Abstract
Mucosal surfaces are under constant bombardment from potentially antigenic particles and so must maintain a balance between homeostasis and inappropriate immune activation and consequent pathology. Epithelial cells play a vital role orchestrating pulmonary homeostasis and defense against pathogens. TGF-β regulates an array of immune responses- both inflammatory and regulatory, however its function is highly location and context dependent. We demonstrate that epithelial derived TGF-β acts as a pro-viral factor suppressing early immune responses during influenza A infection. Mice specifically lacking bronchial epithelial TGF-β1 (epTGFβKO) displayed marked protection from influenza induced weight loss, airway inflammation and pathology. However, protection from influenza induced pathology was not associated with a heightened lymphocytic immune response. In contrast, the kinetics of IFNβ release into the airways was significantly enhanced in epTGFβKO mice compared to control mice, with elevated IFNβ at day 1 in epTGFβKO compared to control mice. This induced a heighted anti-viral state resulting in impaired viral replication in epTGFβKO mice.
Thus, epithelial derived TGF-β acts to suppress early IFNβ responses leading to increased viral burden and pathology. This study demonstrates the importance of the local epithelial micro-environmental niche in shaping initial immune responses to viral infection and controlling host disease.
Introduction
Pulmonary epithelial cells are continually exposed to an inhaled environment comprising billions of innocuous particles as well as potential pathogens. Interactions between epithelial cells of the conducting airways and environmental stimuli are important in the development of pulmonary pathology and perturbations in the level of expression of the pleiotropic cytokine TGF-β and related signaling molecules in these cells appears to be pivotal. Altered expression of the TGF-β signaling molecule smad 2 at the mucosal surface has previously been shown to drive airway remodelling in response to allergen via modulation of innate epithelial derived mediators(1, 2). Epithelial cell derived TGF-β has also been shown to be a critical co factor for enhanced innate lymphoid cell function and generation of allergen mediated allergic airways disease(3). However, the precise role of TGF-β of epithelial origin in shaping the immune response to viruses within the respiratory tract is less clear. Seasonal infections with the respiratory virus influenza A result in significant mortality and morbidity in vulnerable groups as well as substantial healthcare and economic costs in the healthy population(4). Epithelial cells sense infection by pathogens via recognition of pathogen-associated molecular pattern (PAMPs) and damage-associated molecular pattern (DAMPs) molecules. Activation of these pathways induces rapid release of epithelial derived type I (IFNα and IFNβ) and type III interferons (IFNλ) which are vital for antiviral immunity.
IFN signaling in adjacent epithelial cells activates a cascade of IFN stimulated genes (ISGs) resulting in neighboring epithelial cells adopting an ‘anti-viral state’, reducing initial viral replication and subsequent viral spread(5). Rapid induction of the antiviral state is crucial as it reduces viral burden allowing time for development of adaptive immune responses.
Expression of TGF-β is increased in the pulmonary epithelium after infection with influenza(6, 7), but our understanding of the function of this pool of bioactive TGF-β is limited. However, work utilizing rhinovirus infection of human epithelial in vitro culture systems indicate an immune suppressive function for TGF-β(8).
In contrast, we show that mice specifically lacking epithelial TGF-β (epTGFβKO) exhibited marked protection from influenza induced weight loss and airway inflammation concomitant with significantly reduced pathology and viral burden. We describe the very early events at the initial site of infection (the bronchiolar epithelium) that shape and direct the subsequent immune response and place the relationship between TGF-β and interferon as a key determinant of host outcome.
Results
Epithelial TGF-β promotes influenza induced pathology
Mice specifically lacking expression of TGF-β1 in club cells which comprise 80% of bronchiolar epithelial cells were generated by crossing TGF-β1fl/fl mice with CCSP Tet-on CRE mice (Figure 1a)(3). After the mice reached adulthood they were administered a single dose of doxycycline inducing Cre recombinase expression in epithelial club cells and subsequent excision of TGF-β, generating epithelial specific TGF-β knockout mice (epTGFβKO). A detailed description of the generation of these mice and the methods used to establish ablation of epithelial TGF-β was included in Denney et al 2015. Littermates which lack one of the genes essential for excision of TGF-β were utilized as control mice. As previously reported, at steady state epTGFβKO mice displayed no inflammatory pathology(3) in stark contrast to mice lacking global or T cell specific TGF-β or TGF-β signaling(9).
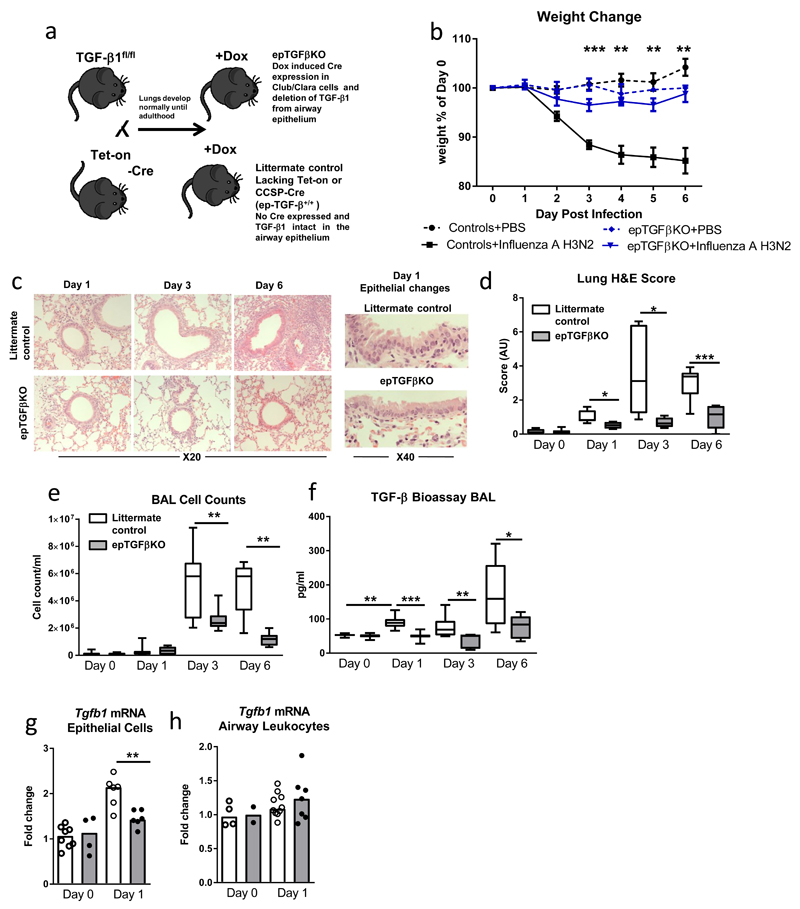
Figure 1. Mice lacking epithelial TGF-β are protected during influenza challenge.
(a) Schematic illustrating breeding scheme and doxycycline (Dox) treatment to generate epithelial TGFβKO mice. (b) Percentage weight loss after influenza infection, stars represent level of significance between infected epTGFβKO and control mice. (c) Hematoxylin and eosin stained lung tissue after infection. (d) Epithelial damage and lung inflammation score. (e) Cells counts in BAL after infection. (f) Bioactive TGF-β1 levels in the airways of influenza infected epTGFβKO and control mice. (g,h) tgfb1 mRNA levels in (g) epithelial cells from tracheal brushings and (h) airway leukocytes at day 1 post infection. Data shown is pooled from two independent experiments with a total of N≥11 mice per group. Box and whisker plots depict the median and IQR and minimum and maximum values. Line graphs are expressed as mean +/- SEM. *p < 0.05, **p < 0.01, and ***p < 0.001.
