Abstract
Rodents have historically been associated with zoonotic pandemics that claimed the lives of large human populations. Appropriate pathogen surveillance initiatives could contribute to early detection of zoonotic infections to prevent future outbreaks. Bordetella species are bacteria known to cause mild to severe respiratory disease in mammals and, some have been described to infect, colonize and spread in rodents. There is a lack of information on the population diversity of bordetellae among Malaysian wild rodents. Here, bordetellae recovered from lung tissues of wild rats were genotypically characterized using 16S rDNA sequencing, MLST and nrdA typing. A novel B. bronchiseptica ST82, closely related to other human-derived isolates, was discovered in three wild rats (n=3) from Terengganu (5.3333° N, 103.1500° E). B. pseudohinzii, a recently identified laboratory mice inhabitant, was also recovered from one rat (n=1). Both bordetellae displayed identical antimicrobial resistance profiles, indicating the close phylogenetic association between them. Genotyping using the 765-bp nrdA locus was shown to be compatible with the MLST-based phylogeny, with the added advantage of being able to genotype non-classical bordetellae. The recovery of B. pseudohinzii from wild rat implied that this bordetellae has a wider host range than previously thought. The findings from this study suggest that bordetellae surveillance among wild rats in Malaysia has to be continued and expanded to other states to ensure early identification of species capable of causing public health disorder.
Keywords: bordetellae, infectious disease, MLST, nrdA, tropical
The increase in human-wildlife interactions resulting from human encroachment into wildlife habitats will undoubtedly lead to increase in the opportunity for transmission of zoonotic diseases [23]. Rodents undeniably, play a central role in transmitting zoonotic diseases as they are the most abundant mammal and can be found in every continent except for Antartica [23]. Historical records have shown that rodents are linked to the spread of plague which drastically reduced human populations in Europe, Africa and Asia [7], highlighting the propensity of rodents to provoke major pandemics. The risks of emerging zoonoses to public health and the economy have provided impetus for pathogen surveillance initiatives, as early detection of zoonotic diseases is key to outbreak prevention.
Members of the bacteria genus Bordetella are closely related and are recognized for causing mild to severe respiratory disease in mammals [30]. The classical bordetellae; B. pertussis, B. parapertussis and B. bronchiseptica can cause respiratory disease in humans and livestocks [4], and receive the most research attention [13]. The other Bordetella subspecies, referred to as non-classical bordetellae, are mostly associated with the environment [13] and rodents [21], albeit more human infections have been recently reported [13]. The potential for inter-species transmission is considerable since a number of classical and non-classical bordetellae possess the ability to colonize and grow in rodents [21].
In Malaysia, there is a dearth of information on the genetic structure of bordetellae populations in wild rodents. Despite its zoonotic potential, no study has been conducted to investigate the diversity and clonal relationships among the bordetellae populations maintained in wild rodents. Reports of whooping cough and other pulmonary symptoms in humans living close to animals infected with B. bronchiseptica suggests that the infections originated from infected animals [12, 27, 32]. Even though there has not been any report of human B. bronchiseptica infection in Malaysia, all these past cases underscore the possibility of zoonotic transmission of bordetellae to humans or to other animals, and these animals can then serve as carrier in the transmission cycle [23]. Detection of B. bronchiseptica from livestock, domestic pets and wild animals in Africa, Asia, Europe and the U.S.A. have been documented, illustrating the global distribution of this species and the diversity of hosts it occupies [14, 25, 27, 33]. B. pseudohinzii also has a relatively wide geographical distribution evident by its recovery in Germany, Japan, Malaysia and the U.S.A., although this bacterium has only been isolated from laboratory-raised rodents [16].
Here, we recovered bordetellae from rodents captured in the wet markets and identified them to subspecies level using molecular tools. Multi locus sequence typing (MLST) and nrdA typing were utilized to determine the phylogenetic relationships among the different bordetellae isolates, which could shed useful insights into factors that shape the genetic structure of bordetellae. Sequence analysis of nrdA, encoding the ribonucleoside-diphosphate reductase alpha chain, has been demonstrated to reliably differentiate species within the Bordetella genus [21, 28]. In addition, the antimicrobial resistance and virulence profiles of the bordetellae isolates were also determined.
MATERIALS AND METHODS
Rodent trappings
Prior approvals were obtained from the local municipal councils to accompany their pest eradication teams to conduct rodent trappings within wet markets in several states in Peninsular Malaysia. Wet markets in four states were included; Johor (1.4927° N, 103.7414° E), Kedah (6.1248° N, 100. 3678° E), Kelantan (6.1168° N, 102.2777° E) and Terengganu (5.3333° N, 103.1500° E) (Fig. 1). Rodent trappings were conducted over a period of 14 months, from November 2014 until December 2015. Steel wire traps (18 × 12 × 28 cm) were placed on floors in the wet market in the evening with baits such as dried fish, bread or peanut butter. The traps were collected early in the morning before the market was opened [3]. All rodents were captured alive and their species were determined before they were euthanized at the field laboratory according to protocols described elsewhere [1]. Lung tissues were harvested and homogenized, followed by inoculation onto Columbia agar according to previously published methods [22]. Cross contamination between tissue specimens from different individual rodents was prevented by washing the surgical tools with alcohol, bleach and water, in between surgical procedures as previously described [20]. This study was approved with the ethics reference no. ISB/31/01/2013/SNMZ (R) by the Institutional Animal Care and Use Committee and did not involve any endangered or protected species.
