Abstract
Zika and dengue viruses belong to the Flavivirus genus, a close group of antigenically related viruses that cause significant arthropod‐transmitted diseases throughout the globe. Although infection by a given flavivirus is thought to confer lifelong protection, some of the patient's antibodies cross‐react with other flaviviruses without cross‐neutralizing. The original antigenic sin phenomenon may amplify such antibodies upon subsequent heterologous flavivirus infection, potentially aggravating disease by antibody‐dependent enhancement (ADE). The most striking example is provided by the four different dengue viruses, where infection by one serotype appears to predispose to more severe disease upon infection by a second one. A similar effect was postulated for sequential infections with Zika and dengue viruses. In this review, we analyze the molecular determinants of the dual antibody response to flavivirus infection or vaccination in humans. We highlight the role of conserved partially cryptic epitopes giving rise to cross‐reacting and poorly neutralizing, ADE‐prone antibodies. We end by proposing a strategy for developing an epitope‐focused vaccine approach to avoid eliciting undesirable antibodies while focusing the immune system on producing protective antibodies only.
Keywords: antibody neutralization, antibody‐dependent enhancement, flavivirus structure, particle heterogeneity, vaccine design
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology; Synthetic Biology & Biotechnology
Glossary
- Ab
antibody
- ADE
antibody‐dependent enhancement
- C
capsid protein
- DC‐SIGN
dendritic cell‐specific ICAM‐grabbing non‐integrin
- EDE
E dimer epitope
- E
envelope protein
- ELISA
enzyme‐linked immunosorbent assay
- EM
electron microscopy
- ER
endoplasmic reticulum
- Fab
antigen binding fragment
- FLE
fusion loop epitope
- gp
glycoprotein
- HI
hemagglutination inhibition
- HIV
human immunodeficiency virus
- IC50
50% inhibitory concentration
- LALA
leucine‐alanine‐leucine‐alanine
- Mab
monoclonal antibody
- M
membrane protein
- NHP
non‐human primates
- prM
precursor of membrane protein
- RNP
ribonucleoprotein
- scFv
single‐chain variable fragment
- sE
soluble envelope protein E
- TAM
Tyro3‐Axl‐Mer
- TGN
trans‐golgi network
- TIM
T‐cell immunoglobulin and mucin domain
- T
triangulation number
Introduction
Flaviviruses are a worldwide threat to public health, as exemplified by the global spread of dengue with an estimated 390 million annual infections 1, the explosive Zika virus epidemics across the Pacific, South and Central America since 2013 2, and the inherent danger of urban yellow fever in Africa and South America 3, 4. The major driver of these epidemics is the virus transmission by peridomestic Aedes aegypti mosquitoes, which constitute the amplification motor in urban viral transmission cycles from human to human 5. The expansion of urban environments with insufficient infrastructure in tropical and subtropical regions makes vector control extremely difficult and provides an ideal ground for the spread of Aedes‐transmitted viruses. In addition to the impact of urbanization, international travel and trade facilitate the introduction of vectors and viruses into new geographical environments, as exemplified by the dissemination of the Asian tiger mosquito Aedes albopictus in the Americas and in Europe 6. Similarly, the omnipresence of Culex mosquitoes and susceptible hosts has led to the expansion of West Nile virus throughout the Americas at the turn of the century 7. Other encephalitogenic flaviviruses such as Japanese encephalitis (also primarily transmitted by Culex mosquitoes) and tick‐borne encephalitis viruses further illustrate the broad range of flavivirus diseases and natural cycles.
Although numerous compounds with anti‐flaviviral activity have been identified (reviewed in reference 8), none has so far been developed for clinical application. Efficient vaccines are available for only a few flaviviruses—yellow fever, Japanese encephalitis, and tick‐borne encephalitis viruses 9, 10, 11. Despite decades of intensive efforts, an efficient vaccine against dengue virus is not available. The vaccine Dengvaxia, currently licensed in certain endemic countries, provides suboptimal protection and is not recommended for children under 9 years of age (reviewed in 12, 13).
We discuss here the structural aspects of the interaction of flavivirus particles with antibodies, in particular those features that relate to virus neutralization and antibody‐mediated mechanisms that may potentially aggravate disease. We do not discuss the antibody response against the non‐structural protein NS1, which can contribute to protection (by mechanisms that are unrelated to particle neutralization), but has also been implicated as a factor in dengue pathogenesis 14. Similarly, the patient's T‐cell response, which is another important aspect of protection and pathogenesis, is also outside the scope the present review.
We recently reviewed structural determinants that may contribute to the broad tropism of flaviviruses 15, which mainly derive from the heterogeneity of the viral particles as a result of their complex morphogenetic pathway. This heterogeneity is particularly important in the case of the dengue viruses. We also analyzed the impact of the dynamic behavior of the envelope protein and its potential for exposing alternative surfaces for the interaction with receptors for entry. Here we discuss those same properties of the flavivirus particle in relation to its interactions with the humoral immune system. We analyze the reasons for the double‐edged sword properties of antibodies against dengue virus—having a protecting role but also a disease enhancing potential—and conclude by providing elements for the design of engineered immunogens that avoid the drawbacks of the dengue virus humoral immune response in humans, while focusing on the induction of protective antibodies only. Such a subunit vaccine approach has the potential of protecting both against dengue and Zika viruses.
Early landmarks in flavivirus research
The name flavivirus derives from latin (flavus = yellow) in relation to yellow fever virus, the type‐species of the genus 16 and, in 1900, the first identified virus infecting humans 17. Yellow fever was also the first disease for which transmission by infected mosquitoes had been already proposed and demonstrated at the end of the 19th century 17, 18. A major breakthrough was the development in the late 1930s of an attenuated strain of the virus, called 17D 19, which is still today one of the most efficient vaccines against any virus 20. The determination of the nucleotide sequence of the yellow fever virus genome in 1985 21 opened the flavivirus molecular biology era. Flaviviruses were then assigned as a genus within a new family 22, separate from the arboviruses in the family Togaviridae. The new classification matched the “group B” arboviruses, which were serologically defined back in the 1950s by Casals and Brown 23 using hemagglutination inhibition assays. In these assays, all the flaviviruses tested exhibited considerable cross‐reactivity, demonstrating that they displayed common antigens. This broad cross‐reactivity contrasted with the neutralization patterns, which were much more specific for the flavivirus against which the antibodies were raised 24.
Flavivirus structural biology
The structure of flavivirus particles
Flaviviruses are small icosahedral enveloped viruses, composed of a positive‐stranded RNA genome (Fig 1A) and only three structural proteins: C (capsid), E (envelope), and prM (precursor of membrane) in immature virions or M (membrane) in mature virions (Fig 1B–E). The E protein mediates cellular attachment and membrane fusion after virus uptake by receptor‐mediated endocytosis. The membrane fusion process is triggered by the acidic pH in the endosomes and is driven by extensive conformational and oligomeric rearrangements of E that lead to the formation of a post‐fusion E trimer (Fig 2). Early landmarks in the structural biology of flaviviruses have been the high‐resolution structure of the envelope protein E of tick‐borne encephalitis virus 25 and of dengue virus type 2 in the pre‐fusion dimer form 26 and in their post‐fusion trimer conformations 27, 28 (Figs 1 and 2); a first cryo‐EM structure resolving the organization of the E dimers at the surface of mature dengue virus serotype 2 particles 29, the X‐ray structure of the prM/E heterodimer of dengue virus 30, and the cryo‐EM structure of the dengue virus serotype 2 immature particle 31. These were followed by the cryo‐EM structures of the mature particles of dengue virus serotypes 2 32 and 4 33 to a resolution of 3.5 Å and 4 Å, respectively. A number of important structures obtained by cryo‐EM were reported recently, such as the structures of Zika virus 34, 35 and Japanese encephalitis virus 36 particles also at around 4 Å resolution. In addition, numerous X‐ray structures of complexes of the E protein (or individual domains) with antibody fragments were reported, as well as cryo‐EM structures of the particles in complex with antibodies, which have been the focus of extensive recent reviews 37, 38, 39, 40.
Figure 1. Flavivirus particle assembly.
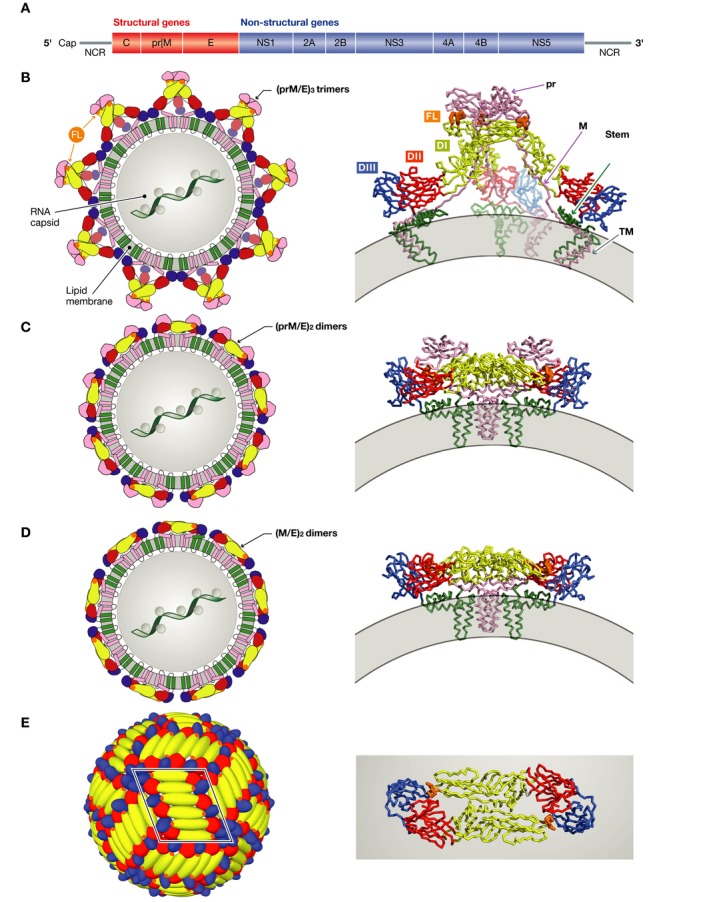
(A) The flavivirus open‐reading frame coding for a single precursor polyprotein. Co‐ and post‐translational proteolytic processing results in the various proteins indicated, with structural and non‐structural proteins in red and blue, respectively. (B–E) Sketches representing the flavivirus particle at different maturation stages (left), with a ribbon representation of the relevant envelope protein complexes on the right. The genomic ribonucleoprotein complex with protein C has not been visualized and is represented here within a central gray circle (in B–D), although its organization is unknown. Protein E is colored according to domains: red, yellow, blue, and green for domains I, II, III, and stem/TM (transmembrane anchor), respectively. The fusion loop is highlighted in orange, and prM/M (including its TM region) is shown in pink. The viral membrane is represented in gray. (B) Left: The immature flavivirus particle as it buds in the ER of the infected cells. Right: A single (prM/E)3 spike is displayed as ribbons (PDB code 4B03). (C) Left: The immature flavivirus particle after exposure to the acidic pH of the trans‐Golgi apparatus, where the trimeric spikes dissociate and the 180 prM/E heterodimers re‐associate into 90 (prM/E)2 dimers. Right: A single (prM/E)2 dimer is shown as ribbons (PDB codes 3C6R and 3JP2). (D) Left: The mature flavivirus particle with 90 (M/E)2 dimers. Right: A single (M/E) 2 dimer is shown in ribbons. (E) Left: Herringbone pattern of E dimers on the surface of mature virus particles, consisting of 30 rafts of three E dimers. One raft is framed in black. Right: Top view of a single E homodimer shown as ribbons.
Figure 2. The fusogenic conformational change of the E protein during cell entry.
