Abstract
Folate receptors (FRα, FRβ and FRγ) are cysteine-rich cell-surface glycoproteins that bind folate with high affinity to mediate cellular uptake of folate. Although expressed at very low levels in most tissues, folate receptors, especially FRα, are expressed at high levels in numerous cancers to meet the folate demand of rapidly dividing cells under low folate conditions1–3. The folate dependency of many tumours has been therapeutically and diagnostically exploited by administration of anti-FRα antibodies, high-affinity antifolates4,5, folate-based imaging agents and folate-conjugated drugs and toxins6–8. To understand how folate binds its receptors, we determined the crystal structure of human FRα in complex with folic acid at 2.8 Å resolution. FRα has a globular structure stabilized by eight disulphide bonds and contains a deep open folate-binding pocket comprised of residues that are conserved in all receptor subtypes. The folate pteroate moiety is buried inside the receptor, whereas its glutamate moiety is solvent-exposed and sticks out of the pocket entrance, allowing it to be conjugated to drugs without adversely affecting FRα binding. The extensive interactions between the receptor and ligand readily explain the high folate-binding affinity of folate receptors and provide a template for designing more specific drugs targeting the folate receptor system.
Folates (vitamin B9) are important one-carbon donors for the synthesis of purines and thymidine—essential components of nucleic acids—and indirectly, via S-adenosyl methionine, for methylation of DNA, proteins and lipids9. Folate deficiency is therefore associated with many diseases, including fetal neural tube defects, cardiovascular disease and cancers10. In adult tissues, folate is mainly taken up by reduced folate carrier, a ubiquitously expressed anion channel that has relatively low folate-binding affinity (Km = 1–10 μM)11. By contrast, high-affinity uptake of the food supplement folic acid (Kd < 1 nM)12 and the physiologically prevalent folate N5-methyltetrahydrofolate (5-mTHF) requires the function of three subtypes of folate receptor (FRα, FRβ and FRγ), which are cysteine-rich glycoproteins that mediate folate uptake through endocytosis. Inside of the cell, the acidic environment of the endosome promotes the release of folate from receptors, which is then transported into the cytoplasm by proton-coupled folate transporter13. The expression of folate receptors is largely restricted to cells important for embryonic development (for example, placenta and neural tubes) and folate resorption (kidney). Among the three FR isoforms, FRα is the most widely expressed, with very low levels in normal tissues, but high expression levels in many tumours14. As such, FRα has become the molecular target for the development of many cancer therapeutics, including anti-FRα antibodies, high-affinity anti-folates, folate-based imaging agents and folate-conjugated drugs and toxins. Despite intense research on the folate structure–activity relationship, the molecular basis for the high-affinity recognition of folates by FRα remains elusive owing to the technical difficulties in expression, purification and crystallization of FRα for structural studies.
To obtain FRα protein for structural studies, we stably expressed human FRα lacking its carboxy-terminal glycophosphatidylinositol anchor as a secreted IgG Fc fusion protein (FRα–Fc) in HEK293 cells. As fully glycosylated fusion protein purified from culture medium yielded poorly diffracting crystals, we reduced crystallization-inhibiting glycosylation heterogeneity by combined treatment with kifunensine and endoglycosidase H, which together reduce complex carbohydrates to single N-acetylglucosamine (NAG) moieties15 (Supplementary Fig. 1a, b). The deglycosylated FRα–Fc had a similar folic acid-binding affinity (~190 pM) to the fully glycosylated protein (Supplementary Fig. 1d) and yielded crystals, which diffracted to 2.8 Å (Supplementary Fig. 1c). We solved the structure by combining the phase information from one Pt derivative and six native S anomalous data sets (see Methods and Supplementary Table 1).
