Abstract
Purpose
Genetically encoded reporters can assist in visualizing biological processes in live organisms and have been proposed for longitudinal and noninvasive tracking of therapeutic cells in deep tissue. Cells can be labeled in situ or ex vivo and followed in live subjects over time. Nevertheless, a major challenge for reporter systems is to identify the cell population that actually expresses an active reporter.
Methods
We have used a nucleoside analog, pyrrolo-2′-deoxy-cytidine, as an imaging probe for the putative reporter gene, Drosophila melanogaster 2′-deoxynucleoside kinase. Bioengineered cells were imaged in vivo in animal models of brain tumor and immunotherapy using chemical exchange saturation transfer MRI. The number of transduced cells was quantified by flow cytometry based on the optical properties of the probe.
Results
We performed a comparative analysis of six different cell lines and demonstrate utility in a mouse model of immunotherapy. The proposed technology can be used to quantify the number of labeled cells in a given region, and moreover is sensitive enough to detect less than 10,000 cells.
Conclusion
This unique technology that enables efficient selection of labeled cells followed by in vivo monitoring with both optical and MRI.
Keywords: Reporter gene, MRI, drosophila melanogaster 2′-deoxynucleoside kinase (Dm-dNK), pyrrolo-2′-deoxycytidine (pyrrolo-dC), fluorescence-activated cell sorting (FACS), dendritic cells (DCs)
INTRODUCTION
Advances in molecular and synthetic biology and recent developments in the field of biomedical imaging have created unique opportunities for visualizing, in real time, biological processes deep in an intact subject using genetically engineered imaging reporters. The diversity of modalities, from fluorescent and bioluminescent imaging to nuclear imaging and MRI, allows multiple choices and numerous alternatives for an appropriate methodology (1). Among these, MRI is unique because it can provide high spatial resolution of anatomical structures, which can be combined with functional information (2).
In MRI, injectable compounds known as contrast agents often are used to enhance the contrast between the tissue of interest and its surrounding tissue. Early studies have demonstrated that MRI contrast agents can be used to detect genetically encoded reporters, such as the human transferrin receptor (3) and β-galactosidase (4). The versatility of MRI contrast mechanisms has now resulted in the engineering of a variety of genetically encoded MRI reporters (5–10), including nonmetallic probes based on chemical exchange saturation transfer (CEST) (11–15).
Several deoxynucleoside kinases have been explored as genetically encoded reporter systems for a wide range of applications in animals (16–20) and humans (21). Because we were interested in generating a reporter that would allow fluorescent cell sorting, followed by both MRI and optical imaging, we sought to exploit a nucleoside kinase that has the ability to phosphorylate natural as well as synthetic, nonnaturally occurring nucleosides. The high homology between the Drosophila melanogaster fly and the human genome has made this one of the most studied organisms, enabling the study of human genetics and diseases in a well-established experimental setup (22). The Drosophila melanogaster 2′-deoxynucleoside kinase (Dm–dNK) (23,24) is an enzyme that phosphorylates all native deoxynucleosides and a wide range of synthetic nucleoside analogs, including fluorescent nucleosides (25–27). This exceptional property has been exploited for gene therapy (28). In this study, we show that a fluorescent nucleoside analog, 2′-deoxycytidine (pyrrolo-dC), generates highly specific CEST MRI contrast, enabling cell sorting in vitro followed by MRI in vivo.
Figure 1 illustrates the working concept of a bimodal, genetically engineered reporter system. A synthetic nucleoside analog (with imaging capabilities) freely crosses the cell membrane, facilitated by nucleoside transporters (29). The Dm-dNK then phosphorylates the probe, with the now negatively charged nucleoside-monophosphates accumulating in the cytoplasm. In wild-type (wt) cells, the probe washes out quickly; thus, only cells expressing the Dm-dNK reporter will retain a fluorescent or MR-detectable label.
FIG. 1.
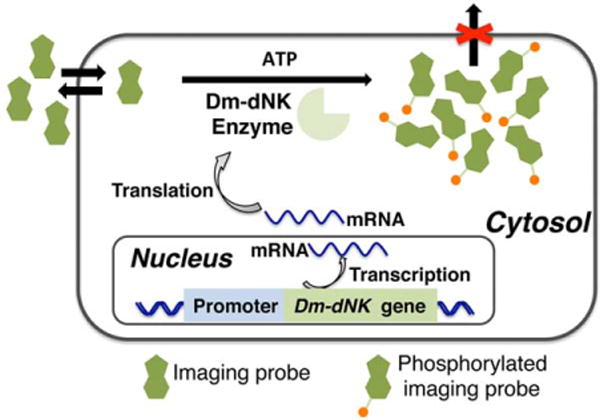
Scheme for imaging Dm-dNK reporter gene expression. The nucleoside analog imaging probe enters the cells and accumulates by phosphorylation only in cells expressing Dm-dNK.Dm-dNK, drosophila melanogaster 2′-deoxynucleoside kinase.
METHODS
Reagents
Pyrrolo-dC is a fluorescent analog of 2′-deoxycytidine commercially available by Berry and Associate (Dexter, MA, USA) (PYA 11090). AddaVax was purchased from InvivoGen (San Diego, CA, USA).
Fluorescence Measurements
Pyrrolo-dC was dissolved in 10 mM phosphate-buffered saline (PBS), and the pH was adjusted to 7.2.
For fluorescent spectra, the excitation and emission profiles of 1 mM pyrrolo-dC were recorded using an RF-5301PC spectrofluorophotometer (Shimadzu, MD, USA).
For plate reader measurements, 100 µL of 20 mM pyrrolo-dC solution and PBS (control) were added to four different wells of a black 96-well plate, and fluorescent measurements were recorded using a 1420 VICTOR3V Multilabel Plate Reader (PerkinElmer Inc., MA, USA) with a setup containing λexcitation = 355 nm and λemission = 460 nm.
Fluorescence of Cells That Accumulated Pyrrolo-dC
For plate-reader measurements, human embryonic kidney 293 (HEK293T) cells were transiently transfected with either pcDNA3.1-HSV1-tk (human herpes simplex virus)-v5– or pcDNA3.1-Dm-dNK-v5–expressing plasmids. Twenty-four hours after transfection, nontransfected wt cells (293wt), and 293 cells transfected with either HSV1-tk (293HSV1-tk) or Dm-dNK (293Dm-dNK) were incubated in the cell culture media containing 4 mM pyrrolo-dC for 3 hours. Following incubation, 7×106 cells of each cell type (293wt, 293HSV1-tk, and 293Dm-dNK) were harvested, washed three times with cold PBS to remove excess pyrrolo-dC, and lysed using the Mammalian Protein Extraction Reagent (M-PER) buffer (Thermo-Fisher Scientific, Waltham, MA, USA). Fluorescence was measured on the cell supernatant, as described above, using a fluorescent multilabel plate reader.
For fluorescence microscopy, HEK 293T cells were transduced with a pLenti-Cytomegalovirus (CMV)-Dm-dNK virus to create a stably expressing 293Dm-dNK cell line. Cells were treated with 5 µg/ml of Blasticidin Thermo Fisher for 10 days to remove all nonexpressing cells; 6×105 cells (293wt and 293Dm-dNK) were plated in six-well plates, and after their attachment, the medium was replaced with cell culture medium containing 4 mM pyrrolo-dC. After 3 hours, cells were washed three times with new medium, and fluorescent images were obtained using a Zeiss fluorescence microscope.