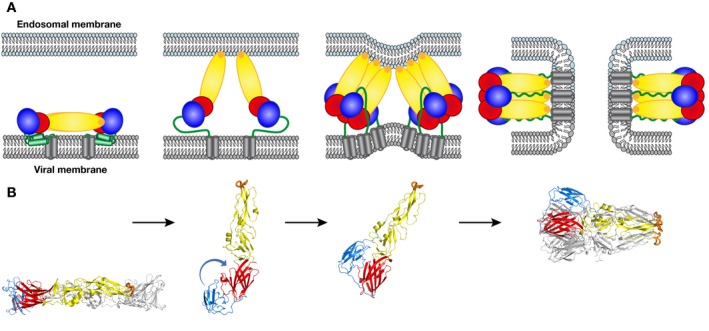
The E protein is colored as in Fig 1. (A) Schematic of the fusion process: A mature E dimer anchored in the viral membrane is represented in the left panel. The dimer dissociates upon exposure to acidic pH in the endosome, inserting the fusion loop into the endosomal membrane (second panel). The aligned E monomers then trimerize, thereby creating a binding site for domain III at the sides of a “core trimer”. Domain III then flips to the sides of the trimer, pulling the stem and TM segments toward the endosomal membrane (third panel). The final, post‐fusion conformation, brings the viral TM segment next to the fusion loop, inducing first hemi‐fusion (i.e., fusion of only the outer leaflets of the two membranes) followed by opening of a fusion pore (fourth panel). The final post‐fusion conformation of E is achieved only after fusion pore formation. (B) 3D structures of the dengue virus 2 E ectodomains (lacking the stem/TM regions) matching the steps indicated in (A) (PDB codes 1OAN and 1OK8 for pre‐fusion and post‐fusion conformations, respectively).
Flavivirus particles and pH sensitivity
The mature particle
The mature particles of flaviviruses all display the same architecture (Fig 1D and E), exposing 180 E proteins organized as dimers in a herringbone pattern. Contrary to what is often quoted, this icosahedral architecture does not conform to a T = 3 surface lattice (which would also yield 180 = T*60 subunits), as the E proteins do not follow the quasi‐equivalence principle 41, meaning that the three E proteins in the icosahedral asymmetric unit display non‐equivalent interactions. This feature is also relevant for antibody recognition, as a given epitope on protein E will be displayed in three different environments on the mature particle.
The fusogenic conformational change
The mature E dimer at the particle surface corresponds to a metastable form of the protein that is sensitive to the mildly acidic environment of the endosomes, reflecting the receptor‐mediated endocytic pathway of flavivirus entry 42. Conformational changes of E have been shown to be triggered already at the pH of early endosomes (6.5–6.0 43) but at least in the case of dengue virus, fusion may also depend on acidic lipids found in late endosomes 44. The pH sensitivity of flavivirus particles leads to their inactivation when exposed to an acidic environment in the absence of target cells 45, 46. Particle inactivation is caused by an exothermic, irreversible conformational change of E 27, 28 triggered upon binding of protons. When this change takes place in an endosome, it catalyzes the fusion reaction between viral and endosomal membranes. This step is essential for releasing the viral genome into the cytoplasm. The fusion mechanism (Fig 2A and B) includes dissociation of the E dimer, which then exposes the fusion loop at the tip of domain II to insert into the inner endosomal membrane leaflet. The two membranes are thereby bridged by the E proteins, which trimerize during this transient extended intermediate conformation. The subsequent re‐location of domain III to the side of the trimer projects the stem and the transmembrane region toward the tip of domain II, forcing the close apposition of viral and endosomal membranes at the same end of the post‐fusion trimer, thereby catalyzing membrane fusion (reviewed in reference 47). The final, stable trimer thus has the three E subunits in the characteristic hairpin conformation (Fig 2A and B, right panels) observed in the post‐fusion forms of all three characterized structural classes of viral fusion proteins 48. The E protein is therefore a “suicidal” catalyst that acts only once, after which it becomes non‐functional and is degraded in the lysosomes.
The acid sensitivity of the flavivirus particle also requires a built‐in mechanism to keep the inactivating conformational change of E from taking place prematurely, because the newly synthesized particles are exposed to acidic pH during their transit through the trans‐Golgi network (TGN) in the course of exocytosis 42. Flavivirus virions therefore assemble in the endoplasmic reticulum (ER) in an immature, non‐infectious form (Fig 1B and C), which is activated for membrane fusion only after proteolytic maturation in the acidic environment of the TGN 30, as described below.
Flavivirus particle assembly
In the infected cell, genome replication takes place within membrane invaginations at the cytosolic side of the ER 49. The newly synthesized genomic RNA associates with the core protein C (Fig 1) to form a ribonucleoprotein (RNP) complex. The role of protein C in this process and the organization of the resulting RNP are still not understood 50. Budding of the RNP across the ER membrane appears to be driven by the viral transmembrane glycoproteins prM and E, which can lead to budding of empty particles in the absence of the RNP (reviewed in 42). The two glycoproteins form heterodimers that in turn associate into (prM/E)3 trimeric spikes projecting into the ER lumen 42. The spikes interact laterally to induce curvature, resulting in formation of a closed icosahedrally symmetrical particle displaying 60 spikes (Fig 1B). These particles, which accumulate in the neutral pH ER lumen to then be transported across the exocytic pathway, are also pH sensitive. As shown for dengue virus, they react to the TGN's acidic environment by undergoing a reversible conformational change. The 60 (prM/E)3 trimers thus convert into 90 (prM/E)2 dimers (Fig 1C), generating the characteristic herringbone‐like arrangement of E, which remains in mature virions 30 (Fig 1E). Through this conformational change, prM exposes a specific cleavage site for furin, a TGN‐resident protease 51, which cleaves prM in the linker between its N‐terminal globular head (containing roughly 80 amino acids, termed pr) and the C‐terminal, transmembrane stub (also about 80 amino acids, termed protein M). This proteolytic step renders the conformational change of the particle irreversible. The pr globular domain remains bound as long as the pH is acidic, keeping E from undergoing the fusogenic conformational change. Its affinity for the E dimers drops substantially at neutral pH and is therefore shed from the virion upon release into the extracellular, neutral pH environment 30, giving rise to activated mature particles that are ready to infect other cells (Fig 1D and E).
Particle heterogeneity and dynamics
Two important main features derive from the complex flavivirus morphogenesis pathway and the metastability of the exposed E dimer, both of which are a requirement for the particle to react to the environmental pH:
Furin processing is often incomplete, resulting in mosaic particles (reviewed in reference 52) as documented for dengue virus 53, 54. The fusion loop and other conserved regions of the E protein are exposed in the immature areas of these particles (Fig 3A and B), with important consequences for the specificity of antibodies induced and their interaction with virus particles.
The reactivity to acid pH also implies that the E dimers in the “activated” form are metastable and display a dynamic behavior (reviewed in reference 55) commonly referred to as virus “breathing”. The kinetics of breathing are strain specific, and—like particle heterogeneity—affect the interaction with antibodies. Antibody binding to transiently exposed epitopes can also induce exposure of the fusion loop to insert into the plasma membrane for cell entry in the absence of a receptor, in a novel ADE mechanism described recently 56, or modulate antibody responses 57. This breathing also allows sporadic binding of circulating antibodies targeting the fusion loop in sub‐neutralizing stoichiometry, leading to entry via Fcγ receptors present in some cells in a classical ADE context.
Figure 3. The fusion loop is exposed in the immature (prM/E)3 spikes.
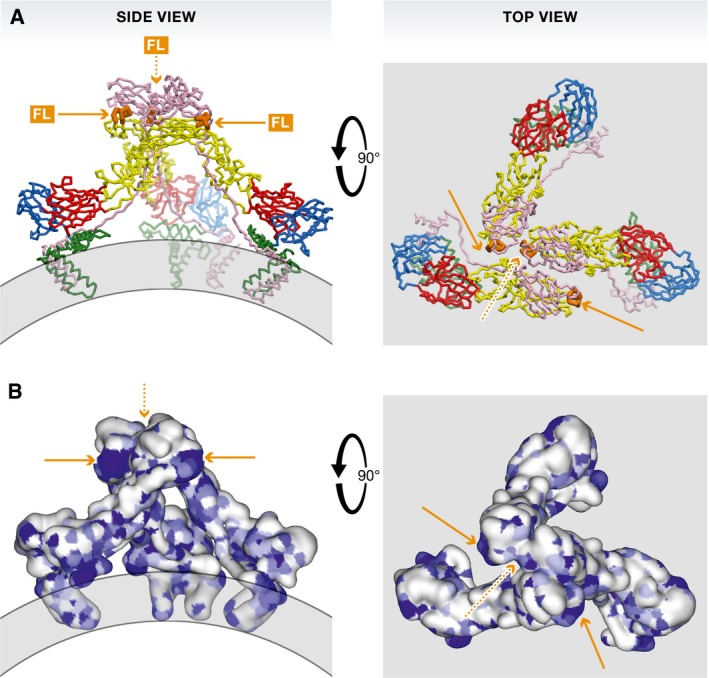
Left panels: side views, right panels: top views. (A) The spike shown in ribbons as in Fig 1. (B) The spike shown in a surface representation colored according to amino acid conservation across all flaviviruses, from white (variable) to dark blue (absolutely conserved). Patches corresponding to the fusion loop are indicated by orange arrows. One of the three fusion loops in the trimer is not exposed (orange dashed arrow).
Both of these properties, particle heterogeneity and dynamic “breathing” behavior, therefore contribute to the biological and antigenic characteristics of flaviviruses. Certain strains of dengue virus serotype 2 were shown to adopt a different, expanded conformation of their mature particles, termed “bumpy” 58, 59 upon incubation at temperatures above 34°C. This change was irreversible, unlike the “breathing” behavior discussed above, suggesting that in those particles the envelope protein was trapped in a local energy minimum such that the particles could be visualized by single particle cryo‐EM with icosahedral averaging. In other strains, this “bumpy” arrangement has not been observed, which may be due simply to the fact that such local minimum energy state is not always available, and the E dimers reversibly move about the local minimum in the herringbone conformation. The highly dynamic behavior also leads to a rapid particle inactivation, which is accelerated with temperature (reviewed in 55).
The possible reasons for flaviviruses to maintain these properties most probably have to do with interactions for entry, as the diversity of circulating forms may allow them to expand their host and tissue tropism, as discussed recently 15. The antigenic complexity conferred by particle heterogeneity and dynamics may also confer them an advantage in the vertebrate hosts by exploiting non‐protective antibody responses to favor subsequent heterologous infections.
The antigenic landscape of flaviviruses
Most of the residues exposed at the external surface of the E dimers in the mature herringbone lattice are not conserved and are specific to each virus (Fig 4A and B). Antibodies directed against this surface therefore potently neutralize the autologous and closely related viruses only. This criterion of cross‐neutralization by polyclonal sera has led to the classification of flaviviruses into serocomplexes 24.
Figure 4. The mature particle exposes a conserved patch.
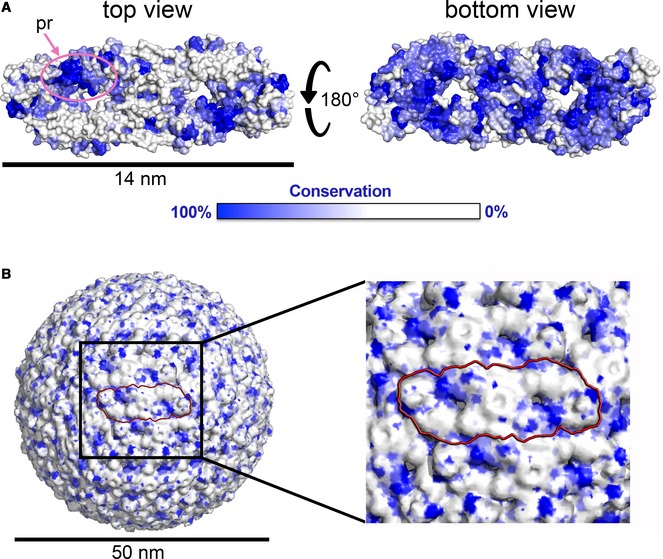
(A) The E dimer in surface representation colored according to amino acid conservation across all flaviviruses, in its top and bottom view. The pr binding site is encircled in pink. This site corresponds to the conserved E dimer epitope (EDE) discussed in the text. Notice the high conservation of the E dimer “underside” in the “bottom” view (right panel), which faces the viral membrane in the particle. (B) The mature flavivirus particle shown in surface representation, colored as in (A). One dimer is highlighted by a red contour. The black box indicates the region zoomed in the right panel.