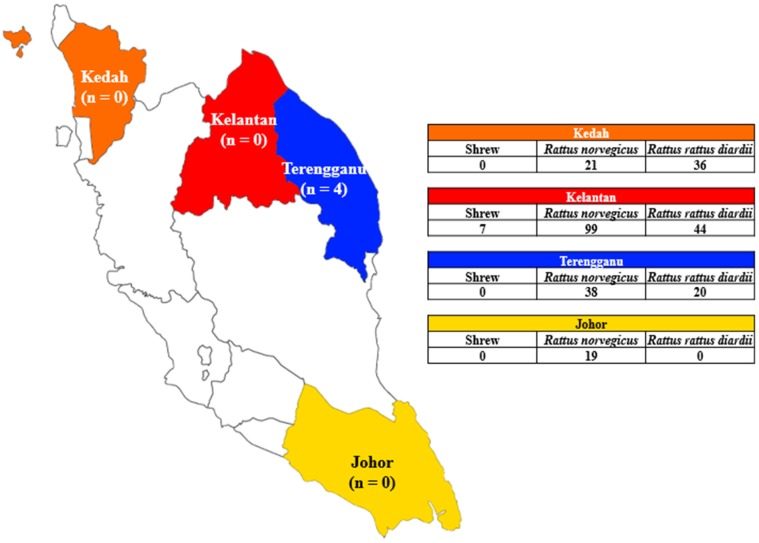
Fig. 1.
Map of Peninsular Malaysia showing the states that were involved in the study; Johor, Kedah, Kelantan and Terengganu, together with the number of bordetellae recovered, indicated below the names of each respective state. The table shows the number of rodents and shrews captured in the individual states.
Antimicrobial susceptibility testing and amplification of virulence genes
All mixed cultures grown from the rodent lung homogenates were sub-cultured until pure cultures were obtained. Chromosomal DNA of all pure-cultured bacteria isolates was extracted using NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. Subsequently, 16S rDNA sequencing [18] was performed to identify the respective isolates’ species identity. Only members of the genus Bordetella were selected for this study and they were stocked in −80°C until further use. Antimicrobial susceptibility profiles were determined using M.I.C.Evaluator Strips (Thermo Fisher Scientific, Waltham, MA, U.S.A.) on Mueller-Hinton agar. The following antibiotics were utilized to determine the minimum inhibitory concentrations (MIC); amikacin, amoxicillin-clavulanic acid, ampicillin, ceftazidime, ciprofloxacin, ceftriaxone, cefotaxime, erythromycin, gentamicin, imipenem and meropenem. Antimicrobial susceptibility results were interpreted according to Clinical and Laboratory Standards Institute breakpoints for Enterobacteriaceae [5, 24]. The presence of the virulence genes; dermonecrotic toxin (dnt), exogenous ferric siderophore receptor (bfrZ) and flagella (fla), were examined using nucleic acid amplification protocols previously described [29].
Multi locus sequence typing and nrdA typing
DNA fragments from seven housekeeping genes (adk, fumC, glyA, tyrB, icd, pepA and pgm) were amplified, trimmed to a standard length and compared with the Bordetella MLST database at http://pubmlst.org/bordetella/ [6]. A novel allele number was assigned to allele sequences that are not available in the database, resulting in the assignment of a novel ST. An additional nrdA typing method [28] was performed on bordetellae which failed MLST characterization [21]. This method was also applied to the other Bordetella isolates and sequences of all nrdA loci were extracted from the database, aligned and phylogenetically analyzed using MEGA 5.2.
Analysis of evolutionary relationships
The evolutionary relationships among bordetellae in this study, were analyzed and presented as a minimum spanning tree using the algorithm, PHYLOViZ [9]. The displayed minimum spanning tree was used to visualize the possible evolutionary relationships between Bordetella isolates based on their MLST allelic profile data. The minimum spanning tree firstly, links STs sharing single locus variants and the subsequent branches connect to STs with more than one locus variation. STs located multiple branches away from the core ST have the most loci variants.
Amplification of the CRISPR-cas locus
A Type II-C CRISPR-Cas system was recently discovered in a newly described bordetellae, B. pseudohinzii [15]. CRISPR-Cas systems have not been identified in any other Bordetella species, making the amplification of genes associated to the Type II-C CRISPR-Cas system a defining feature for the identification of B. pseudohinzii. Nucleic acid amplification of the Type II-C CRISPR associated genes was performed on bordetellae which could not be characterized using MLST [15].
