Abstract
The non-reducing disaccharide trehalose is widely distributed among various organisms. It plays a crucial role as an instant source of energy, being the major blood sugar in insects. In addition, it helps countering abiotic stresses. Trehalose synthesis in insects and other invertebrates is thought to occur via the trehalose-6-phosphate synthase (TPS) and trehalose-6-phosphate phosphatase (TPP) pathways. In many insects, the TPP gene has not been identified, whereas multiple TPS genes that encode proteins harboring TPS/OtsA and TPP/OtsB conserved domains have been found and cloned in the same species. The function of the TPS gene in insects and other invertebrates has not been reviewed in depth, and the available information is quite fragmented. The present review discusses the current understanding of the trehalose synthesis pathway, TPS genetic architecture, biochemistry, physiological function, and potential sensitivity to insecticides. We note the variability in the number of TPS genes in different invertebrate species, consider whether trehalose synthesis may rely only on the TPS gene, and discuss the results of in vitro TPS overexpression experiment. Tissue expression profile and developmental characteristics of the TPS gene indicate that it is important in energy production, growth and development, metamorphosis, stress recovery, chitin synthesis, insect flight, and other biological processes. We highlight the molecular and biochemical properties of insect TPS that make it a suitable target of potential pest control inhibitors. The application of trehalose synthesis inhibitors is a promising direction in insect pest control because vertebrates do not synthesize trehalose; therefore, TPS inhibitors would be relatively safe for humans and higher animals, making them ideal insecticidal agents without off-target effects.
Keywords: trehalose, trehalose-6-phosphate synthase, physiological function, chitin regulation, TPS inhibitor
Trehalose and its function in invertebrates
Trehalose is a non-reducing disaccharide in which two glycosyl moieties are linked together by an α,α-1,1 bond (Elbein et al., 2003; Bansal et al., 2013). It is found ubiquitously as a metabolite in various bacteria, fungi, slime molds, protozoa, plants, and invertebrates (Kern et al., 2012; Tang et al., 2012a,b, 2014a,b, 2016; Lyu et al., 2013). Trehalose functions not only as a reserve carbohydrate, but also as an important stress-protecting molecule in different organisms (Elbein et al., 2003; Pampurova et al., 2014). Trehalose has been shown to serve as a mobile energy source for flight, and its levels in the blood have been reported to control the expenditure of flight energy in insects (Clegg and Evans, 1961; Cui and Xia, 2009). High levels of trehalose are also present in the hemolymph of insects at nonflying stages and in the blood of invertebrates that use lipids for flight energy (Wyatt, 1961). The levels of blood trehalose vary greatly in the developmental history of different species, and in all probability, trehalose has been adapted for diverse functions within the class Insecta (Murphy and Wyatt, 1965). However, trehalose synthesis pathway has not been found in higher animals (mammals) or vertebrates, even though trehalase (TRE) has been reported in the small intestine, digestive system, and other organs of various species, especially in insects and other invertebrates (Richards et al., 2002; Chen and Haddad, 2004).
In the animal kingdom, trehalose was first identified as an important constituent of insect hemolymph in silkworm pupae (Wyatt and Kalf, 1956). Trehalose was then found in concentrations of up to 2% in the hemolymph of the desert locust Schistocerca gregaria (Howden and Kilby, 1956). This sugar is an important soluble carbohydrate and energy reserve in insects (Kandy and Kilby, 1959). It is secrected into the hemolymph of insects at all developmental stages (Matsuda et al., 2015). Trehalose functions as a source of glucose for energy in adult insects during flight and energy-requiring activities; it also serves as an energy source to meet the demands of insect flight muscles and other tissues and is continuously synthesized in the fat body (Evans and Dethier, 1957; Wyatt and Kalf, 1957; Bücher and Klingenberg, 1958; Candy and Kilby, 1961; Becker et al., 1996; Elbein et al., 2003; Chen and Haddad, 2004; Kern et al., 2012; Gao et al., 2014; Shukla et al., 2015). Trehalose serves not only as a reserve carbohydrate but also as an efficient protection factor, playing important roles in the protection of organisms against adverse environmental conditions (Iordachescu and Imai, 2008; Tang et al., 2008; Shukla et al., 2015; Liu et al., 2016). Trehalose is also essential for stress response in various microorganisms, and its inhibition may be a promising antimicrobial strategy as TPS genes are entirely absent in humans (Magalhães et al., 2017). Survival strategies for overwintering insects are determined by biochemical components of their body fluids. Freeze-tolerant and freeze-avoiding insects often accumulate a high level of trehalose that acts as a supercooling agent and cryoprotectant (Storey and Storey, 2012; Wen et al., 2016). During menadione stress, trehalose has been found to be necessary for yeast intracellular functions (Herdeiro et al., 2006), whereas the presence of trehalose on both sides of the lipid bilayer minimized oxidative damage to proteins and lipids (da Costa Morato Nery et al., 2008).
In nematodes, trehalose is usually present at a concentration higher than that of free glucose (Fairbairn, 1958; Dmitryjuk et al., 2009), and has many important functions: it protects cellular structures during stresses such as high osmotic pressure, drying, or freezing; it provides energy as the major circulating sugar; and it is important for egg hatching (Perry, 1989; Behm, 1997; Dmitryjuk and Zółtowska, 2003; Elbein et al., 2003). Nearly all insects maintain high level of trehalose in their hemolymph (Wyatt, 1967; Kramer et al., 1978; Becker et al., 1996; Mariano et al., 2009). Trehalose protects organisms against different environmental stresses, including heat, oxidation, cold, anoxia, or desiccation, because of its unique chemical properties (Crowe et al., 1998; Elbein et al., 2003; Matsuda et al., 2015). In Drosophila larvae desiccated for 10 h at <5% relative humidity, the desiccation-responsive trehalose metabolic pathway was activated in concert with the enzymes TPS and TRE (Shukla et al., 2015). These data indicate that trehalose is a potential marker for anhydrobiosis in Drosophila (Thorat et al., 2012).
As in mammals, insulin-like peptides (Dilps) and a glucagon-like peptide regulate circulating sugar levels in Drosophila (Yasugi et al., 2017). Feeding on dietary sugar immediately changes the levels of the circulating sugar (Ugrankar et al., 2015). Genetic manipulation of the function of Dilps and adipokinetic hormone (Akh) changes trehalose and glucose levels in the circulating hemolymph, which means that mobilization of blood trehalose to glucose is critical for metabolic homeostasis (Rulifson et al., 2002; Gáliková et al., 2015). Flight, feeding, and parasitic infections in insects produce hypertrehalosemia in the hemolymph (Becker et al., 1996; Zółtowska and Lopieniska-Biernat, 2006). These findings further support the notion that trehalose plays a role in the response to several biological functions as a physiological adaptations and as an energy source in insects (Chung, 2008).
In recent years, several approaches have been applied to study the trehalose synthesis genes TPS and TPP. Their special functions in molecular mechanisms underlying different stresses and even in the regulation of chitin synthesis have been described in insects and other invertebrates (Chen et al., 2002, 2003; Tang et al., 2010; Chen and Zhang, 2015; Shi et al., 2016; Xiong et al., 2016; Yang et al., 2017), taking advantage of their ability to adapt to stress conditions (Chen and Haddad, 2004; Qin et al., 2012; Tang et al., 2014b; Guo et al., 2015). On the one hand, an increasing number of TPS genes are being identified and cloned from different insects and other invertebrate species. On the other hand, many insects seem to possess only TPS genes but no TPP gene according to genome sequencing results. In this regard, several following questions arise. How does trehalose synthesis proceed in invertebrates and is there another pathway in addition to the TPS/TPP pathway? Is the presence of just the TPS gene sufficient for trehalose synthesis because the encoded protein has both TPS and TPP domains? Do trehalose synthesis pathways vary between different insects? In this review, we summarize and discuss the current knowledge of the invertebrate trehalose synthesis pathway; the cloning and expression of the underlying genes identified so far; their role in development, stress conditions, and chitin metabolism regulation. We also point out the knowledge gaps that need to be filled, especially regarding future pest control by using inhibitors of trehalose synthesis, considering the absence of TPS in vertebrates.
Genetic architecture
TPS and TPP gene identification, cloning, and evolution
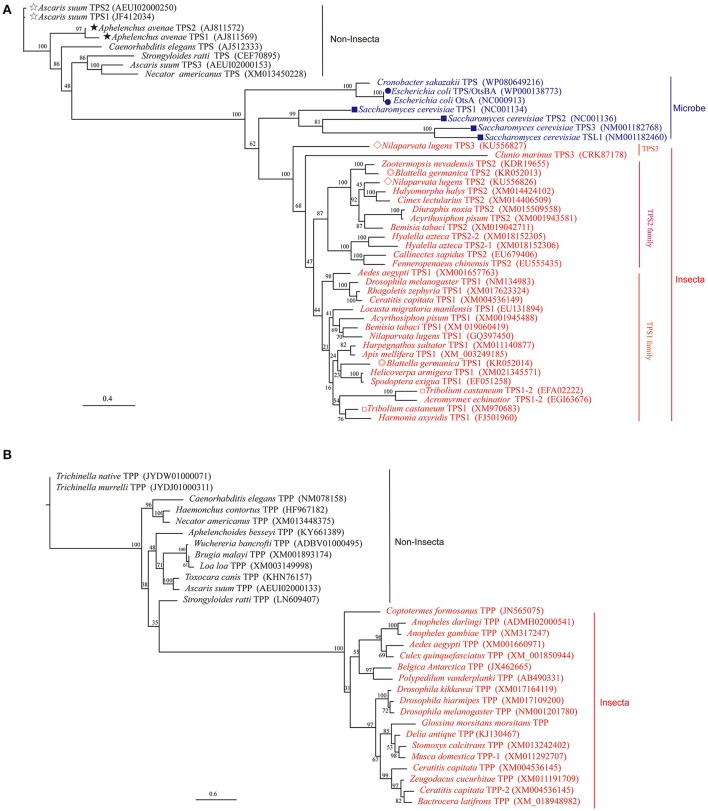
The first insect TPS gene was cloned in Drosophila (Chen et al., 2002; Chen and Haddad, 2004), and the induction of TPS1 gene expression was shown to increase tolerance to anoxia (Chen et al., 2002). Subsequently, insect TPS genes were cloned from Helicoverpa armigera (Xu et al., 2009), Locusta migratoria manilensis (Cui and Xia, 2009), Spodoptera exigua (Tang et al., 2010), Nilaparvata lugens (Chen et al., 2010b), Catantops pinguis (Tang et al., 2011), Ctenocephalides felis (Kern et al., 2012), Harmonia axyridis (Qin et al., 2012), Blattella germanica (Chen and Zhang, 2015), Delia antiqua (Guo et al., 2015), Leptinotarsa decemlineata (Shi et al., 2016), Bactrocera minax (Xiong et al., 2016), and other organisms. Moreover, two TPS genes have been found in B. germanica, Tribolium castaneum, and Aphelenchoides besseyi, and three TPS genes have been found in Ascaris suum and N. lugens (Figure 1A and Table 1, Shen, 2017). In addition, TPS from the Chinese shrimp Fenneropenaeus chinensis has been cloned and reported (Zhang J. et al., 2012), and one full-length cDNA sequence of four structural isoforms of TPS was isolated from the chela muscles of an adult female (Shi and Chung, 2014). Furthermore, three TPS genes have been isolated and sequenced from the muscles of the parasite A. suum (Dmitryjuk et al., 2013; Dmitryjuk and Łopienska-Biernat, 2016).