Over the years, large panels of mostly murine monoclonal antibodies (Mabs) were obtained through immunization in a variety of ways, using recombinant fragments of protein E, or using inactivated virus and also live virus. Later, human Mabs were isolated from vaccinated or naturally infected individuals. The resulting Mabs revealed a broad range of neutralizing potencies, with the most potent Mabs neutralizing in the ng/ml range (reviewed in references 37, 39), indicating that polyclonal sera contain a mixture of antibodies with different degrees of neutralizing activities and cross‐reactivities.
A number of the epitopes of neutralizing Mabs were characterized structurally by X‐ray crystallography of Fab or scFv fragments in complex with the soluble E (sE) protein (i.e., lacking the stem and trans‐membrane regions), and by cryo‐EM of complexes with mature particles. The epitopes identified were generally conformational and comprised discontinuous segments of the E polypeptide chain. Some localized to single domains, such as those encompassing the lateral ridge of domain III—exemplified by the murine antibody E16 against West Nile virus 60. Epitopes have also been identified on domain I, like the epitope of the chimpanzee Mab 5H2 specific for dengue virus serotype 4 61. Other antibodies target the hinge region between domains I and II, such as Mab 1F4 which potently neutralizes dengue virus serotype 1 62. Other Mabs bind more complex assemblies and were found to be dependent on the quaternary structure of E, such as Mab 2D22, which is specific for dengue virus serotype 2 63 and the “EDE” antibodies described below. Finally, several antibodies were found to bind across E dimers arranged in the herringbone pattern, such as the West Nile virus‐specific Mab CR4354 64, the dengue virus serotype 1‐specific Mab 14c10 65, the dengue virus serotype 3‐specific Mab 5J7 66, and the Zika virus‐specific antibody ZIKV‐117 67.
Overall, the surface of flavivirus particles appears to display a continuum of epitopes with different degrees of structural complexity that can induce potently neutralizing antibodies.
EDE antibodies
Although the exposed E protein surface on mature virions is essentially variable and most strongly neutralizing Mabs are virus type‐specific, the E dimers also expose a vulnerable region, termed “EDE” for “E dimer epitope”, which is targeted by antibodies with a broader spectrum of viruses they potently neutralize 68. For instance, EDE antibodies can cross‐neutralize all four serotypes of dengue virus, and some can also potently neutralize Zika virus 69, which does not formally belong to the dengue serocomplex. The EDE is more conserved than other regions at the surface of E (Fig 4A), because it is the binding site of prM during the particle maturation process in the Golgi apparatus of the infected cell (Fig 1). This conserved patch exposes the main chain of β‐strand b at the tip of domain II and is therefore relatively insensitive to side chain alterations on this strand. It also includes the exposed part of the fusion loop, corresponding mainly to the main chain of glycine residues in this area, as the strictly conserved non‐polar side chains are buried in the dimer contact with domain III from the adjacent E subunit. The fusion loop surface is partially protected by an asparagine‐linked glycan on the variable “150 loop” of the opposite E subunit in the dimer. Antibodies targeting this epitope are divided into two subclasses, EDE1 and EDE2, depending on the recognition of the conserved glycan at Asn153 of dengue virus. EDE1 antibodies do not require glycosylation for binding, whereas EDE2 antibodies strongly rely on the glycan for binding. The crystal structures of these antibodies in complex with the E protein showed that the EDE1 Mabs induce disorder of the 150 loop and the glycan, and recognize also the domain III surface that interacts with the fusion loop on the adjacent subunit, partially overlapping the epitope of the dengue virus serotype 2‐specific 2D22 antibody discussed above. The EDE2 antibodies, in contrast, do not interact with domain III. The EDE1 antibodies were shown to also potently neutralize Zika virus, whereas the EDE2 Mabs displayed only intermediate neutralizing potency, mainly because the glycan on the 150 loop in the Zika virus E protein (which is linked to Asn154) is shifted by about 10 Å, as shown in the crystal structure 69, and the binding affinity is therefore substantially reduced.
EDE antibodies bind partially mature particles equally well as fully mature virions
In spite of being incompetent for binding to fully immature dengue virus particles (which do not expose an E dimer at their surface, see Fig 5A), the EDE MAbs were found to bind virions having over 60% uncleaved prM as efficiently as they bind fully mature particles, which expose only dimers at their surface 68. This result can be explained by the fact that in fully immature particles, the 60 (prM/E)3 trimeric spikes form a tightly intertwined lattice, in which each trimer is supported by three adjacent trimers 70, such that there are as many intra‐ as inter‐spike stabilizing interactions (Fig 5A). When one‐third of the prM molecules are cleaved, there are two possibilities: either random cleavage, in which on average each trimeric spike will be destabilized by the absence of one of the three prMs, or non‐random cleavage leaving regions of uncleaved prM on one side of the particle, forming a “spiky patch”, with the rest of the virion displaying the smooth, mature herringbone arrangement. Structural studies on dengue virions have shown this second possibility to actually be the case 53, 54, which is likely a reflection of the membrane‐anchored nature of furin, such that under limiting furin expression, only the side of the virion closest to the TGN membrane will be processed. The (prM/E)3 trimers at the interface between immature and mature patches will lack the stabilizing effect of three adjacent trimers and will more easily flip reversibly to (prM/E)2 dimers, generating a substrate for EDE Mab binding. Because the affinity of the EDE antibodies for the E dimer is very high, they shift the equilibrium toward the dimeric form while displacing the uncleaved prM (Fig 5B).
Figure 5. Differential binding modes of EDE versus FLE antibodies (Abs).
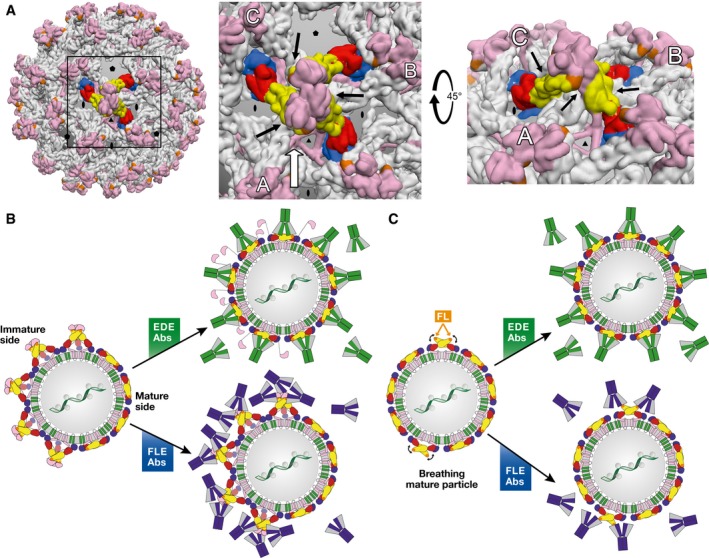
(A) Left panel: Intertwined lattice of (prM/E)3 trimers in the spiky immature particles. A central (prM/E)3 spike is represented with protein E colored by domains as in Fig 1. All other spikes have protein E in gray. prM is displayed in pink throughout. A box highlights the region zoomed in the middle panel. Middle panel: intertwined array of (prM/E)3 spikes, in which the three adjacent trimers (A, B, and C) interact underneath the central one (black arrows), underpinning it. In partially immature spikes, such stabilization is lost for the trimers at the interface between spiky and mature patches. Right panel: View along the white arrow of the middle panel, rotated by 45°. The black arrows point to the contacts between adjacent spikes. (B) Interaction of partially mature particles with EDE (green) and FLE (blue) antibodies. The spikes at the interface between mature and immature patches do not have the underpinning and stabilizing effect of three surrounding spikes shown in (A), and are therefore destabilized at these edge regions. The very high affinity of the EDE antibodies for the E dimer appears to shift the trimer‐dimer equilibrium toward dimers at the interface. Inner trimers become destabilized in turn upon EDE antibody binding, resulting in a domino effect such that the particle ends up fully coated with antibodies, as illustrated in the middle, top panel. FLE antibodies, in contrast, can readily bind to the immature patches, but also to the mature side depending on the extent of breathing (see panel C). (C) Interaction of mature particles with EDE (green) and FLE (blue) antibodies. Particle breathing (indicated by curved black arrows) leads to transient exposure of the fusion loop. EDE antibodies bind readily, regardless of the breathing behavior of the E dimers. FLE antibodies can only bind when the fusion loop is exposed through breathing, allowing particle internalization via Fcγ receptor‐mediated endocytosis before attaining sufficient antibody coating for neutralization depending on the breathing kinetics, giving rise to ADE. This situation can also explain why neutralizing antibodies, if present with ADE‐prone antibodies such as the FLEs, can override ADE by completing the antibody coating, without the need to displace the bound FLE antibody.
Antibodies targeting cryptic epitopes
Several epitopes have been described that would be inaccessible for antibody binding in a compact assembly of E proteins in mature virions (Fig 1E) and were therefore designated cryptic epitopes. The most prominent of these include epitopes at different sites of domain III and the fusion loop epitopes (FLEs).
Domain III epitopes
Examples of partially cryptic epitopes are those of the murine dengue virus‐specific Mabs 1A1D2 71 and 4E11 72, which target the A strand in domain III and can neutralize several serotypes, and also the 2H12 antibody targeting the AB loop of domain III 73. A special case is the dengue virus type 1‐specific antibody E111 74, which targets an entirely buried surface of domain III between E dimers in the herringbone pattern (the CC’ loop in domain III). This is the only antibody described so far that is specific for a cryptic epitope and strongly neutralizing, but this activity is restricted to a laboratory‐adapted strain exhibiting a highly dynamic behavior of the E protein 75. For Zika virus, two similar mouse antibodies, ZV‐48 and ZV‐64, both targeting the CC’ loop, were shown to neutralize only one out of several strains, suggesting a different breathing mode for susceptible strains, which affects the packing of dimers at the particle surface 76.
Fusion loop epitopes
Fusion loop epitopes are defined as an ensemble of epitopes comprising the fusion loop, which is normally buried in the E dimer. Binding to the FLEs therefore requires dimer dissociation. Because the amino acid sequence of the fusion loop is conserved across flaviviruses, antibodies targeting the FLE are broadly flavivirus cross‐reactive. The majority of them specifically recognize the side chain of the strictly conserved Trp101 of the fusion loop, as shown by structural studies 53, 77. FLE antibodies are reactive in immunoblots only under non‐reducing conditions, as several disulfide bonds stabilize the conformation of the fusion loop. The FLEs are exposed in the immature patches (Figs 3 and 5B) of the mosaic particles resulting from incomplete furin maturation 53, 54, in line with the observation that partially mature virus preparations are more sensitive to neutralization by patients serum than fully mature particles (reviewed in reference 78). Consistent with the cryptic nature of FLEs in mature virions, Pierson and colleagues have demonstrated, using West Nile virus, that antibodies targeting the FLEs can bind such particles only upon “breathing” of the E dimer (illustrated in Fig 5C). The breathing rate is strain dependent, as discussed below, and affects the neutralization potential of these antibodies 79.
The extent of breathing and the exposure of the fusion loop are apparently controlled by sequences outside the FLEs. When Serafin and Aaskov obtained escape mutants of the FLE antibody 4G2, the corresponding mutations did not map to the fusion loop, but to residues elsewhere in the E protein 80, which presumably control FLE exposure by affecting the breathing rate. Similarly, the kinetics of breathing were shown to be strain dependent (reviewed in reference 81) and single mutations of the E protein were found to have a strong impact on the overall reactivity of the particle with antibodies targeting cryptic epitopes 75, 82. Although there have not been systematic experiments addressing breathing rates for many strains, it is currently being recognized that laboratory‐adapted dengue virus strains are much more dynamic than clinical strains, exposing cryptic epitopes more easily and behaving differently in neutralization tests 83. The highly dynamic behavior of such strains can also lead to greater instability of virus preparations and more rapid loss of infectivity 79. Such particle instability may not be a disadvantage under the conditions used in cell culture, but is likely to be selected against in naturally circulating viruses, in particular for flaviviruses, which go back and forth between arthropod and vertebrate hosts.