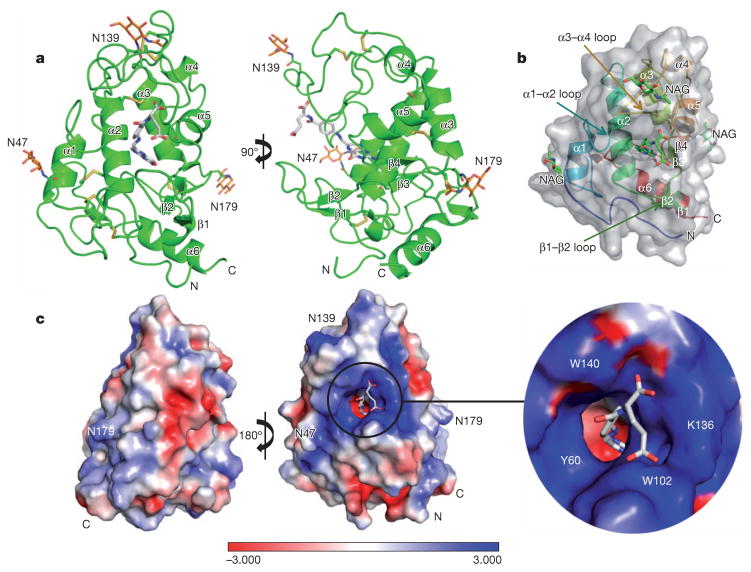
FRα has an overall globular structure, comprising four long α-helices (α1, α2, α3, α6), two short α-helices (α4, α5), four short β-strands (β1–β4) and many loop regions (Fig. 1a, b). The tertiary structure is greatly stabilized by eight disulphide bonds formed by 16 conserved cysteine residues (C15–C43, C35–C83, C44–C87, C67–C153, C74–C124, C113–C187, C117–C167 and C130–C147). FRα has three predicted N-glycosylation sites at N47, N139 and N179. Clear electron density for NAG is observed for N47 and N139, and partial electron density for N179. The overall fold of FRα is similar to that of riboflavin-binding protein (22% sequence identity to FRα)16, with a root mean squared deviation of 1.56 Å for 163 Cα atoms, but the two proteins have very differently shaped ligand pockets and ligand-binding modes (Supplementary Fig. 2).
Figure 1. Structure of FRα bound to folic acid.
a, Two views of the complex, with FRα in green, folic acid in grey, NAG in orange and the disulphide bonds depicted as yellow sticks. The N and C termini are labelled. b, Ribbon diagram of FRα, with folic acid and NAG in green stick presentations, overlaid with the semi-transparent receptor surface. c, Charge distribution surface of FRα with a close-up view of the ligand-binding pocket entrance. Folic acid carbon atoms are coloured grey, nitrogen atoms blue, and oxygen atoms red. A colour-code bar (bottom) shows an electrostatic scale from −3 to +3 eV.
The core domain consists of helices α1, α2, α3 and α5 tied together by four disulphide bridges (C35–C83, C44–C87, C74–C124 and C117–C167; Fig. 1a). The structure of FRα contains a long and open folate-binding pocket, which is formed by α1, α2 and α3 in the back; the amino-terminal loop, β1 and β2 in the bottom; the α1–α2 and α3–α4 loops in the left and top; and α4, α5, β4 and β3 in the right (Fig. 1a, b). Folic acid is oriented with its basic pteroate moiety docked deep inside of the negativelycharged pocket and the two negatively charged carboxyl groups of its glutamate moiety sticking out of the positively charged entrance of the ligand-binding pocket, which is formed by the α1–α2, β1–β2 and α3–α4 loops (Figs 1 and 2b).