Figure 1.
Phylogenetic analysis of insect TPS and TPP based on their amino acid sequences. Full-length amino acid sequences were aligned using Mega 6.0 and to the ML phylogenetic tree (TPS for A; TPP for B) was performed using PhyML with the model of WAG (Guindon et al., 2005). A bootstrap analysis was carried out and the robustness of each cluster was verified in 1,000 replications. Some species have more than two TPS proteins have marked by different form front of species name (A). The monophyly of insect TPP and TPS is well supported, but the monophyly of non-insect and microbe is not supported.
Table 1.
Characteristics of reported invertebrate TPS gene and their encoding protein.
| Organism | TPS Family | GeneBank No. | No. amino acid | References |
|---|---|---|---|---|
| N. lugens | TPS1 | GQ397450 | 807 | Chen et al., 2010a |
| TPS2 | KU556826 | 820 | Yang et al., 2017 | |
| TPS3 | KU556827 | 783 | Shen, 2017 | |
| Sogatella furcifera | TPS1 | JQ743627 | 807 | Zhang D. W. et al., 2012 |
| Diabolocatantops pinguis | TPS1 | GQ389790 | 809 | Tang et al., 2011 |
| Locusta migratoria manilensis | TPS1 | EU131894 | 813 | Cui and Xia, 2009 |
| Antheraea pernyi | TPS1 | KU977454 | 828 | Huang et al., 2016 |
| Ctenocephalides felis | TPS1 | JX025053 | 824 | Kern et al., 2012 |
| Delia antique | TPS1 | JX681124 | 815 | Li et al., 2013 |
| Blattella germanica | TPS1 | KR052013 | 833 | Chen and Zhang, 2015 |
| TPS2 | KR052014 | 822 | Chen and Zhang, 2015 | |
| Leptinotarsa decemlineata | TPS1 | KU756283 | 821 | Shi et al., 2016 |
| Harmonia axyridis | TPS1 | FJ50196 | 805 | Qin et al., 2012 |
| Helicoverpa armigera | TPS1 | XM_021345571 | 826 | Xu et al., 2009 |
| Spodoptera exigua | TPS1 | EF051258 | 826 | Tang et al., 2010 |
| Drosophila melanogaster | TPS1 | NM_134983 | 809 | Chen et al., 2002, 2003; Chen and Haddad, 2004; Matsuda et al., 2015; Thorat et al., 2016 |
| Delia antiqua | TPS1 | JX681124 | 815 | Guo et al., 2015 |
| Ascaris suum | TPS1 | JF412034 | 1,298 | Dmitryjuk et al., 2014 |
| TPS2 | AEUI02000250 | 1,254 | Dmitryjuk et al., 2014 | |
| TPS3 | AEUI02000153 | 1,269 | Dmitryjuk et al., 2014 | |
| Callinectes sapidus | TPS2 | EU679406 | 755 | Chung, 2008 |
| Fenneropenaeus chinensis | TPS2 | EU555435 | 844 | Zhang J. et al., 2012 |
| Aphelenchoides besseyi | TPS1 | KY661388 | 1,250 | Chen Q. et al., 2017 |
| TPS2 | KY661389 | 507 | Chen Q. et al., 2017 | |
| Caenorhabditis elegans | TPS1 | AJ512333 | 1,230 | Pellerone et al., 2003 |
| Bactrocera minax | TPS1 | KU379749 | 814 | Xiong et al., 2016 |
In 2005, gob-1, the first TPP in Caenorhabditis elegans, was identified. Loss-of-function mutations in gob-1 resulted in early larval lethality, which was completely suppressed by the ablation of C. elegans tps-1 and tps-2 genes (Kormish and McGhee, 2005). Furthermore, a TPP gene was identified in Brugia malayi in 2011 (Kushwaha et al., 2011), and its silencing was found to be lethal for the third instar larvae as its in vivo development became impaired (Kushwaha et al., 2012). No more TPP genes were reported in insects between 2011 and 2015. The identification and cloning of a single TPP gene was reported in a diapausing insect, D. antiqua (Guo et al., 2015). Furthermore, single TPP genes from insects have been reported in GenBank (e.g., Coptotermes formosanus [JN565075], Drosophila melanogaster [NM_135269], and Plutella xylostella [XM_011559193]) (Yang et al., 2017). These genes are shorter than TPS and encode proteins containing only the TPP domain, very similar to the TPS protein of the same species, with only some protein sequence differences at the N-terminus (Figure 1B and Table 2).
Table 2.
Some invertebrate TPP protein sequences from NCBI and published article.
| Organism | GeneBank No. | No. amino acid | References |
|---|---|---|---|
| Toxocara canis | KHN76157 | 485 | |
| Aphelenchoides besseyi | KY661389 | 507 | Chen Q. et al., 2017 |
| Brugia malayi | XM_001893174 | 492 | |
| Wuchereria bancrofti | ADBV01000495 | 467 | |
| Ascaris suum | AEUI02000133 | 532 | Dmitryjuk et al., 2014 |
| Haemonchus contortus | HF967182 | 432 | |
| Strongyloides ratti | LN609407 | 451 | |
| Necator americanus | XM_013448375 | 466 | |
| Delia antiqua | KJ130467 | 273 | Guo et al., 2015 |
| Caenorhabditis elegans | NM078158 | 468 | Kormish and McGhee, 2005 |
| Loa loa | XM_003149998 | 254 | |
| Trichinella nativa | JYDW01000071 | 455 | |
| Trichinella murrelli | JYDJ01000311 | 455 | |
| Stomoxys calcitrans | XM_013242402 | 343 | |
| Musca domestica | XM_011292707 | 331 | |
| Ceratitis capitata | XM_004536145 | 323 | |
| Zeugodacus cucurbitae | XM_011191709 | 333 | |
| Drosophila kikkawai | XM_017164119 | 273 | |
| Drosophila biarmipes | XM_017109200 | 296 | |
| Drosophila melanogaster | NM_001201780 | 296 | |
| Bactrocera latifrons | XM_018948982 | 323 | |
| Ceratitis capitata | XM_004536143 | 273 | |
| Belgica antarctica | JX462665 | 303 | |
| Polypedilum vanderplanki | AB490331 | 294 | |
| Anopheles darlingi | ADMH02000541 | 297 | |
| Culex quinquefasciatus | XM_001850944 | 309 | |
| Anopheles gambiae | XM_317247 | 261 | |
| Aedes aegypti | XM_001660971 | 281 | |
| Coptotermes formosanus | JN565075 | 299 |
Although some invertebrates have more than one TPS and TPP gene, two TPS genes from a single insect were first cloned and reported in B. germanica (BgTPS1: KR050213 and BgTPS2: KR050214) (Chen and Zhang, 2015), followed the discovery of two separate TPS genes in N. lugens (TPS1: GQ397450, TPS2: KU556826; Yang et al., 2017). The third TPS (KU556827) was cloned from N. lugens in 2017 (Shen, 2017). All these TPS genes have been found to encode proteins with two conserved TPS and TPP domains with high similarity in their amino acid sequences (Figure 1A, Table 1; Yang et al., 2017). Meanwhile, the use of Illumina RNA-seq technology showed that the beetle Microdera punctipennis may have five potential TPS UniGenes (Lu et al., 2014). However, the exact number of TPS genes in this insect remains unknown. In the evolution of the TPS gene, bacteria and yeasts are likely to be closer to the relatives of insects than to nematodes and other non-insects (Figure 1A). Because a) no TPS gene has been found in higher animals, like mammals and b) most insects only have one TPS gene, which can synthesis trehalose only and has TPS and TPP enzymatic activities (Yoshida et al., 2016). So we hypothesized that the evolution of insect TPS evolved from multiple homologs to a single one. Insect or other invertebrate trehalose synthesis from TPS/TPP pathway maybe evolved to one TPS pathway because of TPS replaced the function of the TPP gene in some species (Figures 1, 2). And this could be the reason for so many insects lacking TPP and most of the known TPP sequences being closely related to the TPS gene sequence. Also it is reported that Drosophila have two TPPs (CG5171 and CG5177), but only CG5171 can dephosphoryte T6P under experimental conditions (Yoshida et al., 2016). Of course, more work needs to be done to clearly distinguish the functions of different TPS genes in the same species and to elucidate whether all of TPS can synthesize trehalose independently when the species possesses only one TPS gene.
Figure 2.
