Abstract
The Potyviridae is the largest family of RNA plant viruses, members of which have single-stranded, positive-sense RNA genomes and flexuous filamentous particles 680–900 nm long and 11–20 nm wide. There are eight genera, distinguished by the host range, genomic features and phylogeny of the member viruses. Genomes range from 8.2 to 11.3 kb, with an average size of 9.7 kb. Most genomes are monopartite but those of members of the genus Bymovirus are bipartite. Some members cause serious disease epidemics in cultivated plants. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Potyviridae, which is available at www.ictv.global/report/potyviridae.
Keywords: Potyviridae, ICTV Report, Taxonomy
Virion
The flexuous, filamentous particles are 680–900 nm long and 11–20 nm wide, with helical symmetry and a pitch of about 3.4 nm (Table 1). Particles of some viruses are longer in the presence of divalent cations than in the presence of EDTA. Virion sedimentation coefficient S20,w is 137–160S; density in CsCl is 1.31 g cm−3; extinction coefficient E0.1 %1cm, 260nm=2.4–2.7. Virions contain a core capsid protein (CP) of 30–47 kDa; the tip of the capsid may contain the virus-encoded proteins genome-linked protein (VPg) and helper-component proteinase (HC-Pro) [1]. Virions contain about 5 % RNA by weight [2]. Virions are moderately immunogenic; there are serological relationships among many members. Some monoclonal antibodies react with most aphid-transmitted potyviruses [3, 4].
Table 1. Characteristics of the family Potyviridae.
| Typical member: | potato virus Y-O (U09509), species Potato virus Y, genus Potyvirus |
|---|---|
| Virion | Non-enveloped, flexuous and filamentous capsid, 680–900 nm long and 11–20 nm in diameter with a single core capsid protein |
| Genome | 8–11 kb of positive-sense, single-stranded, usually monopartite RNA (bipartite in the genus Bymovirus) |
| Replication | Cytoplasmic, initiated in virus replication complexes on membranous vesicles at ER exit sites. Replication initiates at 6K2-induced ER-originated vesicles |
| Translation | Directly from genomic RNA |
| Host range | Plants (all virus genera). Most members are arthropod-borne but those of the genus Bymovirus are transmitted by plasmodiophorids |
| Taxonomy | Currently eight genera containing nearly 200 species |
Genome
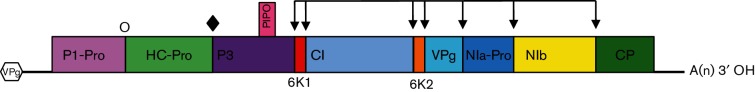
The single-stranded, positive-sense genome ranges from 8.2 kb (members of the species Artichoke latent virus, genus Macluravirus) to 11.3 kb (members of the species Wheat yellow mosaic virus, genus Bymovirus), with an average of around 9.7 kb (Table 2). The genomes have a VPg covalently linked to the 5′ end, and the 3′ terminus is polyadenylated. Most genomes are monopartite but those of members of the genus Bymovirus are bipartite. The genome of members of the species Potato virus Y, the type species of the genus Potyvirus, is organized as described in Fig. 1. The single large ORF of monopartite genomes encodes a single polyprotein that is cleaved into functional proteins at semi-conserved sites by three self-encoded proteases. Bipartite Bymovirus genomes encode two polyproteins that are cleaved by two proteases [5]. A second small ORF, PIPO, is generated by a polymerase slippage mechanism and is expressed as the trans-frame protein P3N-PIPO [6–8]. Coding region order and protein sequences are generally conserved throughout the family, although one of or both the P1 and the HC-Pro N-terminal coding regions may be lacking and a genus-specific or species-specific region may be present instead (Table 2). The coat protein of most isolates of the type species, Potato virus Y, contains 267 amino acids.
Table 2. Characteristics of members of the eight genera and two unassigned species in the family Potyviridae.
