Abstract
Background
DAZAP1 (DAZ Associated Protein 1) was originally identified by a yeast two-hybrid system through its interaction with a putative male infertility factor, DAZ (Deleted in Azoospermia). In vitro, DAZAP1 interacts with both the Y chromosome-encoded DAZ and an autosome-encoded DAZ-like protein, DAZL. DAZAP1 contains two RNA-binding domains (RBDs) and a proline-rich C-terminal portion, and is expressed most abundantly in the testis. To understand the biological function of DAZAP1 and the significance of its interaction with DAZ and DAZL, we isolated and characterized the mouse Dazap1 gene, and studied its expression and the subcellular localization of its protein product.
Results
The human and mouse genes have similar genomic structures and map to syntenic chromosomal regions. The mouse and human DAZAP1 proteins share 98% identity and their sequences are highly similar to the Xenopus orthologue Prrp, especially in the RBDs. Dazap1 is expressed throughout testis development. Western blot detects a single 45 kD DAZAP1 protein that is most abundant in the testis. Although a majority of DAZAP1 is present in the cytoplasmic fraction, they are not associated with polyribosomes.
Conclusions
DAZAP1 is evolutionarily highly conserved. Its predominant expression in testes suggests a role in spermatogenesis. Its subcellular localization indicates that it is not directly involved in mRNA translation.
Background
Spermatogenesis is a complex developmental process in which male germ cells progress through mitotic proliferation, meiotic division and dramatic morphological changes to form mature sperm. This process is vital for the propagation of a species, and involves a large portion of the genome of an organism to ensure the quality and quantity of the final products. It is estimated that mutations in up to 11% of all genes in Drosophila might lead to male sterility [1]. This is likely to be true for humans also, considering the extremely high incidence (4–5%) of infertility in men [2]. Among the genes associated with male infertility is the DAZ (Deleted in Azoospermia) gene family. The family includes the Y-linked DAZ genes that are present only in great apes and old world monkeys [3], and the autosomal DAZL1 (DAZ-like 1) and BOULE genes [4,5] in all mammals. Deletion of the DAZ genes is found in about 10% of infertile males with idiopathic azoospermia [2], and disruption of Dazl1 causes infertility in both male and female mice [6]. Mutations in the DAZ family members of Drosophila[7], C. elegans[8], and Xenopus[9] also affect the fertility in either males, females, or both sexes.
The DAZ gene family encodes RNA binding proteins that are expressed specifically in germ cells. DAZ and DAZL are expressed in the nucleus and cytoplasm of primordial germ cells and spermatogonia, and in the cytoplasm of meiotic spermatocytes [6,10]. BOULE is expressed later, in the cytoplasm of pachytene spermatocytes [5]. Genetic and biochemical studies suggest a role for the DAZ family in the regulation of mRNA translation. Drosophila Boule mutants was defective in the translation of the meiosis-specific CDC25 homologue, Twine [11], and DAZL was found to be associated with polyribosomes in mouse testes [12]. More recently, DAZL was shown both in vitro and in a yeast three-hybrid system to bind specifically to oligo(U) stretches interspersed by G or C residues, including a U-rich segment in the 5' UTR of mouse Cdc25C mRNA [13].
In an attempt to elucidate the function of the DAZ gene family and to understanding the mechanisms of its action, we used a yeast two-hybrid system to isolate two human genes encoding DAZ associated proteins (DAZAPs) [14]. One of them, DAZAP1, is expressed predominantly in testes. It encodes a protein with two RNA binding domains and a proline rich C-terminal portion. The DAZAP1 protein interacted with both DAZ and DAZL in vitro. It also bound to RNA homopolymers. We now report our characterization of the mouse Dazap1 gene and its protein product. The subcellular localization of DAZAP1 suggests that it is not involved directly in mRNA translation.