RESULTS
A total of four Malaysian states were included in this study (Johor, Kedah, Kelantan and Terengganu). Two types of rodent species were caught from the four states; Rattus norvegicus (n=177) and Rattus rattus diardii (n=100), with Kelantan having the most number of captured rats (n=143), followed by Terengganu (n=58), Kedah (n=57) and Johor (n=19). Rattus norvegicus dominated the number of captured rats in Kelantan (n=99), followed by Terengganu (n=38), Johor (n=19) and Kedah (n=21). The most number of Rattus rattus diardii was captured in Kelantan (n=44), followed by Kedah (n=36) and Terengganu (n=20). There was no Rattus rattus diardii trapped in Johor (Fig. 1). Several shrews (n=7) were trapped in Kelantan, however, they were omitted as this species was not included in the animal ethics application.
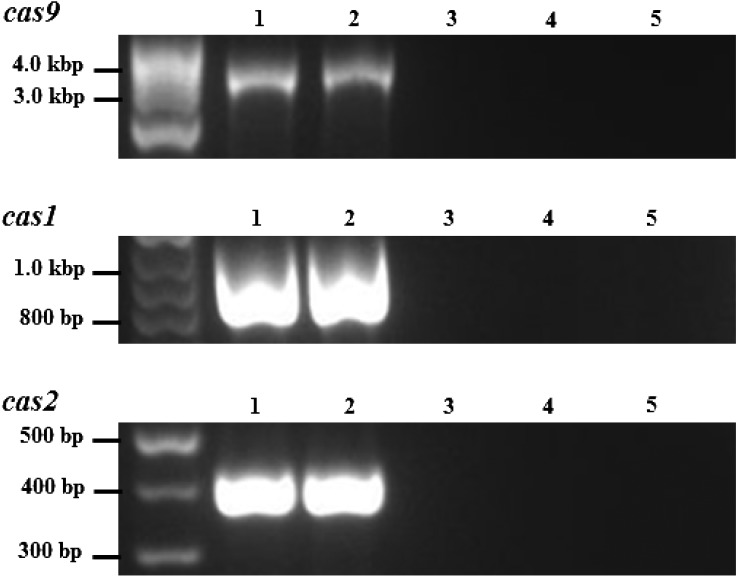
Analyses of the bacterial isolates cultured from rat lung homogenates using 16S rDNA sequencing revealed the isolation of 4 Bordetella isolates, yielding the prevalence of bordetellae at 1.44%. Based on the partial 16S rDNA sequences, the isolates were identified as B. bronchiseptica (n=3) and B. hinzii (n=1) (Table 1). These four bordetellae were isolated from Terengganu rats. Rattus rattus diardii harbored three isolates (2 B. bronchiseptica and 1 B. hinzii), while the other B. bronchiseptica was isolated from the lung of one Rattus norvegicus (Table 1). The B. bronchiseptica isolates (TRE151600, TRE150202 and TRE155202) were subjected to MLST and nrdA typing, whereas only the nrdA typing was performed on isolate TRE152202 (Table 1). Genotyping analyses uncovered that all of the B. bronchiseptica isolates as ST82 (novel ST) possessing nrdA locus 162 (Table 1). Isolate TRE152202 was shown to possess nrdA locus 189, corresponding to Bordetella genogroup 16 (Table 1). Upon obtaining this finding, the extracted DNA of isolate TRE152202 was used for the amplification and sequencing of the CRISPR-cas locus. Amplicons of the cas1, cas2 and cas9 were amplified (Fig. 2) and all displayed identical sequences as the CRISPR-associated genes of B. pseudohinzii HI4681 (accession no. CP016440) and BH370 [19]. Isolate TRE152202 was thus, redesignated as B. pseudohinzii.
Table 1. Antimicrobial susceptibilities, sequence types, nrdA loci and host diversity of bordetellae recovered from wild rodents.