MRI
All CEST MRI experimental protocols were performed and processed, as previously described in detail in published protocols (30,31). Pyrrolo-dC solutions (10 mM PBS, pH = 7.2) were prepared from 1.25 mM to 10 mM and placed in microcapillaries to perform high-throughput CEST MRI experiments using a preclinical 11.7 tesla (T) MRI scanner (vertical Bruker Advance system, Billerica, MA, USA). CEST-weighted images (−10 to+10 ppm, with increments of 0.2 ppm around the water resonance) were acquired after the sample temperature was stabilized at 37 °C with a modified Rapid Acquisition with Relaxation Enhancement (RARE) sequence (repetition time/echo time (TR/TE) = 8,000/9.4 ms; RARE factor 16; 1-mm slice thickness; field of view (FOV) = 14 × 12 mm2; matrix size = 64 × 32; resolution = 0.16 × 0.375 mm2; and number of averages (NA) 2), including a presaturation pulse (variable B1 and a saturation time of 4,000 ms). Pixel-based B0 correction was achieved using experiments performed with the same parameters as above, except for TR = 1,500 ms and B1/tsat = 21 Hz/500 ms, with a sweep range from −1 to +1 ppm (0.1 ppm steps). A clinical 3.0 T MRI scanner (Philips Achieva system, Philips Healthcare, Best, The Netherlands) equipped with a body coil for transmitting and a 32-channel head coil for reception also was used to measure pyrrolo-dC solutions (40 mM in PBS, pH = 7.2) in 5-mm NMR tubes. CEST data were acquired using a spin echo sequence (TR/TE = 12,000/7.1 ms), including a magnetization transfer module with B1 = 2.0 μT and a saturation time (tsat) 4,000 ms. CEST-weighted images were acquired from – 12 to + 12 ppm, in increments of 0.3 ppm around the water resonance, which was assigned as 0 ppm. Other parameters were a slice thickness of 5 mm, a FOV = 200 × 176 mm2, and an in-plane resolution of 1.04 × 1.05 mm2.
CEST MRI of Cells That Accumulated Pyrrolo-dC
Transiently transfected 293Dm-dNK and control 293wt were incubated in a media containing 4-mM pyrrolo-dC. After 3 hours, 2 × 107 cells were lysed using a dual-phase extraction methodology for simultaneous extraction of cellular lipids and water-soluble metabolites, as described elsewhere (32). The dry content was dissolved in 10 mM PBS, pH = 7.2; CEST MRI experiments were acquired using an 11.7 T MRI scanner, as described above.
Liposome Preparation and Characterization
Liposomes were prepared using a standard lipid hydration method, as previously described (33), using the following composition: hydrogenated soy phosphatidylcholine/distearoyl-phosphatidylethanolamine polyethylene glycol conjugate)/cholesterol = 00/00/00, with 15 mg/mL of pyrrolo-dC as the encapsulated imaging agent. Empty liposomes (no pyrrolo-dC in the preparation) were used as controls. The hydrodynamic diameter of the final liposome formulation was found to be 150 to 200 nm. Fluorescence and CEST properties of the liposomes were determined using a fluorescent plate reader and an 11.7 T MRI scanner, as described above.
In Vivo CEST MRI
In vivo CEST MRI experiments were performed on an 11.7 T preclinical scanner (Bruker, Biospec, Billerica, MA, USA), as previously described (30,34). In brief, for the immunotherapy model, C57BL6 female mice were injected with dendritic cells (DCs), as described above. For the intracranial brain tumor xenograft model, 9Lwt and 9LDm-dNK were inoculated (2 × 105 cells/2 μL PBS) bilaterally into the striatum of adult nonobese diabetic/severe combined immunodeficient (NOD-SCID) male mice to generate intracranial tumors in both hemispheres. Seven days after cell transplantation, mice were anesthetized with 1.5% isoflurane, and a full CEST spectrum was acquired for each animal at 1 hour and 2.5 hours after intravenous injection of pyrrolo-dC (200 μL in saline, 150 mg/kg). CEST-weighted images were acquired with a modified RARE pulse sequence (TR/TE = 6,000/35 ms) using a 213 Hz/4,000 ms saturation pulse from −8 to was + 8 ppm around the water resonance, which was assigned to 0 ppm. Pixel-based B0 correction was achieved with a set of experiments using TR = 1,500 ms, B1/tsat = 21 Hz/500 ms, with a sweep range from – 1 to + 1 ppm (0.1 ppm steps). A single 1-mm slice with an FOV of 1.6 × 1.6 cm2 and a 128 × 48 matrix was used. Mean CEST spectra were plotted from a region of interest of each popliteal lymph nodes (PLN) (wt or Dm-dNK), or tumor and normal brain tissue, after B0 correction and ΔMTRasym was defined as [MTRasym (tumor)] − [MTRasym (normal brain tissue)](32), in order to remove magnetization transfer effects.
Cloning, Cell Transfection and Virus Transduction, and Western Blot Analysis
Cloning
The DNA encoding for the Dm-dNK gene was a kind gift from Dr. Stefan Lutz (Department of Chemistry, Emory University, Atlanta, GA, USA). The DNA sequence is similar to NM_079675.3, with the exception that the first seven amino acids (the nuclear localization sequence) were deleted. The DNA encoding for the Dm-dNK was subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) or pLenti6.2/V5-DEST Gateway Vector (Invitrogen), both under the cytomegalovirus promoter and a fused V5-tag. pLenti virus (a pLenti-CMV-Dm-dNK) was produced according to the manufacturer’s instructions.
Cell Transfection and Virus Transduction
Cells were either transfected using lipofectamine 2000 (Ther-moFisher Scientific) or transduced with a pLenti-CMV-Dm-dNK virus. Cells expressing the Dm-dNK were positively selected by adding a cell-type–appropriate concentration of Blasticidin Thermo Fisher.
Western Blot Analysis
Cells were transiently transfected with either pcDNA3.1-HSV1-tk-v5– or pcDNA3.1-Dm-dNK-v5–expressing plasmids or were infected by lentivirus. Twenty-four hours after transfection, nontransfected wt cells and cells transfected with either HSV1-tk or Dm-dNK were lysed using M-PER (ThermoFisher Scientific). For lentivirus transduction, transduced cells were positively selected with the cell-type–appropriate concentration of Blasticidin Thermo Fisher (Invitrogen) for 10 days. Protein extracts were run on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and protein blots were reacted either with Anti-V5 antibody (Invitrogen) or anti b-actin antibody (Sigma-Aldrich) as a loading control.
Fluorescence-Activated Cell Sorting
In Vitro
pLenti-CMV-Dm-dNK was used to transduce six different cell types. The cell lines 293Dm-dNK, human mesenchymal stem cells (hMSC)Dm-dNK, mouse insulinoma cells (bTC6)Dm-dNK, rat glioma (9L)Dm-dNK, human glial-restricted progenitors (hGRP) Dm-dNK, and DCDm-dNK were obtained after 10 days of selection with 5, 10, 5, 2.5, 5, and 1 μg/mL Blasticidin Thermo Fisher, respectively. Nontransduced wt cells were used as a control, that is, 293wt, hMSCwt, βTC6wt, 9Lwt, hGRPwt, and DCwt. One million cells of transduced cells and their corresponding wt controls were incubated with 1 mL of culture media containing 4 mM pyrrolo-dC. After 3 hours of incubation, cells first were washed with PBS and then with 1% fetal bovine serum in PBS, and then were filtered through a 40-µm filter into fluorescence-activated cell-sorting (FACS) tubes.