Results
Characterization of the mouse Dazap1 cDNA
Mouse Dazap1 cDNA clones were isolated by library screening, and the 5' end of the cDNA was isolated by 5' RACE [15]. The near fall length cDNA consists of a 53 bp 5' untranslated region (UTR), an open reading frame for a protein of 405 amino acid residues, and a 362 bp 3' UTR (GenBank Accession No: AF225910). The coding region shares 89% similarity with that of the human orthologue. The 3' UTR sequence is remarkably conserved. It contains three segments of 35 bp, 133 bp and 90 bp that share 85%, 90%, and 97% similarity with segments in the human 3' UTR, respectively. These segments probably contain regulatory elements.
The DAZAP1 protein contains two RNA-binding domains (RBDS) and a C-terminal portion that is rich in proline (Figure 1). It is highly conserved evolutionarily. The mouse and the human proteins differ in 9 amino acids only, with 7 substitutions and two deletions/insertions. The mammalian proteins shares 89% similarity and 81% identity with Xenopus Prrp (for proline-rich RNA binding protein) [16]. The two RBDs are especially highly conserved. They share 98% and 97% similarity and 97% and 92% identity, respectively, between DAZAP1 and Prrp. These proteins may therefore have a similar RNA binding specificity. The C-terminal proline-rich portions of DAZAP1 and Prrp are less conserved (81% similarity and 71% identity). There is an insertion of a 58 bp segment in Prrp cDNA that causes a change of reading frame and results in a shorter Prrp with a different C-terminal end sequence.
Figure 1.
Evolutionary conservation of the DAZAP1 proteins. The amino acid sequences of the human and mouse DAZAP1s and the Xenopus Prrp are compared. The two RNA binding domains are boxed. Differences between the human and the mouse sequences, and between the mouse and Xenopus sequences are marked by #'s and *'s, respectively.
Genomic structure of Dazap1 and chromosomal mapping
Several overlapping lambda clones containing mouse Dazap1 genomic sequences were isolated. The locations of exons were determined by PCR amplification across exon-intron boundaries following by sequencing. All but the first exon were isolated and mapped. The genomic structure of the human DAZAP1 gene was also determined by blasting the human genome sequence at National Center for Biotechnology Information with the human DAZAP1 cDNA sequence. The mouse and the human genes have very similar structures, consisting of 12 exons spanning about 28 kb. All intron insertion sites are conserved (Table 1). The two RBDs are encoded by exons 1–4 and 5–8, respectively.
Table 1.
Exon-intron Organization of the Mouse and Human DAZAP1 genes
| Exon | Exon Size (bp) | Intron insertion sitesa | Intron size (kb) | |||
| Mouse | Human | Mouse | Human | Mouse | Human | |
| 1 | >82 | >137 | 82 | 137 | ? | 9.70 |
| 2 | 41 | 41 | 123 | 178 | 0.40 | 0.66 |
| 3 | 167 | 167 | 290 | 345 | 0.19 | 0.30 |
| 4 | 66 | 66 | 356 | 411 | 2.7 | 2.42 |
| 5 | 108 | 111 | 464 | 522 | 0.7 | 1.09 |
| 6 | 49 | 49 | 513 | 571 | 2.6 | 3.44 |
| 7 | 83 | 83 | 596 | 654 | 2.1 | 2.88 |
| 8 | 154 | 154 | 750 | 808 | 1.2 | 0.97 |
| 9 | 30 | 30 | 780 | 838 | 0.28 | 0.22 |
| 10 | 141 | 141 | 921 | 979 | 0.95 | 2.15 |
| 11 | 177 | 177 | 1098 | 1156 | 2.2 | 2.0 |
| 12 | 535 | 564 | ||||
A pair of PCR primers was designed from Dazap1 intronic sequences that amplified mouse but not hamster genomic sequences. Using a panel of mouse-hamster radiation hybrids, the mouse Dazap1 gene was mapped to chromosome 10 placed 27.84 cR from D10Mit260 (lod > 3.0) (data not shown). This region is syntenic to human 19p13.3 where the human DAZAP1 gene is located [14,17]. It contains no known mutant alleles that are associated with infertility.