| Isolate name (Bordetella species) |
Host | Virulence gene | ST | nrdA locus | MIC (µg/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | AMC | AMP | CAZ | CIP | CRO | CTX | ERY | GEN | IMP | MEM | |||||
| TRE151600 (B. bronchiseptica) |
RR | dnt, bfrZ, fla | 82 | 162 | S (16) | R (256) | R (128) | S (0.5) | S (0.5) | R (8) | R (16) | R (8) | S (1) | S (0.06) | S (0.03) |
| TRE150202 (B. bronchiseptica) |
RR | dnt, bfrZ, fla | 82 | 162 | S (4) | R (128) | R (64) | S (0.06) | S (0.25) | R (16) | R (16) | R (8) | S (2) | S (0.5) | S (0.5) |
| TRE155202 (B. bronchiseptica) |
RN | dnt, bfrZ, fla | 82 | 162 | S (8) | R (256) | R (64) | S (0.5) | S (0.12) | R (16) | R (32) | R (8) | S (1) | S (0.25) | S (0.25) |
| TRE152202 (B. pseudohinzii) |
RR | na | na | 189 | S (8) | R (128) | R (128) | S (0.03) | S (0.25) | R (8) | R (16) | S (0.5) | S (0.5) | S (0.06) | S (0.06) |
RR, Rattus rattus diardii; RN, Rattus norvegicus; dnt, dermonecrotic toxin; bfrZ, exogenous ferric siderophore receptor; fla, flagella; ST, sequence type; na, not available; MIC, minimum inhibitory concentration; AMK, amikacin; AMC, amoxicillin-clavulanic acid; AMP, ampicillin; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CTX, cefotaxime; ERY, erythromycin; GEN, gentamicin; IMP, imipenem; MEM, meropenem; R, resistant; S, sensitive.
Fig. 2.
Nucleic acid amplification products of cas9, cas1 and cas2 of B. pseudohinzii TRE152202. Lane 1, B. pseudohinzii BH370 (positive control); Lane 2, B. pseudohinzii TRE152202; Lane 3, B. bronchiseptica TRE151600; Lane 4, B. bronchiseptica TRE150202; Lane 5, B. bronchiseptica TRE155202.
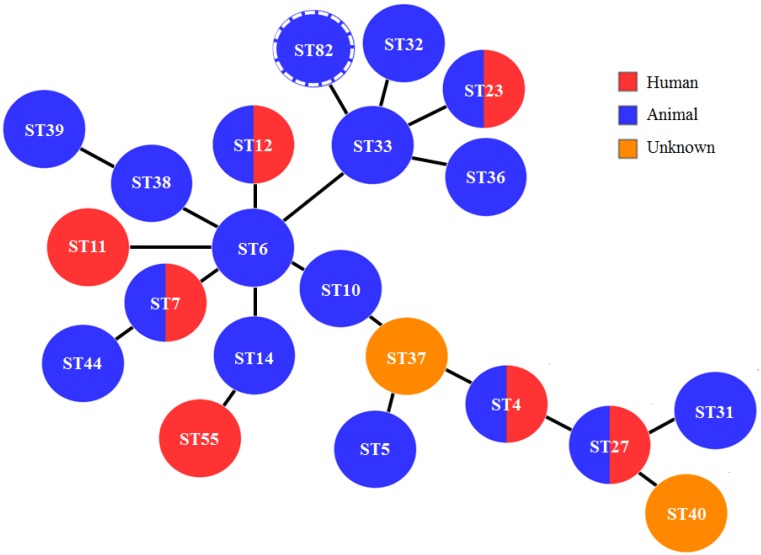
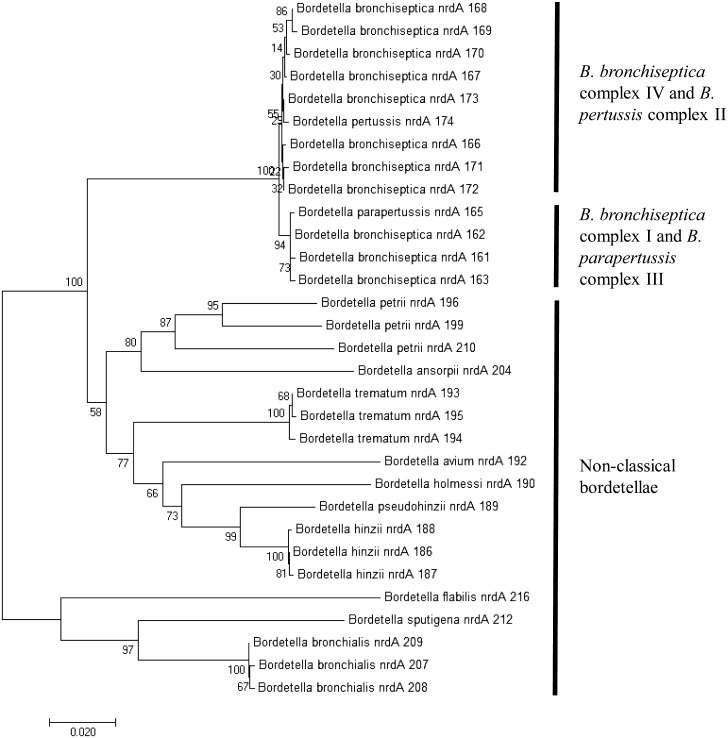
The population structure among B. bronchiseptica isolated in this study was examined by building a minimum spanning tree using an established algorithm, PHYLOViZ. The ST82 isolates were shown to have evolved from the central ST6-lineage (Fig. 3). As depicted in Fig. 3, all of the STs were also members of the B. bronchiseptica complex I, which consists of predominantly animal-associated isolates. Based on the constructed phylogenetic tree using the trimmed 765-bp nrdA gene, we observed that the classical and non-classical bordetellae were distinctively separated into 3 clusters (Fig. 4); one belonging to the classical bordetellae, one belonging to non-classical bordetellae, of which has been partially characterized and one belonging to the recently discovered non-classical bordetellae (B. bronchialis, B. flabilis and B. sputigena). Two sub-clusters were noted among the classical bordetellae; one containing predominantly human isolates (B. bronchiseptica complex IV and B. pertussis complex II) and the other consists of predominantly animal isolates (B. bronchiseptica complex I and B. parapertussis complex III) (Fig. 4). The B. bronchiseptica isolates carrying nrdA locus 162 were clustered together with B. bronchiseptica complex I and B. parapertussis complex III. The B. pseudohinzii isolate carrying nrdA locus 189, however was clustered together with the non-classical bordetellae (Fig. 4).