In Vivo
Ten million DCwt or DCDm-dNK were stained with the fluorescent dye, carboxyfluorescein succinimidyl ester (CFSE), mixed with AddaVax Vaccine Adjuvant (1:1; InvivoGen), and a total volume of 100 µl was subcutaneously administered to the footpad of C57BL6 female mice. Forty-eight hours post-cell injection, mice were injected intravenously with pyrrolo-dC (200 µl in saline, 150 mg/kg body weight). Two hours after intravenous injection, the popliteal lymph nodes (PLNs) were either imaged by MRI or harvested and analyzed by FACS. FACS experiments were performed using an LSRII instrument (BD, Franklin Lakes, NJ, USA) with an ultraviolet laser for excitation and a blue filter for analyzing cells that accumulated the pyrrolo-dC. Data were processed using FlowJo software (TreeStar, Inc., Ashland, OR, USA).
Calculation of Cell Numbers
DCs were prestained with the fluorescent dye CFSE before subcutaneous injection into the mice footpad. Forty-eight hours post-cell injection, mice were injected intravenous with pyrrolo-dC (200 µl in saline, 150 mg/kg body weight). Two hours after intravenous injection, the PLNs were harvested from treated mice, mechanically disrupted to obtain a single-cell suspension, and counted by Trypan blue exclusion to determine the total number of cells per PLN. The total number of CFSE+ pyrrolo-dC+ cells ( , which represents the fraction of DC that took up pyrrolo-dC) was calculated by multiplying the total live cells counted per PLN (NPLN) by the fraction of the cell population (CFSE+ pyrrolo-dC+ cells), as measured by FACS ( ), according to the following formula:
Where error bars are included in the graphs, the standard error of the mean (SEM) represents the error.
RESULTS
Imaging Features of Pyrrolo-dC
The imaging probe pyrrolo-dC was dissolved in PBS in order to evaluate its fluorescent properties. Figure 2a shows the excitation and emission profiles of pyrrolo-dC, indicating an Exmax of excitation and Emmax of emission at 395 and 460 nm, respectively. The Exmax absorbance value obtained for pyrrolo-dC is slightly red-shifted compared to previously reported values (340–350 nm) (26,35), which may be attributable to the solvent. These optical properties enabled fluorescence intensity measurements using a conventional plate reader (Fig. 2b).
FIG. 2.
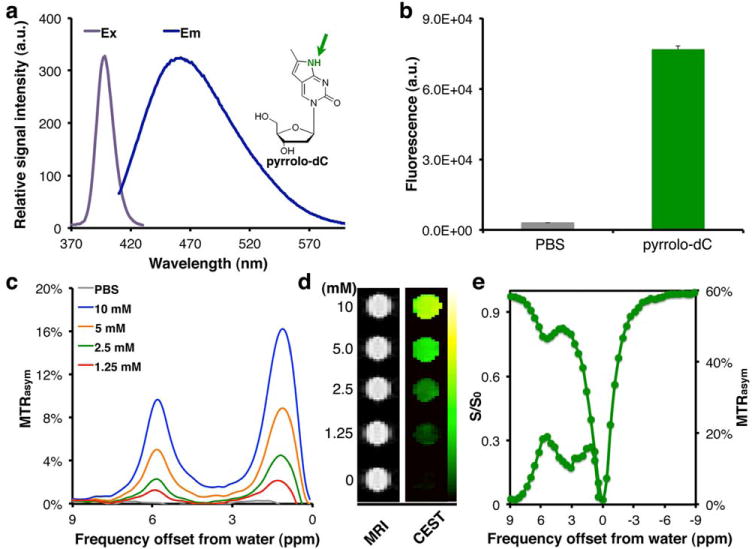
Imaging features of pyrrolo-dC. (a) Ex (gray) and Em (blue) spectra of pyrrolo-dC (inset) in solution. Arrow points to MRI CEST-detectable imino proton. (b) Fluorescent signal from 20 mM pyrrolo-dC. (c) MTRasym plots of pyrrolo-dC in solution. Corresponding (d) MR images (left) and MTRasym maps at 5.8 ppm (right), (e) CEST-spectra and MTRasym plots obtained using a 3T clinical scanner.CEST, chemical exchange saturation transfer; Em, emission; Ex, excitation; PBS, phosphate-buffered saline; pyrrolo-dC, pyrrolo-2′-deoxycytidine; T, tesla.
Figures 2c through 2e depict the CEST MRI features of pyrrolo-dC. In CEST imaging, are selectively saturated followed by their magnetization transfer to the water molecules surrounding (36,37). After transfer of their saturation to bulk water, from which the proton MRI signal is derived, the water signal is reduced; hence, these exchangeable protons are detected indirectly. The NH proton of the pyrrolo-dC generates a well-defined peak when a saturation radio-frequency (RF) pulse is applied at the 5.8 ppm frequency offset (Δω) from the water proton resonance frequency, as observed in the MTRasym plot (Fig. 2c). An additional peak from the hydroxyl (-OH) protons of the deoxyribose moiety of pyrrolo-dC also can be observed; however, these protons have relatively low Δω values and therefore are less favorable for CEST applications. A representative MTRasym map (Fig. 2d) shows the dynamic range of detectable pyrrolo-dC concentrations. This correlation between the pyrrolo-dC concentration and the MTRasym values at Δω = 5.8 ppm is linear (Supporting Fig. S1a). By varying the saturation power (38) (B1), we found the exchange rate (kex) of the pyrrolo-dC-NH exchangeable proton to be 850 s−1 (Supporting Fig. S1b). This relatively slow kex, together with the large chemical shift (Δω = 5.8 ppm), allows the detection of pyrrolo-dC when using MRI scanners operating at low magnetic fields, such as clinical MRI scanners. Using an MRI scanner operating at 3.0 T (128 MHz), for which the kex (850 s−1) is on the same order of magnitude as Δω (742 Hz), a well-defined peak from pyrrolo-dC can be seen in both the CEST spectrum and the MTRasym (Fig. 2e). Finally, to determine the potential of using pyrrolo-dC as an imaging agent in an extensive range of biomedical applications, pyrrolo-dC was encapsulated in liposomes. The pyrrolo-dC–containing liposomes generated a higher CEST contrast at 5.8 ppm and showed higher fluorescence levels compared to the empty-control liposomes (Supporting Figs. S2a–c).
Bimodal in Vitro Imaging of Dm-dNK Expression
Next, mammalian cells were engineered to express the Dm-dNK protein or the HSV1-tk protein as a control (Fig. 3a, inset). HEK 293T cells expressing the nucleoside kinases (HSV1-tk or Dm-dNK) and wt cells were incubated with 4 mM pyrrolo-dC in culture medium. After 3 hours, cells were lysed and the intracellular content was analyzed. As shown in Figure 3a, the cell lysate of the 293Dm-dNK showed a much higher fluorescent signal compared to 293wt (P < 5 × 10−9, Student t test) and 293HSV1-tk (P < 5 10−8, Student t test). For recombinant HSV1-tk expression, a nucleoside kinase that also phosphorylates a wide range of nucleoside analogs, little fluorescence could be detected, most likely due to a low level of pyrrolo-dC phosphorylation and accumulation. These data indicate that pyrrolo-dC has a higher substrate specificity for Dm-dNK compared to HSV1-tk. Fluorescence also was observed in intact cells expressing 293Dm-dNK (Fig. 3b).