To investigate the role of local epithelial TGF-β secretion during the pulmonary response to virus, epTGFβKO mice were infected with (1.2x104 TCID50/50 μl) influenza A (subtype X31 H3N2). Although control mice showed characteristic weight loss associated with influenza infection, we found epTGFβKO mice were strongly protected from weight loss (Figure 1b), and showed only a minimal weight change from mock-infected mice at day 3 post infection. epTGFβKO mice were also protected from pathological changes within lung tissue induced by influenza infection (Figures 1c,d). Indeed, this protection was apparent very early during the disease course. The bronchiolar epithelium of control mice at day 1 post infection displayed more areas of damaged epithelial cells with sections of ruffling and disruption to normal columnar organization than mice lacking epithelial TGF-β. Associated airway inflammation was also markedly reduced in epTGFβKO mice with fewer infiltrating cells at day 3 and 6 post infection compared to infected control mice (Figure 1e).
Influenza infection is known to increase levels of bioactive TGF-β and as mice lacking epithelial TGF-β were protected from early pathological changes we investigated the primary cellular sources of TGF-β in the lung. In control mice, increased levels of bioactive TGF-β were detected in the airway lumen from day 1 post infection (Figure 1f and supplementary figure 1a). In contrast, this early increase in bioavailable TGF-β was not seen in mice lacking epithelial TGF-β suggesting that during the initial immune response to influenza TGF-β is derived from bronchiolar epithelial cells rather than from immune or other stromal cells.
Next, in order to further investigate the potential sources of TGF-β after infection, we determined Tgfb1 mRNA transcript levels at day 1 post infection in both epithelial cells and cells from the airway lumen (Figures 1g,h). Epithelial cells were obtained from tracheal brushings and as expected, showed high relative expression of CCSP (Scgb1a1) transcript, strongly suggesting a club cell enriched fraction (Supplementary Figure 1b). Airway lumen cells comprising resident and recruited leukocytes were obtained by via bronchoalveolar lavage (BAL). At 12hrs post infection airway leukocytes comprise ~95% airway macrophages (Supplementary figure 1c). By 24hrs post infection these cells included roughly equal proportions of (CD11c+SiglecF+) airway macrophages and neutrophils with a minority (<3%) lymphocytes and other cells as determined by flow cytometry (Supplementary Figure 1c).
At 12hrs post infection Tgfb1 mRNA levels were not altered from homeostasis in either the epithelial cell or the airway leukocyte fraction (Supplementary Figure 1d-e). However, the epithelial cell fraction exhibited a significant increase in Tgfb1 mRNA expression from 24hrs after infection compared to mock infected mice, while this surge in transcript abundance was not observed in epTGFβKO mice (Figure 1g). In contrast, airway leukocyte Tgfb1 mRNA transcript levels were not modulated by either infection or ablation of epithelial TGF-β at 24hrs post infection (Figure 1h).
We note that in control mice relative basal Tgfb1 transcript expression was much higher in the airway leukocytes (which comprise mostly airway macrophages) than epithelial cells (Supplementary Figure 1f). However, only epithelial cells displayed increased Tgfb1 mRNA expression in response to infection which was attenuated in epTGFβKO mice suggesting de novo Tgfb1 transcription in bronchial club cells upon infection.
Muted cellular immune response in influenza infected mice lacking epithelial TGF-β
Activation of resident airway macrophages as well as the early and efficient recruitment of other immune cells (neutrophils, infiltrating monocytes/macrophages, NK cells, T cell and ILC1s) to the lung is vital for the generation of an effective anti-viral inflammatory response and ultimately viral clearance. These cell types express TGF-βRII, and TGF-β has been shown to variously activate, recruit, maintain and regulate these cell populations(9).
In keeping with published studies, we found an increase in the number of neutrophils early after infection in the airways (Figure 2a, for gating strategy see Supplementary figure 2).
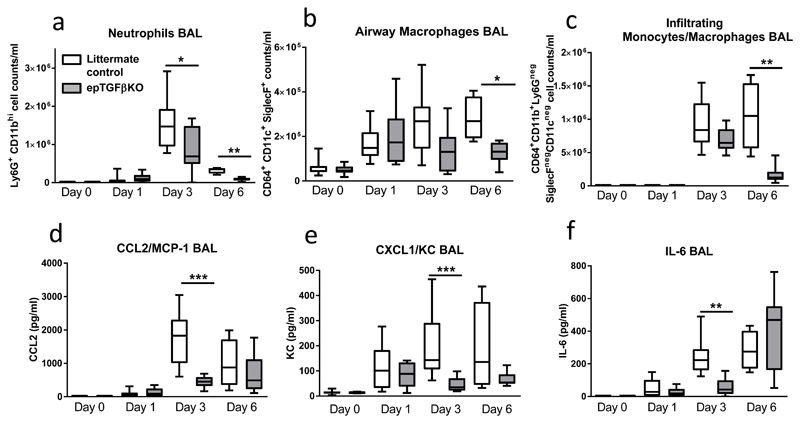
Figure 2. Immune response after influenza infection in epTGFβKO and control mice.
(a) Airway neutrophils (Ly6G+CD11bhi) total count after influenza infection of epTGFβKO and control mice. (b) Airway macrophages (CD64+CD11c+SiglecF+) total count. (c) Airway Infiltrating monocytes and macrophages (CD64+CD11b+Ly6GnegSiglecFnegCD11cneg) total count. (d-f) levels of (d) CCL2/MCP-1, (e) KC/CXCL1 and (f) IL-6 in the BAL of epTGFβKO and control mice at day 0, 1, 3, 6 post influenza infection. Data shown is pooled from two independent experiments with a total of N≥11 mice per group. Box and whisker plots depict the median and IQR and minimum and maximum values. *p < 0.05, **p < 0.01, and ***p < 0.001.
There was a significant reduction in peak neutrophilia (day 3 post infection) in epTGFβKO mice (Figure 2a). The number of CD11c+ Siglec F+ airway macrophages increased in response to influenza infection within 24 hours both in control and epTGFβKO mice (Figure 2b). However, whilst the magnitude of this response was further elevated at days 3 and 6 post infection in control mice there was no additional increase in macrophage numbers in epTGFβKO mice. There was also no differential expression of activation markers (CD11b/MHCII) on airway macrophages between control and epTGFβKO mice at either homeostasis or after infection (Supplementary Figure 3A-B). Infiltrating monocytes and macrophages (SiglecFnegCD11cneg) are potent producers of IFNs and TNF-α and induce inflammation that promotes viral clearance; however they can also cause severe immune pathology(10). Again, we observed a reduction in peak numbers of infiltrating monocyte and macrophages 6 days post influenza challenge in epTGFβKO mice (Figure 2c).
Numbers of IFNγ+ NK cells and IFNγ+ ILC1s increased in response to influenza infection but were not affected by epithelial TGF-β (Supplementary Figures 2c-d. As expected, substantial numbers of IFNγ+ T cells were only induced at day 6 after infection with a reduction in both IFNγ+ CD8 and CD4 T cells seen in epTGFβKO mice, consistent with the diminished immune response to influenza infection observed in these mice (Supplementary Figures 2e-f). IL-17+ CD4 T cells follow similar kinetics as seen with IFN-γ+ CD4 and CD8 T cells with a trend to reduced numbers at day 6 post infection (Supplementary Figures 2g). Similarly, at day 6 post infection, epTGFβKO mice showed a trend towards reduced numbers of Foxp3+ Tregs in the airways but not the lung tissue (Supplementary Figure 3h-i).