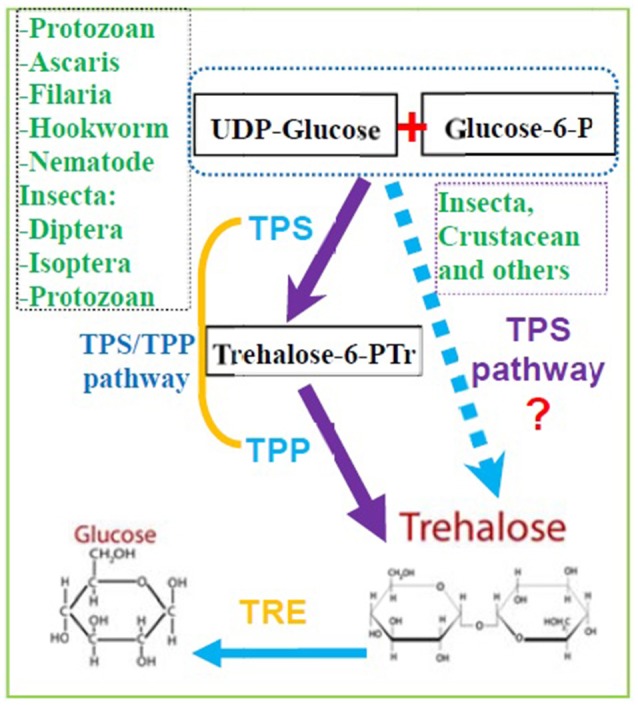
The potential pathway of trehalose synthesis in insects and other invertebrate animals. Trehalose are mainly synthesized by the pathway of TPS and TPP in many kind of invertebrates, but it also be synthesis by TPS only in some species from Insecta and Crustacean. (TPS, Trehalose-6-phosphate synthase; TPP, Trehalose-6-phophate phosphatase; TRE, Trehalase).
TPS gene structure
One study reported that Drosophila has only one TPS gene, and this gene has domains that are conserved when compared with the yeast genes TPS (OtsA in E. coli) and TPP (OtsB in E. coli) (Chen et al., 2002). Later, TPS genes from H. armigera (EU878265) and S. exigua (EF051258) and many insects have been found and cloned. Insect TPS gene encodes an 820–850-aa protein with two conserved domains—TPS and TPP—corresponding to OtsA and OtsB genes in yeast (Xu et al., 2009; Tang et al., 2010, 2014a; Yang et al., 2017). The TPS genes of the blue crab C. sapidus were cloned in 2014 and found to be very similar to those of insects. TPS genes of four different lengths were isolated: TPS-mus-1 (EU910087), TPS-mus-1a (EU910088), TPS-mus-1b (EU910089), and TPS-mus-1c (EU910090) (Shi and Chung, 2014; Yang et al., 2017). TPS-mus-1b and TPS-mus1 contain conserved TPS and TPP structures, whereas TPS-mus-1b and TPS-mus-1c harbor only a TPS conserved domain (Shi and Chung, 2014; Yang et al., 2017).
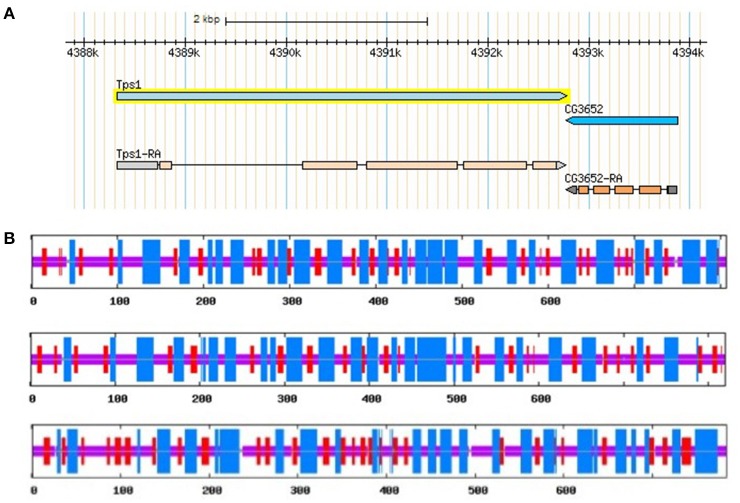
The length of TPS genes is variable among different species. It has been shown that D. melanogaster TPS (DmTPS or Dmtps1) has 5 exones (Figure 3A). However, Anopheles gambiae TPS (AgTPS), Aedes aegypti TPS (AaTPS), Nasonia vitripennis TPS (NvTPS), Apis mellifera TPS (AmTPS) and S. exigua TPS (SeTPS) have 5, 5, 3, 10, 8, and 12 exons, respectively. Comparison between SeTPS and NvTPS showed that they have seven common exon–intron boundaries (Tang et al., 2010). The genomic structure of F. chinensis TPS (FcTPS) comprises three exons and two introns (Zhang J. et al., 2012). Thus, TPS gene structure has been examined in several insect species, in which genomic sequencing has been completed. Therefore, to determine whether TPS alone can synthesize trehalose, more experiments need to be performed, and structures of gene sequences homologous to TPS and TPP have to be analyzed in insect genomes.
Figure 3.
D. melanogaster TPS genomic structure (A) from FlyBase and predicted secondary structures of three TPS proteins of the brown planthopper, including TPS1, TPS2, and TPS3 (B). Blue and red regions represent α-helixes and β-sheets in Figure 3B, respectively.
Biochemistry
TPS and the trehalose synthesis pathway
Trehalose biosynthetic pathway was first identified in Saccharomyces cerevisiae (Cabib and Leloir, 1958; Kern et al., 2012). Initially, insects were thought to synthesize trehalose through the TPS/TPP pathway (Candy and Kilby, 1961), and it was suggested that insects might have the same pathway as that of yeast (Candy and Kilby, 1959, 1961). Now, many studies have reported that there are at least five trehalose biosynthetic pathways in different species (Magalhães et al., 2017). In most invertebrates, including nematodes, TPS/TPP is the predominant trehalose biosynthetic pathway, which involves the following steps (Magalhães et al., 2017): TPS catalyzes the transfer of glucose from UDP-glucose to glucose-6-phosphatase, forming trehalose-6-phosphate (T6P), and TPP dephosphorylates T6P to trehalose (Cabib and Leloir, 1958; Behm, 1997; Elbein et al., 2003; Avonce et al., 2006; Tang et al., 2010; Guo et al., 2015).
The N-terminal TPS (Tre-6-P synthase) domain catalyzes the production of Tre-6-P using glucose 6-phosphate and UDP-glucose as substrates, whereas the C-terminal TPP (Tre-6-P phosphatase) domain then dephosphorylates Tre-6-P, generating trehalose (Matsuda et al., 2015; Yasugi et al., 2017). Expression of Drosophila TPS gene in mammalian HEK-293 cells enables them to synthesize trehalose (Matsuda et al., 2015). In H. armigera, the level of TPS expression corresponds to TPS enzymatic activity as a result of increased trehalose production (Xu et al., 2009). The catalytic activity of H. armigera TPS increased approximately fivefold when it was overexpressed in Bombyx mori hemolymph infected by using a recombinant baculovirus expression system (Xu et al., 2009). Therefore, we believe that some insects can synthesize trehalose by the TPS/TPP pathway, whereas other insects can synthesize trehalose by TPS alone, just as we described that the evolution of insect TPS evolved from multiple homologs into a single one. In addition, the trehalose synthesis enzyme TPS is solely responsible for the de novo syhthesis of trehalose in Drosophia based on the genetic and biochemical evidence (Yoshida et al., 2016). Therefore, a revised trehalose synthesis pathway in insects and other invertebrates is illustrated in Figure 2.
Tissue expression of TPS genes
Because insect fat bodies appear to be important sites for the production of α,α-trehalose, studies of trehalose synthesis have necessarily centered around this organ (Gans et al., 1968), which is analogous to the mammalian liver (Candy and Kilby, 1959; Murphy and Wyatt, 1965; Friedman, 1968). Insects express TPS in the fat body (Cui and Xia, 2009; Xu et al., 2009; Chen et al., 2010b; Tang et al., 2010; Xiong et al., 2016), whereas C. sapidus displays ubiquitous expression of TPS in most tissues examined (Chung, 2008). TPS is expressed in all tissues of adult crabs of both sexes, indicating that these tissues can produce trehalose (Chung, 2008). Furthermore, in L. migratoria manilensis TPS transcripts are expressed in the fat body, midgut, hemolymph, and leg muscle (Cui and Xia, 2009). It has been reported that Drosophila TPP of CG5171 was mainly expressed in the Malpighian tubules and the components of the carcass (Yoshida et al., 2016), so it can't play a role in insect trehalose synthesis because it only works in the fat body.
F. chinensis TPS gene was found to be expressed in various tissues, including the muscles, hemocytes, ovaries, gills, nerves, lymphoid organs, intestine, stomach, heart, and epidermis, with the strongest level observed in the hepatopancreas (Zhang J. et al., 2012). Previous reports showed that L. decemlineata TPS was highly expressed in the fat body, and it was also transcribed in the foregut, hindgut, trachea, ovaries, and testes, indicating that trehalose might be synthesized in these tissues (Shi et al., 2016). Several reports demonstrated expression of the TPS gene in the tissues of foregut and trachea, likely because these two tissues may have been doped with fat body during the extraction process. RT-PCR or northern blot analysis in C. elegans showed mRNA expression of two TPS genes at all stages of C. elegans life cycle (Pellerone et al., 2003; Grewal et al., 2006). B. minax TPS expression was detectable in all developmental stages, with a higher expression level in the final (third) instar larvae (Xiong et al., 2016). In vitro treatment with a lethal dose of ivermectin decreased TPS and TPP activities in the muscle of adult A. suum females compared with those in the control groups (Dmitryjuk et al., 2014).