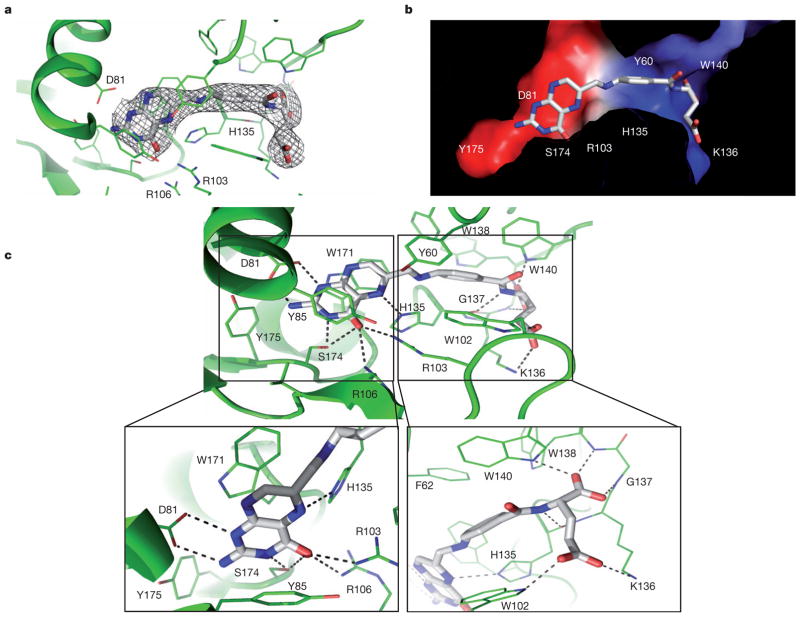
Figure 2. Structural and biochemical analysis of FRα–folic acid interactions.
a, The σA-weighted 2Fo – Fc electron density map for folic acid, shown as a grey mesh. b, The internal charge distribution surface of the binding pocket is shown using the same colour code as in Fig. 1c, with folic acid shown in stick presentation. c, Folic acid-binding network with close-ups of the folic acid head and tail groups. Residues that line the binding pocket are shown in green and folic acid is shown in grey. Hydrogen bonds are indicated by dashed lines.
Clear electron density was observed for folic acid and the surrounding amino acid residues, which allowed for accurate modelling of the ligand and its interacting residues lining the binding pocket (Fig. 2a and Supplementary Fig. 3). The cross-section of the binding pocket reveals the complementary shape and charge between the bound ligand and the receptor (Fig. 2b). Folic acid docks into an extended groove of FRα in the direction roughly perpendicular to the plane formed by helices α1, α2 and α3, with the pterin head group buried inside against the back formed by α1, α2 and α3 (Figs 1a and 2b, c). The interactions around the pteroate moiety contain both hydrogen bonds and hydrophobic interactions. First, the pterin ring is stacked between the parallel side chains of Y85 and W171, and capped by Y175 (Fig. 2c). Second, the hydrophilic pterin ring N and O atoms form a series of hydrogen bonds with receptor residues. Specifically, the pterin N1 and N2 atoms form strong hydrogen bonds with the side-chain carboxyl group of D81, the N3 and O4 atoms with the S174 hydroxyl group, the O4 atom forms two hydrogen bonds with the guanidinium groups of R103 and R106, and the N5 atom forms one hydrogen bond with the H135 side chain (Figs 2c and 3a). Interestingly, folic acid O4 is replaced by an amino group in the antifolates methotrexate and aminopterin, which have reduced affinity for FRα5,17. The amino group would not allow for the formation of hydrogen bonds with R103 and R106 and would sterically clash with the position of R103 (see Fig. 3a) in the folic acid-bound structure, providing a structural rationale for the poor FRα-binding of these two compounds and their preferential uptake by reduced folate carrier.
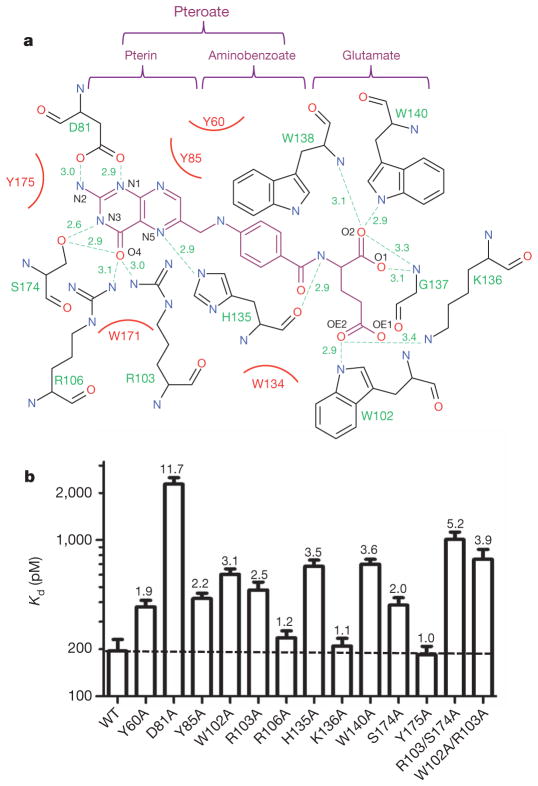
Figure 3. Folic acid affinities of FRα ligand-binding-pocket mutants.