The amino acid sequence conservation of the fusion loop, together with its exposure at acid pH to drive membrane fusion provides a framework to understand the early findings that led to the serological classification of the flaviviruses (then classified as “Group B Arboviruses”) by hemagglutination inhibition (HI) assays 23. For certain viruses, for instance influenza virus, hemagglutination relies on binding of the viral hemagglutinin to sialic acid (the influenza virus receptor) present at the surface of red blood cells 84. In the case of flaviviruses (as well as other arboviruses), hemagglutination requires acid treatment 85, leading to E dimer dissociation and exposure of the fusion loop, which then inserts into the plasma membrane of red blood cells, inducing their agglutination around virus particles. This means that a substantial proportion of antibodies in the sera that are positive in HI tests can be assumed to bind FLEs and thus interfere with membrane insertion, explaining their broad cross‐reactivity.
Immune responses to FLEs
Differences in the dynamics and heterogeneity of flavivirus particles may not only affect the mechanisms of virus neutralization (see below) but may also skew the antibody response toward certain epitopes. This effect appears to be especially prominent in dengue virus infections, in which a majority of antibodies were found to be cross‐reactive and poorly neutralizing, targeting the immunodominant FLEs 86, 87, 88, 89, 90, 91. Importantly, the antibody response to prM was also found to be particularly strong in dengue virus infections 87, 88, 89. These antibodies have no or only marginal neutralizing activity, and their epitopes have been mapped to a single dominant site in dengue prM 92 or to complex quaternary epitopes encompassing surfaces from both prM and E 88, 92. Although the degree of maturity of viruses circulating in dengue virus‐infected patients has not been determined, the strong induction of prM‐specific antibodies may be an indirect indicator for the circulation of partially mature virions. Since the FLEs are accessible in the immature patches of such particles (Fig 3), their presence may contribute significantly to the dominance of FLE antibodies in dengue virus infections 86, 87, 88, 89, 90, 91.
In secondary infections with dengue or other flaviviruses, the dominance of FLE antibodies is likely to be a consequence of the original antigenic sin phenomenon that leads to preferential boosting of pre‐existing cross‐reactive antibodies upon sequential infections or immunizations with antigenically related molecules 93. For instance, a recent longitudinal study following the B‐cell response of Zika virus‐infected individuals with a pre‐existing memory of dengue virus infection showed clear evidence of original antigenic sin in the first few months of the infection, resulting only in cross‐reactive antibodies against dengue and Zika virus, which neutralized Zika virus poorly. But in subsequent months, the same individuals had B cells making potently neutralizing antibodies against Zika virus, which were unrelated to the original ones and were not cross‐reactive, co‐existing with the initial ADE‐prone antibodies 94.
The presence in the patient's serum of poorly neutralizing cross‐reactive antibodies together with strongly neutralizing serotype‐specific antibodies is responsible for the dual activity of the humoral response against flaviviruses: It is important for protection, but in certain instances may also aggravate disease via antibody‐dependent enhancement of the infection, as discussed below.
Interactions with antibodies
Antibody neutralization mechanisms
In order to develop efficient vaccines against closely related flaviviruses such as those in the dengue virus serocomplex, it is important to understand the mechanism of action of the strongly neutralizing antibodies at the molecular level. Such an understanding can provide clues to the correlates with protection. The main target of neutralizing antibodies is the E glycoprotein, present in 180 copies at the particle surface, as described above (Fig 1D). The mechanism of flavivirus neutralization by antibodies was initially studied using West Nile virus, where neutralization was found to follow a “multiple hit” model with a minimum antibody/virus particle stoichiometry 95, in line with the model for virion “coating” by the antibodies proposed earlier for other viruses 96. The minimum stoichiometry for neutralization was shown to depend on the antibody, and for potently neutralizing antibodies, it was found that occupancy of about 30 antibodies per particle was sufficient 95. Of note, for Mab E16, used in these experiments, the maximum number of occupied epitopes by Fab fragments was found to be 120 and not 180 60, 97. The reduced number of accessible epitopes is due to partial occlusion of one of the three epitopes in the icosahedral asymmetric unit, which in this case is also a consequence of the lack of quasi‐equivalence in the lateral interactions between E dimers in the herringbone lattice (Fig 1E).
Neutralization by a “coat” of antibodies around the virus particle can block its infectivity in multiple ways. One of them is by cross‐linking circulating virus particles via the two Fab arms of the antibody, reducing the number of available particles capable to infect cells as the big cross‐linked virus/antibody aggregates appear to be efficiently cleared from the circulation 98. For non‐cross‐linked particles, there are essentially two options for neutralization: interference with receptor binding, and/or blocking the fusogenic conformational change of the E protein once in the endosome, thereby preventing membrane fusion.
Inhibition of the interactions with receptors
In spite of decades of intense investigation, there are still many unknowns remaining in our understanding of flavivirus biology, in particular concerning the entry pathways into relevant cells. A large body of studies on flavivirus receptors have identified protein–protein interactions such as domain III binding to integrins in the encephalotropic mosquito‐borne flaviruses, protein–sugar interactions like the DC‐SIGN lectin with the high‐mannose glycans on the dengue virus E protein, and interactions of phosphatidylserine lipids in the viral membrane with TIM and TAM/Axl lipid receptors in the case of dengue, Zika, West Nile, and yellow fever viruses (reviewed recently in 49, 99). Nevertheless, no single receptor has been identified that accounts for the complete tissue tropism for any flavivirus as of today. As we discussed in a previous review, the flavivirus maturation status depends on the cell in which the particles were assembled, and the resulting heterogeneity in turn allows the particles to employ different entry pathways to infect other cells, potentially using different receptors and attachment factors depending on the particular tissue 15.
An early study with murine antibodies against dengue virus identified that Mabs targeting E domain III were most efficient at blocking adsorption to cultured Vero cells 100, in line with the proposed role of domain III for receptor binding—at least for some flaviviruses and for infection of certain cells 101, 102. A similar finding was obtained for the domain III‐specific Mab 2B8 against dengue virus serotype 2, which interferes with virus attachment to BHK cells 103.
It follows from the apparent heterogeneity of flavivirus attachment factors and entry receptors that determining the neutralization potency of an entry‐blocking antibody in tissue culture, that is, for a given type of cell, may not necessarily reflect the situation in vivo. For instance, if an antibody neutralizes by blocking the interaction with DC‐SIGN for entry into certain cells, the same antibody may not neutralize the virus with respect to other receptors, for example, integrins or the lipid receptors. This phenomenon may partially explain the apparent lack of correlation between protection and neutralization in cell culture observed with the antibodies elicited in patients during the phase II and III dengue vaccine trials with Dengvaxia 104, 105, 106, 107 (see below).
Inhibition of membrane fusion
Studies with polyclonal sera 108 and Mabs (reviewed in reference 37) have indicated that they may confer neutralization at a post‐attachment stage, most likely by interfering with the conformational change required for membrane fusion. Domain III, for instance, undergoes a drastic re‐location during this process, resulting in a hairpin‐like structure of the E protein that defines its post‐fusion conformation (Fig 2). Targeting this domain can therefore also block the conformational change and impair membrane fusion, as elegantly shown for the potently neutralizing Mab E16 directed against West Nile virus 60, 109. Moreover, a cryo‐EM reconstruction of the West Nile virus coated with the Fab fragment of E16 showed that upon acid treatment, the bound Fab locks the particle in an intermediate expanded state, with the E dimers at a higher radial distance from the viral membrane, but unable to proceed to the fusogenic conformational change 110. Similarly, the chimpanzee Mab 5H2 targeting domain I and the linker between domains I and III, which must extend to allow re‐location of domain III, was shown to block fusion of the virus particles with fluorescently labeled liposomes 61.
Some of the most potently neutralizing Mabs are directed against domain III 111, 112. While such antibodies constitute a substantial proportion of the murine antibody response 113, 114, they make up only a minor fraction in human antibody responses against dengue virus 88, 89. Their frequency, however, may increase in individuals undergoing sequential flavivirus infections 94, 111. As described above, many potently neutralizing antibodies were found to recognize quaternary epitopes exposed at the surface of mature virions (reviewed recently in 81, 115, 116). These antibodies often cross‐link the E dimers at the particle surface and would interfere with the conformational change to drive fusion 66, 67, 117. An important prerequisite for their neutralizing activity is that their binding affinity remains high in the acidic environment of the endosome, as shown for Mab C10 and its interaction with Zika virus 117. The mechanism of neutralization by fusion inhibition can be considered universal and not subject to variations in cell receptor usage for infection of different tissues. The contribution of fusion inhibition to overall neutralization, however, may be overestimated in certain instances, because of possible pitfalls in the post‐attachment assays usually used for such analyses 66.
It is worth noting that antibodies capable of blocking fusion may also interfere with receptor binding, such that the particle never reaches the endosome. Such a dual activity is best illustrated by the domain III‐specific Mab E16, which inhibits both virus attachment and fusion, as discussed above. In several instances, the neutralization titers of Mabs were higher in pre‐attachment than in post‐attachment assay formats 67, 100, 103, suggesting that both mechanisms may contribute to the overall neutralization. These antibodies can therefore provide a double level of protection, such that any particle that managed to interact with a receptor in spite of being bound by the antibody, may still be blocked at the second step of entry, within the endosome.
Antibody‐dependent enhancement
A number of studies have shown that antibodies can not only inhibit but also enhance flavivirus infection by providing an entry pathway involving Fcγ receptors. This phenomenon was clearly shown in vitro for several flaviviruses using cultured cells bearing Fcγ receptors that are refractory to infection unless antibodies are present (reviewed in 115, 118, 119. The available evidence further indicates that any antibody that can bind flavivirus particles without neutralizing will cause ADE, even potently neutralizing antibodies when present at sub‐neutralizing concentrations 95. But antibodies targeting prM and the fusion loop of E appear to play an especially important role in ADE 87, 88, 92, 120. Both types of antibodies were shown to be dominant components of the humoral response of patients following a dengue virus infection 87, 88, to have low or no neutralizing potency, and to be strongly infection enhancing in vitro 120 and in vivo 121.
Epidemiological observations provide indirect evidence for a possible role of ADE on the course of natural dengue infections in humans. Infants born to dengue‐immune mothers were shown to have a higher risk of developing severe forms of dengue disease when infected in a time window in which the maternal antibodies have dropped below the neutralization threshold 122. To what extent ADE caused by sub‐neutralizing concentrations of maternal antibodies is responsible for disease progression in these cases is still controversial 123, 124. The first experimental in vivo model of enhanced dengue disease severity through maternally acquired heterotypic dengue antibodies was developed only recently 125 and confirmed that this mechanism results in more severe disease in mice.
In addition to potentially detrimental effects of maternally transmitted antibodies, epidemiological studies have also shown that the vast majority of severe dengue disease in children and adults occurs in secondary infections with a different serotype, indicating that pre‐existing heterologous immunity may in certain instances aggravate the infection outcome 122, 126. After a primary infection, the patients appear to be transiently protected from heterotypic infection, but this effect vanishes over time to render the individuals more vulnerable to severe forms of the disease upon subsequent heterotypic infection 127. A recent study of a long‐term pediatric cohort in Nicaragua revealed that the risk of severe dengue was correlated with pre‐existing dengue antibody titers and was significantly higher when the titers were low 128. A similar disease enhancement phenomenon of heterologous dengue infections had also been shown experimentally, four decades ago already, with non‐human primates that received antibodies against dengue virus from human immune serum 129.
More recently, cross‐enhancement was also shown between Zika and dengue viruses, both in vitro 130, 131 and in vivo using mouse models 112, 132. In their analysis of Zika virus infections in mice, Bardina et al 132 observed that antibody‐dependent disease enhancement was not only caused by human sera from dengue but also from West Nile virus infections, albeit to lower extents and frequencies. An extension of the phenomenon beyond dengue and Zika viruses was also suggested by a recent study showing that subjects with cross‐reactive antibody titers from a prior inactivated Japanese encephalitis vaccination had a prolonged yellow fever vaccine viremia upon yellow fever vaccination 130. In this case, antibody‐enhanced infection of the attenuated 17D yellow fever vaccine strain had a beneficial effect by inducing higher titers of neutralizing antibodies against yellow fever virus in these vaccinees.
To what extent cross‐enhancement contributes to the pathogenesis of Zika or dengue infections in humans is still unresolved. So far, studies in rhesus macaques did not find an influence of previous infections with dengue or yellow fever viruses on Zika virus pathogenesis 133, 134, although in vitro ADE was clearly demonstrable with the sera from these animals. Possible effects of pre‐existing flavivirus immunity on the teratogenic potential of Zika virus infections have not yet been analyzed in non‐human primates (NHPs). Nevertheless, a recent study in rhesus macaques provides the first in vivo evidence that dengue infections can be significantly enhanced by pre‐existing immunity to Zika virus 135.