FIG. 3.
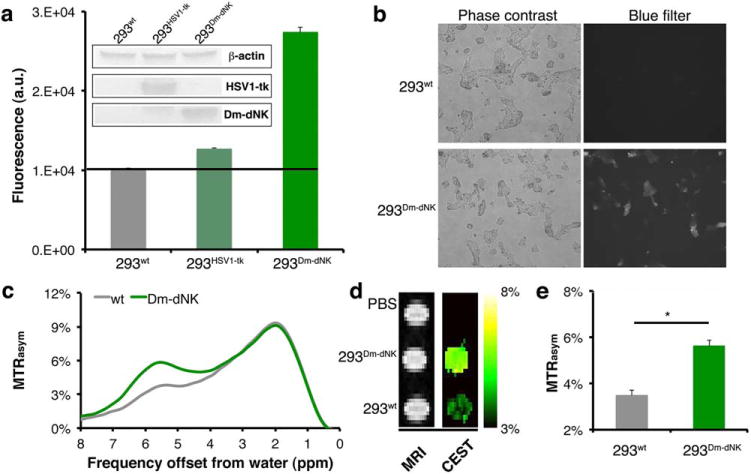
In vitro imaging of Dm-dNK expression. (a) Emitted fluorescence from cell lysate after 3 hours incubation with 4 mM pyrrolo-dC. Shown are HEK 293T cells (293wt) and cells that were transiently transfected with either HSV1-tk (293HSV1-tk) or Dm-dNK (293Dm-dNK) (n = 4). Inset shows Western blot analysis of cell extracts using an anti-V5 tag antibody for staining of HSV1-tk and Dm-dNK expression, with anti-β-actin as a control. (b) Microscopic images of 293wt and 293Dm-dNK incubated for 3 hours with pyrrolo-dC. (c-e) CEST MRI of cell lysates (293wt and 293Dm-dNK) incubated for 3 hours with 4 mM of pyrrolo-dC. (c) Representative MTRasym plot (293wt, gray; and 293Dm-dNK, green). (d) MR images of phantoms without a saturation pulse (right) and MTRasym maps (radiofrequency saturation at Δω = 5.8 ppm). (e) Averaged MTRasym values (with standard error of mean) from maps in d (n = 3).CEST, chemical exchange saturation transfer; Dm-dNK, drosophila melanogaster 2′-deoxynucleoside kinase; HSV1-tk, herpes simplex virus thymidine kinase type 1; PBS, phosphate-buffered saline; pyrrolo-dC, pyrrolo-2′-deoxycytidine; wt, wild type.
CEST experiments were subsequently performed on cell lysates after 3 hours of incubation in media containing pyrrolo-dC. As expected, a well-defined peak at Δω = 5.8 ppm could be observed for the MTRasym plot obtained from 293Dm-dNK (Fig. 3c). The CEST maps shown in Figure 3d and the obtained CEST contrast was significantly higher for 293Dm-dNK compared to 293wt controls (Fig. 3e) (P = 0.0004).
Flow Cytometry
Unless a fluorescent reporter is used, one of the major challenges for reporter systems is to identify the cell population that express an active reporter. One of the advantages of the Dm-dNK system is that pyrrolo-dC also can be detected by fluorescence. FACS analysis was performed on five different mammalian cell types. In all examined cells, that is, HEK293T cells, hMSC, bTC6, 9L cells, and hGRP, higher fluorescence was observed for Dm-dNK–expressing cells compared to wt cells after incubation with pyrrolo-dC (Fig. 4), indicating retention of pyrrolo-dC. As expected, in all examined cells the histogram representing the Dm-dNK–expressing cells (blue histograms) was further right-shifted (higher blue fluorescence) compared to control cells (red histograms). Although the fluorescence intensity and the relative population of cells that can be sorted using blue fluorescence is different for each examined cell type, a well-defined and highly fluorescent cell population could be easily observed for all cells. These findings demonstrate the advantage of Dm-dNK/pyrrolo-dC for cell sorting without the need for an additional fluorescent reporter (either fused or coexpressed).
FIG. 4.
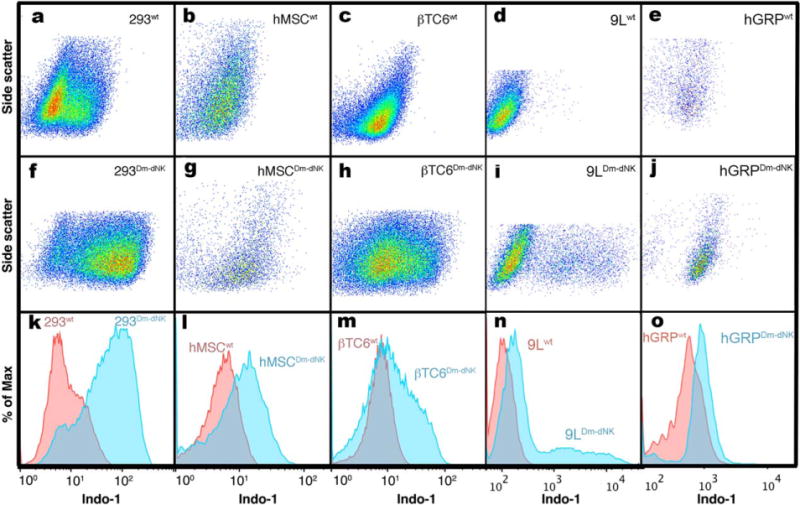
Flow cytometry analysis of wt- and Dm-dNK–transduced mammalian cell lines incubated with 4 mM pyrrolo-dC. Following incubation with 4 mM pyrrolo-dC, cells were analyzed via flow cytometry for pyrrolo-dC retention, which can be detected in the Indo-1 channel. HEK 293 (a, f, k), hMSC (b, g, l), βTC6 (c, h, m), 9L (d, i, n), and hGRP (e, j, o). Scatter plots (a-j) of side scatter versus Indo-1 (blue) channel of wt- (a-e) and Dm-dNK–expressing (f-j) cells. FACS histograms (k-o) display relative blue fluorescence of wt- (red histogram) and Dm-dNK–expressing cells (blue histogram) for each of the examined cell lines (a and f, b and g, c and h, d and i, e and j). Increased blue fluorescence corresponds to increased retention of pyrrolo-dC.9L, rat glioma; βTC6, mouse insulinoma cells; Dm-dNK, drosophila melanogaster 2′-deoxynucleoside kinase; FACS, fluorescence-activated cell sorting; HEK 293, human embryonic kidney 293; hGRP, human glial-restricted progenitors; hMSC, human mesenchymal stem cells; HSV1-tk, herpes simplex virus thymidine kinase type 1; pyrrolo-dC, pyrrolo-2′-deoxycytidine; wt, wild type.
Quantifying Labeled Cells In Vivo
After demonstrating that pyrrolo-dC accumulates exclusively in Dm-dNK–expressing cells, and that this accumulation can be detected both with optical (fluorescence) and MR (CEST) imaging in vitro, we examined the feasibility of monitoring the cells labeled with Dm-dNK/pyrrolo-dC in vivo. To that end, we used a well-established (39,40) mouse model relevant to immunotherapy— dendritic cell cancer vaccines (Fig 5a)—tracking them from the site of injection (the footpad) to the popliteal lymph node.