Insect development and TPS gene expression
In D. melanogaster, P element mutagenesis experiments showed that TPS gene disruption is lethal at early larval stages (Chen et al., 2002, 2003; Chen and Haddad, 2004). TPS mutant Drosophila larvae exhibited diet-dependent growth and survival phenotypes when they lacked hemolymph trehalose (Matsuda et al., 2015). Those findings confirmed the assumed crucial functions of TPS synthesis in insects (Becker et al., 1996). In D. antiqua, differential expression of TPS and TPP shared similar trends among summer- and winter-diapausing pupae populations, and their enzyme activities were consistent with the expression levels of corresponding genes (Guo et al., 2015). In C. sapidus, trehalose concentrations showed a bimodal pattern, and it exhibited two peaks at early ecdysis and post ecdysis, indicating that C. sapidus consumes energy from trehalose during the molting process (Chung, 2008). The changes in trehalose content and TPS activity in H. armigera hemolymph showed a similar trend during larval-pupal development of diapause and non-diapause programming (Xu et al., 2009).
Physiological function
Diversity of TPS genes and their functions
TPS plays a key role in the perception of carbohydrate availability and carbohydrate metabolism (Jin et al., 2016) in insects, other invertebrates, as well as in plants (Gao et al., 2014). TPS is considered a cytoplasmic protein with two functionally distinct catalytic domains (Elbein et al., 2003; Matsuda et al., 2015). Drosophila TPS gene was cloned and studied at early 2000 (Chen et al., 2002, 2003). Overexpression of D. melanogaster TPS in mammalian cells (HEK-293) made them capable of trehalose synthesis (Chen et al., 2003). In N. lugens, three TPS genes were cloned, and their protein secondary structures showed similar structures and composition of α-helix, β-sheet, and random coil (Table 3, Figure 3B, Shen, 2017). Thus, different TPS genes can synthesize trehalose, but it is unclear if different TPS genes vary in their function and genomic structure within the same species. Currently, at least three other insect species (N. lugens, B. germanica, and T. castaneum) have more than two TPS genes (Chen and Zhang, 2015; Yang et al., 2017).
Table 3.
The prediction of the secondary structure of TPS in brown planthopper.
| Item | α-helix(%) | β-sheet(%) | Radom coil(%) |
|---|---|---|---|
| TPS1 | 40.27 | 12.76 | 45.35 |
| TPS2 | 36.59 | 13.29 | 49.39 |
| TPS3 | 35.50 | 18.01 | 46.49 |
In addition, TPS1 and TPS2 enzymes have been identified in C. elegans (Pellerone et al., 2003), as well as in the anhydrobiotic nematode Aphelenchus avenae (Goyal et al., 2005; Kormish and McGhee, 2005). Simultaneous RNA interference (RNAi) targeting of both TPS1 and TPS2 in wild-type C. elegans lowered trehalose levels to 7% of control levels (Pellerone et al., 2003; Kormish and McGhee, 2005). Nonetheless, on the background of age-1 mutant, RNAi of TPS1 and TPS2 greatly decreased C. elegans resistance to osmotic shock (Kormish and McGhee, 2005). Meanwhile, there are several instances when multiple TPS genes have been found in the same insect species, and these TPS genes could have different functions in trehalose synthesis (Chen and Zhang, 2015; Yang et al., 2017). Further research on the distinct roles of different TPS genes is warranted.
Role of TPS in regulating sugar metabolism
It has been reported that larvae lacking trehalose exhibit diet-dependent phenotypes relating to growth and survival in the genetically tractable organism of Drosophila (Matsuda et al., 2015). Moreover, a lack of TPS can cause an accumulation of trehalose that is lethal during the pupal period, as well as results in a significant reduction in circulating glucose and the larvae exhibit a high lethality after desiccation stress (Yoshida et al., 2016). Temporary and simultaneous knockdown of both TPS genes in C. elegans by RNAi resulted in a 90% decline in trehalose levels but no obvious phenotype was observed (Pellerone et al., 2003; Cui and Xia, 2009). In the crustacean Artemia franciscana, a fraction of trehalose is quickly mobilized as an energy source, whereas the remainder serves as a substrate for glycogen and glycerol synthesis when dormancy is broken (Collins and Clegg, 2004; Argüelles, 2014). Members of the phylum Apicomplexa, a group of protists evolutionarily close to dinoflagellates and ciliates, synthesize trehalose through the biosynthetic pathway similar to that in plants and fungi (Yu et al., 2010; Argüelles, 2014). In nematodes, e.g., in Anisakis simplex, glycogen and trehalose metabolism plays a key role in supporting life processes (Łopienska-Biernat et al., 2015). Because TPP has a high affinity for trehalose-6-phosphate and the later hydrolyzes quickly to trehalose, TPS activity is an important limiting factor in trehalose synthesis (Behm, 1997). Two TPS genes with very high resemblance to the tps2 gene of C. elegans were also identified in A. avenae, but the expression of a gene similar to C. elegans tps1 has not yet been confirmed (Łopienska-Biernat et al., 2015).
Drosophila larvae were shown to be unable to synthesize trehalose when dTPS1 transcript levels were decreased by the ubiquitous daGAL4-driven expression of the dTPS1-RNAi transgene (Thorat et al., 2016). This result highlighted the significance of trehalose in the regulation of desiccation-responsive redox homeostasis (Thorat et al., 2016). The result on the function of TPS further demonstrated that the regulation of trehalose metabolism is essential for normal development, body water homeostasis, and desiccation tolerance in Drosophila (Yoshida et al., 2016). Dietary trehalose has also been shown to be directly transported to the hemolymph from the larval gut in insects (Shi et al., 2016), because feeding of trehalose dramatically increased the in vivo trehalose pools in D. melanogaster larvae treated with DmTPS RNAi (Thorat et al., 2016). In addition, the knockdown of LdTPS delayed larval development, strongly reduced hemolymph monosaccharides in the fat body, and potentiated sugar absorption in the larval gut of L. decemlineata (Shi et al., 2016). Trehalose can be maintained at a high level while glucose is broken down and used shortly after food intake (Ugrankar et al., 2015; Yasugi et al., 2017). In this condition, the production of trehalose from diet appears to be critical for buffering the fluctuation of sugar levels in the body and for producing trehalose in fat body on a long-term basis (Yasugi et al., 2017). Trehalose is the main hemolymph sugar, and its metabolism plays a pivotal role in systemic energy homeostasis based on the requirement for dietary sugar when both TRE and TPS1 are mutated (Yasugi et al., 2017).
TPS functions during stress conditions
During 18-h starvation, the maximum distance by which Harmonia axyridis moved initially increased and then decreased with time and falling levels of trehalose and glycogen as well as with the reduction in TPS expression. This indicates that insects need to consume trehalose to search for food (Tang et al., 2014b; Shi et al., 2017). The Arctic collembolan Onychiurus arcticus can survive winter temperatures of −25°C by increasing trehalose concentrations, decreasing glycogen reserves, and reducing TRE activity as temperature decreases. Meanwhile, TPP activity peaks at 0°C (Montiel et al., 1998). TPS induction in Schizosaccharomyces pombe transformed with TPS gene increased intracellular trehalose levels and the increase correlated with increased tolerance to heat shock and other stresses (Soto et al., 1999). Furthermore, human primary fibroblasts transformed using a recombinant adenovirus vector to express the trehalose biosynthetic enzymes encoded by OtsA and OtsB genes from Escherichia coli., which produced increased amounts of trehalose with increasing multiplicities of infection (Guo et al., 2000). In addition, elevated trehalose levels in mammalian cells transfected with the Drosophila TPS gene were reported to protect the cells from hypoxic injury (Chen et al., 2002, 2003; Chen and Haddad, 2004).
In Polypedilum vanderplanki, one of the mechanisms of the tolerance to extreme conditions is that the larvae can rapidly accumulate trehalose to the levels up to 18% of dry body mass (Watanabe et al., 2002; Chen and Haddad, 2004). In 2009, Xu et al. reported TPS activity regulates the changes in trehalose content during H. armigera larval-pupal development, and that this is the reason of a significantly higher trehalose concentration in diapausing insects than in non-diapausing insects (Xu et al., 2009). Furthermore, it has been reported that trehalose concentrations were lower in summer- and winter-diapausing pupa at the initial phase, but then, they increased gradually and peaked during the maintenance phase (Guo et al., 2015). The concentration then declined in the quiescence phase, indicating that trehalose metabolism plays an important role through the expression of TPS, TPP, and TRE genes (Guo et al., 2015). In overwintering mountain pine beetle larvae, TPS levels are high in the autumn proteome, whereas in the spring proteome, they are significantly lower. This observation supports the hypothesis that trehalose is produced for survival during cold periods (Bonnett et al., 2012). TPS has also been found to possess anti-stress functions and play putative roles in physiological adaptation to environmental stress in Bactrocera dorsalis (Yang et al., 2014) and C. sapidus (Yednock and Neigel, 2014).
Anastrepha ludens larvae developed a protection mechanism based on the synthesis of trehalose by TPS to achieve greater survivability to stress caused by hydrostatic pressure (Vargas-Ortiz et al., 2013). Starvation and the injection of dsSeHTF—an Akh-like hypertrehalosemic factor—can significantly decrease TPS expression level (Park and Kim, 2017). Although no conspicuous phenotype changes were observed after TRE and TPS genes were silenced individually or simultaneously in the nematode A. besseyi, its survival under hypertonic osmotic pressure decreased significantly and the recovery was delayed. Thus, trehalose metabolism genes, including TPS and TRE, play an important role in osmobiosis regulation in a time/season-dependent fashion (Chen Q. et al., 2017).
Regulation of chitin metabolism by TPS
In silkworm larvae, trehalose has been reported to be a source of carbon for chitin synthesis during the new cuticle production and molting stages (Duchateau-Bosson et al., 1963). Trehalose is also considered a major substrate for chitin synthesis (Shi et al., 2016; Xiong et al., 2016). In insects, ecdysis, i.e., shedding of the cuticle at the end of a larval stadium, only occurs when ecdysteroid returns to a low level after its peak titer in the hemolymph (Steele, 2016). In Periplaneta americana, ecdysis is strongly correlated with the increase in trehalose and glucose concentrations in the hemolymph (Steele, 2016), suggesting a causal relationship between both events. 20-Hydroxyecdysone has been shown to induce the expression of BmTPS and three other genes in the chitin biosynthesis pathway, including TRE, glucose-6-phosphate isomerase (G6PI), and chitin synthase (CHS) (Xiong et al., 2016). TRE is the first gene in the chitin synthesis pathway (Tang et al., 2008, Zhang et al., 2011), and it regulates insect chitin synthesis and degradation (Tang et al., 2016; Zhao et al., 2016). Figure 4 illustrates how TPS in the chitin synthesis pathway regulates insect molting (Chen Q. W. et al., 2017; Yang et al., 2017).