Expression of Dazap1
Northern analyses of adult mouse tissues showed the presence of two Dazap1 transcripts of 1.75 kb and 2.4 kb, respectively (Figure 2). Only the shorter transcript has been isolated in cDNA clones. Dazap1 was expressed most abundantly in the testis, much less in liver, heart and brain, and even less in other tissues. This pattern of expression is similar to that of the human DAZAP1 (14). RT-PCR analyses showed that Dazap1 mRNA was already present in fetal testes at embryonic day 15, similar to Dazl1 mRNA (Figure 3). The expression of both Dazl1 and Dazap1 persisted throughout testes development, in both the prenatal and postnatal periods. Dazl1 and Dazap1 transcripts were also present in the testes of Wv/Wv mutant mice which contained diminished number of germ cells [18]. However, only Dazap1 was expressed in a mouse germ cell line GCl-spg [19] and a Sertoli cell line MT4. The results suggest that Dazap1 is expressed in both somatic and germ cells in the testis.
Figure 2.
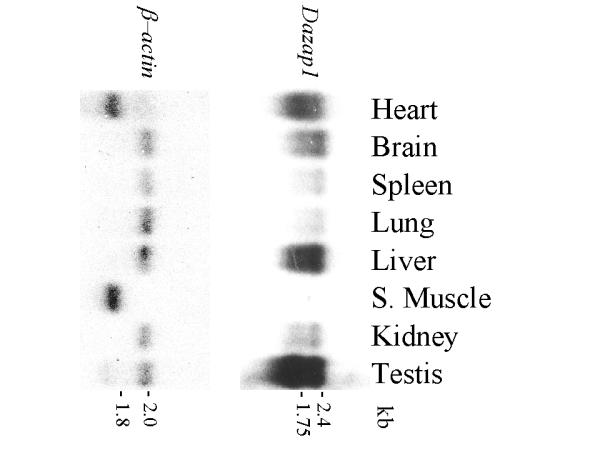
Expression of Dazap1 in adult mouse tissues. A mouse multiple-tissue Northern blot was hybridized with a Dazap1 cDNA probe, stripped, and rehybridized with a β-actin probe. Dazap1 is expressed most abundantly in the testis.
Figure 3.
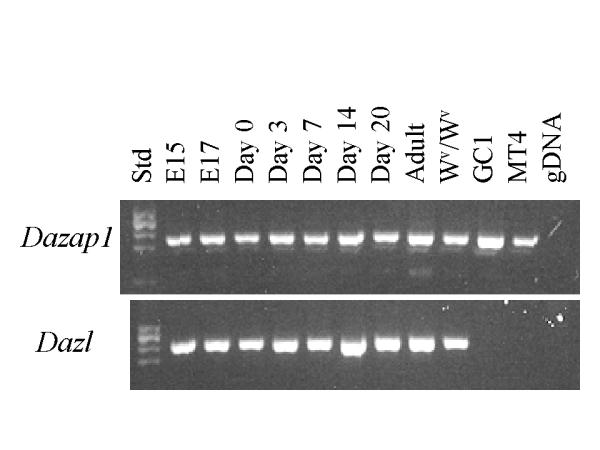
Developmental expression of Dazap1 and Dazl in mouse testes. RT-PCR was performed on total testicular RNAs isolated from day 15 (El 5) and day 17 (El 7) embryos, new born mice (Day 0), and mice at various days after birth. Wv/Wv testes contain diminished germ cell population due to a mutated W (White spotted) gene. GC1 and MT4 are mouse germ cell and Sertoli cell lines, respectively, and gDNA is mouse genomic DNA. The PCR primers span over introns and produce much larger (if any) fragments from genomic DNA.