Next, we examined the levels of key immune mediators (CCL2/MCP-1, CXCL1/ KC and IL-6) that could explain the differential cell numbers of inflammatory cells in the airways of infected epTGFβKO and control mice. CCL2/MCP-1 attracts monocytes, dendritic cells and memory T cells, CXCL1/KC has neutrophil chemoattractant activity and IL-6 is essential for neutrophil recruitment and function(11). In control mice these mediators all followed a similar pattern: peaking at day 3 and remaining elevated at day 6 (Figure 2d-f). There was a comparable induction of these mediators at day 1 in both epTGFβKO and control mice. However, at days 3 and 6 there was no further increase in the levels of CCL2 and KC in epTGFβKO mice and concentrations were significantly reduced at day 3 compared to control mice. Secretion of IL-6 was also significantly reduced in epTGFβKO mice at day 3 although by day 6 levels were comparable with control mice. The peak in IL-6 levels in the epTGFβKO mice at day 6 post infection is likely derived from a combination of recruited immune cell populations as well as structural cells (including epithelial cells)(11, 12). IL-1β levels followed a similar pattern to IL-6 (Supplementary Figure 3j). In contrast to pro-inflammatory cytokines, IL-10 was not detected in the airways and in the lung tissue IL-10 levels were similar in both epTGFβKO and control mice (Supplementary Figure 3k).
Together these data suggest a generally muted inflammatory response to influenza infection in epTGFβKO mice rather than reduced recruitment or activation of specific leukocyte populations in the absence of epithelial cell-derived TGF-β. It is therefore likely that these immune changes are a consequence of earlier events rather than the initial drivers of a protective mechanism.
Ablating epithelial TGF-β expression does not protect mice from RSV driven pathology
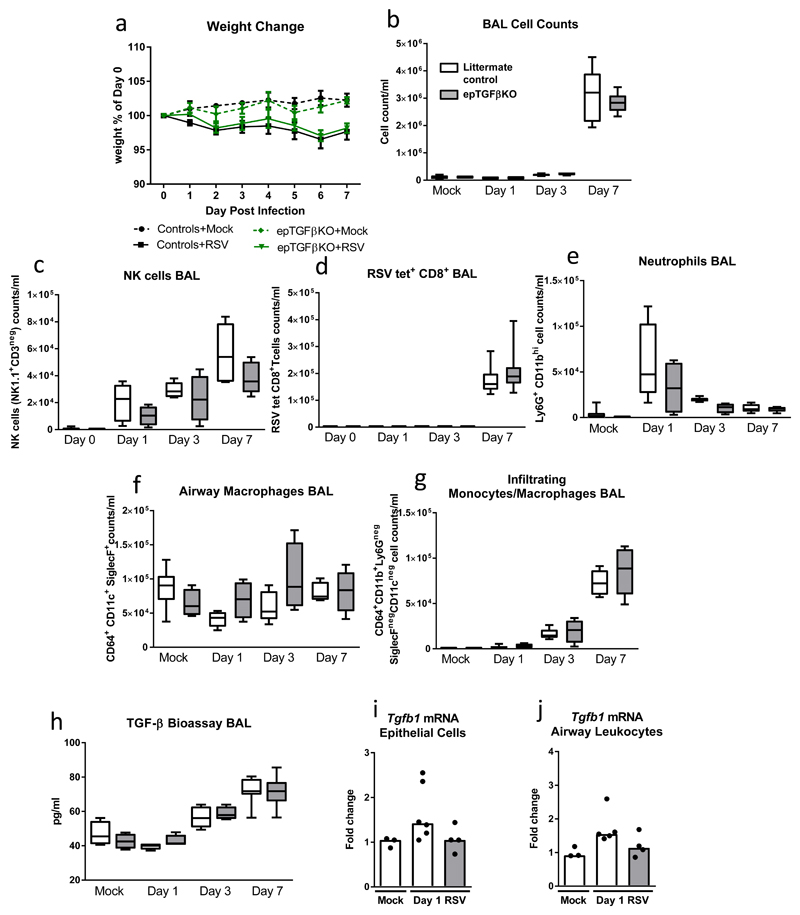
Next, we questioned if this protection from pathology associated with the loss of epithelial TGF-β was restricted to influenza A infection, which is known to cleave inactive TGF-β and displays distinct tropism for bronchial epithelium or was indicative of a more general association with viral immunity. Therefore, we infected mice with respiratory syncytial virus (RSV) which preferably infects alveolar epithelial cells and lacks the neuraminidase protein that mediates TGF-β activation by influenza virus(13). In stark contrast to influenza infection, epTGFβKO and control mice showed indistinguishable weight loss and airway inflammation following RSV infection (Figures 3a,b). In the airways, frequency of RSV tetramer+ CD8+T cells, NK cells, neutrophils, airway macrophages and inflammatory monocyte/macrophages, were equivalent in epTGFβKO mice and control (Figures 3c-g). Unlike influenza, RSV infection did not lead to increased levels of bioactive TGF-β in the airways at day 1 post infection and at all time points levels were similar in epTGFβKO and control mice (Figure 3h). In keeping with this data, Tgfb1 mRNA levels were not altered after RSV infection in either epithelial cells or airway leukocytes (Figure 3,j). These data indicate the key importance to host outcome of both the site of viral infection as well as the capacity of the pathogen to alter the immediate immune environment by promoting the generation of bioactive TGF-β.
Figure 3. Mice lacking epithelial TGF-β are not protected during RSV challenge.
(a) Percentage weight loss after RSV infection (1x106 FFU) in epTGFβKO and control mice. (b) Cells counts in BAL with mock infection and day 1, 3 and 7 after RSV infection. (c) RSV tet+ CD8 T cells (CD8+CD3+) in the airways. (d) Number of NK cells (NK1.1+CD3neg) in the airways. (e) Airway neutrophils (Ly6G+CD11bhi) total count after influenza infection of epTGFβKO and control mice. (f) Airway macrophages (CD64+CD11c+SiglecF+) total count. (g) Airway infiltrating monocytes and macrophages (CD64+CD11b+Ly6GnegSiglec FnegCD11cneg) total count. (h) Bioactive TGF-β1 levels in the airways of RSV infected epTGFβKO and control mice. (i,j) tgfb1 mRNA levels in (i) epithelial cells from tracheal brushings and (j) airway leukocytes at day 1 post infection. N≥7 mice per group. Box and whisker plots depict the median and IQR and minimum and maximum values. Line graphs are expressed as mean +/- SEM.