FIG. 5.
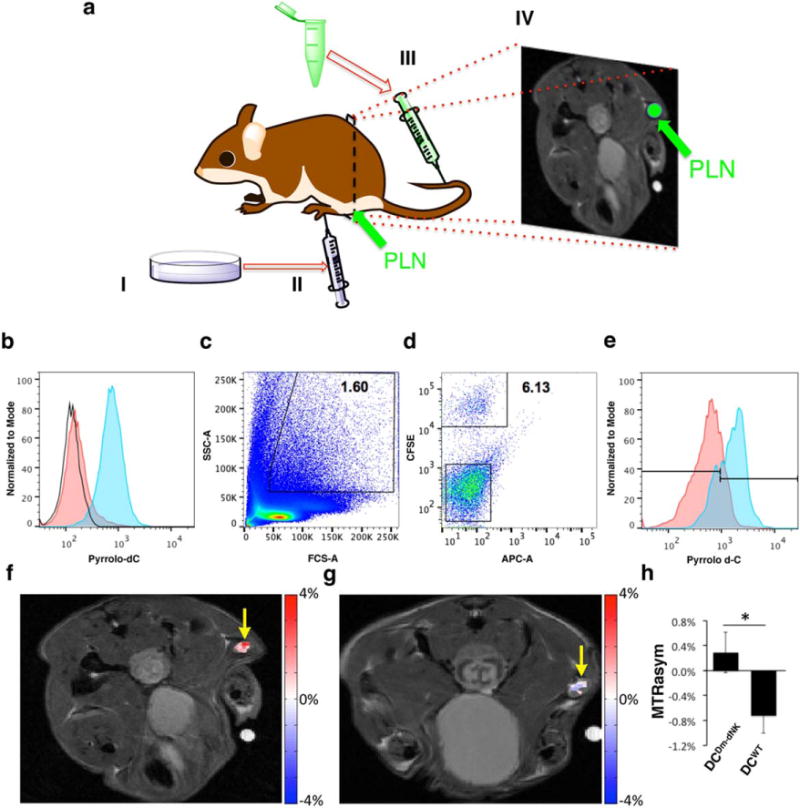
(a) Illustration of the experimental procedure. Ten million DCwt or DCDm-dNK activated with AddaVax Vaccine Adjuvant (InvivoGen, San Diego, CA, USA (1:1) were administered subcutaneously to the footpad of mice. Pyrrolo-dC was administered intravenously, 48 hours post-cell transplantation; and 2 hours postinjection, the PLNs were analyzed either by MRI or by FACS. (b) DCwt and DCDm-dNK were incubated in vitro with 4 mM pyrrolo-dC. Histogram displays the relative fluorescence of control DCwt+Pyrrolo-dC (red) and DCDm-dNK+Pyrrolo-dC (blue). Increased blue fluorescence corresponds to increased retention of pyrrolo-dC. No fluorescence was observed in the DCwt without pyrrolo-dC (empty histograms). (c-e) Ten million DCDm-dNK stained with CFSE (CFSE+ DCDm-dNK) were processed and injected into mice as described in (a), and PLNs were harvested and analyzed by FACS. The myeloid cell population was gated based on forward-scatter and side-scatter measure of granularity and size, respectively, (c) and the pyrrolo-dC uptake of the CFSE+ DCDm-dNK cells (d and e; blue) was compared to the CFSE− cells (APC-A channel is empty) (d and e; red). (f-h) Following the procedure described in (a), mice were imaged by MRI. CEST MRI map overlaid on anatomical MRI shows pyrrolo-dC accumulation in the PLN (arrow) of a mouse injected with DCDm-dNK (f; n = 5), but not with nontransduced DCwt (g; n = 5). (h) Average MTRasym values at 5.8 ppm plots of DCwt and DCDm-dNK (mean ± standared error of the mean; n = 5 mice per group), Student t test, unpaired two-tailed, P value = 0.046.CEST, chemical exchange saturation transfer; CFSE, carboxyfluorescein succinimidyl ester; dC, dendritic cells; Dm-dNK, drosophila melanogaster 2′-deoxynucleoside kinase; FACS, fluorescence-activated cell sorting; PLN, popliteal lymph nodes; pyrrolo-dC, pyrrolo-2′-deoxycytidine; SSC, side scatter; wt, wild type.
Mouse dendritic cells (DC2.4) were transduced with a lentivirus encoding for the Dm-dNK reporter gene. Dendritic cells expressing the Dm-dNK (DCDm-dNK) were selected with 1 µg/mL Blasticidin Thermo Fisher For 10 days. FACS analysis was performed on DCwt and DCDm-dNK after 3 hours of incubation with 4 mM pyrrolo-dC. DCwt without pyrrolo-dC served as an additional control. As shown in Figure 5b, higher fluorescence was observed for the DCDm-dNK (96.8%) compared to DCwt (13.2%) following incubation with pyrrolo-dC, indicating retention of pyroolo-dC. No fluorescence was observed in the DCwt that were not incubated with pyrrolo-dC.
Next, DCDm-dNK were stained with the fluorescent dye, CFSE, to facilitate tracking of the cells, and 107 DCDm-dNK CFSE+ with or without AddaVax Vaccine Adjuvant (1:1; InvivoGen) were subcutaneously administered to the footpad of C57BL6 female mice (Figs. 5a, c–e). Forty-eight hours post-cell injection, mice were injected intravenously with pyrrolo-dC (150 mg/kg body weight; the same concentration used for luciferin in bioluminescent imaging) and the PLNs were dissected and analyzed by FACS. As expected, the AddaVax Vaccine Adjuvant (InvivoGen) increased the number of migrating DCs into the PLNs (data not shown). Figures 5c through 5e demonstrate pyrrolo-dC accumulation in migrating DCDm-dNK CFSE+ (77.05 ± 0.65%), compared to CFSE− cells (intrinsic PLN-residing cells), which showed only a basal level of pyrrolo-dC accumulation (22.95 ± 0.65%). Based on these FACS analyses, we calculated that less than 10,000 migrating DCDm-dNK accumulated the fluorescent pyrrolo-dC (8155 ± 1077 cells per node; n = 3).
In Vivo Cell Tracking with Dm-dNK/Pyrrolo-dC
Ten million of either DCwt or DCDm-dNK mixed with Adda-Vax Vaccine Adjuvant (InvivoGen) were administered subcutaneously to the footpad of mice. Forty-eight hours post-cell injection, mice were injected intravenously with pyrrolo-dC (150 mg/kg body weight), and the mice (n = 5 per each group) were imaged using an 11.7 T MRI scanner. Two hours post–pyrrolo-dC injection, sufficient accumulation was observed in the PLN of mice injected with DCDm-dNK (Fig. 5f), compared to the DCwt-injected mice (Fig. 5g). As shown in Figure 5h, at the 2-hour time point, the DCDm-dNK showed higher CEST contrast (MTRasym = 0.29% ± 0.32) compared to the control DCwt (MTRasym =− 0.72 ± 0.28) at the frequency offset of the imino exchangeable proton of the pyrrolo-dC (Δω =5.8 ppm), with P values of 0.046. Moreover, a two-tailed Mann-Whitney U Test was used to determine if there was a significant difference between the medians of these two groups. This test is the nonparametric alternative to the t test and does not make any assumptions about the normality of the data. Indeed, there was a significant (P = 0.0216) difference between the MTRasym of these two groups (Supporting Fig. S5).