Figure 4.
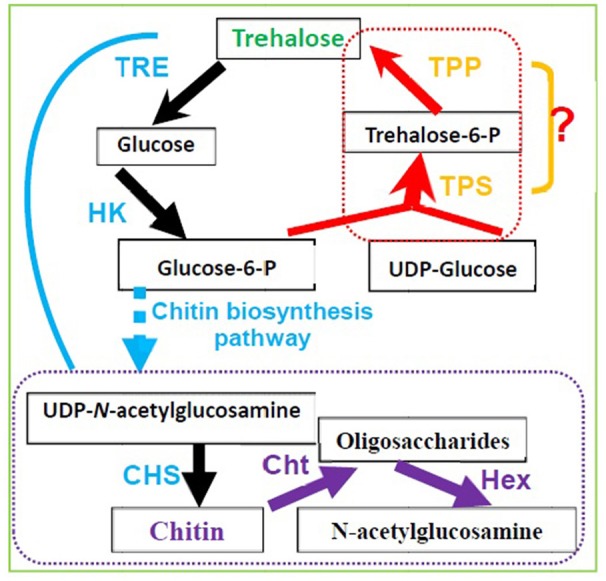
Trehalose metabolism and its relevance to chitin biosynthesis and degradation in insects and invertebrate animals. (TRE, Trehalase; HK, Hexokinase; CHS, Chitin synthase; Cht, Chitinase; Hex, β-N-acetylhexosamindase or β-N-acetyl-D-Hexosamindase; TPS, Trehalose-6-phosphate synthase; TPP, Trehalose-6-phophate phosphatase).
S. exigua complete the process of molting and die when TPS is knocked down by RNAi (Tang et al., 2010): the decrease in trehalose content causes larval and pupal lethality. In L. decemlineata, when LdTPS was knocked down by RNAi, surviving insects consumed a greater amount of foliage; accumulated more glycogen, lipid, and proline; and gained a larger body mass with a lower amount of chitin than did control insects (Shi et al., 2016). Moreover, TPS activity and trehalose content decreased significantly when dsRNA was injected into third -instar larvae, successfully silencing the transcription of BmTPS in B. minax and inhibiting the expression of three key genes in the chitin biosynthesis pathway. Furthermore, this treatment was associated with 52% mortality rate and the appearance of abnormal phenotypes (Xiong et al., 2016). In N. lugens, three phenotypes, namely molting deformity, molting and wing deformity, and wing deformity, occurred when the expression of TPS1 or TPS2 was decreased significantly by RNAi, along with 30% mortality (Chen et al., 2010b; Yang et al., 2017) and a significant decrease in trehalose content (Zhang et al., 2017). In addition, the expression of chitinase genes and chitin content decreased significantly, after that mainipulation, suggesting that the chitin metabolism balance is disrupted upon TPS gene knockdown (Chen Q. W. et al., 2017; Shen, 2017; Yang et al., 2017).
An increasing number of key enzymes and proteins of crop insects are being identified as candidates for RNAi-based gene silencing (Kola et al., 2015; Joga et al., 2016; Reisenman et al., 2016; Kolliopoulou et al., 2017). In a study by Shi et al. (2016), in TPS RNAi group, the chitin content in the body and epidermis decreased significantly, compared with that in the control group, from the third day to the eighth day of the life cycle. Moreover, a rescue bioassay revealed that trehalose feeding increased the survival of TPS RNAi hypomorphs and partially recovered chitin content.
Potential target for insecticides
The non-reducing disaccharide trehalose is absent in vertebrates, and, in particular, in mammals (Argüelles, 2014). This physiological difference might provide clues regarding the evolutionary branching of invertebrates and vertebrates (Argüelles, 2014). An increasing number of studies have shown significant mortality in insects when the trehalose balance is blocked (Chen et al., 2010a; Tang et al., 2010), further supporting the notion that TPS enzyme inhibition might be a viable insecticidal mechanism (Kern et al., 2012). However, until recently, no attempts to use inhibitors of insect TPS have been undertaken. In 2012, 4-substituted 2,6-diamino-3,5-dicyano-4H-thiopyrans were applied at potential inhibitory concentrations on insect TPS and highlighted as potential lead compounds for the development of insecticides (Kern et al., 2012). TPP is suggested to be a promising target for the development of antibacterial, antifungal, and antihelmintic therapeutics (Liu et al., 2017). The World Health Organization has included B. malayi TPP enzyme in the priority list of prospective antifilarial drug targets for lymphatic filariasis (Ho et al., 1992).
Studies on some potent inhibitors of insect TREs such as trehazolin (Ando et al., 1991, 1995), validoxyamine-A (Asano et al., 1990), and its derivative validamycin, have suggested that these compounds can act as insecticides by interfering with trehalose utilization in flight muscles, wing buds, cuticle, nervous system, and other body parts (Kono et al., 1999; Wegener et al., 2003, 2010; Tang et al., 2017). Application of TPS RNAi constructs via injection into S. exigua larvae (Tang et al., 2010) or via feeding into N. lugens larvae (Chen et al., 2010b) led to significant mortality in these insect species, further supporting the notion that TPS enzyme inhibition might be a viable insecticidal mechanism (Kern et al., 2012). Aryl d-glucopyranoside 6-sulfate prototypes are expected to find future applications for the development of tailored second-generation T6PP inhibitors (Liu et al., 2017). Interfering with trehalose biosynthesis could also be an insecticidal approach, making the trehalose biosynthesis enzyme TPS a potential drug target for pest control (Kern et al., 2012). A considerable body research over the recent years has demonstrated that TPS is indispensable for larval-pupal metamorphosis and that it is a suitable target to control insect and helminth pests by inhibiting the trehalose synthesis pathway (Tang et al., 2010; Xiong et al., 2016; Yang et al., 2017).
To conclude, TPS genes have been identified so far in hundreds of insect and other invertebrate species. We have reviewed the current understanding of the evolutionary and physiological significance of trehalose. In future studies, different trehalose synthesis pathways, distinct functions of multiple TPS genes, and sensitivity of TPS proteins to potential pest control inhibitors should be investigated in depth.
Author contributions
Conceived and manuscript structure design: SW, S-GW, S-YC, and BT. Current articles collection and trehalose metabolism genes' analysis: SW, H-JW, and BT. Sequence data analysis and figure drawing: BT and J-YZ. Wrote the paper: S-YC and BT.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFD0201000), the National Natural Science Foundation of China (Grant No. 31672081 and 31371996), Beijing Technology Program (Grant No. D171100001617003), Beijing Key Laboratory of Environment Friendly Management on Fruit Diseases and Pests in North China (BZ0432) and Youth Scientific Research Fund of Beijing Academy of Agricultural and Forestry Science (Grant No. QNJJ201725).