a, Interaction map of folic acid with ligand-binding-pocket residues. The folic acid chemical structure is shown in magenta, pocket residues in black and hydrogen bonds as green dashed lines with bond distances (Å) indicated. Hydrophobic interactions are presented as curved red lines. The pteroate and glutamate moieties of folic acid are indicated above the map. b, Folic acid affinities of wild-type and mutant FRα proteins as measured by [3H]-folic acid binding assay (see Supplementary Figs 7 and 8 for binding isotherms). The numbers on top of the bars indicate the fold decrease in affinity (increase in Kd) relative to wild-type FRα. Error bars indicate s.d. (n –2).
The folic acid aminobenzoate is stabilized by hydrophobic interactions with Y60, W102 and W134, which line the middle of the long ligand-binding pocket (Fig. 3a). Extensive interactions are also observed for the glutamate group, which engages six hydrogen bonds, contributed by the side chains of W102, K136 and W140, as well as by backbone interactions with H135, G137 and W138 (Figs 2c and 3a). Most residues involved in ligand binding are identical among different subtypes of FR regardless of their origins (Supplementary Figs 4 and 5), indicating that the observed folate-binding interactions are probably conserved in all three different receptor subtypes. In addition, the most physiologically prevalent folate, 5-mTHF, can be easily docked into the FRα ligand-binding pocket in a mode very similar to that of folic acid, suggesting that the fundamental mechanism of folate recognition is conserved (Supplementary Fig. 6).
To validate the structure observations, we examined the ligand-binding affinities of FRα mutants that have alanine mutations in the key folate-contacting residues. The W171A mutation abolished the expression of the receptor (Supplementary Fig. 7a), suggesting that this residue is critical for protein stability. All other mutants expressed relatively well and were purified to determine their folate-binding affinity by radioligand-binding assay (Supplementary Figs 7b and 8b). Whereas wild-type FRα bound to [3H]-folic acid with a Kd of ~0.19 nM, replacement of D81 decreased affinity by more than one order of magnitude, consistent with the strong interaction of the aspartate carboxyl oxygens with the pterin N1 and N2 nitrogens, and indicating that this interaction is a key contributor to high-affinity ligand binding. By contrast, mutations of Y175, K136 and R106 (bond lengths ≥3.1 Å) have little effect, and mutations of all other ligand-binding residues (hydrogen bonds ≤3.0 Å) have only moderate effects on folic acid binding (affinity deceases of ≤3.6-fold), which are approximately additive for the double mutants R103A/S174A and W102A/R103A. This extensive interaction network therefore makes FRα–folic acid binding remarkably resistant to single amino acid substitutions (Fig. 3b and Supplementary Figs 7b and 8b). Together, the structural and mutational analyses present a structural rationale for the absolute requirement of the pterin group for anchoring folate in the binding pocket of the receptor and for the availability of the glutamate group for conjugation with drugs and imaging reagents18, without adversely affecting the interactions between receptor and ligand.
In summary, many cancers highly express FRα, which has therefore become an important target for receptor-mediated chemotherapy. How FR binds to folate and folate-conjugated drugs, however, has remained unknown. The FRα–folic acid complex structure illustrates how the receptor assumes a deep folate-binding pocket that is formed by conserved residues across all receptor subtypes and provides detailed insights into how folic acid interacts with its receptors. Together, these observations establish a rational foundation for designing specific drugs targeting the folate receptor system.