Neutralizing antibodies can override ADE
Several studies have shown that potently neutralizing antibodies can block the activity of enhancing antibodies, both in vitro and in vivo 112, 121, 131. Such an ADE blocking activity is displayed by some but not all strongly neutralizing antibodies, as revealed by an analysis of antibodies engineered with a mutation at the site recognized by the Fcγ receptors (LALA mutants 136) in a mouse model of antibody‐enhanced lethal dengue virus serotype 2 infections 121. In this case, inhibition of disease enhancement was related to the capacity of the blocking antibody to out‐compete the enhancing antibodies for binding to the virion, and this concept formed the basis for the development of an in vitro suppression‐of‐enhancement assay 121. It is, however, not necessary that the neutralizing antibody out‐competes ADE‐prone antibodies bound to cryptic epitopes to override ADE. It is sufficient to complete the antibody coating of the particle (Fig 5B) by binding the remaining available sites on the virion that are left unhindered by the already bound enhancing antibodies, without displacing them.
After Fcγ receptor‐mediated virus uptake in the form of immune complexes, only antibodies capable of preventing endosomal fusion will be able to inhibit infection. This mechanism of neutralization requires stability of the virus‐antibody complex at acid pH, a property usually reflected by bell‐shaped curves of in vitro ADE analyses, that is, enhancement at low antibody concentrations but neutralization at high concentrations. Some antibodies that are potently neutralizing in Fcγ receptor‐negative cells, however, cause ADE also at high concentrations. Examples are the EDE2 MAbs A11 and B7, which neutralize Zika virus infection of Vero cells with IC50s in the nanomolar range 69, but which also enhance infection of Fcγ receptor + U937 cells at concentrations over 2 logs higher 131. The Zika virus‐specific human Mab ZKA 230 112 is another example of a potently neutralizing antibody that enhances infection also at high concentrations. In these cases, it is likely that the antibody binding does not withstand the acidic pH in the endosomes, and the neutralization mechanism is restricted to blocking attachment.
Considerations for in vitro neutralization as correlate for protection
The occurrence of breakthrough infections after vaccination with the recently licensed dengue vaccine 12, 13 in individuals who had positive titers of neutralizing antibodies against the infecting serotype 137 raised concerns about the value of in vitro neutralization as correlate for in vivo protection 106, 138. A lack of correlation has also been suggested by previous studies of secondary dengue infections in children despite the presence of neutralizing antibodies against the infecting serotype 123.
Although correlation between neutralizing antibody titers against dengue virus and protection from symptomatic infection was observed by following a longitudinal cohort of patients 139, clinically overt disease despite the presence of neutralizing antibodies against the re‐infecting serotype was also apparent in this study. For the vaccines against yellow fever, Japanese encephalitis, and tick‐borne encephalitis, it is generally accepted that the responses measured by in vitro neutralization correlate with protection 140, but specific data on neutralizing antibody titers at the time of breakthrough infections—similar to those generated in the course of the Dengvaxia field trials 106—are not available. It is therefore currently not possible to establish an unambiguous comparison of this parameter between the various vaccines.
Most virus neutralization measurements—especially with dengue viruses—employ laboratory strains that have undergone multiple passages and have adapted for better growth in tissue culture. The higher degree of particle dynamics of the laboratory strains compared to primary isolates discussed above 33, 34, 58, 59 can also affect the results of neutralization and/or ADE assays because of differences in the exposure of certain epitopes, such as the FLEs 82. In a very interesting study, Chaichana et al 83 demonstrated that human dengue post‐infection sera strongly neutralized the primary virus isolates obtained from the same patient, but failed to neutralize laboratory‐adapted strains. The opposite was seen in ADE assays, that is, strong ADE with laboratory‐adapted strains but no ADE with the primary virus isolates. The antigenic heterogeneity of dengue virus serotypes observed by Katzelnick et al 141 is also a reflection of strong quantitative differences in the neutralization of different virus strains by the same serum sample. Whether these discrepancies are caused by differences in breathing or by antigenic differences between strains remains to be further clarified.
Using in vitro neutralization as correlate for in vivo protection is further challenged by variability in the extent of prM cleavage, which is a virus strain‐specific property but also a cell‐dependent variable that can affect antigenic reactivity (see above) and therefore affect the results of in vitro neutralization. The problem can be partly overcome by the use of cell lines that overexpress furin and thus lead to the production of homogeneous mature virus preparations 142. Whether such a standardized setting, however, reflects the situation in vivo remains to be elucidated.
Because of the discrepancies of in vitro neutralization and protection observed in the Dengvaxia study 106 and the potential impact of ADE in the pathogenesis of dengue and possibly other flavivirus infections, the use of Fcγ receptor‐bearing cell lines for in vitro neutralization has been proposed as an alternative for yielding better correlates for protection in humans 143. These cells can be tuned to express a set of Fcγ receptors closely mimicking those on natural target cells such as primary monocytes and are likely to measure primarily the capacity of antibodies to inhibit viral membrane fusion in the endosome. The induction of these antibodies may be desirable for any flavivirus vaccine (see below “Implications for immunogen design”) because of the universality of the inhibition of membrane fusion as a mechanism of virus neutralization in all cell types. Antibodies that are unable to inhibit fusion but neutralize by other mechanisms, however, may not be detected by such assays, and a comparative analysis of dengue post‐vaccination sera in Fcγ receptor‐negative and Fcγ receptor‐positive cells would be required to really inform about the quality of the correlation of these two settings. Careful analyses will be necessary to evaluate whether the proposed assay formats can be standardized to a similar degree as the conventional neutralization tests.
The many variables influencing the effects of antibody interactions with heterogeneous and dynamic dengue virus particles circulating in patients make the correlation of in vitro neutralization with in vivo protection a topic of intensive discussion among stakeholders in the area of dengue vaccines. Outstanding questions in our understanding of protective or detrimental immune responses to dengue virus infections and vaccinations were recently the focus of a “Summit on Dengue Immune Correlates of Protection” and are excellently summarized by Katzelnick et al 144. It is apparent that differences in the nature of vaccines as well as the status of flavivirus immunity at the time of vaccination will affect the specifics of immune responses and make the correlation between protection and in vitro parameters even more difficult. Assay standardization is one of the keys to further improvement in this area, but can only complement future research into existing imponderables of immune responses to dengue virus infections or vaccination and their impact on protection from disease.
Use of antibodies for passive immunization
Potently neutralizing Mabs were shown to be protective in several animal models of Zika and dengue virus infections when administered before or even a short period after infection (reviewed in 37, 39, 145). These animal studies are encouraging for developing therapeutic human antibodies to be used in pre‐ and post‐exposure treatments. A difficulty is that the time point of infection is usually unknown, and the relatively short half‐life of antibodies as well as high costs are further limiting factors for large‐scale and long‐term antibody‐based immunization practices. Nevertheless, such strategies may be of value in specific instances when protection is needed for a certain period of time only, for instance to protect against Zika virus infection during pregnancy. Recent data from a mouse model have shown that an EDE antibody (containing the LALA mutation to prevent FcγR interactions and ADE effects 121, 146, 147) is able to protect adult mice from Zika and dengue virus‐induced disease and also to prevent fetal infection with Zika virus 148. This dual protection is consistent with the cross‐neutralization patterns observed with EDE antibodies (see Section “EDE Antibodies” above) and would be an important benefit for application in pregnant women, considering that most of Zika virus infections have occurred in countries endemic for dengue.
A possible concern could be the development of neutralization escape mutants, which are readily selected when viruses replicate in the presence of single Mabs. The problem may be solved by the use of cocktails of potently neutralizing Mabs that are specific for different epitopes and thus prevent ready escape from neutralization. In this context, an innovative concept of antibody engineering has been presented by Wang et al 149, who constructed a recombinant tetravalent symmetric antibody out of two strongly Zika virus‐neutralizing human Mabs, one specific for domain III and one for domain II. Tetravalency was achieved by fusing the Fab of the domain III‐Mab to the N‐terminus of the domain II‐Mab through a flexible linker. The tetravalent antibody was further engineered with a LALA mutation to prevent ADE. This chimeric antibody was highly protective against Zika virus infection in a mouse model, neutralized escape mutants generated by each of the parent antibodies individually, and did not allow escape mutants to grow during eight passages in vitro. In conclusion, rapid progress in the generation and engineering of human antibodies is being made which may offer valuable opportunities for passive immunotherapies in specific situations.
Implications for dengue immunogen design
Put together, the available data converge to indicate that, for efficient flavivirus neutralization, the epitope's accessibility is as crucial as the binding affinity of the antibody. Only when these two conditions are met, particle coating by the antibody can readily reach the stoichiometry threshold required for neutralization. Moreover, when affinity is high but accessibility is poor, the ADE potential is high. Antibodies that are strongly neutralizing thus target readily accessible epitopes on the E protein at the virion surface, whereas poorly neutralizing/ADE‐prone antibodies target cryptic epitopes. As the virion‐exposed E surface is mostly variable, and the buried E surfaces are mostly conserved, it follows that the strongly neutralizing antibodies are mostly serotype specific, while the broadly cross‐reactive antibodies are poorly neutralizing and ADE prone. In the case of dengue virus, the exception to these general principles are the EDE antibodies, which are cross‐reactive as they bind to several flaviviruses—although not as broad as the FLE antibodies, which bind to the E protein of essentially all medically relevant flaviviruses—yet the EDE Mabs potently neutralize the virus particles to which they bind.
Current vaccine approaches against dengue virus have not considered the implications of partial maturation and particle dynamics of the immunogen. A number of vaccines use a mixture of four live‐attenuated chimeric viruses, containing the prM‐E genome portion of the four serotypes of dengue virus 150, but the properties of the particular dengue virus strain from which the prM/E segments were derived have not been analyzed, neither with respect to their breathing behavior nor to their maturation status. This is particularly important as the furin cleavage site in dengue viruses appears to have been selected to be suboptimal 15, resulting in the production of mostly partially mature particles, which expose the FLEs and prM (Fig 3), and therefore induce ADE‐prone antibodies. A proper selection of strains with optimal furin cleavage sites in prM and lowest possible “breathing” behavior of protein E would provide important advantages. Nevertheless, in a live vaccine the E protein must remain metastable to be able to induce membrane fusion for entry—so that a certain level of breathing must remain. In addition, complete prM processing does not depend only on an optimal cleavage site but also on the levels of furin present in the cells of the various tissues in which the vaccine replicates, which are out of the experimenter's control.
It is possible, in contrast, to control these properties experimentally in the context of the development of a subunit vaccine. In such an approach, (i) the E protein can be engineered to eliminate breathing and (ii) prM can be eliminated altogether. A number of studies have explored vaccines that rely on virus subunits, like DNA immunisation with plasmids coding for domain III of the Envelope protein of dengue virus 151, or the use of particles formed by the hepatitis B core antigen exposing domain III of the the dengue virus 152 and of the Zika virus E protein 153. Also, an mRNA vaccine platform encoding a mutated prM/E gene of Zika virus was reported, in which the fusion loop was mutated to avoid eliciting antibodies targeting the FLEs 154. The drawback of this approach is that, as the fusion loop is important for the E dimer interactions, most mutations will interfere with dimer formation, exposing other conserved regions of the E monomer normally buried in the dimer (Fig 4). Furthermore, antibodies targeting quaternary epitopes, which are among the most strongly neutralizing, will not be elicited—the same is true for the particles exposing only domain III mentioned above.