Ablation of epithelial TGF-β reduced live viral titre
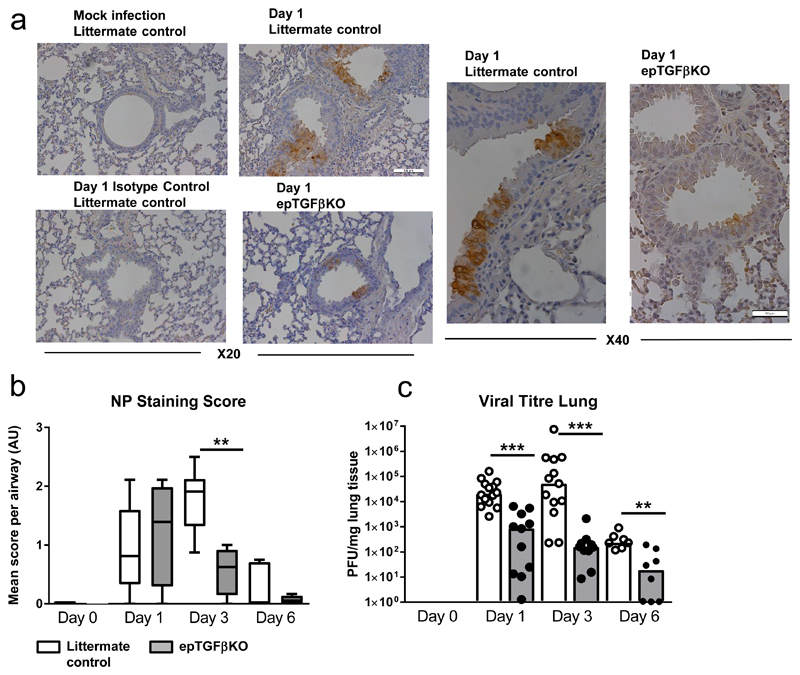
Since epTGFβKO mice exhibited a strong, early protection from the pathogenic effects of influenza infection we examined whether the virus was able to infect bronchial epithelial cells in these mice. Influenza A initially infects club cells in the epithelium before subsequent infection of both structural and immune cells (including macrophages and DCs)(14). We surveyed the location of viral particles within the lung at early time points after infection to determine primary infection sites. In keeping with published studies, infected cells were initially restricted to the bronchial epithelium and viral protein staining was observed in both control mice and those lacking epithelial TGF-β (Figure 4a), consistent with both control and TGF-β-deficient epithelium being permissive to the initial entry of IAV particles. Quantification of NP stained airways also showed reduced NP staining at day 3 post infection in epTGFβKO mice compared to control mice (Figure 4b). In order to further examine viral tropism we determined the levels of viral RNA in epithelial cells and airway leukocytes finding that at 12hrs and 1 day post infection X31 RNA levels are highest in epithelial cells (Supplementary Figure 4a). Although staining for NP protein and qPCR for viral RNA allows identification of infected cells and viral tropism, neither technique can quantitatively determine the live viral burden.
Figure 4. Reduced Viral titre in mice lacking epithelial TGF-β.
(a) NP influenza protein immuno-histochemistry staining in the bronchial airways of epTGFβKO and control mice at day 1 after influenza infection (20X and inset X40). (b) NP staining quantification in the lung. (c) Viral titer in lung tissue by plaque assay in MDCK cells. Data shown is pooled from two independent experiments with a total of N≥11 mice per group. Bar chart depict the median. ***p < 0.001.
Therefore, we determined viral titre in whole lung homogenate by plaque forming unit (PFU) assay(15). As expected, virus was detectable at day 1 post infection and PFU peaked at day 3 in control mice (Figure 4c). In contrast, there was a significant reduction in PFU in epTGFβKO mice at both day 1 and day 3 compared to control mice (Figure 4c). Therefore, mice lacking epithelial TGF-β had a substantially diminished early viral burden as well as a reduced peak viral load. These differences in viral titre at day 1 post infection suggest an altered early anti-viral response in mice lacking epithelial TGF-β and likely accounts for the reduced immunopathology seen from day 3 onwards.
Differential Interferon responses mediate immune protection in mice lacking epithelial TGF-β
Detection of viral infection, induction of a rapid immune response and the appropriate curtailment of that response to prevent immune pathology are key steps in effective host protective immunity. Mice lacking epithelial TGF-β have reduced viral burden and are protected from influenza induced weight loss, inflammation and pathological changes. However, contrary to expectation key mediators and effector cell populations involved in viral clearance/limiting viral infection were either equivalent or reduced in these mice. Therefore, we explored more closely the mechanisms that are associated with the initial epithelial response to viral infection namely, the type I and III interferon response.
Firstly, we examined expression of IFNλ protein, finding no significant induction of IFNλ above homeostatic levels in the airways at day 1 post infection in either control or epTGFβKO mice that would reflect the difference in early epithelial damage or viral titre (Figure 5a). In accordance with published work, control infected mice displayed substantially increased IFNλ levels at day 3 post infection which by day 6 had reduced to levels similar to baseline(16). However, in epTGFβKO mice contrary to expectation the magnitude of peak day 3 response was greatly reduced. We found that IFNλ was the predominant IFN released after infection and was present at much higher levels than type I IFNs (Figures 5a-c), in keeping with published work(16). However, the kinetics of its release were too slow to drive the early immune protection seen in epTGFβKO mice and thus it is unlikely to mediate the early epithelial anti-viral response and subsequent protection from influenza induced pathology observed in mice lacking epithelial TGF-β.
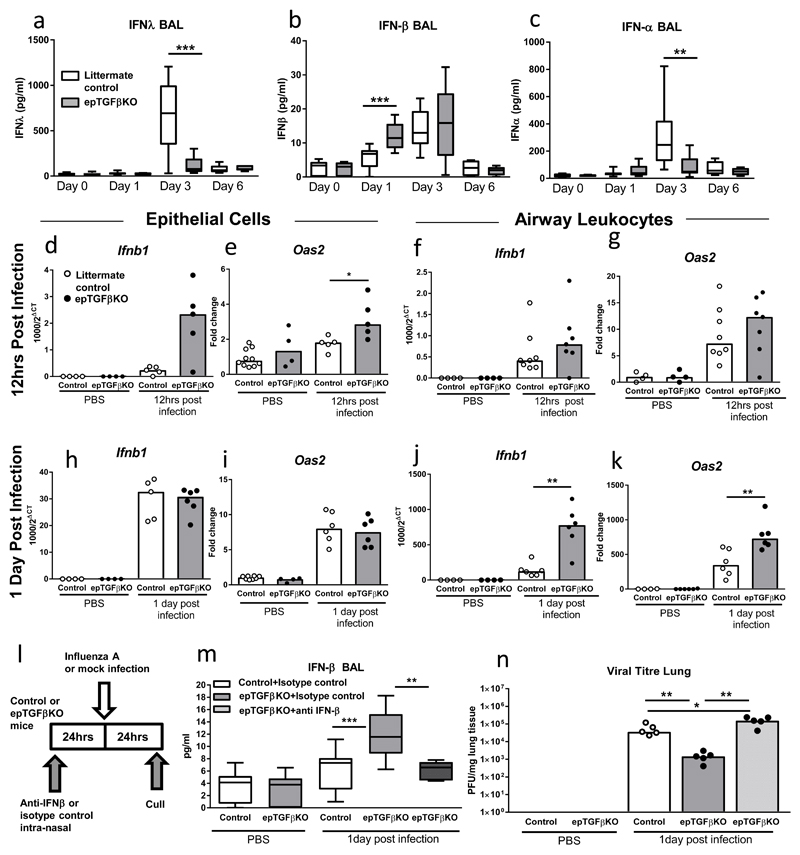
Figure 5. Enhanced early IFNβ response in epTGFβKO mice.
(a-c) Concentration of (a) IFNλ, (b) IFNβ and (c) IFNα in the airways of epTGFβKO mice and control mice after influenza infection. Limits of detection were 15.6pg/ml, 7.25 pg/ml and 7.48 pg/ml respectively. (d,e) mRNA levels of (d) Ifnb1 and (e) Oas2 in epithelial cells 12hrs after infection. (f,g) mRNA levels of (f) Ifnb1 and (g) Oas2 in airway leukocytes 12hrs after infection. (h,i) mRNA levels of (h) Ifnb1 and (i) Oas2 in epithelial cells 1 day after infection. (j-k) mRNA levels of (j) Ifnb1 and (k) Oas2 in airway leukocytes 1 day after infection. (l) Schematic showing anti-IFN-β treatment and influenza infection schedule. (m) Concentration IFNβ airways of epTGFβKO mice and control mice after anti-IFN-β (or isotype control) treatment and influenza infection. (n) Viral titres in lung tissue of anti-IFN-β (or isotype control) treatment epTGFβKO mice and control mice infected with influenza. Data shown is representative of two independent experiments with a minimum of n=5 per group. Box and whisker plots depict the median and IQR and minimum and maximum values and bar charts depict the median. **p < 0.01, and ***p < 0.001.