To study the expression of the DAZAP1 protein, two antibodies against mouse DAZAP1 were generated. The anti DAZAP1-C antibody was raised against a recombinant protein containing the C-terminal proline-rich portion, and the anti DAZAP1-P antibody was raised against an oligopeptide containing the last 19 amino acid residue at the C-terminus. Both antibodies recognized in vitro synthesized DAZAP1 in an immunoprecipitation assay (data not shown). Western blotting of mouse tissue extracts detected a 45 kD protein that was present most abundantly in the testis, and to a lesser degree in spleen, liver, lung and brain (Figure 4). The protein was also present in the ovary. The expression of DAZAP1 during germ cell development paralleled that of DAZL (Figure 5). It is present at a low level in the testes of 6 days old mice which contained only primitive type A spermatogonia. The expression of DAZAP1 increased afterward, as the testes contained increasing number of proliferating and meiotic germ cells.
Figure 4.
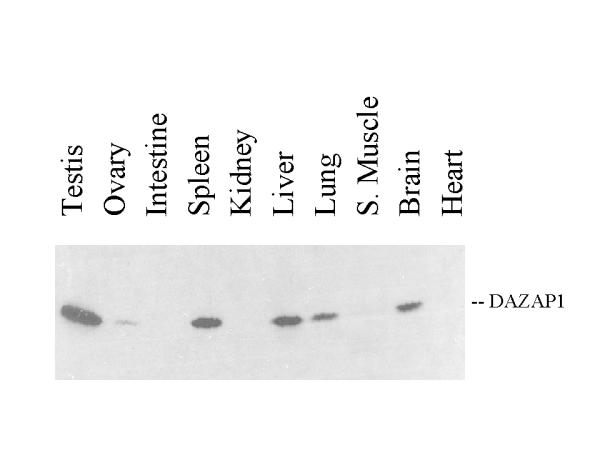
Expression of the DAZAP1 protein in adult mouse tissues. Equal amounts of total protein from various tissue extracts were applied to a 10% SDS-polyacrylamide gel and western blotted with the anti-DAZAP1-P antibody.
Figure 5.
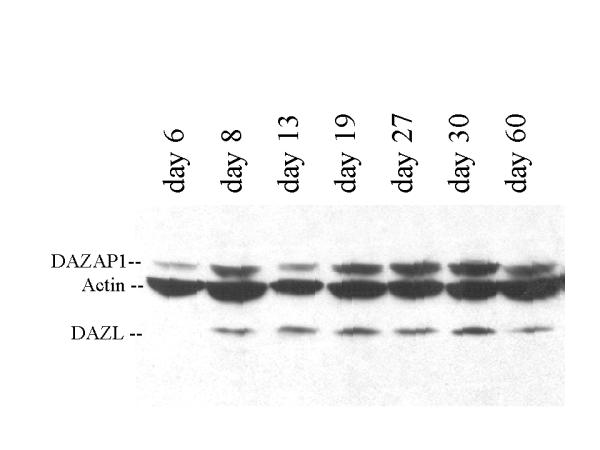
Western blot analyses of the expression of DAZAP1 and DAZL in mouse testes during postnatal development.
Subcellular localization of DAZAP1
Our previous fractionation of mouse testis extracts showed that most DAZL were present in the post mitochondrial fraction, and some of them were associated with polyribosomes [12]. Similar analyses showed that a majority of DAZAP1 in adult mouse testes was also present in the cytoplasmic fraction (data not shown). However, sucrose gradient analyses of the post- mitochondria fraction showed that, unlike DAZL, DAZAP1 did not co-sediment with polyribosomes (Figure 6).
Figure 6.
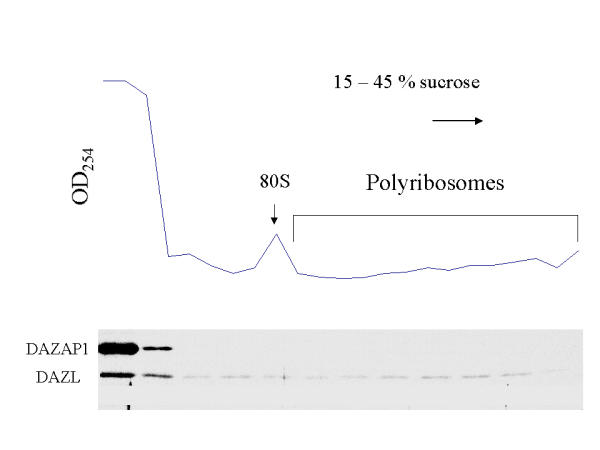
Sucrose gradient analyses shows that DAZAP1 is not associated with polyribosomes. The post-mitochondrial supernatant of mouse testis extracts was analyzed on a 15–45% sucrose gradient. Sedimentation was from left to right. The presence of DAZAP1 and DAZL in each fractions was analyzed by Western blotting.