Recent studies have also shown that cysteine residues can be engineered at strategic locations on E to make inter‐subunit disulfide bonds and stabilize the dimer 155, 156. These engineered disulfide bridges prevent the dissociation of the dimer into monomers and the exposure of the FLEs, which would occur at low concentrations. The crystal structures of these covalently linked sE dimers were indistinguishable from wild‐type dimers 155. Two different cysteine mutants were generated: The first approach involved one mutation at the center of the dimer (A259C in dengue virus serotype 2), such that the engineered C259 faces itself at the twofold molecular axes of the sE dimer to make a disulfide bond. The second approach involved a double mutation—L107C and T313C in dengue virus serotype 2—resulting in two cysteines facing each other across the dimer interface, where the fusion loop (which contains residue 107) contacts domain III of the adjacent subunit. Importantly, in ELISA experiments using immobilized sE, the EDE antibodies bound efficiently to the cysteine mutants and not to wild‐type E—which is immobilized essentially as monomer, therefore not exposing dimer‐dependent epitopes. But the FLE antibodies bound efficiently to both wild‐type E and the A259C mutant, indicating that even with a covalently stabilized dimer there is enough breathing of the E protein about the central disulfide bond to allow binding of FLE Mabs. This mutant also retained a certain degree of interaction with liposomes, as assayed by co‐flotation in density gradients at acid pH, consistent with exposure of the fusion loop. The double cysteine mutant, in contrast, did not react with most of the FLE antibodies and was unable to co‐float with liposomes, demonstrating that it is possible to generate recombinant immunogens that stably display the EDE but do not expose the fusion loop 155.
An issue remains about exposure of the dimer's “underside”, which is highly conserved (Fig 4A, right panel) and would be exposed if isolated stabilized dimers were used as immunogens. The ideal scaffold would be to present the E dimer as on the virion, assembled into a herringbone pattern of 90 covalently linked dimers, which would not expose the underside. How to obtain these particles is not trivial, however, as the herringbone lattice is formed only after the initial budding of immature particles with 60 spikes of prM/E heterodimers (Fig 1). But it should be possible to find conditions to drive a similar assembly, or to use other scaffolds to present correctly stabilized dimers and masking their conserved side. An alternative would be to re‐surface the underside of the sE dimer, similar to the re‐surfaced HIV gp120 that has been used to isolate antibodies targeting the CD4 binding site 157.
The above approach should lead to the induction of strongly neutralizing antibodies that target the variable, exposed side of the E dimer. Because of the relative conservation of the EDE, which is also exposed by the stabilized dimers, an adequate priming and boosting strategy using two different E protein immunogens that only have the EDE in common (such as using the E protein from Zika virus and from any of the four dengue virus serotypes) should focus the immune system into producing antibodies targeting the EDE. In addition, the N‐linked glycan on the 150 loop could be engineered out to avoid inducing EDE2 antibodies, which require the glycan to bind and were shown to induce ADE against Zika virus. Such engineered immunogens should result in broad protection while avoiding induction of unwanted antibodies with ADE potential. Because the presence of strongly neutralizing antibodies has been shown to override ADE (see above) 89, 112, 121, 131, this vaccine approach should be valid not only for flavivirus naïve individuals, but also in endemic countries where previous dengue infections may have induced ADE‐prone antibodies in the population. In other words, it should be possible to use the positive side of the original antigenic sin phenomenon, by boosting cross‐reactive antibodies targeting the EDE while avoiding to boost those against the FLEs. The identification of the EDE has therefore opened a new window of opportunity for next‐generation, EDE‐focused subunit vaccine design with a broad impact.
Conclusions and outlook
The advances made in the structural biology of flaviviruses have led to unprecedented insight into the interactions of viruses with antibodies, and into the effects these interactions can have on the different steps of the cell entry process. Especially in the case of dengue viruses, the much‐debated Janus‐faced nature of antibody responses—beneficial and protective on the one hand but potentially harmful on the other (causing ADE of infection in certain constellations of sequential infections)—has hampered the development of effective dengue vaccines for decades. In this context, it is worth noting that the manufacturer of the sole dengue vaccine on the market (Dengvaxia) recently released a statement (www.sanofipasteur.com/en/Documents/PDF/PR/20171130_Sanofi-Updates-Info-on-Dengue-Vaccine_EN.pdf) and that a 6‐year study revealed more severe dengue disease in vaccinated individuals who were naïve to dengue virus at the time of vaccination, indicating that the vaccine was protective only in individuals with prior dengue infections 158. In the last years, the strong suspicion that cross‐reactive antibodies induced by previous dengue infections may have contributed to the observed pathogenesis of Zika virus infections—most of which have occurred in dengue hyperendemic regions—has brought the issue of antibody cross‐reactivity without cross‐neutralization to the frontline. High‐resolution analyses of the antigenic determinants and detailed structural features of the epitopes, in combination with functional studies of the corresponding antibodies, have dramatically increased our mechanistic understanding of the virus neutralization process and the antibody‐mediated infection enhancement phenomena. Efforts to put this information into practice to design next‐generation vaccines, focusing the antibody response on the most protective antigenic sites, are underway, but remain challenging (see Box 1).
Box 1: In need of answers.
-
How heterogeneous are naturally circulating flavivirus particles?
Studies so far were performed with viruses grown in cell culture and revealed heterogeneities with respect to prM cleavage as well as viral envelope dynamics, both of which can affect the induction of and interaction with antibodies. Structural analyses of viruses circulating in humans are challenging but will be critical for understanding flavivirus immune responses.
-
What entry mechanisms are used in different tissues of infected humans? What receptors are involved?
Current data on flavivirus receptors are diverse and inconclusive as to their role in the infection of different tissues and the organotropism of different flaviviruses. Similar to the interaction with antibodies, structural heterogeneity and dynamics can provide a number of interaction partners for cellular molecules that may function as attachment and/or entry receptors in different tissues and in different hosts.
-
How important is the ADE effect in Zika virus infections?
Epidemiological evidence indicates that the risk of severe forms of dengue disease is substantially higher in sequential infections with heterologous dengue virus serotypes than after primary infections and this finding has been linked to ADE. In vitro and in vivo data suggest that there may also be an association between pre‐existing dengue immunity and enhancement of Zika virus infections. Conclusive answers to this question will be important for understanding the epidemiology of Zika disease and for informing vaccine design. An important, related question is what would be the impact of the immune status induced by a Zika vaccine during subsequent dengue infections.
-
Can neutralization assays be improved to become a better correlate of protection?
Studies with dengue post‐vaccination sera have revealed discrepancies between in vitro neutralization of a given serotype and protection from natural infection with the same serotype. Research into better defining in vitro correlates of protection is of utmost importance for the evaluation of immune responses to different kinds of vaccines and their regulatory approval.
-
Can a dengue or Zika vaccine be designed to avoid inducing ADE‐prone antibodies?
Studies with monoclonal antibodies have provided detailed structural insights into the characteristics of sites in the viral envelope that can induce potently neutralizing and protective antibodies and those that induce ADE‐prone antibodies. Exploiting this information is expected to inform the design of novel immunogens and immunization schedules to maximize vaccine‐induced protection.
The recent progress in the area of flavivirus structural biology has also made clear that there are still fundamental gaps in our understanding of the flavivirus entry process and the way it is inhibited or enhanced by antibodies. Properties such as the heterogeneity of the viral particles due to incomplete proteolytic maturation, and the highly dynamic behavior of the virions, have been demonstrated with viruses grown in vitro. This situation can affect the correlation between antibody‐mediated virus neutralization in vitro and protection in vivo. It is possible that structural heterogeneity and dynamic behavior are key features required for flaviviruses to replicate efficiently in phylogenetically distant hosts such as arthropods and vertebrates. Understanding how these properties can affect antibody responses and protection in vivo will certainly form important topics of future research and inform the development of effective vaccines (Box 1). But the lessons derived from the ensemble of available studies already provide new leads toward the goal of next‐generation immunogens that selectively target the “bright” side of the immune response, that is, protection from disease, in such a way that it dominates over potentially harmful effects. In particular, the gained knowledge that it is crucial to avoid exposure to the immune system of conserved cryptic elements of the envelope protein, or exposure of elements exposed only in immature flavivirus particles, paves the way to novel approaches for vaccine design—in particular to protect against dengue disease—with brighter prospects to provide better protection to the general population.
Conflict of interest
MCV and FAR have a patent on “anti‐dengue vaccine and antibodies” (Number WO2016/012800 in Great Britain, EP3172229 in the European Union) and have filed another one on a super vaccine Dengue/Zika (deposit number GB1610162.8).
Acknowledgements
FAR, MCV, and MD acknowledge support from the French ANR (grant ANR‐13‐ISV8‐0002‐01), Institut Pasteur, CNRS, and the LABEX IBEID. FXH and KS acknowledge support from the Austrian Science Fund FWF.
See the Glossary for abbreviations used in this article.
Contributor Information
Félix A Rey, Email: felix.rey@pasteur.fr.
Franz X Heinz, Email: franz.x.heinz@meduniwien.ac.at.
References
- 1. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O et al (2013) The global distribution and burden of dengue. Nature 496: 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weaver SC, Costa F, Garcia‐Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi P‐Y, Vasilakis N (2016) Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO (2017) Yellow fever in Africa and the Americas, 2016. Wkly Epidemiol Rec 92: 442–452 [PubMed] [Google Scholar]
- 4. ECDC (2017) Rapid risk assessment: outbreak of yellow fever in Brazil. First update, 13 April 2017. Stockholm: ECDC; [Google Scholar]
- 5. Wilder‐Smith A, Gubler DJ, Weaver SC, Monath TP, Heymann DL, Scott TW (2017) Epidemic arboviral diseases: priorities for research and public health. Lancet Infect Dis 17: e101–e106 [DOI] [PubMed] [Google Scholar]
- 6. Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W et al (2015) The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife 4: e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roehrig JT (2013) West Nile virus in the United States – a historical perspective. Viruses 5: 3088–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boldescu V, Behnam MAM, Vasilakis N, Klein CD (2017) Broad‐spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov 16: 565–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staples JE, Monath TP, Gershman MD, Barrett ADT (2017) Chapter 63 ‐ Yellow fever vaccines In Plotkin's vaccines, Plotkin SA, Orenstein WA, Offit PA, Edwards KM. (eds), pp 1181–1265. Amsterdam, The Netherlands: Elsevier B.V., RELX Group; [Google Scholar]
- 10. Halstead SB, Hills SL, Dubischar K (2017) Chapter 33 ‐ Japanese encephalitis vaccines In Plotkin's vaccines, Plotkin SA, Orenstein WA, Offit PA, Edwards KM. (eds), pp 511–548. Amsterdam, The Netherlands: Elsevier B.V., RELX Group; [Google Scholar]
- 11. Hombach J, Barrett ADT, Kollaritsch H (2017) Chapter 59 ‐ Tickborne encephalitis vaccines In Plotkin's vaccines, Plotkin SA, Orenstein WA, Offit PA, Edwards KM. (eds), pp 1080–1094. Amsterdam, The Netherlands: Elsevier B.V., RELX Group; [Google Scholar]
- 12. Rothman AL, Ennis FA (2016) Dengue vaccine: the need, the challenges, and progress. J Infect Dis 214: 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guy B, Noriega F, Ochiai RL, L'Azou M, Delore V, Skipetrova A, Verdier F, Coudeville L, Savarino S, Jackson N (2017) A recombinant live attenuated tetravalent vaccine for the prevention of dengue. Expert Rev Vaccines 16: 1–13 [DOI] [PubMed] [Google Scholar]
- 14. Watterson D, Modhiran N, Young PR (2016) The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antiviral Res 130: 7–18 [DOI] [PubMed] [Google Scholar]
- 15. Rey FA, Stiasny K, Heinz FX (2017) Flavivirus structural heterogeneity: implications for cell entry. Curr Opin Virol 24: 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pierson TC, Diamond MS (2013) Flaviviruses In Fields virology, Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (eds), pp 747–794. Philadelphia: Lippincott. Williams & Wilkins; [Google Scholar]