Fig. 3.
Minimum spanning tree of B. bronchiseptica complex I, constructed using PHYLOViZ. Each circle represents individual STs and different colors indicate the host at which B. bronchiseptica was recovered. ST and hosts data were recovered from the Bordetella MLST database at http://pubmlst.org/bordetella/. The novel ST82 was indicated by a circle with dashed lines.
Fig. 4.
Neighbor-joining tree based on nrdA loci of classical and non-classical bordetellae. nrdA locus numbers are indicated behind the bordetellae species names. The nrdA locus numbers were extracted from the Bordetella MLST database at http://pubmlst.org/bordetella/. Bordetellae isolates which have not been classified to the species level were excluded from the study. Separation of complexes I-IV in the classical bordetellae group based on MLST data was consistent with the nrdA sequence analyses. All non-classical bordetellae were clustered separately.
Antimicrobial susceptibility testing performed on the Bordetella isolates showed that they had almost similar antimicrobial profiles. Resistance was displayed towards amoxicillin-clavulanic acid, ampicillin, ceftriaxone and cefotaxime. All of the Bordetella isolates were susceptible to amikacin, ceftazidime, ciprofloxacin, gentamicin, imipenem and meropenem (Table 1). The only observed antimicrobial disparity was on erythromycin, by which the B. bronchiseptica isolates displayed resistance while B. pseudohinzii was susceptible (Table 1). Additionally, the B. bronchiseptica isolates carried the virulence genes; dnt, bfrZ and fla, while B. pseudohinzii did not carry any (Table 1).
DISCUSSION
Even though rats were implicated as the host for B. bronchiseptica [2], the prevalence of this species in the Malaysian wild rat population is unknown. The overall prevalence of bordetellae in rodents from the four Malaysian states was 1.44% (4/277), slightly higher as compared to the findings by Jiyipong et al. (2013) where only one bordetellae was cultured out of 206 rodent specimens yielding a prevalence of 0.49% [17]. This was also consistent with the reported low prevalence of B. bronchiseptica carriers in rodent populations [2] and a well-fed population of rats in the market would have developed stronger immunological defenses, reducing B. bronchiseptica infection risk [8]. The isolation of bordetellae from the lung tissues of Rattus norvegicus and Rattus rattus diardii suggests that these two rodent species were competent hosts and indicated the propensity of this bacterial genus to the respiratory tract, similar to other reports [4, 30]. Only rats from the state of Terengganu harbored bordetellae and this was to be expected due to the low prevalence of this bacterium in the host [2] coupled with the limited number of study sites. The absence of Rattus rattus diardii in Johor probably reflected the sampling limitation in this study whereby trappings were conducted for only one night and rat species with low abundance was not captured. Identification of bordetellae using a three-pronged approach (16S rDNA sequencing, nrda typing and MLST) ensures resolution to the species level, at least for the classical bordetellae [21], owing to the close phylogenetic relationships within the genus [28]. It has been reported that rodents are unnatural hosts for B. bronchiseptica although they can be colonized and shed the bacteria [30]. Rats have been shown to demonstrate strong resistance towards clinical infection by B. bronchiseptica, however they become asymptomatic carrier for life after exposure [2], similar to our observation of the seemingly healthy captured rats. B. bronchiseptica can remain within the respiratory epithelial cells to evade the host’s immune system [10] contributing to life-long disease persistence in infected rats.