METHODS
Protein expression and purification
The human FRα (residues 23–234) complementary DNA excluding the secretion signal peptide (residues 1–22) and glyco-phosphatidylinositol anchor signal peptide (residue 235–257) was expressed as a human IgG Fc fusion protein from the expression vector pcDNA6. This construct also contained a murine Igκ leader sequence at the N terminus to allow target protein secretion into media supernatant, a thrombin cleavage site between FRα and Fc, and a His6 tag after the Fc tag. For small-scale expression, HEK293 cells were transiently transfected with the FRα–Fc DNA. Media supernatants were collected after 4 days and dialysed against 20 mM Tris, pH 8.0, 0.15 M NaCl, 5% glycerol before nickel-nitrilotriacetic acid (Ni-NTA) chromatography. For large-scale purification, a stable HEK293 cell line expressing FRα–Fc was established by selection of HEK293 cells transiently transfected with FRα–Fc DNA in the presence of 10 μg ml−1 blasticidin (Invitrogen). Single colonies were grown in 24-well plates and expression of secreted FRα–Fc fusion protein in cell media supernatants was examined by biolayer interferometry using an Octet Red instrument (ForteBio) and by immunoblot analysis.
For large-scale purifications, a stable clone was maintained in 500 ml of DMEM supplemented with 5% fetal bovine serum, 20 mM HEPES, 5 μM kifunensine and 200 μM folic acid in one-litre roller bottles at 37 °C. Two litres of conditioned media were collected, concentrated to 400 ml and dialysed against buffer C (25 mM Tris, pH 8.0, 150 mM NaCl, 1 μM folic acid) at 4 °C overnight before loading on a 50-ml Ni-chelating Sepharose column (GE Healthcare). The column was washed with 300 ml buffer A (25 mM Tris, pH 8.0, 150 mM NaCl, 25 mM imidazole, 10% glycerol, 1 μM folic acid) and eluted with buffer A plus 500 mM imidazole. Peak fractions were pooled, digested with thrombin at a 1:1,000 mass ratio during overnight dialysis against buffer C at 4 °C to remove imidazole, and loaded on a 5-ml Ni-chelating Sepharose column (GE Healthcare) to remove the Fc His6 tag. The flow-through was collected, adjusted to pH 5.6 and deglycosylated with endoglycosidase Hf (New England Biolabs). Deglycosylated protein was finally separated by Sephadex S-200 gel filtration in 25 mM Tris, pH 8.0, 200 mM ammonium acetate, 1 mM EDTA and 1 μM folic acid. The protein eluted from the gel-filtration column at a volume corresponding to the size of a monomer at a purity >95% as judged by SDS–PAGE (Supplementary Fig. 1).
Crystallization
Purified FRα protein was concentrated to about 7 mg ml−1 before crystallization trials. Initial screening identified that polyethylene glycol (PEG) is favourable for crystal formation. Optimization trays using PEG were set up manually using the hanging drop method at 20 °C. Needle-shaped crystals were obtained, which diffracted X-rays to about 9–10 Å. To reduce glycosylation, FRα protein was expressed in the presence of 5 μM kifunensine (GlycoSyn)15 and purified FRα protein was further deglycosylated with endoglycosidase Hf (New England Biolabs) (Supplementary Fig. 1). Crystals were grown at 20 °C in hanging drops containing 1.5 μl of the purified protein and 1 μl of well solution (0.1 M MES, pH 6.5, 12% (v/v) PEG 2000, 0.15 M potassium sodium tartrate). Crystals appeared within 5–6 days and grew to a dimension of ~250 μm in length with a hexagonal shape by day 14. These crystals diffracted to 2.8 Å at the Advanced Photon Source (APS) synchrotron, Life Sciences Collaborative Access Team (LS-CAT).