References
- Ando O., Nakajima M., Kifune M., Fang H., Tanzawa K. (1995). Trehazolin, a slow tight-binding inhibitor of silkworm trehalase. Biochim. Biophys. Acta 1244, 295–302. 10.1016/0304-4165(95)00029-B [DOI] [PubMed] [Google Scholar]
- Ando O., Satake H., Itoi K., Sato A., Nakajima M., Takahashi S., et al. (1991). Trehazolin, a new trehalase inhibitor. J. Antibiot. 44, 1165–1168. 10.7164/antibiotics.44.1165 [DOI] [PubMed] [Google Scholar]
- Argüelles J. C. (2014). Why can't vertebrates synthesize trehalose? J. Mol. Evol. 79, 111–116. 10.1007/s00239-014-9645-9 [DOI] [PubMed] [Google Scholar]
- Asano N., Takeuchi M., Kameda Y., Matsui K., Kono Y. (1990). Trehalase inhibitors, validoxylamine A and related compounds as insecticides. J. Antibiot. 4, 722–726. 10.7164/antibiotics.43.722 [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G. (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109. 10.1186/1471-2148-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R., Mian M. A., Mittapalli O., Michel A. P. (2013). Molecular characterization and expression analysis of soluble trehalase gene in Aphis glycines, a migratory pest of soybean. Bull. Entomol. Res. 103, 286–295. 10.1017/S0007485312000697 [DOI] [PubMed] [Google Scholar]
- Becker A., Schlöder P., Steele J. E., Wegener G. (1996). The regulation of trehalose metabolism in insects. Experientia 52, 433–439. 10.1007/BF01919312 [DOI] [PubMed] [Google Scholar]
- Behm C. A. (1997). The role of trehalose in the physiology of nematodes. Int. J. Parasitol. 27, 215–229. 10.1016/S0020-7519(96)00151-8 [DOI] [PubMed] [Google Scholar]
- Bonnett T. R., Robert J. A., Pitt C., Fraser J. D., Keeling C. I., Bohlmann J., et al. (2012). Global and comparative proteomic profiling of overwintering and developing mountain pine beetle, Dendroctonus ponderosae (Coleoptera: Curculionidae), larvae. Insect Biochem. Mol. Biol. 42, 890–901. 10.1016/j.ibmb.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Bücher Th., Klingenberg M. (1958). Wege des Wasserstoffs in der lebendigen Organisation. Angew. Chem. 70, 552–570. 10.1002/ange.19580701707 [DOI] [Google Scholar]
- Cabib E., Leloir L. F. (1958). The biosynthesis of trehalose phosphate. J. Biol. Chem. 231, 259–275. [PubMed] [Google Scholar]
- Candy D. J., Kilby B. A. (1959). Site and mode of trehalose biosynthesis in the locust. Nature 183, 1594–1595. 10.1038/1831594a0 [DOI] [PubMed] [Google Scholar]
- Candy D. J., Kilby B. A. (1961). The biosynthesis of trehalose in the locust fat body. Biochem. J. 78, 531–536. 10.1042/bj0780531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Tang B., Chen H. X., Yao Q., Huang X. F., Chen J., et al. (2010a). Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference. PLoS ONE 5:e10133. 10.1371/journal.pone.0010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang D. W. (2015). Molecular cloning, tissue distribution and temperature-induced expression of two trehalose-6-phosphate synthase genes in Blattella germanica (Blattodea: Blattellidae). Acta Entomol. Sin. 58, 1046–1053. [Google Scholar]
- Chen J., Zhang D. W., Yao Q., Zhang J. Q., Dong X. L., Tian H. G., et al. (2010b). Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 19, 777–786. 10.1111/j.1365-2583.2010.01038.x [DOI] [PubMed] [Google Scholar]
- Chen Q., Behar K. L., Xu T., Fan C., Haddad G. G. (2003). Expression of Drosophila trehalose-phosphate synthase in HEK-293 cells increases hypoxia tolerance. J. Biol. Chem. 278, 49113–49118. 10.1074/jbc.M308652200 [DOI] [PubMed] [Google Scholar]
- Chen Q., Haddad G. G. (2004). Role of trehalose phosphate synthase and trehalose during hypoxia: from flies to mammals. J. Exp. Biol. 207, 3125–3129. 10.1242/jeb.01133 [DOI] [PubMed] [Google Scholar]
- Chen Q., Li D., Wang F., Zhang R., Ling Y. (2017). Trehalose metabolism genes of Aphelenchoides besseyi (Nematoda: Aphelenchoididae) in hypertonic osmotic pressure survival. Biol. Open 6, 664–672. 10.1242/bio.023267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ma E., Behar K. L., Xu T., Haddad G. G. (2002). Role of trehalose phosphate synthase in anoxia tolerance and development in Drosophila melanogaster. J. Biol. Chem. 277, 3274–3279. 10.1074/jbc.M109479200 [DOI] [PubMed] [Google Scholar]
- Chen Q. W., Jin S., Zhang L., Shen Q. D., Wei P., Wei C. M., et al. (2017). Regulatory functions of trehalose-6-phosphate synthase in the chitin biosynthesis pathway in Tribolium castaneum (Coleoptera: Tenebrionidae) revealed by RNA interference. B. Entomol. Res. 18, 1–12. 10.1017/S000748531700089X [DOI] [PubMed] [Google Scholar]
- Chung J. S. (2008). A trehalose-6-phosphate synthase gene of the hemocytes of the blue crab, Callinectes sapidus: cloning, the expression, its enzyme activity and relationship to hemolymph trehalose levels. Salin. Syst. 4:18. 10.1186/1746-1448-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg J. S., Evans D. R. (1961). Blood trehalose and flight metabolism in the blowfly. Science 134, 54–55. 10.1126/science.134.3471.54 [DOI] [PubMed] [Google Scholar]
- Collins C. H., Clegg J. S. (2004). A small heat-shock protein, p26, from the crustacean Artemia franciscana protects mammalian cells (Cos-1) against oxidative damage. Cell Biol. Int. 28, 449–455. 10.1016/j.cellbi.2004.03.014 [DOI] [PubMed] [Google Scholar]
- Crowe J. H., Carpenter J. F., Crowe L. M. (1998). The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60, 73–103. 10.1146/annurev.physiol.60.1.73 [DOI] [PubMed] [Google Scholar]
- Cui S. Y., Xia Y. X. (2009). Isolation and characterization of the trehalose-6-phosphate synthase gene from Locusta migratoria manilensis. Insect Sci. 16, 287–295. 10.1111/j.1744-7917.2009.01268.x [DOI] [Google Scholar]
- da Costa Morato Nery D., da Silva C. G., Mariani D., Fernandes P. N., Pereira M. D., Panek A. D., et al. (2008). The role of trehalose and its transporter in protection against reactive oxygen species. Biochim. Biophys. Acta 1780, 1408–1411. 10.1016/j.bbagen.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Dmitryjuk M., Łopienska-Biernat E. (2016). The gene expression and the activity of enzyme synthesis of trehalose during development of Ascaris suum (Nematoda) eggs. Invertebr. Reprod. Dev. 60, 95–102. 10.1080/07924259.2016.1160000 [DOI] [Google Scholar]
- Dmitryjuk M., Łopienska-Biernat E., Farjan M. (2009). The level of sugars and synthesis of trehalose in Ascaris suum tissues. J. Helminthol. 83, 237–243. 10.1017/S0022149X08165178 [DOI] [PubMed] [Google Scholar]
- Dmitryjuk M., Łopienska-Biernat E., Zaobidna E. A. (2014). The in vitro effect of ivermectin on the activity of trehaose synthesis pathway enzymes and their mRNA expression in the muscle of adult female Ascaris suum (Nematoda). Sci. World J. 2014:936560. 10.1155/2014/936560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitryjuk M., Dopieralska M., Łopienska-Biernat E., Fraczek R. J. (2013). Purification and partial biochemical-genetic characterization of trehalose 6-phosphate synthase from muscles of adult female Ascaris suum. J. Helminthol. 87, 212–221. 10.1017/S0022149X12000259 [DOI] [PubMed] [Google Scholar]
- Dmitryjuk M., Zółtowska K. (2003). Purification and characterization of acid trehalase from muscle of Ascaris suum (Nematoda). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 61–69. 10.1016/S1096-4959(03)00170-2 [DOI] [PubMed] [Google Scholar]
- Duchateau-Bosson G., Jeuniaux C., Florkin M. (1963). Contributions to the biochemistry of the silkworm. XXVII. Trehalose, trehalase and molting. Arch. Int. Physiol. Biochim. 71, 566–576. [DOI] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R−27R. 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- Evans D. R., Dethier V. G. (1957). The regulation of taste thresholds for sugars in the blowfly. J. Insect Physiol. 1, 3–17. 10.1016/0022-1910(57)90019-7 [DOI] [Google Scholar]
- Fairbairn D. (1958). Trehalose and glucose in helminths and other invertebrates. Can. J. Zool. 36, 787–795. 10.1139/z58-065 [DOI] [Google Scholar]
- Friedman S. (1968). Trehalose regulation of glucose-6-phosphate hydrolysis in blowfly extracts. Science 159, 110–111. 10.1126/science.159.3810.110 [DOI] [PubMed] [Google Scholar]
- Gáliková M., Diesner M., Klepsatel P., Hehlert P., Xu Y., Bickmeyer I., et al. (2015). Energy homeostasis control in Drosophila adipokinetic hormone mutants. Genetics 201, 665–683. 10.1534/genetics.115.178897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans H., Subramanian V., Tan B. H. (1968). Selective phagocytosis: a new concept in protein catabolism. Science 159, 107–110. 10.1126/science.159.3810.107 [DOI] [PubMed] [Google Scholar]
- Gao Y., Jiang Y., Liu Q., Wang R., Liu X., Liu B. (2014). Enzymatic and regulatory properties of the trehalose-6-phosphate synthase from the thermoacidophilic archaeon Thermoplasma acidophilum. Biochimie 101, 215–220. 10.1016/j.biochi.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Goyal K., Browne J. A., Burnell A. M., Tunnacliffe A. (2005). Dehydration-induced tps gene transcripts from an anhydrobiotic nematode contain novel spliced leaders and encode atypical GT-20 family proteins. Biochimie 87, 565–574. 10.1016/j.biochi.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Grewal P. S., Bornstein-Forst S., Burnell A. M., Glazer I., Jagdale G. B. (2006). Physiological, genetic, and molecular mechanisms of chemoreception, thermobiosis, and anhydrobiosis in entomopathogenic nematodes. Biol. Control. 38, 54–65. 10.1016/j.biocontrol.2005.09.004 [DOI] [Google Scholar]