| Genus | Type species | Genome organization | Genome size range (kb) |
Host range | Vectors | Notable features (Alk-1, activin receptor-like kinase-1; HC-Pro, helper-component protease; NTR, non-translated region; P1, protein one; RNAi, RNA-interference) |
|---|---|---|---|---|---|---|
|
Brambyvirus
(1 species) |
Blackberry virus Y | Monopartite | 10.8 | Rubus species | Unknown | Alk1 domain encoded in a very large P1 coding region. HC-Pro lacks motifs for genome amplification and systemic movement. |
|
Bymovirus
(6 species) |
Barley yellow mosaic virus | Bipartite | RNA1 : 7.2–7.6 RNA2 : 2.2–3.6 |
Gramineae | Polymyxa graminis | Members lack P1 and HC-Pro coding regions. RNA2 encodes an HC-Pro-like protein unique to bymoviruses. |
|
Ipomovirus
(6 species) |
Sweet potato mild mottle virus | Monopartite | 9.0–10.8 | Wide | Whitefly (Bemisia tabaci) | Some members lack the P1 and/or HC-Pro coding regions and encode a P1b protein instead, performing a role in host RNAi suppression. |
|
Macluravirus
(8 species) |
Maclura mosaic virus | Monopartite | 8.2 | Wide | Aphids | No P1 coding region and the HC-Pro region is shorter than in potyviruses. |
|
Poacevirus
(3 species) |
Triticum mosaic virus | Monopartite | 9.7–10.2 | Gramineae and Orchidaceae | Wheat curl mite (TriMV) | Unusually long 5′ NTR with 12 translation initiation codons and three ORFs. |
|
Potyvirus
(160 species) |
Potato virus Y | Monopartite | 9.4–11.0 | Wide | Aphids | |
|
Rymovirus
(3 species) |
Ryegrass mosaic virus | Monopartite | 9.4–9.5 | Gramineae | Eriophyid mites | |
|
Tritimovirus
(6 species) |
Wheat streak mosaic virus | Monopartite | 9.2–9.6 | Gramineae | Eriophyid mites | P1 protein rather than HC-Pro serves as a suppressor of gene silencing. |
| Unassigned | Rose yellow mosaic virus | Monopartite | 9.5 | Rosa sp. | ||
| Unassigned | Spartina mottle virus | Unknown | Unknown | Gramineae | Unknown |
Fig. 1.
Genome organization of a typical member of the genus Potyvirus. Viruses of other genera may differ as described in Table 2. VPg, viral protein genome-linked; P1-Pro, protein 1 protease; HC-Pro, helper component protease; P3, protein 3; PIPO, pretty interesting Potyviridae ORF; 6K, six kilodalton peptide; CI, cytoplasmic inclusion; NIa-Pro, nuclear inclusion A protease; NIb, nuclear inclusion B RNA-dependent RNA polymerase; CP, coat protein. Cleavage sites of P1-Pro (O), HC-Pro (♦) and NIa-Pro (↓) are indicated.
Replication
Viruses are transmitted horizontally by arthropods or plasmodiophorids; some are transmitted vertically in seed. Some members cause serious disease epidemics in cultivated plants. Members of a few species infect over 30 plant families, but most infect one or a few host species or families [9] (Table 2).
Taxonomy
The family is divided into eight genera, the members of which are distinguished by host range, genomic features and phylogeny (Table 2). The species demarcation criteria, based upon the large ORF or its protein product, are generally accepted as <76 % nucleotide identity and <82 % amino acid identity. If the complete ORF sequence is not available, similar criteria can be used for the coat protein coding region and its product. The corresponding thresholds for species demarcation using nucleotide identity values for other coding regions range from 58 % (P1 coding region) to 74–78 % (other regions), although these ranges are exceeded in some cases [10].
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/potyviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Michael J. Adams, Donald B. Smith, Richard J. Orton and Hélène Sanfaçon.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Torrance L, Andreev IA, Gabrenaite-Verhovskaya R, Cowan G, Mäkinen K, et al. An unusual structure at one end of potato potyvirus particles. J Mol Biol. 2006;357:1–8. doi: 10.1016/j.jmb.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Adams MJ, Zerbini FM, French R, Rabenstein F, Stenger DC, et al. Potyviridae. In: King AM, editor. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. London, UK: Elsevier; 2011. (editor) [Google Scholar]
- 3.Shukla DD, Ward CW. Identification and classification of potyviruses on the basis of coat protein sequence data and serology. Brief review. Arch Virol. 1989;106:171–200. doi: 10.1007/BF01313952. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Moya JJ, Valli A, Garcia JA. Potyviridae. Chichester UK: John Wiley & Sons; 2009. [Google Scholar]
- 5.Adams MJ, Antoniw JF, Beaudoin F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol. 2005;6:471–487. doi: 10.1111/j.1364-3703.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung BY, Miller WA, Atkins JF, Firth AE. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA. 2008;105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015;16:995–1004. doi: 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, et al. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol. 2015;89:6965–6967. doi: 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla DD, Ward CW, Brunt AA. The Potyviridae. UK: CAB International; 1994. [Google Scholar]
- 10.Adams MJ, Antoniw JF, Fauquet CM. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol. 2005;150:459–479. doi: 10.1007/s00705-004-0440-6. [DOI] [PubMed] [Google Scholar]