Data collection and structure determination
Crystals were transferred to well solution with 20% (v/v) ethylene glycol as a cryoprotectant before flash freezing in liquid nitrogen. Data collection was performed at sector 21-ID-D (LS-CAT) of the APS synchrotron using single native crystals and the diffraction data were processed with HKL2000 (ref. 23). On the basis of Matthew’s coefficient calculation, the crystals have an unusually large unit cell with an estimate of 8–10 molecules per asymmetric unit. Initial structure determination by molecular replacement using riboflavin-binding protein (which shares 22% sequence identity with FRα) as a search model failed to yield any correct solution. To solve the phase problem, a heavy-atom derivative was prepared by soaking the native FRα crystals with a Pt salt before data collection. Also, six data sets of native FRα were collected at a wavelength of 1.77 Å to measure the S anomalous signal to aid in structure determination. These six data sets were processed using XDS24, combined using Pointless, and merged using Scala of the CCP4 suite25 as previously described26. Merging multiple data sets increased the S anomalous signal and redundancy of the data, but also led to an increase of the merging R-factor26. Initial phases were established by using the SHELX program19 with Pt-soaked derivative data and native data (Supplementary Table 1). Fifteen Pt atoms were found by SHELXD with a CC/CCweak score of 31.4/17.1 (CC is the correlation coefficient between Ecalc and Eobs for all data and CCweak is the correlation coefficient for 30% of reflections that were not used during the dual-space refinement). Subsequent phasing using SHELXE generated a contrast score of 0.8 and connectivity of 0.79 for the correct hand solution. Density modification for the initial electron density map was performed using DM20. A crude model was built automatically using the CCP4 program buccaneer and improved by manual building using Coot21. Phases were further improved by using the S anomalous data and a total of 29 S atoms were found based on the anomalous difference Fourier using the Phenix program27. The initial FRα model was manually adjusted on the basis of the electron density map using the riboflavin-binding protein structure as a reference and the improved model allowed accurate location of eight molecules in one asymmetric unit by molecular replacement (Supplementary Fig. 9). The models were refined against the native data with eight-fold non-crystallographic symmetry restraints using the Refmac program of CCP4 (ref. 22). The densities for folic acid became clear after several rounds of model adjustments and refinements and eight molecules of folate were built into the model. The final model was refined to an R factor of 0.206 and an Rfree factor of 0.256 (Supplementary Table 1). The Ramachandran statistics are 87% in the favoured regions, 12.5% in additional allowed regions and 0.5% in generously allowed regions.
Mutagenesis
Site-directed mutagenesis was carried out using the QuickChange method (Stratagene). Mutations and all plasmid constructs were confirmed by DNA sequencing.
Radioligand-binding assay
The binding affinity of each FRα mutant was determined by saturation radioligand-binding assay. 40 nM of each mutant in 100 μl binding buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Triton X-100) was immobilized in the wells of a protein G-coated 96-well plate (Thermo Scientific) for 40 min. Endogenous ligand was stripped with 100 μl stripping buffer (25 mM acetate acid, pH 3.5, 150 mM NaCl, 0.1% Triton X-100) for 1 min as described previously28. After neutralizing and washing with 200 μl binding buffer, proteins were incubated for 40 min with 100 μl binding buffer supplemented with the indicated concentrations of [3H]-folic acid (Moravek Biochemicals). FRα-bound [3H]-folic acid was determined by scintillation counting following removal of unbound ligand by two 100-μl washes with binding buffer. Kd was determined by nonlinear regression using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Y. Jones for the pHL-Fc plasmid and H. L. Monaco for providing the chicken riboflavin-binding protein coordinates. The atomic coordinates have been deposited in the Protein Data Bank with accession codes listed in Supplementary Table 1. We thank staff members of the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under contract no. DE-AC02-06CH11357. This work was supported by the Jay and Betty Van Andel Foundation, and work by the Yong, Xu and Melcher laboratories is supported by the American Asthma Foundation, Ministry of Science and Technology (China) grants 2012ZX09301001-005 and 2012CB910403, Amway (China), by National Institutes of Health grants R01 DK071662 (H.E.X.) and R01 GM102545 (K.M.), and by the National Research Foundation Singapore under its Clinician Scientist Award NMRC/CSA/026/2011 (E.-L.Y.). C.C. is recipient of the NUS Graduate School for Integrative Sciences and Engineering Scholarship.
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Supplementary Information is available in the online version of the paper.