In contrast, IFNβ was substantially increased at day 1 post infection in the airways of epTGFβKO mice compared to both mock infected and day 1 post infection control mice, with levels remaining elevated at day 3 post infection (Figure 5b). Indeed, in control mice IFNβ levels were not detected until day 3 post infection. We also found that expression of IFNβ in the airways inversely correlated with viral titre at day 1 post infection (p=0.0018, spearman correlation, r value -0.6032 and Supplementary figure 4b). Hence, in mice lacking epithelial TGF-β, IFNβ kinetics in the airways were substantially altered, with a more rapid induction of the peak response and sustained elevation.
However, IFNα the other key type I interferon family member did not exhibit the pattern of expression seen with IFNβ (Figure 5c). There was no significant induction of IFNα levels at day 1 post infection between epTGFβKO and control mice. By day 3, IFNα levels were substantially higher in control mice than the epTGFβKO mice, a comparable trend to that seen with IFNλ. These data suggest that IFNβ expression is differentially regulated compared to IFNλ and IFNα. Thereafter, we investigated the cellular source of IFNβ after infection by examining expression patterns of Ifnb1 in both epithelial cells and airway leukocytes. As early as 12hrs post infection Ifnb1 gene expression was increased in epithelial cells from epTGFβKO mice (Figure 5d). This observation correlates with a significant induction of the interferon stimulated gene (ISG) Oas2 (which activates latent RNAses and aids viral degradation/inhibits viral replication), implying the rapid implementation of an antiviral state in epithelial cells lacking TGF-β expression (Figure 5e). We questioned whether this adoption of an antiviral state was restricted to epithelial cells or if airway leukocytes (which comprise ~95% airway macrophages at 12hrs post infection, Supplementary figure 1c) also showed a heighted antiviral state. Expression of Ifnb1 was also increased in airway leukocytes from epTGFβKO mice although the fold induction relative to infected control was less than that observed in epithelial cells (Figure 5f). Oas2 expression in airway leukocytes also followed a similar pattern (Figure 5g and Supplementary Figure 4c).
At 1 day post infection, both control and epTGFβKO epithelial cells exhibited a marked increase in expression levels of Ifnb1 and Oas2 compared to uninfected mice (Figures 5h,i). However, by this time point there was no difference in Ifnb1 and Oas2 transcript levels between infected control and epTGFβKO epithelial cells; consistent with a rapid process in which the differential Ifnb1 and Oas2 gene expression at 12hrs post infection precedes the elevated airway IFNβ protein and reduced pulmonary viral titre observed at 1 day post infection. In contrast, at day 1 post infection, there was a significant increase in Ifnb1 and Oas2 transcript levels epTGFβKO in airway leukocytes compared to control (Figures 5j,k).
This pattern of increased ISG transcript expression in airway leukocytes from mice lacking epithelial TGF-β was observed in other key ISGs- Irf7 (an interferon regulatory transcription factor that positively regulates the type 1 interferon response) and Ifitm3 (inhibits viral entry to host cytoplasm via altered cholesterol homeostasis) (Supplementary Figures 4e-f). These early changes in Ifnb1 and ISG gene expression patterns between control and epTGFβKO are consistent with differences in both IFNβ protein in the airways and viral titre at 1 day post infection.
We also examined the early IFN response in RSV infected mice. In keeping with published studies(17, 18), we find RSV induced IFNβ peaked at day 1 post infection there was however, no difference in IFNβ levels between TGFβKO and control mice (Supplementary Figure 5). Unlike influenza, Ifnb1 mRNA was not induced in epithelial cells at day 1 post RSV infection (Supplementary Figure 5b). Airway leukocytes did exhibit increased Ifnb1 expression at day 1 post RSV infection however, there was no difference in Ifnb1 levels between TGFβKO and control mice (Supplementary Figure 5c). OAS2 was induced in both epithelial cells and airway leukocytes at day 1 post infection but was not differentially expressed in TGFβKO and control mice (Supplementary Figure 5d-e). Therefore, the kinetics of IFNβ induction in RSV are distinct from Influenza. In this context, ablating epithelial TGF-β in RSV infection has little effect on the IFNβ pathways and ultimately on disease pathology.
In order to establish the central role of early induction of IFNβ in protection of TGFβKO mice from influenza infection, we examined the effect of blocking IFNβ during the initial stages of infection. Mice were treated with anti-IFNβ (or isotype control) intranasally 1 day prior to infection and culled at 1 day post infection (Figure 5l). At day 1 post infection, epTGFβKO mice treated with isotype control had a significant induction of airway IFNβ and this was abrogated in epTGFβKO mice treated with anti-IFNβ (Figure 5m). Concomitant with reduced IFNβ levels, viral titre in epTGFβKO mice treated with anti-IFN-β was significantly increased compared to influenza infected isotype control treated mice (Figure 5n). Therefore, the early induction of IFNβ in the airways and generation of a heighted antiviral state is critical to the protection from disease pathology observed in mice lacking epithelial TGF-β.
Discussion
The pulmonary epithelium plays a pivotal role in both the maintenance of immune homeostasis and defense against pathogens in the lung. The detection of viral infection, induction of a rapid immune response and the appropriate curtailment of that response to prevent immune pathology are key steps in effective host protection, allowing effective viral clearance while minimizing immunopathology(19). We have determined that influenza infection rapidly triggers production and activation of TGF-β from CCSP+ epithelial cells, which acts to constrain early IFNβ production and impair adoption of a pulmonary antiviral state, leading to unrestrained viral replication in the host and increased immunopathology (described schematically Figure 6). Importantly, local antibody blockade of IFNβ reversed the protective effect of epithelial TGF-β ablation on pulmonary viral load, supporting the conclusion that the pro-viral effect of epithelial TGF-β is reliant on negative regulation of IFNβ. We describe this novel function for epithelial TGF-β in vivo, identifying a link between two cytokines with highly location and context dependent functions. In this setting, epithelial TGF-β functions as a pro-viral factor by restraining the local antiviral IFNβ response enabling influenza A to temporarily evade host defenses and favor its own replication and survival.
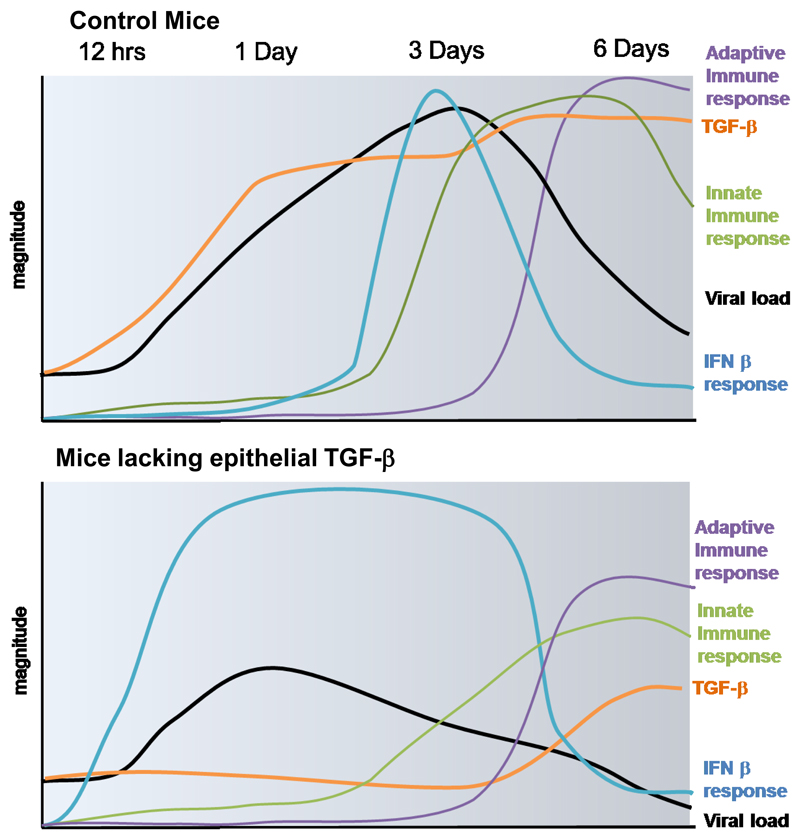
Figure 6. Schematic illustrating timing and magnitude of key immune events in the lung after influenza infection in mice lacking epithelial TGF-β expression and control mice.
