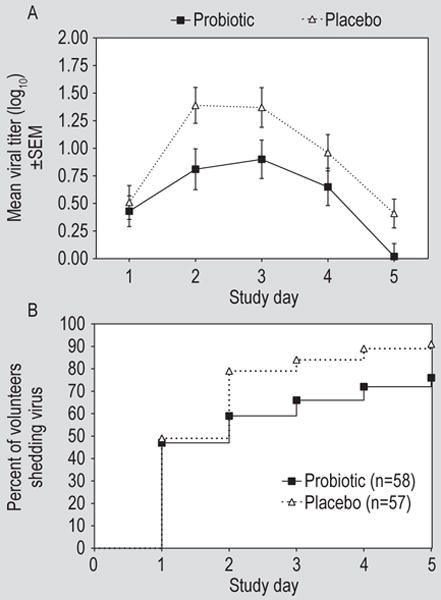
(A) Viral titres in nasal lavage from subjects treated with probiotic or placebo by day after rhinovirus challenge (P=0.03, ANOVA, for probiotic vs placebo) and (B) the cumulative proportion of subjects shedding virus in nasal secretions by day after virus challenge (P=0.02 logrank test, for probiotic vs placebo). SEM = standard error of the mean.
Effect of probiotic on lower respiratory tract inflammation
There was a statistically significant increase in the concentration of eNO in response to rhinovirus infection in both the probiotic and placebo arms with no significant difference between the two groups (Figure 5A). A post-hoc analysis revealed a modest correlation between the peak eNO concentration after virus challenge and total cough symptom score (Figure 5B; rs=0.44, P<0.001, Spearman rank correlation).
Figure 5.
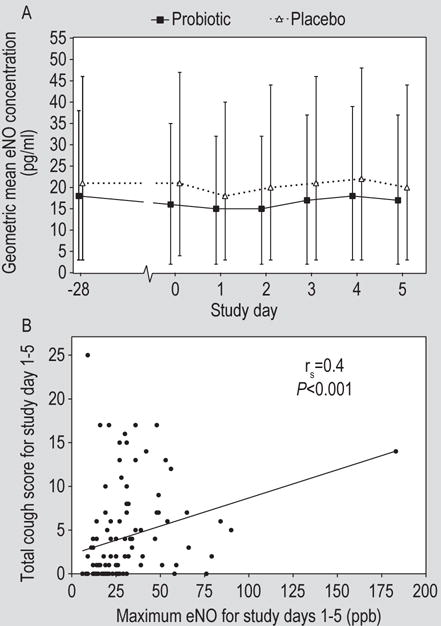
(A) Exhaled nitric oxide (eNO) concentrations in volunteers treated with probiotic or placebo. (B) Correlation between peak eNO concentration and total cough symptom score (rs=0.44, P<0.0001, Spearman rank correlation).
Effect of probiotic treatment on symptom scores
There was no effect of probiotic on symptom severity in this study. The mean (± standard error of the mean) respiratory symptom score for the five days after virus challenge was 9.2 (0.9) in the probiotic group compared to 8.0 (0.6) in the placebo group. Comparison of total or daily nasal symptom scores or total or daily rhinorrhoea or nasal obstruction symptom scores revealed no significant differences between the probiotic and placebo treatment groups (Figure 6).
Figure 6.
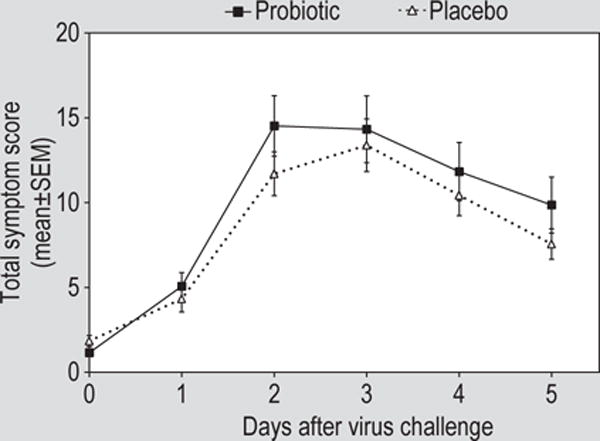
Total symptom scores by study day in volunteers treated with probiotic or placebo. Rhinovirus challenge was on day 0. SEM = standard error of the mean.