- 17. Reed W, Carroll J, Agramonte A, Lazear JW (1900) The etiology of yellow fever–a preliminary note. Public Health Pap Rep 26: 37–53 [PMC free article] [PubMed] [Google Scholar]
- 18. Finlay CJ (1881) The mosquito hypothetically considered as the agent of transmission of yellow fever. 1881. New Orleans Med Surg J 18: 601–616 [PubMed] [Google Scholar]
- 19. Theiler M, Smith HH (1937) The use of yellow fever virus modified by in vitro cultivation for human immunization. J Exp Med 65: 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collins ND, Barrett AD (2017) Live attenuated yellow fever 17D vaccine: a legacy vaccine still controlling outbreaks in modern day. Curr Infect Dis Rep 19: 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH (1985) Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229: 726–733 [DOI] [PubMed] [Google Scholar]
- 22. Westaway EG, Brinton MA, Gaidamovich S, Horzinek MC, Igarashi A, Kaariainen L, Lvov DK, Porterfield JS, Russell PK, Trent DW (1985) Flaviviridae. Intervirology 24: 183–192 [DOI] [PubMed] [Google Scholar]
- 23. Casals J, Brown LV (1954) Hemagglutination with arthropod‐borne viruses. J Exp Med 99: 429–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE (1989) Antigenic relationships between flaviviruses as determined by cross‐neutralization tests with polyclonal antisera. J Gen Virol 70(Pt 1): 37–43 [DOI] [PubMed] [Google Scholar]
- 25. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC (1995) The envelope glycoprotein from tick‐borne encephalitis virus at 2 A resolution. Nature 375: 291–298 [DOI] [PubMed] [Google Scholar]
- 26. Modis Y, Ogata S, Clements D, Harrison SC (2003) A ligand‐binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA 100: 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA (2004) Structure of a flavivirus envelope glycoprotein in its low‐pH‐induced membrane fusion conformation. EMBO J 23: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Modis Y, Ogata S, Clements D, Harrison SC (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427: 313–319 [DOI] [PubMed] [Google Scholar]
- 29. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG et al (2002) Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG (2008) The flavivirus precursor membrane‐envelope protein complex: structure and maturation. Science 319: 1830–1834 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG (2003) Structures of immature flavivirus particles. EMBO J 22: 2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH (2013) Cryo‐EM structure of the mature dengue virus at 3.5‐A resolution. Nat Struct Mol Biol 20: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kostyuchenko VA, Chew PL, Ng TS, Lok SM (2014) Near‐atomic resolution cryo‐electron microscopic structure of dengue serotype 4 virus. J Virol 88: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kostyuchenko VA, Lim EXY, Zhang S, Fibriansah G, Ng T‐S, Ooi JSG, Shi J, Lok S‐M (2016) Structure of the thermally stable Zika virus. Nature 533: 425–428 [DOI] [PubMed] [Google Scholar]
- 35. Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ (2016) The 3.8 Å resolution cryo‐EM structure of Zika virus. Science 352: 467–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X, Li SH, Zhu L, Nian QG, Yuan S, Gao Q, Hu Z, Ye Q, Li XF, Xie DY et al (2017) Near‐atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat Commun 8: 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fibriansah G, Lok SM (2016) The development of therapeutic antibodies against dengue virus. Antiviral Res 128: 7–19 [DOI] [PubMed] [Google Scholar]
- 38. Dai L, Wang Q, Qi J, Shi Y, Yan J, Gao GF (2016) Molecular basis of antibody‐mediated neutralization and protection against flavivirus. IUBMB Life 68: 783–791 [DOI] [PubMed] [Google Scholar]
- 39. Heinz FX, Stiasny K (2017) The antigenic structure of Zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol Mol Biol Rev 81: e00055‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Priyamvada L, Hudson W, Ahmed R, Wrammert J (2017) Humoral cross‐reactivity between Zika and dengue viruses: implications for protection and pathology. Emerg Microbes Infect 6: e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caspar DL, Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 27: 1–24 [DOI] [PubMed] [Google Scholar]
- 42. Lindenbach BD, Murray CL, Thiel HJ, Rice CM (2013) Flaviviridae In Fields virology, Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (eds), pp 712–746. Philadelphia: Lippincott. Williams & Wilkins; [Google Scholar]
- 43. Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX (1995) Oligomeric rearrangement of tick‐borne encephalitis virus envelope proteins induced by an acidic pH. J Virol 69: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zaitseva E, Yang S‐T, Melikov K, Pourmal S, Chernomordik LV (2010) Dengue virus ensures its fusion in late endosomes using compartment‐specific lipids. PLoS Pathog 6: e1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corver J, Ortiz A, Allison SL, Schalich J, Heinz FX, Wilschut J (2000) Membrane fusion activity of tick‐borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269: 37–46 [DOI] [PubMed] [Google Scholar]
- 46. Stiasny K, Allison SL, Schalich J, Heinz FX (2002) Membrane interactions of the tick‐borne encephalitis virus fusion protein E at low pH. J Virol 76: 3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrison SC (2015) Viral membrane fusion. Virology 479–480: 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Igonet S, Rey FA (2012) SnapShot: viral and eukaryotic protein fusogens. Cell 151: 1634 [DOI] [PubMed] [Google Scholar]
- 49. Acosta EG, Kumar A, Bartenschlager R (2014) Revisiting dengue virus‐host cell interaction: new insights into molecular and cellular virology. Adv Virus Res 88: 1–109 [DOI] [PubMed] [Google Scholar]
- 50. Byk LA, Gamarnik AV (2016) Properties and functions of the dengue virus capsid protein. Annu Rev Virol 3: 263–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garten W, Hallenberger S, Ortmann D, Schafer W, Vey M, Angliker H, Shaw E, Klenk HD (1994) Processing of viral glycoproteins by the subtilisin‐like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76: 217–225 [DOI] [PubMed] [Google Scholar]
- 52. Pierson TC, Diamond MS (2012) Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol 2: 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR et al (2009) Structural basis for the preferential recognition of immature flaviviruses by a fusion‐loop antibody. EMBO J 28: 3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, Sittisombut N, Kuhn RJ, Steven AC, Rossmann MG (2011) Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep 12: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuhn RJ, Dowd KA, Beth Post C, Pierson TC (2015) Shake, rattle, and roll: impact of the dynamics of flavivirus particles on their interactions with the host. Virology 479–480: 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haslwanter D, Blaas D, Heinz FX, Stiasny K (2017) A novel mechanism of antibody‐mediated enhancement of flavivirus infection. PLoS Pathog 13: e1006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsouchnikas G, Zlatkovic J, Jarmer J, Strauß J, Vratskikh O, Kundi M, Stiasny K, Heinz FX (2015) Immunization with immune complexes modulates the fine specificity of antibody responses to a flavivirus antigen. J Virol 89: 7970–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG (2013) Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci USA 110: 6795–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM (2013) Structural changes in dengue virus when exposed to a temperature of 37°C. J Virol 87: 7585–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH (2005) Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437: 764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cockburn JJ, Navarro Sanchez ME, Goncalvez AP, Zaitseva E, Stura EA, Kikuti CM, Duquerroy S, Dussart P, Chernomordik LV, Lai CJ et al (2012) Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J 31: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A et al (2014) A potent anti‐dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med 6: 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM et al (2015) Cryo‐EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG (2010) Neutralization of West Nile virus by cross‐linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci USA 107: 18950–18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS et al (2012) The structural basis for serotype‐specific neutralization of dengue virus by a human antibody. Sci Transl Med 4: 139ra83 [DOI] [PubMed] [Google Scholar]
- 66. Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE et al (2015) A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun 6: 6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS et al (2017) A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat Commun 8: 14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T et al (2015) A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16: 170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Barba‐Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon‐Loriere E, Sakuntabhai A, Cao‐Lormeau VM, Haouz A et al (2016) Structural basis of potent Zika‐dengue virus antibody cross‐neutralization. Nature 536: 48–53 [DOI] [PubMed] [Google Scholar]
- 70. Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM (2013) Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol 87: 7700–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi‐Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT et al (2008) Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol 15: 312–317 [DOI] [PubMed] [Google Scholar]
- 72. Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA (2012) Mechanism of dengue virus broad cross‐neutralization by a monoclonal antibody. Structure 20: 303–314 [DOI] [PubMed] [Google Scholar]
- 73. Midgley CM, Flanagan A, Tran HB, Dejnirattisai W, Chawansuntati K, Jumnainsong A, Wongwiwat W, Duangchinda T, Mongkolsapaya J, Grimes JM et al (2012) Structural analysis of a dengue cross‐reactive antibody complexed with envelope domain III reveals the molecular basis of cross‐reactivity. J Immunol 188: 4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH (2012) Structural basis of differential neutralization of DENV‐1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog 8: e1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dowd KA, DeMaso CR, Pierson TC (2015) Genotypic differences in dengue virus neutralization are explained by a single amino acid mutation that modulates virus breathing. MBio 6: e01559‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS et al (2016) Structural basis of zika virus‐specific antibody protection. Cell 166: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, Zhang Y, Yuan Y, Song H, Haywood J et al (2016) Structures of the zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19: 696–704 [DOI] [PubMed] [Google Scholar]
- 78. Pierson TC, Diamond MS (2015) A game of numbers. Prog Mol Biol Transl Sci 129: 141–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mukherjee S, Lin T‐Y, Dowd KA, Manhart CJ, Pierson TC (2011) The infectivity of prM‐containing partially mature west Nile virus does not require the activity of cellular furin‐like proteases. J Virol 85: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Serafin IL, Aaskov JG (2001) Identification of epitopes on the envelope (E) protein of dengue 2 and dengue 3 viruses using monoclonal antibodies. Arch Virol 146: 2469–2479 [DOI] [PubMed] [Google Scholar]
- 81. Lok S‐M (2016) The interplay of dengue virus morphological diversity and human antibodies. Trends Microbiol 24: 284–293 [DOI] [PubMed] [Google Scholar]
- 82. Goo L, VanBlargan LA, Dowd KA, Diamond MS, Pierson TC (2017) A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog 13: e1006178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chaichana P, Okabayashi T, Puiprom O, Sasayama M, Sasaki T, Yamashita A, Ramasoota P, Kurosu T, Ikuta K (2014) Low levels of antibody‐dependent enhancement in vitro using viruses and plasma from dengue patients. PLoS ONE 9: e92173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Skehel JJ, Wiley DC (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69: 531–569 [DOI] [PubMed] [Google Scholar]
- 85. Porterfield JS (1954) The haemagglutination‐inhibition test in the diagnosis of yellow fever in man. Trans R Soc Trop Med Hyg 48: 261–266 [DOI] [PubMed] [Google Scholar]
- 86. Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK (2008) Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross‐reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol 82: 6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S et al (2010) Cross‐reacting antibodies enhance dengue virus infection in humans. Science 328: 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi‐Petty S, Navarro‐Sanchez E, Young PR, de Silva AM et al (2010) The human immune response to Dengue virus is dominated by highly cross‐reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8: 271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE Jr (2012) Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86: 2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE Jr (2014) Isolation of dengue virus‐specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 88: 12233–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu M, Züst R, Toh YX, Pfaff JM, Kahle KM, Davidson E, Doranz BJ, Velumani S, Tukijan F, Wang C‐I et al (2016) Protective capacity of the human anamnestic antibody response during acute dengue virus infection. J Virol 90: 11122–11131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Smith SA, Nivarthi UK, de Alwis R, Kose N, Sapparapu G, Bombardi R, Kahle KM, Pfaff JM, Lieberman S, Doranz BJ et al (2016) Dengue virus prM‐specific human monoclonal antibodies with virus replication‐enhancing properties recognize a single immunodominant antigenic site. J Virol 90: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rothman AL (2011) Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 11: 532–543 [DOI] [PubMed] [Google Scholar]