Club cell derived TGF-β is rapidly released into the airways after influenza infection, acting to suppress early IFNβ responses from both epithelium and airway leucocytes (a population consisting predominantly of macrophages). Hence, pulmonary viral load rises and although the viral infection is ultimately cleared by a robust immune response this is at the cost of host immunopathology. In the absence of club cell derived TGF-β, IFNβ is promptly induced in both epithelial cells and airway leukocytes impairing viral fitness, reducing viral load and leading to muted innate and adaptive immune responses, protecting the host from both viral and immune driven pathology.
This early TGFβ - IFNβ axis, beginning at the initial site of infection, the bronchial epithelium, critically shapes the outcome of influenza infection. While others have previously reported the activation of TGF-β by influenza (6, 20, 21), we describe for the first time a disease-modifying role for a specific cellular source of TGF-β1 in influenza infection. This relationship between epithelial-derived TGF-β and IFNβ underscores the importance of local and appropriate immune responses in determining the outcomes of infection for the host.
We describe the potential for differential patterns of expression of IFNβ compared to both IFNα and IFNλ, consistent with a report that type I and III IFNs are independently controlled during influenza infection(16). Additionally, our data reinforce the concept that, despite overlapping activation pathways, there is a hierarchal relationship between type I and III IFNs, with IFNβ acting prior to IFNα and IFNλ induction(22, 23). Accordingly, we detected evidence of enhanced epithelial IFNβ activity in epTGFβKO mice as early as 12hrs post-infection, which quickly spread to an enhanced pulmonary antiviral state detectable in airway leukocytes at 24hrs, coincident with decreased lung viral load. In contrast, no enhancement of IFNα levels were detected in BAL of epTGFβKO mice at this time point and IFNλ was undetectable in BAL before 72hrs post-infection.
The preference for type I versus type III IFN production by epithelial cells appears organ specific. Gut epithelial cells exclusively producing type III IFNs(24), while pulmonary epithelial cells secrete both type I and III IFNs upon infection(25). Our study supports the theory that although abundant, both IFNα and IFNλ play a subordinate role in influenza infection, with IFNβ acting as the pivotal inducer/mediator of ISG activation and the antiviral response at the earliest stages of infection(26). Indeed, Wack et al show that small differences in early interferon induction can have dramatic influences on host survival(27). Notably, the levels of IFNs we have observed in the current study are comparable to those reported to lead to differential survival outcomes.
In contrast to the protective effects of enhanced IFNβ production in our model, Wack et al show a pathogenic role for increased type 1 IFN production in the 129 mouse strain after influenza infection. However, while IFNβ levels at 1 day post infection were enhanced in epTGFβKO mice, peak levels did not exceed those of controls at any of the time-points tested, indicating that the timing, as well as context of IFNβ release is critical to its effects during influenza infection.
TGF-β has previously been suggested, by in vitro studies, to act as a modulator of IFNβ during infection(8). Utilizing human epithelial cells infected with rhinovirus Bedke et al showed that epithelial TGF-β influences IFNβ production(8). Exogenous TGF-β increased viral replication and decreased IFNβ protein secretion, while conversely, blocking TGF-β had reduced viral titre and increased IFNβ levels. We demonstrate for the first time that epithelial TGF-β suppresses IFNβ production in vivo, revealing interactions between the epithelial cytokines and the antiviral response in airway leukocytes that dictate the consequences of mucosal infection.
A very recent study utilizing mice deficient in β6 integrin (and therefore impaired activation of TGF-β) also showed improved survival following influenza infection(21). This protective phenotype could be ablated via the addition of exogenous TGF-β. In this model lower levels of TGF-β at steady state lead to constitutively activated airway macrophages with increased CD11b expression and IFN responsiveness. In contrast, protection in our model was independent of differential expression of activation markers (CD11b/HMCII) on airway macrophages between control and epTGFβKO mice at either homeostasis or after infection. Cumulatively, this suggests that there are differential outcomes to ablating integrin mediated activation of TGF-β in the lung versus specific deletion of TGF-β from the club cells of the conducting airway epithelium, which we show to be the source of bioactive TGF-β released into the airway lumen early in influenza infection.
Several previous studies suggest that the timing of TGF-β release and/or activation is absolutely crucial to host outcome with a variety of regimens and interpretations explored. Conflicting results are reported when TGF-β was blocked using pan TGF-β antibodies. One study showed little effect of TGF-β blockade on key markers of influenza pathology(28) while in another substantially increased mortality was observed(20). However, in the latter study, pathogenic effects of pan-TGF-β blockade were only reported from 6 days post-infection, strongly suggesting a dependence on the role of TGF-β in regulation of T cell responses(28) in contrast to our rapid effect on viral load. Similarly, Furuya et al show protection of mice from influenza infection after induction of allergic airways disease to be at least partially TGF-β-mediated(29). However this protection was independent of viral load and was likely mediated by the suppressive effects of TGF-β on immune cells.
When viewed in the context of these published findings, our data show that ablation of the cell-specific source of TGF-β1 released rapidly upon infection has drastically different consequences to global blockade throughout infection.
We suggest that in addition to timing, the cellular source of bioactive TGF-β is critical in determining host outcome. It has been proposed that the increase in bioavailable TGF-β after influenza infection arises via direct enzymatic activation of the pool of latent TGF-β(6, 20). However, we note a substantial increase in mRNA levels of Tgfb1 in epithelial cells from infected control mice, which is absent in epTGFβKO mice, suggesting that de novo protein synthesis from CCSP+ epithelial cells also occurs. It is pertinent that the induction of Tgfb1 mRNA is restricted to bronchial epithelial cells, which are the early site of viral attachment and infection. This location specific effect of TGF-β is underscored by the lack of protection of epTGFβKO from RSV, in which no induction of TGF-β mRNA or bioactive protein was observed rapidly after infection and no enhancement of the early antiviral IFNβ response was observed. This clear distinction between the effects of club cell TGF-β ablation on different respiratory viral infections may be due to tropism of RSV for alveolar rather than bronchial epithelial cells, in which TGF-β expression is intact in our model(3), or the inability of RSV to activate latent TGF-β, which is a feature of multiple influenza strains(20).
The hijacking of TGF-β regulatory pathways either through enzymatic activation of host latent TGF-β or via pathogen derived molecules that mimic TGF-β and induce receptor signaling is an evolutionary adaption utilized by a large number of unrelated pathogens both in in vivo models and human disease(30). In humans, understanding precisely how TGF-β functions in the micro-environmental niche at the site of infection is, naturally, fraught with technical difficulties. However, this study clearly demonstrates a role for early epithelial TGF-β as a local immune regulatory factor with capacity for therapeutic modulation and stresses the importance of the airway epithelium in controlling pulmonary immune response.