Discussion
RNA-binding proteins have been found to participate in many cellular functions, including RNA transcription, pre-mRNA processing, mRNA transport, localization, translation and stability [20]. A role for the DAZ family in the regulation of mRNA translation is supported by lines of circumstantial evidence, including the association of DAZL with polyribosomes [12]. The absence of DAZAP1 from polyribosomes indicates that it is not directly involved in protein synthesis. This finding is different from two RNA-binding proteins, FXR1P and FXR2P, that were identified through their interaction with another polysomal-associated RNA-binding protein, the fragile X mental retardation protein [21]. Both FXR1P and FXR2P are associated with the polyribosomes [22].
The significance of the interaction between DAZAP1 and DAZL/DAZ remains to be defined. These proteins may act together to facilitate the expression of a set of genes in germ cells. For example, DAZAP1 could be involved in the transport of the mRNAs of the target genes of DAZL. Alternatively, DAZL and DAZAP1 may act antagonistically to regulate the timing and the level of expression. Such an antagonistic interaction between two interacting RNA-binding proteins is exemplified by the neuron-specific nuclear RNA-binding protein, Nova-1. Nova-1 regulates the alternative splicing of the pre-mRNAs encoding neuronal inhibitory glycine receptor α2 (GlyR α2) [23]. The ability of Nova-1 to activate exon selection in neurons is antagonized by a second RNA-binding protein, brPTB (brain-enriched polypyrimidine tract-binding protein), which interacts with Nova-1 and inhibits its function [24]. DAZAP1 could function in a similar manner by binding to DAZL and inhibiting its function. Comparing the phenotypes of Dazl1 and Dazap1 single and double knock-out mice may provide some clues to the significance of their interaction. Dazl1 knock-out mice have already been generated and studied [6]. The spermatogenic defect in the male becomes apparent only after day 7 post partum when the germ cells are committing to meiosis (H. Cooke, personal communication). The genomic structure of Dazap1, delineated here, should facilitate the generating of Dazap1 null mutation.
DAZAP1 was shown to bind RNA homopolymers in vitro, with a preference for poly U and poly G. Its natural substrates have not been identified. Recently, the Xenopus orthologue of DAZAP1, Prrp, was identified and characterized [16]. Prrp binds to a 340 nt sequence in the 3' UTR of Xenopus Vg1 mRNA. This Vg1 localization element (VLE) is sufficient for the migration and clustering of Vg1 mRNA to the vegetal cortex of mature oocyte. Prrp also interacts through its proline-rich domain with two microfilament-associated proteins profilin and Mena, which may facilitate the anchoring of Vg1 mRNA to the vegetal cortex. The Vg1 RNA encodes a member of the transforming growth factor-β family that is required for generating dorsal mesoderm at the blastula stage of Xenopus embryogenesis [25]. Sequence conservation between the RBDs of DAZAP1 and Prrp suggests that these proteins may bind to similar RNA sequences. However, a BLAST search of the GenBank for the 340 nt VLE sequence failed to identify any mammalian sequences with significant homology. Further mapping of the RNA sequence within VLE that binds Prrp, and possibly DAZAP1, may help to identify the natural substrates of DAZAP1.
Conclusions
DAZAP1 is an evolutionarily conserved RNA-binding protein. It is present at variable levels in many tissues. Its predominant expression in testes suggests a role in spermatogenesis. In mouse testes, DAZAP1 was found both in the nuclei and in the cytoplasm. Its absence from polyribosomes indicates that it is not directly involved in mRNA translation.