These findings indicate the feasibility of imaging DCs migration into the draining PLNs with MRI. The combination of pyrrolo-dC with Dm-dNK, together with MRI and FACS, create a unique approach that enables quantification of the number of cells emitting a signal.
In addition, the intracranial xenograft tumor model, 9LDm-dNK, showed higher CEST contrast 2.5 hours post–pyrrolo-dC injection compared to the control 9Lwt tumor (3). Importantly, when the mean ΔMTRasym values of each tumor type were plotted (Supporting Figs. S3 and S4), a well-defined peak was revealed at the frequency offset corresponding to the NH exchangeable proton of the pyrrolo-dC (Δω = 5.8 ppm), showing specificity with significant P values < 0.05 (Student t test). Interestingly, the 9L (9Lwt or 9LDm-dNK) cells that took up the lowest amount of the pyrrolo-dC in vitro compared to other cell lines showed a very moderate uptake in vivo. We hypothesize that a drug-resistance mechanism that is typical for glioblastoma prevented accumulation.
DISCUSSION
Immunotherapy works by empowering each patient’s individual immune system to target, detect, and destroy cancer cells, and thus offers new hope for the treatment of otherwise incurable cancer. There have been many attempts to incorporate molecular imaging into immunotherapy, and specifically into tracking dendritic cells as well as other antigen-presenting cells. DCs can identify various tumor antigens and serve as essential antigen-presenting cells that stimulate helper and killer T cells, and consequently induce an immune response against tumor antigens by the host [41]. Although immunotherapeutic strategies with DCs have been applied to the treatment of diverse cancer models, their outcomes are only partially effective due to the inability to monitor the behavior of DCs in vivo (42). Therefore, there is a great need for noninvasive imaging tools that can accurately monitor the fate of DCs and track their migration to lymphoid organs. MRI-labeling of DCs with iron oxides has been used in the clinic and has demonstrated the importance of the ability to identify precisely the injection site (43). Similarly, several studies have been performed to monitor activated antigen-presenting DCs in draining lymph nodes and to optimize cell-mediated immune responses to antitumor vaccination (39, 40). Fluorine (19F) MRI also has been used to label DCs in a mouse model (44) and in humans (45). Nevertheless, despite those recent breakthroughs in noninvasive cell tracking, there still is an unmet need to monitor the status of the cells and to distinguish viable DCs from cells that have died and released iron oxide or the perfluorocarbon tracer. One approach is to use genetically encoded reporters. Unlike synthetic imaging probes that report on both live and dead cells and persist after cell death, leading to false-positive results (46), reporter genes can be used to image only live and functioning cells. Therefore, they are widely used, and HSV1-tk, which is a gene similar to Dm-dNK, already has been used in the clinic (21).
A major challenge is to measure directly the levels of the reporter/probe in the cells before transplantation of targeted cells. As can be seen in Figure 4, not all the Dm-dNK–transduced cells accumulate the same amounts of imaging probe. This could lead to transplantation of cells with a very low potential of being tracked. One such example is the 9L cells in this study, which showed poor probe uptake compared to other cell lines, both in vitro and in vivo. This point emphasizes the importance of the prescreening technology. Such a method can help in the future to prevent using inappropriate model systems, information that cannot be obtained without a probe with optical properties or even when the reporter gene is fused to a fluorescent protein. Thus, a robust method for prescreening cells is critical. Several attempts to develop a fusion protein that contains the green fluorescent protein, expressed in conjunction with the reporter gene, have been reported (47,48). However, in cases where there is a discrepancy between the accumulation of the probe and the protein expression level, such as in cells that developed drug resistance mechanisms, a system in which MRI is directly coupled to the optical output may be an advantage.
The ability to sort cells that accumulate fluorescent probe also will allow high-throughput screening following mutagenesis (eg, directed evolution) to gain better conversion of the substrate by the enzyme. Such a methodology can dramatically improve the activity of Dm-dNK toward the desired substrate (26). It also was shown that mutated HSV1-tk (49) or mutated human deoxycytidine kinase (17), selected from libraries of mutants, had higher selectivity for radiolabeled positron emission tomography (PET) probes and better capabilities for imaging of gene expression. In addition, directed evolution was used to improve the sensitivity and specificity of MRI-based protein sensors (50).
The fluorescent properties of pyrrolo-dC are sufficient for detection using a conventional fluorescent apparatus, such as a plate reader and flow cytometry. Recent technological developments and the extensive interest in developing a fluorescent nucleoside (51,52), mostly for investigating DNA structure, damage, and repair mechanisms, have contributed much to our understanding of the optical properties of synthetic nucleoside analogs. Consequently, new synthetic nucleosides with red-shifted fluorescence will allow better light penetration and will improve the optical resolution and the sensitivity for the detection of cellular events in living subjects, similar to developments reported for fluorescent dyes (53) and proteins (54).
The main advantages of diamagnetic CEST sensors is that their exchangeable protons have a large Δω and are characterized by: 1) minimal contribution of endogenous CEST contrast, 2) lower effect of direct saturation of the water signal, and 3) better detection using MRI scanners operating at low magnetic fields. One condition that should be fulfilled for a good CEST probe is that the exchange rate of its exchangeable proton should be in the slow-to-intermediate exchange range (ie, kex ≤ Δω) (37). Because Δω depends on the magnetic field (B0), exchangeable protons that resonate at large Δω, such as the NH of pyrrolo-dC, also can be monitored with MRI scanners operating at low B0, as in the case of clinical MRI scanners. Although pyrrolo-dC displays a well-defined peak when a clinical MRI scanner is used, it is important to mention that the current clinical scanners are not designed to image small animals, both in terms of specific absorption rate (SAR) and RF coils that were built for humans or large samples; therefore, the majority of CEST data in this study were acquired using a preclinical 11.7 T scanner.
Here, we decided to use an immunotherapy model and image DCs localization into draining PLNs. Although the contrast difference between wt- and Dm-dNK–expressing cells is on the order of a few percent (Fig. 5), it is not uncommon for MRI. In fact, in the field of functional MRI (fMRI), which is a well-accepted MRI technique, the change in the blood oxygen-level–dependent (BOLD) contrast is on the same order of magnitude. In molecular MRI, it was shown that a < 0.5% change in contrast may be used for the in vivo study of biological setups using newly developed probes (55). The accuracy and the significance of the contrast are determined by statistical tests. This also is true in the case of the PLN model that we describe. Although the difference in contrast is small, it allows the detection of a low number of cells (less than 105 cells per node, based on in vivo measurements using flow cytometry), which is remarkable for MRI. Therefore, this is a highly sensitive method for the detection of a very low number of cells in vivo, which can be matched with high precision to anatomical resolution images.
CONCLUSION
The recent studies and developments for spatially monitoring reporter gene expression noninvasively in the deep tissue of an intact subject have transformed MRI into a major and important player in the field of cellular and molecular imaging. The combination of pyrrolo-dC (a substrate) with Dm-dNK (an enzyme) permits bimodal in vivo monitoring of reporter gene expression. What makes this system unique is that the signal from both optical imaging and MRI stems from the same probe, and they share the same gene.