The B. bronchiseptica isolates recovered from Rattus norvegicus and Rattus rattus diardii (Table 1) displayed identical genotype (ST82; nrdA locus 162), suggesting inter-species B. bronchiseptica transmission between rats in that particular area. This contradicts the earlier suggestion that B. bronchiseptica transmission does not occur between wild type host [30], although colonization of hosts can be affected by the different physiological responses between mice and rats [20] and/or B. bronchiseptica strain variations [4]. Alternatively, B. bronchiseptica recovered from rats in this study could be transmitted by other animals in the market, as the bacterium is highly contagious among poultry and livestock [30]. This species can also survive in the environment and on inanimate surfaces [2, 30], possibly aiding the spread and transfer to other mammals. nrdA typing identified isolate TRE152202 as Bordetella genogroup 16 possessing nrdA locus 189 after the inability of 16S rDNA sequencing and MLST to resolve its species identity (Table 1). We have previously isolated a similar bordetellae with nrdA locus 189 [21] and this isolate was recently classified as B. pseudohinzii [19]. This species can be differentiated from the other bordetellae by the amplification of the Type II-C CRISPR-Cas system (Fig. 2), exclusively found in B. pseudohinzii [15]. Initially found only in laboratory mice [15, 16, 19, 21], we have now established that B. pseudohinzii also reside in the lung of wild rats, although the clinical significance remains unclear.
The nrdA-based phylogeny [28] (Fig. 4) displayed a congruent clustering of Bordetella complexes with the MLST-based phylogeny. The first cluster comprised B. bronchiseptica complex IV and B. pertussis complex II, and the second cluster comprised B. bronchiseptica complex I and B. parapertussis complex III. The bordetellae population structure inferred from nrdA typing concurred with the MLST-based phylogeny, affirming that B. parapertussis and B. pertussis evolved from B. bronchiseptica complex I and IV, respectively [6]. In addition, the non-classical bordetellae formed a separate cluster, demonstrating excellent Bordetella subspecies demarcation using the nrdA-based phylogeny [28] (Fig. 4).
Based on the MLST Bordetella phylogeny (Fig. 3), the novel ST82 clustered together with other clones from complex I [6], which included isolates from humans and animals. A high possibility of zoonotic transmission can be anticipated with ST82 isolates as they are closely related to other STs (ST4, ST7, ST11, ST12, ST23, ST27 and ST55) (Fig. 3) which were recovered from humans [4, 6, 27]. Transmission from infected animals to humans can occur through inhalation of aerosols or by contact with respiratory secretions [10]. Animals may not show apparent clinical symptoms of B. bronchiseptica infection however, human-adapted strains may modulate their virulence differently and lead to respiratory illnesses [4], augmenting the risk to humans. ST27 from a household cat, for example, was strongly suspected to be the source of B. bronchiceptica infection in a child [27] and a B. bronchiseptica strain originating from companion animals was suggested to infect and, transmit between humans who were in close contact with each other [14]. Human to human transmission, hence, is possible via contact with respiratory droplets [10].
Antimicrobial susceptibility testing revealed that B. bronchiseptica and B. pseudohinzii had almost similar antimicrobial profiles, reflecting the close phylogenetic relationships between the two bordetellae. The only antimicrobial susceptibility variance between B. bronchiseptica and B. pseudohinzii was observed using the macrolide; erythromycin, where it was ineffective against B. bronchiseptica although it was reported to be effective for the treatment of most Bordetella clinical infections [31], including B. pseudohinzii in this study. Resistance to erythromycin could be an innate phenotype of B. bronchiseptica as this was also observed among B. bronchiseptica isolates from pigs with respiratory diseases in China [33]. The anti-pseudomonal cephalosporin; ceftazidime, was effective for the eradication of B. bronchiseptica and B. pseudohinzii, similar to other reports [21, 32]. The isolates were however, susceptible to the third generation cephalosporins, cefotaxime and ceftriaxone, in agreement with a previous case of suspected B. bronchiseptica transmission from a kitten [26] and B. pseudohinzii BH370 recovered from ICR mouse [21]. Both bordetellae appeared to be susceptible to aminoglycosides (amikacin and gentamicin), carbapenem (imipenem and meropenem) and fluoroquinolone (ciprofloxacin), comparable to B. bronchiseptica human infections [11, 14, 32] and B. pseudohinzii BH370 [21]. In accordance with a previous study on human clinical isolates [32], our B. bronchiseptica isolates displayed identical resistance towards ampicillin and amoxicillin-clavulanic acid. B. pseudohinzii BH370 [21], 8-296-03T [16] and TRE152202 (this study) exhibited analogous resistance to ampicillin and even though 8-296-03T was not tested against amoxicillin-clavulanic acid [16], it was expected to display resistance similar to BH370 and TRE152202, suggesting that the difference in hosts did not influence antimicrobial properties. The nucleic acid amplification of dnt, bfrZ and fla from B. bronchiseptica but not B. pseudohinzii may reflect the roles of these genes in host colonization [29], catering to the broad host range for B. bronchiseptica.