Adverse events
Adverse events were recorded for all subjects who were randomised to treatment (95 probiotic, 95 placebo). Gastrointestinal adverse events (13 occurrences in 9 subjects) were reported only in the probiotic group (P<0.01, Fisher exact test). There were no differences in the occurrence of other adverse events between the treatment groups. Adverse events in 5 subjects (4 gastrointestinal and 1 respiratory) were judged to be possibly or probably related to the study interventions.
4. Discussion
This study suggests that ingestion of Bl-04 alters nasal lavage CXCL8 levels at baseline and has a modest effect on the inflammatory response to rhinovirus infection. There also appears to be an effect on rhinovirus replication manifest by decreased shedding of virus in the nasal secretions. There was no effect of the probiotic on symptoms or on the incidence of infection assessed by isolation of virus and/or seroconversion.
Systematic reviews have documented increasing evidence that ingestion of probiotics may be useful for reducing the risk of viral upper respiratory illness (Hao et al., 2015). The published studies report a variety of different probiotics, study populations, dosages, supplementation schedules and endpoints (Lehtoranta et al., 2014), but do not address strain or species specificity of the probiotics on immune function (Hill et al., 2014). In healthy adults B. lactis Bl-04, the probiotic strain used in this study, was associated with a significant reduction in the occurrence of upper respiratory tract illness (West et al., 2014b). Studies of innate immune function in blood specimens from a subset of volunteers revealed no alterations in systemic innate immunity that would explain the observed effect on the incidence of illness (West et al., 2014a).
The experimental rhinovirus challenge model has been used for many years for the study of rhinovirus pathogenesis and treatment. This model is particularly well-suited for the goals of our study. The ability to control the baseline characteristics of the volunteers and give a uniform viral challenge precisely timed in relation to the administration of the probiotic and collection of specimens provides an opportunity to characterise the interaction between the probiotic, the infection and host responses under controlled conditions in human volunteers. While the study was conducted under Good Clinical Practice guidelines and generally follows the CONSORT guidelines for reporting, this study was not intended or designed to establish the clinical efficacy of the probiotic.
The results of our study suggest that ingestion of Bl-04 can modify the inflammatory response in the nose following rhinovirus infection. We chose to study the CXCL8 response to infection as the primary outcome variable based on previous studies that demonstrate a consistent increase in nasal lavage concentrations of CXCL8 and a modest correlation between the increase in CXCL8 from baseline and symptom severity (Barrett et al., 2006; Turner et al., 1998). We found that the nasal CXCL8 response to the infection was modestly reduced by Bl-04. The decrease in CXCL8 concentrations in both the probiotic and placebo groups after starting the study product is unexplained but the volunteers in the Bl-04 group had significantly higher nasal lavage CXCL8 concentrations at day 0 than the placebo group. Studies in animals suggest that probiotics administered directly to the nasal mucosa may induce a mild inflammatory response that limits the ability of the host to mount an inflammatory response to a virus challenge (Garcia-Crespo et al., 2013; Youn et al., 2012). A previous study in the human rhinovirus challenge model also reported that volunteers who were infected but not ill had higher CXCL8 concentrations in the baseline nasal lavage than those volunteers who developed illness in association with their infection (Zhu et al., 1997). Similarly, our observations are consistent with a hypothesis that ingestion of the probiotic is associated with an increased CXCL8 response in the nose at baseline that modifies the response to infection. The modest magnitude of the effect on the CXCL8 response and the fact that symptom scores were not affected; however, suggest that this may not be sufficient to explain the observed effect of probiotic on common cold incidence in the natural setting. Also, in spite of the effect of probiotic on the innate inflammatory responses in the nose, there was no effect on lower respiratory inflammation, as measured by eNO, or on adaptive responses, assessed by development of type-specific neutralising antibody, to the virus.