Methods
Animals, infections and blocking antibody treatment
Mice on a C57BL/6 background carrying floxed alleles for Tgfb1(31) (JaTgfb1tm2.1Doe/J, Jackson Laboratory) were crossed with mice expressing Cre recombinase under the control of the rat CCSP promoter CCSP-rtTA/tetO-Cre(32). Cre expression was induced by a single intraperitoneal injection of 2 mg doxycycline (DOX) (Sigma-Aldrich) resulting in excision of the Tgfb1 gene (epTGFβKO)(3). Littermates carrying two floxed Tgfb1 alleles but lacking either the CCSP-rtTA or tetO-Cre alleles were used as control mice. One week after DOX administration, mice were infected with 1.2x104 TICD50 of X31 (a kind gift from Andreas Wack, Crick Institute) in 50μl PBS, or mock infected with 50μl PBS. Alternatively, mice were infected with 1x106 focus-forming unit (FFU) RSV in 100μl (strain A2 (ATCC) was grown in Hep-2 cells as described previously(11) or mock infected with Hep-2 supernatant. In some experiments, mice were administered 100 μg of hamster anti-mouse IFNβ (clone MIB-5E9.1) or isotype control (clone HTK888, both Biolegend) in 100μl volume intranasally, 24 hours prior to infection. Mice were housed in specific pathogen free conditions, given food and water ad libitum and used for experiments at 7-12 weeks of age. All procedures were conducted in accordance with the Animals (Scientific Procedures) Act 1986.
Cell and Organ processing
BAL was obtained by washing the airways three times with a volume of 400μl PBS. After centrifugation the cell pellet was resuspended in 500μl of complete media (RPMI, 10%FCS, 2mM L-glutamine, 100U/ml penicillin/streptomycin) (GIBCO, Life Technologies) and the supernatant stored at -80°C for further analysis. The left lobe was finely chopped and digested with 15 mg/ml collagenase (Type D; Roche Diagnostics) and 25μg/ml DNase (Type 1; Roche Diagnostics) in 4ml complete media for 1hr at 37°C with agitation. Lung was then passed through a 70μm sieve (BD Bioscience), cells were washed, red blood cells lysed and the cell pellet resuspended in 1ml complete media. Airway epithelial cells were obtained by tracheal brushing using 4mm interdental brushes (TePe).
Viral titre
Influenza virus titres in the lungs were determined by plaque assay on MDCK cells(33). The inferior lobe was homogenised in 1ml DMEM containing 100U/ml penicillin/streptomycin (GIBCO, Life Technologies). Briefly, serial dilutions of lung homogenate were incubated with near confluent monolayers of MDCK cells for 1hr at 37°C, the monolayer was washed and overlaid with plaque overlay media. After 72 hours, the overlay media was removed and cells were fixed, stained with crystal violet and the plaques counted.
Flow cytometry
Intracellular expression of cytokines was assessed after cells were stimulated with PMA (Sigma-Aldrich)/ionomycin (Emdchemicals) in the presence of Brefeldin A (Sigma-Aldrich) for 3.5hr at 37°C.
After stimulation cells were washed and Fc receptors blocked. Extracellular antigens were stained in 5% FCS/1% BSA in PBS for 30 min at 4°C. Cells were then washed and fixed. Where necessary, cells were permeabilized using Fix/Perm kit (eBioscience) and stained for intracellular antigens. All antibodies were obtained from eBioscience except ICOS, CD11c, CD11b, Ly6G, CD64, (Biolegend) and SiglecF (BD Biosciences). Biotinylated Db RSV M2187-195 monomers were kindly provided by the NIH Tetramer Core Facility (Atlanta, GA). They were multimerized using PE-conjugated streptavidin (Molecular Probes, Life Technologies) as described on the NIH TCF website (tetramer.yerkes.emory.edu/support). Data were acquired on an LSRII Fortessa (BD Biosciences) and analysed using FACSDiva (BD Biosciences) and FlowJo (Treestar) software.
Histological scoring
The right superior lobe was inflated and fixed in BNF prior to wax imbedding and sectioning. Haematoxylin and eosin stained lung sections were scored blindly for epithelial damage, inflammation and lung infiltrates. More than 10 airways and vessels were scored in each lung section. Epithelial damage was scored 0 (normal) 1 (small areas of abnormality) 2 (more than 50% abnormal), 3 (areas of epithelial detachment up to 20%), 4 (areas of epithelial detachment up to 50%) and 5 (more than 50% of the epithelium denuded). Airway and vessel inflammation- 0 (normal), 1 (cell infiltrate/cuffing around the vessel only), 2 (cell infiltrate around the vessel and airways), 3 (double layer of cell infiltrate around the vessel and airways), 4 (multiple layers of cell infiltrate around the vessel and airways) and 5 (large areas of infiltrate).
Immunohistochemistry and scoring of influenza infection
Paraffin embedded lung sections were subjected to antigen retrieval in boiling citrate buffer and stained with polyclonal goat anti-influenza NP antibody or goat IgG isotype control (Abcam), followed by biotin-conjugated donkey anti-goat secondary antibody (Jackson Immunoresearch). Staining was developed using the Vectastain ABC kit and DAB substrate (Vector Laboratories) as per manufacturer’s instructions. The extent of bronchiolar NP staining was semi-quantitatively analyzed by a blinded investigator, using the following scoring system. The mean of all airways in one section (≥10) was reported for each mouse.0, No NP staining. 1, 1-25 % of epithelial cells in airway stained. 2, 26-50 % of epithelial cells in airway stained. 3, 51-75 % of epithelial cells in airway stained. 4, 51-75 % of epithelial cells in airway stained. An additional +1 score was added where one or more patches of dense staining was observed, giving a maximum score of 5 per bronchiole.
Measurement of Cytokines
The right middle lobe was homogenized at 50mg/ml in HBSS (GIBCO) containing 1 protease inhibitor tablet per 50ml (Roche Diagnostics) and centrifuged at 800×g for 20 min at 4°C. Paired antibodies for murine IFNβ (PBL Assay Science), IFNλ, KC, MCP-1 (R&D Systems) and IFNα, IL-6, IL-1β (eBioscience) were used in standardized sandwich ELISAs. Bioactive TGF-β in BAL was measured by bioassay as previously described(34).
Real-Time PCR
Airway leukocytes and epithelial brushing cells were lysed in RLT buffer and passed through QIAShredder columns, before extracting RNA using the RNeasy Micro Kit (all QIAGEN). ≥20 ng of total RNA was converted into cDNA using the GoSCRIPT™ Reverse Transcription System (Promega). Real-time PCR reactions for mouse genes were performed using Taqman Fast Advanced Master Mix with TaqMan primer/probe sets for murine Oas2, Irf7, Ifitm3, Tgfb1, Scgb1a1 (CCSP) and Ifnb1 plus housekeeping genes Gapdh and Hprt (all Thermo Fisher Scientific). Real-time PCR for influenza virus was performed using Fast SYBR Green Master Mix (Thermo Fisher Scientific) with forward and reverse primers for X31 (forward: TGAGTCTTCTAACCGAGGTC, reverse: GGTCTTGTCTTTAGCCATTCC) and housekeeping genes Gapdh (forward: TCCCACTCTTCCACCTTCGA, reverse: AGTTGGGATAGGGCCTCTCTT) and Actb (forward: CTAAGGCCAACCGTGAAAAG, reverse: ACCAGAGGCATACAGGGACA). All real-time PCR was performed on a Viia7 instrument and threshold cycle (CT) values determined from duplicate reactions using Viia 7 sofware (Thermo Fisher Scientific). As appropriate, results were expressed as either relative gene expression [1000/2(CT of target gene- CT of housekeeping genes)] or fold change in relative expression from a control group.