Supplementary Material
Fig. S1. a. Average MTRasym values obtained at 5.8 ppm as a function of pyrrolo-dC concentration. Dashed line represents a linear fit (R2=0.99902) of all experimental points (0–10 mM pyrrolo-dC). b. Average MTRasym values at the 5.8 ppm offset for 10 mM pyrrolo-dC solution as a function of saturation pulse power. All experiments were performed at 37°C, on an 11.7 T scanner, B1=4000 ms, B1=4.7 μT (200 Hz).
Fig. S2. Liposomal nanocariers of pyrrolo-dC. CEST MRI and optical properties, as obtained from PBS, empty liposomes, and liposomes loaded with pyrrolo-dC. a. MTRasym plots, b. MTRasym maps at 5.8 ppm, and c. fluorescence.
Fig. S3. a. Representative in vivo MTRasym maps of intracerebrally implanted 9L cells overlaid on anatomical MR images, showing the CEST signal distribution at Δω=5.8 ppm at one hour (left) and at 2.5 hours (right) post i.v. injection of pyrrolo-dC. b. Average ΔMTRasym plot of 9Lwt (gray) and 9LDm-dNK (green) images. Inset represents the range of ΔMTRasym values at 6.8 ppm > Δω > 4.8 ppm (mean ± SEM; n=5 mice). In all experiments, pyrrolo-dC was injected i.v. at 150 mg/kg.
Fig. S4. CEST signal distribution in tumor: Shown are the distribution of ΔMTRasym values within the tumor ROI (from n=5 mice) at Δω=5.8 ppm at one hour a. and at 2.5 hours b. after injection of pyrrolo-dC. Inset in (b) shows the positive ΔMTRasym values on the histograms.
Fig. S5. A dot plot of the individual MTRasym values for this graph as well as the corresponding box-and-whisker plot with the median for each group shown by a red line (0.11% and −0.51% for DCDm-dNK and DCwt respectively). We conducted a two-tailed Mann-Whitney U Test to determine if there was a significant difference between the medians of these two groups. This test is the nonparametric alternative to the t-test and does not make any assumptions about the normality of the data. There was a significant (p=0.0216) difference between the MTRasym of these two groups.
Acknowledgments
The Dm-dNK plasmid was kindly provided by Dr. Stefan Lutz from the Department of Chemistry at Emory University in Atlanta, Georgia, USA. The authors wish to thank Dr. Gayane Yenokyan from the Johns Hopkins School of Public Health Biostatistics Core, Baltimore, Maryland, USA, for the statistical analysis, and Ms. Mary McAllister for her assistance in editing the manuscript.
Supported by grants from the Maryland Stem Cell Research Foundation MSCRFII-0042, MSCRFF-0103-00, and NIH grant NS079288.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article.
References
- 1.Kooreman NG, Ransohoff JD, Wu JC. Tracking gene and cell fate for therapeutic gain. Nat Mater. 2014;13:106–109. doi: 10.1038/nmat3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duyn JH, Koretsky AP. Novel frontiers in ultra-structural and molecular MRI of the brain. Curr Opin Neurol. 2011;24:386–393. doi: 10.1097/WCO.0b013e328348972a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. In vivo magnetic resonance imaging of trans-gene expression. Nat Med. 2000;6:351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 4.Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 5.Genove G, Demarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11:450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 6.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Kodibagkar VD, Yu JX, Mason RP. 19F-NMR detection of lacZ gene expression via the enzymic hydrolysis of 2-fluoro-4-nitrophenyl beta-D-galactopyranoside in vivo in PC3 prostate tumor xenografts in the mouse. FASEB J. 2007;21:2014–2019. doi: 10.1096/fj.06-7366lsf. [DOI] [PubMed] [Google Scholar]
- 8.Bartelle BB, Berrios-Otero CA, Rodriguez JJ, Friedland AE, Aristizabal O, Turnbull DH. Novel genetic approach for in vivo vascular imaging in mice. Circ Res. 2012;110:938–947. doi: 10.1161/CIRCRESAHA.111.254375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrick PS, Hammersley J, Loizou L, et al. Dual-modality gene reporter for in vivo imaging. Proc Natl Acad Sci U S A. 2014;111:415–420. doi: 10.1073/pnas.1319000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro MG, Ramirez RM, Sperling LJ, Sun G, Sun J, Pines A, Schaffer DV, Bajaj VS. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem. 2014;6:629–634. doi: 10.1038/nchem.1934. [DOI] [PubMed] [Google Scholar]
- 11.Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr, Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25:217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Liang Y, Bar-Shir A, et al. Monitoring enzyme activity using a diamagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. J Am Chem Soc. 2011;133:16326–16329. doi: 10.1021/ja204701x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Shir A, Liu G, Liang Y, et al. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J Am Chem Soc. 2013;135:1617–1624. doi: 10.1021/ja312353e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar-Shir A, Liu G, Chan KW, et al. Human protamine-1 as an MRI reporter gene based on chemical exchange. ACS Chem Biol. 2014;9:134–138. doi: 10.1021/cb400617q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Shir A, Liang Y, Chan KW, Gilad AA, Bulte JW. Supercharged green fluorescent proteins as bimodal reporter genes for CEST MRI and optical imaging. Chem Commun (Camb) 2015;51:4869–4871. doi: 10.1039/c4cc10195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Narsinh KH, Lan F, et al. Early stem cell engraftment predicts late cardiac functional recovery: preclinical insights from molecular imaging. Circ Cardiovasc Imaging. 2012;5:481–490. doi: 10.1161/CIRCIMAGING.111.969329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Likar Y, Zurita J, Dobrenkov K, Shenker L, Cai S, Neschadim A, Medin JA, Sadelain M, Hricak H, Ponomarev V. A new pyrimidine-specific reporter gene: a mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. J Nucl Med. 2010;51:1395–1403. doi: 10.2967/jnumed.109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Zhang S, Rabinovich B, et al. Human CD34 + cells in experimental myocardial infarction: long-term survival, sustained functional improvement, and mechanism of action. Circ Res. 2010;106:1904–1911. doi: 10.1161/CIRCRESAHA.110.221762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massoud TF, Paulmurugan R, Gambhir SS. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med. 2010;16:921–926. doi: 10.1038/nm.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhang HE, Gabrielson KL, Laterra J, Fisher PB, Pomper MG. Tumor-specific imaging through progression elevated gene-3 promoter-driven gene expression. Nat Med. 2011;17:123–129. doi: 10.1038/nm.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaghoubi SS, Jensen MC, Satyamurthy N, Budhiraja S, Paik D, Czernin J, Gambhir SS. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munch-Petersen B, Piskur J, Sondergaard L. Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J Biol Chem. 1998;273:3926–3931. doi: 10.1074/jbc.273.7.3926. [DOI] [PubMed] [Google Scholar]