Materials and methods
Isolation of mouse Dazap1 cDNA clones
Dazap1 cDNA clones were isolated from a mouse testis cDNA library (#937308, Stratagene, La Jolla, CA) using a human DAZAP1 cDNA as a probe. The 5' end of the cDNA was isolated by 5' RACE [15], using prdap11 (ttgcgggccatatccttg, #749–732) as the primer for cDNA synthesis from mouse testis RNA, and prdap37 (ttgttgccacgtgggcg, #734–718) and an adaptor primer as the primers for PCR amplification. The PCR products were cloned into a TA cloning vector pCR2.1-TOPO (Invitrogen, Carlsbad CA). Dazap1 clones were identified by colony hybridization and sequenced. The 5' RACE clone with the longest 5' UTR region and the cDNA clone P21 were ligated together through a unique PmlI site at # 722 to generate a cDNA clone (Dazap1-C) with a near full-length insert.
Chromosomal mapping of Dazap1
Dazap1 genomic clones were isolated from a mouse 129SV genomic library (#946305, Stragagene), and sequences flanking each exons were determined. PCR primers (prdap25: cacctccaggatgtgttagc and prdazp26:gtcaccaagggtgtctgaag) were designed from intronic sequences flanking Dazap1 exon 8. These primers amplified a 271 bp fragment from mouse but not hamster genomic DNA. DNA samples of a panel of 100 radiation hybrids containing mouse chromosome fragments in a hamster background were purchased from Research Genetics (Huntsville, AL). The presence of mouse Dazap1 in the radiation hybrids was determined by PCR and the results were sent to the MIT server http://www.gonome.wi.mit.edu/mouse_rh/index.hlml for computerized physical mapping of the gene.
Expression of Dazap1 transcripts
Northern hybridization was carried out according to standard procedures [26] using a mouse Multiple Tissue Northern Blot #7762–1 from Clontech (Palo Alto, CA). The blot was hybridized sequentially with DAZAP1 and β-actin cDNA probes, with stripping of the bound probes in between.
Reverse transcription-polymerase chain reaction (RT-PCR) was carried out as preciously described using an annealing temperature of 54°C [27]. The primers were prdap35: agctcagggagtacttcaaga and prdap24 :ggagcttgattcttgctgtcc for Dazap1 which generated a product of 211 bp, and prdaz71: atcgaactggtgtgtcgaagg and prdaz72: ggaggctgcatgtaagtctca for Dazl1 which generated a product of 245 bp. Both primer pairs annealed across intron insertion sites.
Generation of anti-DAZAP1 antibodies
Antibodies were generated against both a recombinant protein produced in E. coli and an oligopeptide synthesized in vitro. The insert of a Dazap1 cDNA clone P21, which encoded the C-terminal portion of DAZAP1 (starting from aa #197), was cloned in-frame into the EcoRI/XhoI sites of an expression vector pET32b (Novagen, Madison, WI). Sequences at the junctions were verified by DNA sequencing. Milligrams of fusion proteins between thioredoxin and DAZAP1 were prepared and purified on His-Bind metal chelation resins (Novagen, Madison, WI). The proteins were mixed with Freund's adjuvant and injected into rabbits to generate the anti-DAZAPl-C antibody. An oligopeptide containing the last 19 ammo acid residues of the mouse DAZAP1 was synthesized in vitro using the services of Bethyl Laboratories (Montgomery, TX). The peptide was conjugated to KLH as carrier and injected into a goat. The anti-DAZAP1-P antibody thus produced was purified on an affinity column containing the oligopeptide antigen.
Western blotting
Mouse tissues were homogenized in the RIPA lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% DOC, 0.1% SDS, 50 mM Tris, pH 8.0) at a concentration of 0.2 g tissue per ml of buffer. The homogenized samples were cleared of debris by centrifugation at 10,000 × g for 10 minutes. Protein concentration of the tissue extracts was determined by the Bradford method using the Bio-Rad Protein Assay system (Bio-Rad, Hercules, CA). About 50 μg of tissue extracts were separated on 10% SDS-polyacrylamide gels and blotted with either the anti-DAZAP1-C antibody (at a 1/2,000 dilution) or the anti-DAZAP1-P antibody (at a 1/5,000 dilution). After incubation with horseradish peroxidase-conjugated secondary antibodies, the binding of antibodies was detected using the ECL Western Blotting System (Amersham Pharmacia Biotech, Piscataway, NJ).