Statistical Analysis
All data were analyzed with Prism 7 (GraphPad). Box and whisker plots depict medians and IQRs. Line graphs show means ± SEM. Bars on scatter plots show medians and dots indicate data points from individual mice. Non-parametric data were analyzed using Mann-Whitney U test. Correlations used a Spearman test. Significance was defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary Material
Supplementary Material is linked to the online version of the paper at http://www.nature.com/mi
Acknowledgments
The authors thank the staff of the Mary Lyon Centre (MLC) at Harwell for performing mouse re-derivation and animal husbandry support, Lorraine Lawrence for histological sectioning, Jessica Rowley and Jane Srivastava of the Imperial College Core Flow Cytometry facility for assistance. We also thank Andreas Wack (The Francis Crick Institute) for his kind gift of influenza virus stocks, James Harker and Chloe Pyle for guidance with plaque assays and RSV infections. This work was supported by the Welcome Trust (grants 087618/Z/08/Z and 107059/Z/15/Z).
Footnotes
Author Contributions
LD wrote the first manuscript draft. LD and WB performed the majority of the experiments and revised the manuscript. LGG performed some of the analysis and revised the manuscript. CML is the principle investigator who conceived the study and edited the manuscript.
Disclosure
The authors have nothing to disclose.
The authors have no conflict of interest to declare.
References
- 1.Gregory LG, Mathie SA, Walker SA, Pegorier S, Jones CP, Lloyd CM. Overexpression of Smad2 drives house dust mite-mediated airway remodeling and airway hyperresponsiveness via activin and IL-25. American journal of respiratory and critical care medicine. 2010;182:143–154. doi: 10.1164/rccm.200905-0725OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregory LG, Jones CP, Walker SA, Sawant D, Gowers KH, Campbell GA, McKenzie AN, Lloyd CM. IL-25 drives remodelling in allergic airways disease induced by house dust mite. Thorax. 2013;68:82–90. doi: 10.1136/thoraxjnl-2012-202003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denney L, Byrne AJ, Shea TJ, Buckley JS, Pease JE, Herledan GM, Walker SA, Gregory LG, Lloyd CM. Pulmonary Epithelial Cell-Derived Cytokine TGF-beta1 Is a Critical Cofactor for Enhanced Innate Lymphoid Cell Function. Immunity. 2015;43:945–958. doi: 10.1016/j.immuni.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oslund KL, Baumgarth N. Influenza-induced innate immunity: regulators of viral replication, respiratory tract pathology & adaptive immunity. Future Virol. 2011;6:951–962. doi: 10.2217/fvl.11.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz-Cherry S, Hinshaw VS. Influenza virus neuraminidase activates latent transforming growth factor beta. Journal of virology. 1996;70:8624–8629. doi: 10.1128/jvi.70.12.8624-8629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs JD, Ornoff DM, Igo HA, Zeng JY, Imani F. Cell cycle arrest by transforming growth factor beta1 enhances replication of respiratory syncytial virus in lung epithelial cells. Journal of virology. 2009;83:12424–12431. doi: 10.1128/JVI.00806-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedke N, Sammut D, Green B, Kehagia V, Dennison P, Jenkins G, Tatler A, Howarth PH, Holgate ST, Davies DE. Transforming growth factor-beta promotes rhinovirus replication in bronchial epithelial cells by suppressing the innate immune response. PLoS One. 2012;7:e44580. doi: 10.1371/journal.pone.0044580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Travis MA, Sheppard D. TGF-beta activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, Ho LP. Pivotal Advance: Invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukoc Biol. 2012;91:357–368. doi: 10.1189/jlb.0411184. [DOI] [PubMed] [Google Scholar]
- 11.Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, Bunn J, Suratt BT, Haynes L, Rincon M. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 2012;5:258–266. doi: 10.1038/mi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clinical microbiology reviews. 2011;24:210–229. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormier SA, You D, Honnegowda S. The use of a neonatal mouse model to study respiratory syncytial virus infections. Expert review of anti-infective therapy. 2010;8:1371–1380. doi: 10.1586/eri.10.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baer A, Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp. 2014:e52065. doi: 10.3791/52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, Durbin JE. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. Journal of virology. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jewell NA, Vaghefi N, Mertz SE, Akter P, Peebles RS, Jr, Bakaletz LO, Durbin RK, Flano E, Durbin JE. Differential type I interferon induction by respiratory syncytial virus and influenza a virus in vivo. J Virol. 2007;81:9790–9800. doi: 10.1128/JVI.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remot A, Descamps D, Jouneau L, Laubreton D, Dubuquoy C, Bouet S, Lecardonnel J, Rebours E, Petit-Camurdan A, Riffault S. Flt3 ligand improves the innate response to respiratory syncytial virus and limits lung disease upon RSV reexposure in neonate mice. European journal of immunology. 2016;46:874–884. doi: 10.1002/eji.201545929. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson CM, Turpin EA, Moser LA, O'Brien KB, Cline TD, Jones JC, Tumpey TM, Katz JM, Kelley LA, Gauldie J, Schultz-Cherry S. Transforming growth factor-beta: activation by neuraminidase and role in highly pathogenic H5N1 influenza pathogenesis. PLoS pathogens. 2010;6:e1001136. doi: 10.1371/journal.ppat.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meliopoulos VA, Van de Velde LA, Van de Velde NC, Karlsson EA, Neale G, Vogel P, Guy C, Sharma S, Duan S, Surman SL, Jones BG, et al. An Epithelial Integrin Regulates the Amplitude of Protective Lung Interferon Responses against Multiple Respiratory Pathogens. PLoS pathogens. 2016;12:e1005804. doi: 10.1371/journal.ppat.1005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazear HM, Nice TJ, Diamond MS. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS pathogens. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS pathogens. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mordstein M, Michiels T, Staeheli P. What have we learned from the IL28 receptor knockout mouse? Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2010;30:579–584. doi: 10.1089/jir.2010.0061. [DOI] [PubMed] [Google Scholar]
- 27.Davidson S, Crotta S, McCabe TM, Wack A. Pathogenic potential of interferon alphabeta in acute influenza infection. Nat Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods PS, Tazi MF, Chesarino NM, Amer AO, Davis IC. TGF-beta-induced IL-6 prevents development of acute lung injury in influenza A virus-infected F508del CFTR-heterozygous mice. Am J Physiol Lung Cell Mol Physiol. 2015;308:L1136–1144. doi: 10.1152/ajplung.00078.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuya Y, Furuya AK, Roberts S, Sanfilippo AM, Salmon SL, Metzger DW. Prevention of Influenza Virus-Induced Immunopathology by TGF-beta Produced during Allergic Asthma. PLoS pathogens. 2015;11:e1005180. doi: 10.1371/journal.ppat.1005180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston CJ, Smyth DJ, Dresser DW, Maizels RM. TGF-beta in tolerance, development and regulation of immunity. Cellular immunology. 2016;299:14–22. doi: 10.1016/j.cellimm.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azhar M, Yin M, Bommireddy R, Duffy JJ, Yang J, Pawlowski SA, Boivin GP, Engle SJ, Sanford LP, Grisham C, Singh RR, et al. Generation of mice with a conditional allele for transforming growth factor beta 1 gene. Genesis. 2009;47:423–431. doi: 10.1002/dvg.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perl AK, Zhang L, Whitsett JA. Conditional expression of genes in the respiratory epithelium in transgenic mice: cautionary notes and toward building a better mouse trap. American journal of respiratory cell and molecular biology. 2009;40:1–3. doi: 10.1165/rcmb.2008-0011ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matrosovich M, Matrosovich T, Garten W, Klenk HD. New low-viscosity overlay medium for viral plaque assays. Virology journal. 2006;3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesseur I, Zou K, Berber E, Zhang H, Wyss-Coray T. Highly sensitive and specific bioassay for measuring bioactive TGF-beta. BMC cell biology. 2006;7:15. doi: 10.1186/1471-2121-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.