- 94. Rogers TF, Goodwin EC, Briney B, Sok D, Beutler N, Strubel A, Nedellec R, Le K, Brown ME, Burton DR et al (2017) Zika virus activates de novo and cross‐reactive memory B cell responses in dengue‐experienced donors. Sci Immunol 2: pii: eaan6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, Fremont DH, Diamond MS (2007) The stoichiometry of antibody‐mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Burton DR, Saphire EO, Parren PW (2001) A model for neutralization of viruses based on antibody coating of the virion surface. Curr Top Microbiol Immunol 260: 109–143 [DOI] [PubMed] [Google Scholar]
- 97. Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG (2006) West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci USA 103: 12400–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chan KR, Zhang SL, Tan HC, Chan YK, Chow A, Lim AP, Vasudevan SG, Hanson BJ, Ooi EE (2011) Ligation of Fc gamma receptor IIB inhibits antibody‐dependent enhancement of dengue virus infection. Proc Natl Acad Sci USA 108: 12479–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cruz‐Oliveira C, Freire JM, Conceicao TM, Higa LM, Castanho MA, Da Poian AT (2015) Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol Rev 39: 155–170 [DOI] [PubMed] [Google Scholar]
- 100. Crill WD, Roehrig JT (2001) Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol 75: 7769–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bhardwaj S, Holbrook M, Shope RE, Barrett AD, Watowich SJ (2001) Biophysical characterization and vector‐specific antagonist activity of domain III of the tick‐borne flavivirus envelope protein. J Virol 75: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML (2005) Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol 86: 405–412 [DOI] [PubMed] [Google Scholar]
- 103. Deng YQ, Dai JX, Ji GH, Jiang T, Wang HJ, Yang HO, Tan WL, Liu R, Yu M, Ge BX et al (2011) A broadly flavivirus cross‐neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS ONE 6: e16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R et al (2014) Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer‐masked, placebo‐controlled trial. Lancet 384: 1358–1365 [DOI] [PubMed] [Google Scholar]
- 105. Hss AS, Koh MT, Tan KK, Chan LG, Zhou L, Bouckenooghe A, Crevat D, Hutagalung Y (2013) Safety and immunogenicity of a tetravalent dengue vaccine in healthy children aged 2‐11 years in Malaysia: a randomized, placebo‐controlled, Phase III study. Vaccine 31: 5814–5821 [DOI] [PubMed] [Google Scholar]
- 106. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA et al (2012) Protective efficacy of the recombinant, live‐attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380: 1559–1567 [DOI] [PubMed] [Google Scholar]
- 107. Villar L, Dayan GH, Arredondo‐Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales‐Ramirez JO, Carrasquilla G et al (2015) Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 372: 113–123 [DOI] [PubMed] [Google Scholar]
- 108. Gollins SW, Porterfield JS (1986) A new mechanism for the neutralization of enveloped viruses by antiviral antibody. Nature 321: 244–246 [DOI] [PubMed] [Google Scholar]
- 109. Thompson BS, Moesker B, Smit JM, Wilschut J, Diamond MS, Fremont DH (2009) A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog 5: e1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG (2009) Capturing a flavivirus pre‐fusion intermediate. PLoS Pathog 5: e1000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, Schaefer‐Babajew D, Avila‐Rios S, Nogueira L, Patel R et al (2017) Recurrent potent human neutralizing antibodies to zika virus in Brazil and Mexico. Cell 169:597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F et al (2016) Specificity, cross‐reactivity, and function of antibodies elicited by Zika virus infection. Science 353: 823–826 [DOI] [PubMed] [Google Scholar]
- 113. Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS (2007) Induction of epitope‐specific neutralizing antibodies against west Nile virus. J Virol 81: 11828–11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Williams KL, Wahala WMPB, Orozco S, de Silva AM, Harris E (2012) Antibodies targeting dengue virus envelope domain III are not required for serotype‐specific protection or prevention of enhancement in vivo. Virology 429: 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Acosta EG, Bartenschlager R (2016) Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development. Expert Rev Vaccines 15: 467–482 [DOI] [PubMed] [Google Scholar]
- 116. VanBlargan LA, Goo L, Pierson TC (2016) Deconstructing the antiviral neutralizing‐antibody response: implications for vaccine development and immunity. Microbiol Mol Biol Rev 80: 989–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang S, Kostyuchenko VA, Ng T‐S, Lim X‐N, Ooi JSG, Lambert S, Tan TY, Widman DG, Shi J, Baric RS et al (2016) Neutralization mechanism of a highly potent antibody against Zika virus. Nat Commun 7: 13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Porterfield JS (1986) Antibody‐dependent enhancement of viral infectivity. Adv Virus Res 31: 335–355 [DOI] [PubMed] [Google Scholar]
- 119. Dowd KA, Pierson TC (2011) Antibody‐mediated neutralization of flaviviruses: a reductionist view. Virology 411: 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. de Alwis R, Williams KL, Schmid MA, Lai CY, Patel B, Smith SA, Crowe JE, Wang WK, Harris E, de Silva AM (2014) Dengue viruses are enhanced by distinct populations of serotype cross‐reactive antibodies in human immune sera. PLoS Pathog 10: e1004386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Williams KL, Sukupolvi‐Petty S, Beltramello M, Johnson S, Sallusto F, Lanzavecchia A, Diamond MS, Harris E (2013) Therapeutic efficacy of antibodies lacking Fcgamma receptor binding against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies [corrected]. PLoS Pathog 9: e1003157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Halstead SB (2003) Neutralization and antibody‐dependent enhancement of dengue viruses. Adv Virus Res 60: 421–467 [DOI] [PubMed] [Google Scholar]
- 123. Laoprasopwattana K, Libraty DH, Endy TP, Nisalak A, Chunsuttiwat S, Vaughn DW, Reed G, Ennis FA, Rothman AL, Green S (2005) Dengue virus (DV) enhancing antibody activity in preillness plasma does not predict subsequent disease severity or viremia in secondary DV infection. J Infect Dis 192: 510–519 [DOI] [PubMed] [Google Scholar]
- 124. Libraty DH, Acosta LP, Tallo V, Segubre‐Mercado E, Bautista A, Potts JA, Jarman RG, Yoon I‐K, Gibbons RV, Brion JD et al (2009) A prospective nested case‐control study of dengue in infants: rethinking and refining the antibody‐dependent enhancement dengue hemorrhagic fever model. PLoS Med 6: e1000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ng JKW, Zhang SL, Tan HC, Yan B, Maria Martinez Gomez J, Tan WY, Lam JH, Tan GKX, Ooi EE, Alonso S (2014) First experimental in vivo model of enhanced dengue disease severity through maternally acquired heterotypic dengue antibodies. PLoS Pathog 10: e1004031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB (1984) Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol 120: 653–669 [DOI] [PubMed] [Google Scholar]
- 127. Guzman MG, Alvarez M, Halstead SB (2013) Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody‐dependent enhancement of infection. Arch Virol 158: 1445–1459 [DOI] [PubMed] [Google Scholar]
- 128. Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E (2017) Antibody‐dependent enhancement of severe dengue disease in humans. Science 358: 929–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Halstead SB (1979) In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis 140: 527–533 [DOI] [PubMed] [Google Scholar]
- 130. Chan KR, Wang X, Saron WA, Gan ES, Tan HC, Mok DZ, Zhang SL, Lee YH, Liang C, Wijaya L et al (2016) Cross‐reactive antibodies enhance live attenuated virus infection for increased immunogenicity. Nat Microbiol 19: 16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba‐Spaeth G, Duangchinda T, Sakuntabhai A, Cao‐Lormeau VM, Malasit P, Rey FA et al (2016) Dengue virus sero‐cross‐reactivity drives antibody‐dependent enhancement of infection with zika virus. Nat Immunol 17: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D et al (2017) Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. McCracken MK, Gromowski GD, Friberg HL, Lin X, Abbink P, De La Barrera R, Eckles KH, Garver LS, Boyd M, Jetton D et al (2017) Impact of prior flavivirus immunity on Zika virus infection in rhesus macaques. PLoS Pathog 13: e1006487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pantoja P, Perez‐Guzman EX, Rodriguez IV, White LJ, Gonzalez O, Serrano C, Giavedoni L, Hodara V, Cruz L, Arana T et al (2017) Zika virus pathogenesis in rhesus macaques is unaffected by pre‐existing immunity to dengue virus. Nat Commun 8: 15674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. George J, Valiant WG, Mattapallil MJ, Walker M, Huang YS, Vanlandingham DL, Misamore J, Greenhouse J, Weiss DE, Verthelyi D et al (2017) Prior exposure to zika virus significantly enhances peak dengue‐2 viremia in rhesus macaques. Sci Rep 7: 10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hezareh M, Hessell AJ, Jensen RC, van de Winkel JG, Parren PW (2001) Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J Virol 75: 12161–12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sirivichayakul C, Sabchareon A, Limkittikul K, Yoksan S (2014) Plaque reduction neutralization antibody test does not accurately predict protection against dengue infection in Ratchaburi cohort, Thailand. Virol J 11: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. de Silva AM, Harris E (2017) Which dengue vaccine approach is the most promising, and should we be concerned about enhanced disease after vaccination?: The path to a dengue vaccine: Learning from human natural dengue infection studies and vaccine trials. Cold Spring Harb Perspect Biol a029371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Katzelnick LC, Montoya M, Gresh L, Balmaseda A, Harris E (2016) Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc Natl Acad Sci USA 113: 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Pierson TC, Diamond MS (2008) Molecular mechanisms of antibody‐mediated neutralisation of flavivirus infection. Expert Rev Mol Med 10: e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Katzelnick LC, Fonville JM, Gromowski GD, Bustos Arriaga J, Green A, James SL, Lau L, Montoya M, Wang C, VanBlargan LA et al (2015) Dengue viruses cluster antigenically but not as discrete serotypes. Science 349: 1338–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mukherjee S, Sirohi D, Dowd KA, Chen Z, Diamond MS, Kuhn RJ, Pierson TC (2016) Enhancing dengue virus maturation using a stable furin over‐expressing cell line. Virology 497: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gan ES, Ting DHR, Chan KR (2017) The mechanistic role of antibodies to dengue virus in protection and disease pathogenesis. Expert Rev Anti Infect Ther 15: 111–119 [DOI] [PubMed] [Google Scholar]
- 144. Katzelnick LC, Harris E (2017) Immune correlates of protection for dengue: State of the art and research agenda. Vaccine 35: 4659–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Morrison TE, Diamond MS (2017) Animal models of zika virus infection, pathogenesis, and immunity. J Virol 91: e00009‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wines BD, Powell MS, Parren PWHI, Barnes N, Hogarth PM (2000) The IgG Fc contains distinct Fc receptor (FcR) binding sites: The leukocyte receptors FcγRI and FcγRIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J Immunol 164: 5313 [DOI] [PubMed] [Google Scholar]
- 147. Hessell AJ, Hangartner L, Hunter M, Havenith CEG, Beurskens FJ, Bakker JM, Lanigan CMS, Landucci G, Forthal DN, Parren PWHI et al (2007) Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449: 101 [DOI] [PubMed] [Google Scholar]
- 148. Fernandez E, Dejnirattisai W, Cao B, Scheaffer SM, Supasa P, Wongwiwat W, Esakky P, Drury A, Mongkolsapaya J, Moley KH et al (2017) Human antibodies to the dengue virus E‐dimer epitope have therapeutic activity against Zika virus infection. Nat Immunol 18: 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wang J, Bardelli M, Espinosa DA, Pedotti M, Ng T‐S, Bianchi S, Simonelli L, Lim EXY, Foglierini M, Zatta F et al (2017) A human bi‐specific antibody against Zika virus with high therapeutic potential. Cell 171:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pierson TC, Diamond MS (2014) Vaccine development as a means to control dengue virus pathogenesis: Do we know enough? Annu Rev Virol 1: 375–398 [DOI] [PubMed] [Google Scholar]
- 151. Poggianella M, Slon Campos JL, Chan KR, Tan HC, Bestagno M, Ooi EE, Burrone OR (2015) Dengue E protein domain III‐based DNA immunisation induces strong antibody responses to all four viral serotypes. PLoS Negl Trop Dis 9: e0003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Khetarpal N, Poddar A, Nemani SK, Dhar N, Patil A, Negi P, Perween A, Viswanathan R, Lünsdorf H, Tyagi P et al (2013) Dengue‐specific subviral nanoparticles: design, creation and characterization. J Nanobiotechnology 11: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Yang M, Lai H, Sun H, Chen Q (2017) Virus‐like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci Rep 7: 7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, Julander JG, Tang WW, Shresta S, Pierson TC et al (2017) Modified mRNA vaccines protect against Zika virus infection. Cell 168: 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rouvinski A, Dejnirattisai W, Guardado‐Calvo P, Vaney M‐C, Sharma A, Duquerroy S, Supasa P, Wongwiwat W, Haouz A, Barba‐Spaeth G et al (2017) Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat Commun 8: 15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Slon Campos JL, Marchese S, Rana J, Mossenta M, Poggianella M, Bestagno M, Burrone OR (2017) Temperature‐dependent folding allows stable dimerization of secretory and virus‐associated E proteins of Dengue and Zika viruses in mammalian cells. Sci Rep 7: 966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L et al (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV‐1. Science 329: 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Cohen J (2017) If you haven't had dengue infection, don't use our vaccine, drug company warns. Science https://doi.org/10.1126/science.aar6371 [Google Scholar]