There was a suggestion in our study that the Bl-04 may have reduced virus shedding in nasal secretions. Although there was not a significant effect of the tested probiotic on the proportion of volunteers who became infected, there was a significant effect on the quantitative virus titre in nasal lavage. Our study was not able to define the mechanism of this apparent antiviral effect and this observation will require confirmation in future studies. It is noteworthy that probiotics have been shown to induce expression of antiviral responses in vitro and in animal models (Kitazawa and Villena, 2014; Weiss et al., 2010). The role of probiotic modulation of antiviral defence in the human host remains to be explored.
The potential for prevention of viral respiratory infection and illness by probiotic supplementation warrants further investigations. While studies of common cold illnesses in the natural setting have provided evidence of a beneficial effect, the effects have generally been modest and vary among different study populations and the probiotic strains tested. Our study suggests that ingestion of Bl-04 may modulate innate host responses in the nose and may impact virus replication in humans. Although these effects were modest and seem insufficient to explain reduction in illness reported in the natural setting, the results do suggest the utility of further investigations of the effect of specific probiotics on innate immune function in the human host. Whether different preparations or doses or intranasal administration of Bl-04 would enhance the effect of probiotics on rhinovirus infection and illness remains to be determined.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Wesley Morovic for designing the Bl-04 primers. The authors’ contributions were as follows: Funding (SL, MJL), Study design (RBT, SL, MJL), human subjects interaction (RBT), laboratory assays (RBT, JAW, LB, JWS, LMM, MJL), data analysis (RBT, JTP), manuscript preparation and review (all). This work was supported by Danisco Sweeteners Oy, Kantvik, Finland (part of Dupont Nutrition and Health). In addition, LMM was supported by an immunology training grant (NIH T32 AI007496); JAW receives support from NIH/NIAID and NIH/NIAMS; LB is supported by NIH grants AI120055. AI1057438 and AI100799.
Footnotes
Supplementary material
Supplementary material can be found online at https://doi.org/10.3920/BM2016.0160.
S1. Complete inclusion/exclusion criteria.
S2. Complete statistical plan.
Conflict of interest
RBT, JAW, LB, JWS, JTP, and LMM received research funding from the sponsor for the conduct of this study. SL and MJL were employees of the study sponsor at the time of the study.
References
- Anonymous. Recommendations for standardized procedures for the on-line and off-line measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide in adults and children-1999. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, July 1999. American Journal of Respiratory and Critical Care Medicine. 1999;160:2104–2117. doi: 10.1164/ajrccm.160.6.ats8-99. [DOI] [PubMed] [Google Scholar]
- Barrett B, Brown R, Mundt M, Safdar N, Dye L, Maberry R, Alt J. The Wisconsin upper respiratory symptom survey is responsive, reliable, and valid. Journal of Clinical Epidemiology. 2005;58:609–617. doi: 10.1016/j.jclinepi.2004.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett B, Brown R, Voland R, Maberry R, Turner R. Relations among questionnaire and laboratory measures of rhinovirus infection. European Respiratory Journal. 2006;28:358–363. doi: 10.1183/09031936.06.00002606. [DOI] [PubMed] [Google Scholar]