- 24.Johansson M, van Rompay AR, Degreve B, Balzarini J, Karlsson A. Cloning and characterization of the multisubstrate deoxyribonucleoside kinase of Drosophila melanogaster. J Biol Chem. 1999;274:23814–23819. doi: 10.1074/jbc.274.34.23814. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Soni PB, Liu L, Zhang X, Liotta DC, Lutz S. Synthesis of fluorescent nucleoside analogs as probes for 2′-deoxyribonucleoside kinases. Bioorg Med Chem Lett. 2010;20:841–843. doi: 10.1016/j.bmcl.2009.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Li Y, Liotta D, Lutz S. Directed evolution of an orthogonal nucleoside analog kinase via fluorescence-activated cell sorting. Nucleic Acids Res. 2009;37:4472–4481. doi: 10.1093/nar/gkp400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Murphy P, Baker D, Lutz S. Computational design of orthogonal nucleoside kinases. Chem Commun (Camb) 2010;46:8803–8805. doi: 10.1039/c0cc02961k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan S, Zhou X, Paredes JA, Kuiper RV, Curbo S, Karlsson A. Transgene expression of Drosophila melanogaster nucleoside kinase reverses mitochondrial thymidine kinase 2 deficiency. J Biol Chem. 2013;288:5072–5079. doi: 10.1074/jbc.M112.437152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Sun X, Smith KM, et al. Studies of nucleoside transporters using novel autofluorescent nucleoside probes. Biochemistry. 2006;45:1087–1098. doi: 10.1021/bi0520535. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Moake M, Har-el Y-e, et al. In vivo multicolor molecular MR imaging using diamagnetic chemical exchange saturation transfer liposomes. Magn Reson Med. 2011;67:1106–1113. doi: 10.1002/mrm.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar-Shir A, Liu G, Greenberg MM, Bulte JWM, Gilad AA. Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat Protocols. 2013;8:2380–2391. doi: 10.1038/nprot.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 33.Kato Y, Artemov D. Monitoring of release of cargo from nanocarriers by MRI/MR spectroscopy (MRS): significance of T2/T2* effect of iron particles. Magn Reson Med. 2009;61:1059–1065. doi: 10.1002/mrm.21939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minn I, Bar-Shir A, Yarlagadda K, Bulte JW, Fisher PB, Wang H, Gilad AA, Pomper MG. Tumor-specific expression and detection of a CEST reporter gene. Magn Reson Med. 2015;74:544–549. doi: 10.1002/mrm.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noe MS, Rios AC, Tor Y. Design, synthesis, and spectroscopic properties of extended and fused pyrrolo-dC and pyrrolo-C analogs. Org Lett. 2012;14:3150–3153. doi: 10.1021/ol3012327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn Reson Med. 2011;65:927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;43:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 38.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JW, van Zijl PC. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): Ph calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55:836–847. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadayakkara DK, Korrer MJ, Bulte JW, Levitsky HI. Paradoxical decrease in the capture and lymph node delivery of cancer vaccine antigen induced by a TLR4 agonist as visualized by dual-mode imaging. Cancer Res. 2015;75:51–61. doi: 10.1158/0008-5472.CAN-14-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long CM, van Laarhoven HW, Bulte JW, Levitsky HI. Magnetovaccination as a novel method to assess and quantify dendritic cell tumor antigen capture and delivery to lymph nodes. Cancer Res. 2009;69:3180–3187. doi: 10.1158/0008-5472.CAN-08-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HW, Yoon SY, Singh TD, et al. Tracking of dendritic cell migration into lymph nodes using molecular imaging with sodium iodide symporter and enhanced firefly luciferase genes. Sci Rep. 2015;5:9865. doi: 10.1038/srep09865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ardavin C, Amigorena S, Reis e Sousa C. Dendritic cells: immunobiology and cancer immunotherapy. Immunity. 2004;20:17–23. doi: 10.1016/s1074-7613(03)00352-2. [DOI] [PubMed] [Google Scholar]
- 43.de Vries IJ, Lesterhuis WJ, Barentsz JO, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol. 2005;23:1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]
- 44.Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat Biotechnol. 2005;23:983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 45.Ahrens ET, Helfer BM, O’Hanlon CF, Schirda C. Clinical cell therapy imaging using a perfluorocarbon tracer and fluorine-19 MRI. Magn Reson Med. 2014;72:1696–1701. doi: 10.1002/mrm.25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med. 2011;65:564–574. doi: 10.1002/mrm.22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponomarev V, Doubrovin M, Serganova I, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 48.Sun N, Lee A, Wu JC. Long term non-invasive imaging of embryonic stem cells using reporter genes. Nat Protoc. 2009;4:1192–1201. doi: 10.1038/nprot.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR, Iyer M, Namavari M, Phelps ME, Herschman HR. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci U S A. 2000;97:2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapiro MG, Westmeyer GG, Romero PA, Szablowski JO, Kuster B, Shah A, Otey CR, Langer R, Arnold FH, Jasanoff A. Directed evolution of a magnetic resonance imaging contrast agent for noninvasive imaging of dopamine. Nat Biotechnol. 2010;28:264–270. doi: 10.1038/nbt.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teo YN, Kool ET. DNA-multichromophore systems. Chem Rev. 2012;112:4221–4245. doi: 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinkeldam RW, Greco NJ, Tor Y. Fluorescent analogs of biomolecular building blocks: design, properties, and applications. Chem Rev. 2010;110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi HS, Gibbs SL, Lee JH, et al. Targeted zwitterionic near-infrared fluorophores for improved optical imaging. Nat Biotechnol. 2013;31:148–153. doi: 10.1038/nbt.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee T, Cai LX, Lelyveld VS, Hai A, Jasanoff A. Molecular-level functional magnetic resonance imaging of dopaminergic signaling. Science. 2014;344:533–535. doi: 10.1126/science.1249380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. a. Average MTRasym values obtained at 5.8 ppm as a function of pyrrolo-dC concentration. Dashed line represents a linear fit (R2=0.99902) of all experimental points (0–10 mM pyrrolo-dC). b. Average MTRasym values at the 5.8 ppm offset for 10 mM pyrrolo-dC solution as a function of saturation pulse power. All experiments were performed at 37°C, on an 11.7 T scanner, B1=4000 ms, B1=4.7 μT (200 Hz).
Fig. S2. Liposomal nanocariers of pyrrolo-dC. CEST MRI and optical properties, as obtained from PBS, empty liposomes, and liposomes loaded with pyrrolo-dC. a. MTRasym plots, b. MTRasym maps at 5.8 ppm, and c. fluorescence.
Fig. S3. a. Representative in vivo MTRasym maps of intracerebrally implanted 9L cells overlaid on anatomical MR images, showing the CEST signal distribution at Δω=5.8 ppm at one hour (left) and at 2.5 hours (right) post i.v. injection of pyrrolo-dC. b. Average ΔMTRasym plot of 9Lwt (gray) and 9LDm-dNK (green) images. Inset represents the range of ΔMTRasym values at 6.8 ppm > Δω > 4.8 ppm (mean ± SEM; n=5 mice). In all experiments, pyrrolo-dC was injected i.v. at 150 mg/kg.
Fig. S4. CEST signal distribution in tumor: Shown are the distribution of ΔMTRasym values within the tumor ROI (from n=5 mice) at Δω=5.8 ppm at one hour a. and at 2.5 hours b. after injection of pyrrolo-dC. Inset in (b) shows the positive ΔMTRasym values on the histograms.
Fig. S5. A dot plot of the individual MTRasym values for this graph as well as the corresponding box-and-whisker plot with the median for each group shown by a red line (0.11% and −0.51% for DCDm-dNK and DCwt respectively). We conducted a two-tailed Mann-Whitney U Test to determine if there was a significant difference between the medians of these two groups. This test is the nonparametric alternative to the t-test and does not make any assumptions about the normality of the data. There was a significant (p=0.0216) difference between the MTRasym of these two groups.