Fractionation of mouse testicular extracts
Adult mouse testes were homogenized in a buffer containing 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, 40 U/ml of Rnasin ribonuclase inhibitor (Promega, Madison, WI), and a mixture of 10 protease inhibitors provided in the Protease Inhibitors Set (Roche Molecular Biochemicals, Indianapolis, IN). Homogenates were centrifuged at 1,000 × g for 10 minutes to pellet cell debris and nuclei. After an additional centrifugation at 10,000 × g for 10 minutes to pellet the mitochondria, aliquots of the supernatant were applied to 15–45% sucrose gradients in 20 mM Tris, 100 mM KCl and 5 mM MgCl2 and centrifuged in a Beckman SW41 rotor at 39,000 rpm for 2 hours at 4°C. Fractions of 0.5 ml were collected from the bottom of the tubes and analyzed by western blotting.
Acknowledgments
Acknowledgments
We thank Gary Kuo for his involvement in the construction of Dazap1 expression vectors, and Ron Swerdloff's group for helpful discussion. The work was supported by grants from the National Institutes of Health (HD28009 and HD36347). Y. Vera was supported by an NIH grant (GM56902) on Initiative for Minority Student Development.
Contributor Information
Tiane Dai, Email: tianedai@hotmail.com.
Yanira Vera, Email: yvera717@aol.com.
Eduardo C Salido, Email: esalido@ull.es.
Pauline H Yen, Email: pyen@rei.edu.
References
- Hackstein JHP, Hochstenbach R, Pearson PL. Towards an understanding of the genetics of human male infertility. TIG. 2000;16:565–572. doi: 10.1016/S0168-9525(00)02140-5. [DOI] [PubMed] [Google Scholar]
- De Kretser DM. Male infertility. Lancet. 1997;349:787–790. doi: 10.1016/S0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee T-Y, Salo P, Allagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page DC. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Cooke HJ, Lee M, Kerr S, Ruggiu M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- Xu EY, Moore FL, Pera RAR. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. PNAS. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse DAZLA gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Eberhar CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Reijo RA, Dorfman DM, Slee R, Renshaw AA, Laughlin KR, Cooke H, Page DC. DAZ family proteins exist through male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Daz1 orthologue Boule. Nat Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Daz1 protein. Nucl Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S, Dai T, Roettger S, Schempp W, Salido EC, Yen PY. Identification of two novel proteins that interact with germ-cell specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. PNAS. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Jiang C, Kroll TT, Huber PW. proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 2001;20:2315–2325. doi: 10.1093/emboj/20.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- Fleischman RA. From white spots to stem cells: The role of the Kit receptor in mammalian development. Trends Genet. 1993;9:285–290. doi: 10.1016/0168-9525(93)90015-A. [DOI] [PubMed] [Google Scholar]
- Hofmann MC, Narisawa S, Hess RA, Millan JL. Immortalization of germ cells and somatic testicular cells using the SV40 large T antigen. Exp Cell Res. 1992;201:417–435. doi: 10.1016/0014-4827(92)90291-F. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. RNA-binding proteins as regulators of geneexpression. Cur Opin Genet Develop. 1997;7:345–353. doi: 10.1016/S0959-437X(97)80148-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, O'Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR2 proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/S0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. PNAS. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson GH, Melton DA. Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-G. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual 2nd ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1989.
- Salido EC, Yen PH, Mohandas TK, Shapiro LJ. Expression of the X-inactivation-associated gene XIST during spermatogenesis. Nat Genet. 1992;2:196–199. doi: 10.1038/ng1192-196. [DOI] [PubMed] [Google Scholar]