- Esposito S, Rigante D, Principi N. Do children’s upper respiratory tract infections benefit from probiotics? BMC Infectious Diseases. 2014;14:194. doi: 10.1186/1471-2334-14-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Crespo KE, Chan CC, Gabryszewski SJ, Percopo CM, Rigaux P, Dyer KD, Domachowske JB, Rosenberg HF. Lactobacillus priming of the respiratory tract: heterologous immunity and protection against lethal pneumovirus infection. Antiviral Research. 2013;97:270–279. doi: 10.1016/j.antiviral.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwaltney JM, Jr, Colonno RJ, Hamparian VV, Turner RB. Rhinoviruses. In: Schmidt NJ, Emmons RW, editors. Diagnostic procedures for viral rickettsial and chlamydial infections. American Public Health Association; Washington, DC, USA: 1989. pp. 579–614. [Google Scholar]
- Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database of Systematic Reviews. 2015;2:CD006895. doi: 10.1002/14651858.CD006895.pub3. [DOI] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Reviews Gastroenterology and Hepatology. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Kitazawa H, Villena J. Modulation of respiratory TLR3-anti-viral response by probiotic microorganisms: lessons learned from Lactobacillus rhamnosus CRL1505. Frontiers in Immunology. 2014;5:201. doi: 10.3389/fimmu.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtoranta L, Pitkäranta A, Korpela R. Probiotics in respiratory virus infections. European Journal of Clinical Microbiology and Infectious Diseases. 2014;33:1289–1302. doi: 10.1007/s10096-014-2086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang IK, Iwasaki A. Control of antiviral immunity by pattern recognition and the microbiome. Immunological Reviews. 2012;245:209–226. doi: 10.1111/j.1600-065X.2011.01073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud D. Upper airway viral infections. Pulmonary Pharmacology and Therapeutics. 2008;21:468–473. doi: 10.1016/j.pupt.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RB, Wecker MT, Pohl G, Witek TJ, McNally E, St George R, Winther B, Hayden FG. Efficacy of tremacamra, a soluble intercellular adhesion molecule 1, for experimental rhinovirus infection. Journal of the American Medical Association. 1999;281:1797–1804. doi: 10.1001/jama.281.19.1797. [DOI] [PubMed] [Google Scholar]
- Turner RB, Weingand KW, Yeh CH, Leedy D. Association between nasal secretion interleukin-8 concentration and symptom severity in experimental rhinovirus colds. Clinical Infectious Diseases. 1998;26:840–846. doi: 10.1086/513922. [DOI] [PubMed] [Google Scholar]
- Weiss G, Rasmussen S, Zeuthen LH, Nielsen BN, Jarmer H, Jespersen L, Frokiaer H. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology. 2010;131:268–281. doi: 10.1111/j.1365-2567.2010.03301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NP, Horn PL, Barrett S, Warren HS, Lehtinen MJ, Koerbin G, Brun M, Pyne DB, Lahtinen SJ, Fricker PA, Cripps AW. Supplementation with a single and double strain probiotic on the innate immune system for respiratory illness. ESPEN, Clinical Nutrition. 2014a;9:e178–e184. [Google Scholar]
- West NP, Horn PL, Pyne DB, Gebski VJ, Lahtinen SJ, Fricker PA, Cripps AW. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clinical Nutrition. 2014b;33:581–587. doi: 10.1016/j.clnu.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Winkler P, Ghadimi D, Schrezenmeir J, Kraehenbuhl JP. Molecular and cellular basis of microflora-host interactions. Journal of Nutrition. 2007;137:756S–772S. doi: 10.1093/jn/137.3.756S. [DOI] [PubMed] [Google Scholar]
- Youn HN, Lee DH, Lee YN, Park JK, Yuk SS, Yang SY, Lee HJ, Woo SH, Kim HM, Lee JB, Park SY, Choi IS, Song CS. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Research. 2012;93:138–143. doi: 10.1016/j.antiviral.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Tang W, Gwaltney JM, Jr, Wu Y, Elias JA. Rhinovirus stimulation of interleukin-8 in vivo and in vitro: role of NF-kappaB. American Journal of Physiology – Lung Cellular and Molecular Physiology. 1997;273:L814–L824. doi: 10.1152/ajplung.1997.273.4.L814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