- Guindon S., Lethiec F., Duroux P., Gascuel O. (2005). PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557–W559. 10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N., Puhlev I., Brown D. R., Mansbridge J., Levine F. (2000). Trehalose expression confers desiccation tolerance on human cells. Nat. Biotechnol. 18, 168–171. 10.1038/72616 [DOI] [PubMed] [Google Scholar]
- Guo Q., Hao Y. J., Li Y., Zhang Y. J., Ren S., Si F. L., et al. (2015). Gene cloning, characterization and expression and enzymatic activities related to trehalose metabolism during diapauses of the onion maggot Delia antique (Diptera: Anthomyiidae). Gene 565, 106–115. 10.1016/j.gene.2015.03.070 [DOI] [PubMed] [Google Scholar]
- Herdeiro R. S., Pereira M. D., Panek A. D., Eleutherio E. C. (2006). Trehalose protects Saccharomyces cerevisiae from lipid peroxidation during oxidative stress. Biochim. Biophys. Acta 1760, 340–346. 10.1016/j.bbagen.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Ho N. F., Geary T. G., Barsuhn C. L., Sims S. M., Thompson D. P. (1992). Mechanistic studies in the transcuticular delivery of antiparasitic drugs II: ex vivo/in vitro correlation of solute transport by Ascaris suum. Mol. Biochem. Parasitol. 52, 1–13. 10.1016/0166-6851(92)90031-E [DOI] [PubMed] [Google Scholar]
- Howden G. F., Kilby B. A. (1956). Trehalose and trehalase in the locust. Chem. Ind. 1453–1454. [Google Scholar]
- Huang L., Sun L. Z., Wang Y., Ru Y. T., Muhammad I. R. F. A. N., Jiang Y. R., et al. (2016). Changes in the expression of trehalose-6-phosphate synthase gene in Antheraea pernyi (Lepidoptera: Saturniidae) during pupal diapause termination. Acta Entomol. Sin. 59, 938–947. 10.16380/j.kcxb.2016.09.003 [DOI] [Google Scholar]
- Iordachescu M., Imai R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50, 1223–1229. 10.1111/j.1744-7909.2008.00736.x [DOI] [PubMed] [Google Scholar]
- Jin Q., Hu X., Li X., Wang B., Wang Y., Jiang H., et al. (2016). Genome-wide identification and evolution analysis of trehalose-6-phosphate synthase gene family in Nelumbo nucifera. Fron. Plant Sci. 7:1445. 10.3389/fpls.2016.01445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joga M. R., Zotti M. J., Smagghe G., Christiaens O. (2016). RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7:553. 10.3389/fphys.2016.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandy D. J., Kilby B. A. (1959). Site and mode of trehalose biosynthesis in the locust. Nature 183, 1594–1595. [DOI] [PubMed] [Google Scholar]
- Kern C., Wolf C., Bender F., Berger M., Noack S., Schmalz S., et al. (2012). Trehalose-6-phosphate synthase from the cat flea Cetenocephalides felis and Drosophila melanogaster: gene identification, cloning, heterologous functional expression and identification of inhibitors by high throughput screening. Insect Mol. Biol. 21, 456–471. 10.1111/j.1365-2583.2012.01151.x [DOI] [PubMed] [Google Scholar]
- Kola V. S., Renuka P., Madhav M. S., Mangrauthia S. K. (2015). Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front. Physiol. 6:119. 10.3389/fphys.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolliopoulou A., Taning C. N. T., Smagghe G., Swevers L. (2017). Viral delivery of dsRNA for control of insect agricultural pests and vectors of human disease: prospects and challenges. Front. Physiol. 8:399. 10.3389/fphys.2017.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y., Tanaka S., Hasegawa E., Takahashi M., Nishina M., Matsushita K. (1999). The role of trehalose in lipid mobilization of Locusta migratoria: effects of a trehalase inhibitor, validoxylamine A, on flight activity and hemolymph trehalose and lipid concentrations. Entomol. Sci. 2, 449–456. [Google Scholar]
- Kormish J. D., McGhee J. D. (2005). The C. elegans lethal gut-obstructed gog-1 gene is trehalose-6-phosphate phosphatase. Dev. Biol. 287, 35–47. 10.1016/j.ydbio.2005.08.027 [DOI] [PubMed] [Google Scholar]
- Kramer K. J., Speirs R. D., Childs C. N. (1978). A method for separation of trehalose from insect hemolymph. Anal. Biochem. 86, 692–696. 10.1016/0003-2697(78)90796-0 [DOI] [PubMed] [Google Scholar]
- Kushwaha S., Singh P. K., Rana A. K., Misra-Bhattacharya S. (2011). Cloning, expression, purification and kinetics of trehalose-6-phosphate phosphatase of filarial parasite Brugia malayi. Acta Trop. 119, 151–159. 10.1016/j.actatropica.2011.05.008 [DOI] [PubMed] [Google Scholar]
- Kushwaha S., Singh P. K., Shahab M., Pathak M., Bhattacharya S. M. (2012). In vitro silencing of Brugia malayi trehalose-6-phosphate phosphatase impairs embryogenesis and in vivo development of infective larvae in jirds. PLoS Negl. Trop. Dis. 6:e1770. 10.1371/journal.pntd.0001770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hao Y. J., Zhang Y. J., Si F. L., Chen B. (2013). Cloning, bioinformatic analysis and diapauses-related expression of trehalose-6-phosphate synthase gene from the onion maggot, Delia antique (Diptera: Anthomyiidae). Acta Entomol. Sin. 56, 329–338. [Google Scholar]
- Liu C., Dunaway-Mariano D., Mariano P. S. (2017). Rational design of reversible inhibitors for trehalose 6-phosphate phosphatases. Eur. J. Med. Chem. 128, 274–286. 10.1016/j.ejmech.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Liu J. H., Shang X. D., Liu J. Y., Tan Q. (2016). Changes in trehalose content, enzyme activity and gene expression related to trehalose metabolism in Flammulina velutipes under heat shock. Microbiology (Reading. Engl). 162, 1274–1285. 10.1099/mic.0.000324 [DOI] [PubMed] [Google Scholar]
- Lu X., Li J., Yang J., Liu X., Ma J. (2014). De novo transcriptome of the desert beetle Microdera punctipennis (Coleoptera: Tenebrionidae) using illumine RNA-seq technology. Mol. Biol. Rep. 41, 7293–7303. 10.1007/s11033-014-3615-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J. I., Min S. R., Lee J. H., Lim Y. H., Kim J. K., Bae C. H., et al. (2013). Overexpression of a 6-phosphate synthase/phosphatase fusion gene enhances tolerance and photosynthesis during drought and salt stress without growth aberrations in tomato. Plant Cell Tiss Org. 112, 257–262. 10.1007/s11240-012-0225-7 [DOI] [Google Scholar]
- Łopienska-Biernat E., Zaobidna E. A., Dmitryjuk M. (2015). Expression of genes encoding the enzymes for glycogen and trehalose metabolism in L3 and L4 larvae of Anisakis simplex. J. Parasitol. Res. 2015:438145. 10.1155/2015/438145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães R. S., De Lima K. C., de Almeida D. S., De Mesquita J. F., Eleutherio E. C. (2017). Trehalose-6-phosphate as a potential lead candidate for the development of Tps1 inhibitors: insights from the trehalose biosynthesis pathway in diverse yeast species. Appl. Biochem. Biotechnol. 181, 914–924. 10.1007/s12010-016-2258-6 [DOI] [PubMed] [Google Scholar]
- Mariano A. C., Santos R., Gonzalez M. S., Feder D., Machado E. A., Pascarelli B., et al. (2009). Synthesis and mobilization of glycogen and trehalose in adult male Rhodnius prolixus. Arch. Insect Biochem. Physiol. 72, 1–15. 10.1002/arch.20319 [DOI] [PubMed] [Google Scholar]
- Matsuda H., Yamada T., Yoshida M., Nishimura T. (2015). Flies without trehalose. J. Biol. Chem. 290, 1244–1255. 10.1074/jbc.M114.619411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel P. O., Grubor-Lajsic G., Worland M. R. (1998). Partial desiccation induced by sub-zero temperatures as a component of the survival strategy of the Arctic collembolan Onychiurus arcticus (Tullberg). J. Insect Physiol. 44, 211–219. 10.1016/S0022-1910(97)00166-2 [DOI] [PubMed] [Google Scholar]
- Murphy T. A., Wyatt G. R. (1965). The enzymes of glycogen and trehalose synthesis in silk moth fat body. J. Biol. Chem. 240, 1500–1508. [PubMed] [Google Scholar]
- Pampurova S., Verschooten K., Avonce N., Van Dijck P. (2014). Functional screening of a cDNA library from the desiccation-tolerant plant Selaginella lepidophylla in yeast mutants identifies trehalose biosynthesis genes of plant and microbial origin. J. PlantRes. 127, 803–813. 10.1007/s10265-014-0663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Kim Y. (2017). Identification of a hypertrehalosemic factor in Spodoptera exigua. Arch. Insect Biochem. Physiol. 95:e21386. 10.1002/arch.21386 [DOI] [PubMed] [Google Scholar]
- Pellerone F. I., Archer S. K., Behm C. A., Grant W. N., Lacey M. J., Somerville A. C. (2003). Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes. Int. J. Parasitol. 33, 1195–1206. 10.1016/S0020-7519(03)00173-5 [DOI] [PubMed] [Google Scholar]
- Perry R. N. (1989). Dormancy and hatching of nematode eggs. Parasitol. Today (Regul. Ed). 5, 377–383. 10.1016/0169-4758(89)90299-8 [DOI] [PubMed] [Google Scholar]
- Qin Z., Wang S., Wei P., Xu C. D., Tang B., Zhang F. (2012). Molecular cloning and cold-induced expression of trehalose-6-phosphate synthase gene in Harmonia axyridis (Coleoptera: Coccinellidae). Acta Entomol. Sin. 55, 651–658. [Google Scholar]
- Reisenman C. E., Lei H., Guerenstein P. G. (2016). Neuroethology of olfactory-guided behavior and its potential application in the control of harmful insects. Front. Physiol. 7:271. 10.3389/fphys.2016.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A. B., Krakowka S., Dexter L. B., Schmid H., Wolterbeek A. P., Waalkens-Berendsen D. H., et al. (2002). Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem. Toxicol. 40, 871–898. 10.1016/S0278-6915(02)00011-X [DOI] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 296, 1118–1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Shen Q. D. (2017). Genetic Characteristics, Functional Identification and Regulation Analysis of Trehalose-6-Phosphate Synthase 3 in Nilaparvata lugens. MSc Thesis, Hangzhou Normal University, Hangzhou.