Considered together, the much overlooked bordetellae surveillance despite the low prevalence among wild rats in Malaysia is a cause for concern as bordetellae such as B. bronchiseptica can be highly contagious in nature [30], persistent [12] and has a wide host range [2]. The persistence of this species in the host predisposes it to progressively acquire antimicrobial resistance [11] that could exacerbate pre-existing clinical conditions [27]. Upon browsing the Bordetella MLST database at http://pubmlst.org/bordetella/, we noticed that the novel B. bronchiseptica ST82 was the sole complex I representative from Asia, reflecting the neglected state of research on this species. The significant finding of B. pseudohinzii in wild rat is important as this species was previously thought to specifically infect laboratory mice [16]. Although the clinical significance remains unknown and prevalence is low, the potential for pathogen transmission from the wild to the laboratory is possible and this could have serious implications for studies using these infected animals. It is therefore important to study the genetic composition and prevalence of bordetellae in urban rats for comparison with other geographical regions to ensure early identification of pathogens capable of causing public health disorder.
Acknowledgments
This work was supported by the University of Malaya Research Grant (RP016C-14AFR and RU005-2017) and the High Impact Research-Ministry of Higher Education Grant (E000013-20001).
REFERENCES
- 1.Alias S. N., Sahimin N., Edah M. A., Mohd-Zain S. N.2014. Epidemiology of blood parasitic infections in the urban rat population in peninsular Malaysia. Trop. Biomed. 31: 230–240. [PubMed] [Google Scholar]
- 2.Bemis D. A., Shek W. R., Clifford C. B.2003. Bordetella bronchiseptica infection of rats and mice. Comp. Med. 53: 11–20. [PubMed] [Google Scholar]
- 3.Benacer D., Mohd Zain S. N., Amran F., Galloway R. L., Thong K. L.2013. Isolation and molecular characterization of Leptospira interrogans and Leptospira borgpetersenii isolates from the urban rat populations of Kuala Lumpur, Malaysia. Am. J. Trop. Med. Hyg. 88: 704–709. doi: 10.4269/ajtmh.12-0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buboltz A. M., Nicholson T. L., Weyrich L. S., Harvill E. T.2009. Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica. Infect. Immun. 77: 3969–3977. doi: 10.1128/IAI.01362-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2016. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Sixth Informational (Supplement): M100–S26. [Google Scholar]
- 6.Diavatopoulos D. A., Cummings C. A., Schouls L. M., Brinig M. M., Relman D. A., Mooi F. R.2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1: e45. doi: 10.1371/journal.ppat.0010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt M., Raoult D.2016. Molecular history of plague. Clin. Microbiol. Infect. 22: 911–915. doi: 10.1016/j.cmi.2016.08.031 [DOI] [PubMed] [Google Scholar]
- 8.Forbes K. M., Henttonen H., Hirvelä-Koski V., Kipar A., Mappes T., Stuart P., Huitu O.2015. Food provisioning alters infection dynamics in populations of a wild rodent. Proc. Biol. Sci. 282: 20151939. doi: 10.1098/rspb.2015.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francisco A. P., Vaz C., Monteiro P. T., Melo-Cristino J., Ramirez M., Carriço J. A.2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13: 87. doi: 10.1186/1471-2105-13-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-de-la-Fuente C., Guzmán L., Cano M. E., Agüero J., Sanjuán C., Rodríguez C., Aguirre A., Martínez-Martínez L.2015. Microbiological and clinical aspects of respiratory infections associated with Bordetella bronchiseptica. Diagn. Microbiol. Infect. Dis. 82: 20–25. doi: 10.1016/j.diagmicrobio.2015.01.011 [DOI] [PubMed] [Google Scholar]
- 11.Goldberg J. D., Kamboj M., Ford R., Kiehn T. E., Gilhuley K., Perales M. A.2009. ‘Kennel cough’ in a patient following allogeneic hematopoietic stem cell transplant. Bone Marrow Transplant. 44: 381–382. doi: 10.1038/bmt.2009.22 [DOI] [PubMed] [Google Scholar]
- 12.Gueirard P., Weber C., Le Coustumier A., Guiso N.1995. Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J. Clin. Microbiol. 33: 2002–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiso N., Hegerle N.2014. Other Bordetellas, lessons for and from pertussis vaccines. Expert Rev. Vaccines 13: 1125–1133. doi: 10.1586/14760584.2014.942221 [DOI] [PubMed] [Google Scholar]