Author Contributions E.-L.Y., J.L., J.K., H.E.X. and K.M. conceived the project and designed research. C.C., J.K., X.E.Z., W.Y. and J.S.B. performed research. C.C., J.K., H.E.X. and K.M. wrote the paper with contributions from all authors.
The structure of FRα bound to folic acid has been deposited in the Protein Data Bank under the accession code 4LRH.
References
- 1.Kelemen LE. The role of folate receptor α in cancer development, progression and treatment: cause, consequence or innocent bystander? Int J Cancer. 2006;119:243–250. doi: 10.1002/ijc.21712. [DOI] [PubMed] [Google Scholar]
- 2.Kane MA, et al. Influence on immunoreactive folate-binding proteins of extracellular folate concentration in cultured human-cells. J Clin Invest. 1988;81:1398–1406. doi: 10.1172/JCI113469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsue H, et al. Folate receptor allows cells to grow in low concentrations of 5-methyltetrahydrofolate. Proc Natl Acad Sci USA. 1992;89:6006–6009. doi: 10.1073/pnas.89.13.6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuire JJ. Anticancer antifolates: current status and future directions. Curr Pharm Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- 5.Deng YJ, et al. Synthesis and biological activity of a novel series of 6-substituted thieno 2,3-d pyrimidine antifolate inhibitors of purine biosynthesis with selectivity for high affinity folate receptors over the reduced folate carrier and proton-coupled folate transporter for cellular entry. J Med Chem. 2009;52:2940–2951. doi: 10.1021/jm8011323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leamon CP, Reddy JA. Folate-targeted chemotherapy. Adv Drug Deliv Rev. 2004;56:1127–1141. doi: 10.1016/j.addr.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Leamon CP, et al. Preclinical antitumor activity of a novel folate-targeted dual drug conjugate. Mol Pharm. 2007;4:659–667. doi: 10.1021/mp070049c. [DOI] [PubMed] [Google Scholar]
- 8.Reddy JA, et al. Preclinical evaluation of EC145, a folate-vinca alkaloid conjugate. Cancer Res. 2007;67:4434–4442. doi: 10.1158/0008-5472.CAN-07-0033. [DOI] [PubMed] [Google Scholar]
- 9.Bailey LB, Gregory JF. Folate metabolism and requirements. J Nutr. 1999;129:779–782. doi: 10.1093/jn/129.4.779. [DOI] [PubMed] [Google Scholar]
- 10.Stover PJ. Physiology of folate and vitamin B12 in health and disease. Nutr Rev. 2004;62:S3–S12. doi: 10.1111/j.1753-4887.2004.tb00070.x. discussion S13. [DOI] [PubMed] [Google Scholar]
- 11.Zhao R, Matherly LH, Goldman ID. Membrane transporters and folate homeostasis: intestinal absorption and transport into systemic compartments and tissues. Exp Rev Mol Med. 2009;11:e4. doi: 10.1017/S1462399409000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antony AC. The biological chemistry of folate receptors. Blood. 1992;79:2807–2820. [PubMed] [Google Scholar]
- 13.Zhao R, et al. A role for the proton-coupled folate transporter (PCFT-SLC46A1) in folate receptor-mediated endocytosis. J Biol Chem. 2009;284:4267–4274. doi: 10.1074/jbc.M807665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elnakat H, Ratnam M. Distribution, functionality and gene regulation of folate receptor isoforms: implications in targeted therapy. Adv Drug Deliv Rev. 2004;56:1067–1084. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Chang VT, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monaco HL. Crystal structure of chicken riboflavin-binding protein. EMBO J. 1997;16:1475–1483. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratnam M, Freisheim J. In: Folic Acid Metabolism in Health and Disease. Picciano MF, editor. Wiley; 1990. pp. 91–120. [Google Scholar]
- 18.Leamon CP, DePrince RB, Hendren RW. Folate-mediated drug delivery: effect of alternative conjugation chemistry. J Drug Target. 1999;7:157–169. doi: 10.3109/10611869909085499. [DOI] [PubMed] [Google Scholar]
- 19.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowtan K. dm: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker N, et al. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338:284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.