- Shi J. F., Xu Q. Y., Sun Q. K., Meng Q. W., Mu L. L., Guo W. C., et al. (2016). Physiological roles of trehalose in Leptinotarsa larvae revealed by RNA interference of trehalose-6-phosphate synthase and trehalase genes. Insect Biochem. Mol. Biol. 77, 52–68. 10.1016/j.ibmb.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Shi Q., Chung J. S. (2014). Trehalose metabolism in the blue crab Callinectes sapidus: isolation of multiple structural cDNA isoforms of trehalose-6-phosphate synthase and their expression in muscles. Gene 536, 105–113. 10.1016/j.gene.2013.11.070 [DOI] [PubMed] [Google Scholar]
- Shi Z.-K., Wang S., Wang S.-G., Zhang L., Xu Y.-X., Guo X.-J., et al. (2017). Effects of starvation on the carbohydrate metabolism in Harmonia axyridis (Pallas). Biol Open 6, 1096–1103. 10.1242/bio.025189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla E., Thorat L. J., Nath B. B., Gaikwad S. M. (2015). Insect trehalase: physiological significance and potential applications. Glycobiology 25, 357–567. 10.1093/glycob/cwu125 [DOI] [PubMed] [Google Scholar]
- Soto T., Fernandez J., Vicente-Soler J., Cansado J., Gacto M. (1999). Accumulation of trehalose by overexpression of tps1, coding for trehalose-6-phosphate synthase, causes increased resistance to multiple stresses in the fission yeast Schizosaccharomyces pombe. Appl. Environ. Microbiol. 65, 2020–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. E. (2016). Evidence that ecdysis in the larval cockroach, Periplaneta americana L. is triggered by an increase in the concentration of hemolymph sugar. Arch. Insect Biochem. Physiol. 92, 159–172. 10.1002/arch.21323 [DOI] [PubMed] [Google Scholar]
- Storey K. B., Storey J. M. (2012). Insect cold hardiness: metabolic, gene, and protein adaptation. Can. J. Zool. 90, 456–475. 10.1139/z2012-011 [DOI] [Google Scholar]
- Tang B., Chen J., Yao Q., Pan Z. Q., Xu W. H., Wang S. G., et al. (2010). Characterization of a trehalose-6-phosphate synthase gene from Spodoptera exigua and its function identification through RNA interference. J. Insect Physiol. 56, 813–821. 10.1016/j.jinsphys.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Tang B., Chen X. F., Liu Y., Tian H. G., Liu J., Hu J., et al. (2008). Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Mol. Biol. 9:51. 10.1186/1471-2199-9-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Qin Z., Shi Z. K., Wang S., Guo X. J., Wang S. G., et al. (2014b). Trehalase in Harmonia axyridis (Coleoptera: Coccinellidae): effects on beetle locomotory activity and the correlation with trehalose metabolism under starvation conditions. Appl. Entomol. Zool. 49, 255–264. 10.1007/s13355-014-0244-4 [DOI] [Google Scholar]
- Tang B., Wei P., Chen J., Wang S. G., Zhang W. Q. (2012a). Progress in gene features and functions of insect trehalases. Acta Entomol. Sin. 55, 1315–1321. [Google Scholar]
- Tang B., Wei P., Zhao L. N., Shi Z. K., Shen Q. D., Yang M. M., et al. (2016). Knockdown of five trehalase genes using RNA interference regulates the gene expression of the chitin biosynthesis pathways in Tribolium castaneum. BMC Biotechnol. 16:67. 10.1186/s12896-016-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Xu Q. Y., Zhao L. N., Wang S. G., Zhang F. (2014a). Progress in research on the characteristics and functions of trehalose and the TPS gene in insect. Chin. J. Appl. Entomol. 51, 1397–1405. [Google Scholar]
- Tang B., Xu Q., Zou Q., Fang Q., Wang S. G., Ye G. Y. (2012b). Sequencing and characterization of glycogen synthase and glycogen phosphorylase genes from Spodoptera exigua and analysis of their function in starvation and excessive sugar intake. Arch. Insect Biochem. Physiol. 80, 42–62. 10.1002/arch.21027 [DOI] [PubMed] [Google Scholar]
- Tang B., Yang M., Shen Q., Xu Y., Wang H., Wang S. (2017). Suppressing the activity of trehalase with validamycin disrupts the trehalose and chitin biosynthesis pathways in rice brown planthopper, Nilaparvata lugens. Pestic. Biochem. Physiol. 137, 81–90. 10.1016/j.pestbp.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Tang B., Zheng H. Z., Xu Q., Zou Q., Wang G. J., Zhang F., et al. (2011). Cloning and pattern of expression of trehalose-6-phosphate synthase cDNA from Catantops pinguis (Orthoptera: Catantopidae). Eur. J. Entomol. 108, 355–363. 10.14411/eje.2011.044 [DOI] [Google Scholar]
- Thorat L. J., Gaikwad S. M., Nath B. B. (2012). Trehalose as an indicator of desiccation stress in Drosophila melanogaster larvae: a potential marker of anhydrobiosis. Biochem. Biophys. Res. Commun. 419, 638–642. 10.1016/j.bbrc.2012.02.065 [DOI] [PubMed] [Google Scholar]
- Thorat L., Mani K. P., Thangaraj P., Chatterjee S., Nath B. B. (2016). Downregulation of dTps1 in Drosophila melanogaster larvae confirms involvement of trehalose in redox regulation following desiccation. Cell Stress Chaperones 21, 285–294. 10.1007/s12192-015-0658-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R., Berglund E., Akdemir F., Tran C., Kim M. S., Noh J., et al. (2015). Drosophila glucome screening identifies Ck1alpha as a regulator of mammalian glucose metabolism. Nat. Commun. 6:7102. 10.1038/ncomms8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Ortiz M. A., Quintana-Castro R., Oliart-Ros R. M., De la Cruz-Medina J., Ramírez de León J. A., Garcia H. S. (2013). High hydrostatic pressure induces synthesis of heat-shock proteins and trehalose-6-phosphate synthase in Anastrepha ludens larvae. Arch. Insect Biochem. Physiol. 82, 196–212. 10.1002/arch.21085 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kikawada T., Minagawa N., Yukuhiro F., Okuda T. (2002). Mechanism allowing an insect to survive comolete dehydration and extreme temperatures. J. Exp. Biol. 205, 2799–2802. [DOI] [PubMed] [Google Scholar]
- Wegener G., Macho C., Schlöder P., Kamp G., Ando O. (2010). Long-term effects of the trehalase inhibitor trehazolin on trehalase activity in locust flight muscle. J. Exp. Biol. 213, 3852–3857. 10.1242/jeb.042028 [DOI] [PubMed] [Google Scholar]
- Wegener G., Tschiedel V., Schlöder P., Ando O. (2003). The toxic and lethal effects of the trehalase inhibitor trehazolin in locusts are caused by hypoglycemia. J. Exp. Biol. 206, 1233–1240. 10.1242/jeb.00217 [DOI] [PubMed] [Google Scholar]
- Wen X., Wang S., Duman J. G., Arifin J. F., Juwita V., Goddard W. A., III, et al. (2016). Antifreeze proteins govern the precipitation of trehalose in a freezing-avoiding insect at low temperature. Proc. Natl. Acad. Sci. U.S.A. 113, 6683–6688. 10.1073/pnas.1601519113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt G. R. (1961). The biochemistry of insect hemolymph. Annu. Rev. Entomol. 6, 75–102. 10.1146/annurev.en.06.010161.000451 [DOI] [Google Scholar]
- Wyatt G. R. (1967). The biochemistry of sugars and polysaccharides in insects. Adv. Insect Physiol. 4, 287–360. 10.1016/S0065-2806(08)60210-6 [DOI] [Google Scholar]
- Wyatt G. R., Kalf G. F. (1956). Trehalose in insect. Fed. Proc. 15:388. [Google Scholar]
- Wyatt G. R., Kalf G. F. (1957). The chemistry of insect hemolymph. II. Trehalose and other carbohydrates. J. Gen. Phsiol. 40, 833–847. 10.1085/jgp.40.6.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong K. C., Wang J., Li J. H., Deng Y. Q., Pu P., Fan H., et al. (2016). RNA interference of a trehalose-6-phosphate synthase gene reveals its role during larval-pupal metamorphosis in Bactrocera minax (Diptera: Tephritidae). J. Insect Physiol. 91–92, 84–92. 10.1016/j.jinsphys.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Xu J., Bao B., Zhang Z. F., Yi Y. Z., Xu W. H. (2009). Identification of a novel gene encoding the trehalose phosphate synthase in the cotton bollworm, Helicoverpa armigera. Glycobiology 19, 250–257. 10.1093/glycob/cwn127 [DOI] [PubMed] [Google Scholar]
- Yang M. M., Zhao L. N., Shen Q. D., Xie G. Q., Wang S. G., Tang B. (2017). Knockdown of two trehalose-6-phosphate synthases severely affects chitin metabolism gene expression in the rice brown planthopper Nilaparvata lugens. Pest Manag. Sci. 73, 206–216. 10.1002/ps.4287 [DOI] [PubMed] [Google Scholar]
- Yang W. J., Yuan G. R., Cong L., Xie Y. F., Wang J. J. (2014). De novo cloning and annotation of genes associated with immunity, detoxification and energy metabolism from the fat body of the oriental fruit fly, Bactrocera dorsais. PLoS ONE 9:e94470. 10.1371/journal.pone.0094470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T., Yamada T., Nishimura T. (2017). Adaptation to dietary conditions by trehalose metabolism in Drosophila. Sci. Rep. 7:1619. 10.1038/s41598-017-01754-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yednock B. K., Neigel J. E. (2014). Detecting selection in the blue crab, Callinectes sapidus, using DNA sequence data from multiple nuclear protein-coding genes. PLoS ONE 9:e99081. 10.1371/journal.pone.0099081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Matsuda H., Kubo H., Nishimura T. (2016). Molecular characterization of Tps1 and Treh genes in Drosophila and their role in body water homeostasis. Sci. Rep. 6:30582. 10.1038/srep30582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhang H., Zhu G. (2010). Plant-type trehaloe synthetic pathway in Cryptosporidium and some other apicomplexans. PLoS ONE 5:e12593. 10.1371/journal.pone.0012593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. W., Chen J., Guo Y. S. (2012). Cloning and sequence Analysis of trehalose phosphate synthase gene from Sogatell furifera. Heilongjiang Agricul. Sci. 5, 14–19. [Google Scholar]
- Zhang J., Wang J., Li F., Sun Y., Yang C., Xiang J. (2012). A trehalose-6-phosphate synthase gene from Chinese shrimp, Fenneropenaeus chinensis. Mol. Biol. Rep. 39, 10219–10225. 10.1007/s11033-012-1897-0 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang H., Chen J., Shen Q., Wang S., Xu H., et al. (2017). Glycogen phosphorylase and glycogen synthase: gene cloning and expression analysis reveal their role in trehalose metabolism in the Brown planthopper, Nilaparvata lugens Stål (Hemiptera: Delphacidae). J. Insect. Sci. 17:42. 10.1093/jisesa/iex015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Q., Chen X. F., Tang B., Tian H. G., Chen J., Yao Q. (2011). Insect chitin biosynthesis and its regulation. Chin. J. Appl. Entomol. 48, 475–479. [Google Scholar]
- Zhao L. N., Yang M. M., Shen Q., Liu X., Shi Z., Wang S., et al. (2016). Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference. Sci. Rep. 6:27841. 10.1038/srep27841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zółtowska K., Lopieniska-Biernat E. (2006). Content of glycogen and trehalose and activity of α-amylase and trehalase in Galleria mellonella larvae infected with entomophthogenic nematodes Steinemema affinis and S. feltiae. Wiad. Parazytol. 52, 103–107. [PubMed] [Google Scholar]