- 14.Huebner E. S., Christman B., Dummer S., Tang Y. W., Goodman S.2006. Hospital-acquired Bordetella bronchiseptica infection following hematopoietic stem cell transplantation. J. Clin. Microbiol. 44: 2581–2583. doi: 10.1128/JCM.00510-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov Y. V., Shariat N., Register K. B., Linz B., Rivera I., Hu K., Dudley E. G., Harvill E. T.2015. A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 16: 863. doi: 10.1186/s12864-015-2028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov Y. V., Linz B., Register K. B., Newman J. D., Taylor D. L., Boschert K. R., Le Guyon S., Wilson E. F., Brinkac L. M., Sanka R., Greco S. C., Klender P. M., Losada L., Harvill E. T.2016. Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int. J. Syst. Evol. Microbiol. 66: 5452–5459. doi: 10.1099/ijsem.0.001540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiyipong T., Morand S., Jittapalapong S., Raoult D., Rolain J. M.2013. Bordetella hinzii in rodents, Southeast Asia. Emerg. Infect. Dis. 19: 502–503. doi: 10.3201/eid1903.120987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loong S. K., Khor C. S., Jafar F. L., AbuBakar S.2016b. Utility of 16S rDNA sequencing for identification of rare pathogenic bacteria. J. Clin. Lab. Anal. 30: 1056–1060. doi: 10.1002/jcla.21980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loong S. K., Tan K. K., Sulaiman S., Wong P. F., AbuBakar S.2017. Draft genome of Bordetella pseudohinzii BH370 isolated from trachea and lung tissues of a laboratory mouse. Genom. Data 12: 69–70. doi: 10.1016/j.gdata.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loong S. K., Mahfodz N. H., Che Mat Seri N. A., Mohamad Wali H. A., Abd Gani S. A., Wong P. F., AbuBakar S.2016c. Genetic characterization of commensal Escherichia coli isolated from laboratory rodents. Springerplus 5: 1035. doi: 10.1186/s40064-016-2745-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loong S. K., Mahfodz N. H., Wali H. A., Talib S. A., Nasrah S. N., Wong P. F., Abubakar S.2016d. Molecular and antimicrobial analyses of non-classical Bordetella isolated from a laboratory mouse. J. Vet. Med. Sci. 78: 715–717. doi: 10.1292/jvms.15-0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loong S. K., Johari J., Che Mat Seri N. A. A., Abdulrazak O., Douadi B., Ahmad Nasrah S. N., Mohd Zain S. N., AbuBakar S.2016a. Isolation and identification of an emerging pathogen, Kocuria marina, from Rattus rattus diardii. Trop. Biomed. 33: 589–593. [PubMed] [Google Scholar]
- 23.Meerburg B. G., Singleton G. R., Kijlstra A.2009. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 35: 221–270. doi: 10.1080/10408410902989837 [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2002. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals. Approved Standard-Second Addition: M31-A2. National Committee for Clinical Laboratory Standards, Wayne. [Google Scholar]
- 25.Ngom A., Boulanger D., Ndiaye T., Mboup S., Bada-Alambedji R., Simondon F., Ayih-Akakpo A. J.2006. Domestic animals as carriers of Bordetella species in Senegal. Vector Borne Zoonotic Dis. 6: 179–182. doi: 10.1089/vbz.2006.6.179 [DOI] [PubMed] [Google Scholar]
- 26.Redelman-Sidi G., Grommes C., Papanicolaou G.2011. Kitten-transmitted Bordetella bronchiseptica infection in a patient receiving temozolomide for glioblastoma. J. Neurooncol. 102: 335–339. doi: 10.1007/s11060-010-0322-6 [DOI] [PubMed] [Google Scholar]
- 27.Register K. B., Sukumar N., Palavecino E. L., Rubin B. K., Deora R.2012. Bordetella bronchiseptica in a paediatric cystic fibrosis patient: possible transmission from a household cat. Zoonoses Public Health 59: 246–250. doi: 10.1111/j.1863-2378.2011.01446.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spilker T., Leber A. L., Marcon M. J., Newton D. W., Darrah R., Vandamme P., Lipuma J. J.2014. A simplified sequence-based identification scheme for Bordetella reveals several putative novel species. J. Clin. Microbiol. 52: 674–677. doi: 10.1128/JCM.02572-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stępniewska K., Urbaniak K., Markowska-Daniel I.2014. Phenotypic and genotypic characterization of Bordetella bronchiseptica strains isolated from pigs in Poland. Pol. J. Vet. Sci. 17: 71–77. [DOI] [PubMed] [Google Scholar]
- 30.Trainor E. A., Nicholson T. L., Merkel T. J.2015. Bordetella pertussis transmission. Pathog. Dis. 73: ftv068. doi: 10.1093/femspd/ftv068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernli D., Emonet S., Schrenzel J., Harbarth S.2011. Evaluation of eight cases of confirmed Bordetella bronchiseptica infection and colonization over a 15-year period. Clin. Microbiol. Infect. 17: 201–203. doi: 10.1111/j.1469-0691.2010.03258.x [DOI] [PubMed] [Google Scholar]
- 32.Woolfrey B. F., Moody J. A.1991. Human infections associated with Bordetella bronchiseptica. Clin. Microbiol. Rev. 4: 243–255. doi: 10.1128/CMR.4.3.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z., Xue Y., Wang C., Ding K., Wu B., He Q., Cheng X., Chen H.2011. Antimicrobial susceptibility of Bordetella bronchiseptica isolates from pigs with respiratory diseases on farms in China. J. Vet. Med. Sci. 73: 103–106. doi: 10.1292/jvms.10-0184 [DOI] [PubMed] [Google Scholar]