ABSTRACT
Local translation is critical for diverse aspects of neuronal function, including mediating responses of elongating axons to guidance cues and other signaling molecules. A major determinant of the protein synthetic capacity of axons and growth cones is the specific set of mRNAs that are trafficked to these sites. However, recently it has become clear that the axonal transcriptome can also be shaped by local RNA degradation mechanisms, such as nonsense-mediated decay. Here we show that Staufen1-mediated decay can also occur within axons and mediate degradation of specific axonal transcripts. We show that Staufen1 and Upf1, which function together in Staufen1-mediated decay, are localized in growth cones. Selective depletion of Staufen1 from neurons results in a complex pattern of transcriptional alterations, with a subset of transcripts showing increased expression and increased RNA half-life consistent with their regulation by Staufen1-mediated decay. Additionally, we show certain transcripts, such as Rac1, are regulated by Staufen1 within axons and growth cones. The functional significance of Staufen1 in growth cones is supported by morphological alterations in growth cones following Staufen1 knockdown. Together these data point to Staufen1-mediated decay as a novel mechanism to control mRNA expression levels in axons and growth cones through local RNA degradation.
KEYWORDS: mRNA degradation, mRNA stability, local translation, Staufen1, growth cones, axons
Introduction
During embryonic development, local translation is a major mechanism in neurons that controls axon guidance.1-3 Axons that are elongating towards their target cells contain ribosomes and mRNAs, which are locally translated in response to specific guidance cues.1,2,4 Local translation affects the protein composition of growth cones, thereby affecting the responses to guidance cues. Thus, local translation has critical roles for the development of the nervous system.
Although local translation is important for axonal growth and guidance, little is known regarding how mRNA levels are regulated in axons. Numerous studies have focused on specific trafficking elements in mRNAs that mark them for transport.5,6 Although trafficking determines which mRNAs can be present in axons and growth cones, once the mRNAs are localized, their expression levels will be determined by their half-life. Therefore, the pathways that control mRNA degradation in axons and dendrites could ultimately determine the composition of the local mRNA transcriptome, and therefore determine the protein synthesis potential at local translation sites.
We previously demonstrated that “local degradation” has important roles in controlling the levels of local protein synthesis and ultimately determining axon guidance behavior in neurons.7 We found that Upf1 and Smg1, two proteins that promote nonsense-mediated mRNA decay (NMD), are readily detected in mouse commissural axons and growth cones.7 These proteins target specific mRNAs for degradation during axon pathfinding.7 One of these transcripts is Robo3.2, an NMD target mRNA encoding a receptor involved in axon guidance. Robo3.2 mRNA is translationally silent until the growth cone encounters translation-promoting signals at the spinal midline, resulting in its translation and degradation.7 The idea that localized NMD can influence mRNA expression levels has been extended to Arc transcripts in dendrites of hippocampal neurons.8 Thus, one pathway that controls mRNA levels in axons is the NMD pathway.
Although NMD is important for regulating Robo3.2 mRNA levels, it is unlikely that this pathway would regulate a large subset of transcripts. NMD requires the presence of specific transcript features that recruit the NMD machinery. For example, Robo3.2 contains a retained intron7 that causes the ribosome to encounter a stop codon before its typical location in mRNAs, which is the final exon. However, very few transcripts contain retained introns which would make them NMD targets.9,10 Since endogenous NMD targets are relatively rare, it is possible that the high levels of Upf1 in diverse types of axons7 could have functions other than NMD.
Notably, Upf1 also participates in the Staufen1-mediated mRNA decay (SMD) pathway.11-14 Staufen1 is a multifunctional double-stranded RNA-binding protein.15,16 Like NMD, SMD depends on active translation but involves ribosome-mediated detection of Staufen1 binding to the target mRNA. Notably, Staufen1 has been found in axons where its function is linked to the transport of mRNA-containing ribonucleoprotein particles (RNPs) along microtubules within dendrites and axons.17-20 Depletion of functional Staufen1 leads to defects in dendrites and synapse development in hippocampal neurons.21 Since Staufen1 is localized to axons, it is possible that SMD may function in axons to regulate the local transcriptome.
Here we show that Staufen1 contributes to the regulation of mRNA expression levels in axons. Using knockdown approaches, we find that a subset of mRNAs show increased expression levels and increased mRNA half-life upon Staufen1 depletion. We also find that some of these transcripts are localized to axons, and within axons, the mRNA half-life is extended upon Staufen1 depletion, suggesting that Staufen1 controls the stability and expression levels of specific mRNAs in axons. Notably, we find that Staufen1 exists in morphologically distinct granules in axons compared to growth cones, with Staufen1 in growth cones appearing at sites that are in proximity to Upf1, suggesting that these proteins interact to mediate SMD. The importance of Staufen1 and potentially SMD is supported by our finding that Staufen1-deficient neurons exhibit marked abnormalities in growth cone morphology. Together, these findings suggest that axonal growth cones are sites of SMD, and demonstrate that the axonal transcriptome is shaped, in part, by the activity of the SMD pathway.
Materials and methods
DRG isolation and culture
DRG culture was performed as described previously.22 In brief, timed pregnant mice was euthanized using CO2 and E13 embryos were harvested. The embryos were then collected into a 10 cm petri dish containing L-15 media (Gibco) and the DRG were extracted from the spinal cord and incubated with 0.1% trypsin and 500 μg/ml collagenase for approximately 15 minutes. The tissues were then triturated using 18G and 23G needles. DRG neurons were counted and seeded onto a poly-D-lysine (PDL, Gibco) and mouse laminin (Trevigen)-coated plates. DRG neurons were cultured in 1x Neurobasal medium (Gibco) supplemented with 2% B27 (Gibco), 1% L-glutamine (Invitrogen), 1% penicillin-streptomycin (Gibco) and 50 ng/ml NGF (Harlan Bioproducts). To eliminate non-neuronal cells, the DRG neuronal cultures were treated with 20 μM 5-FU (Sigma) and 5 μM aphidicoline (EMD Millipore).
Immunofluorescence
DRG neurons were fixed using 4% formaldehyde on day 4 or 6, as indicated and immunofluorescence was performed as described previously.23,24 To stain for F-actin, the CytoPainter F-actin staining kit (Abcam) was used. The neurons were then washed thoroughly and mounted onto glass slides using ProLong Antifade Diamond Mountant (Invitrogen). Neurons images were captured with Zeiss LSM880.
Generating lentivirus
293LTV cells were cultured in 1x DMEM supplemented with 10% FBS (Invitrogen), 1% L-glutamine (Invitrogen), 1% penicillin-streptomycin (Gibco), and 1x non-essential amino acids (NEAA, Gibco). The night before lentiviral plasmid transfection, the cells were split onto 10 cm petri dishes to achieve 50–60% cellular confluency at the time of the transfection. Lentiviral plasmids used were pΔ8.9, pMD2.G, and pRRL. The pRRL plasmid contains shRNA against lacZ (control) or Staufen1 and a GFP construct. The cells were transfected using Calcium Phosphate Transfection Kit (Invitrogen) following the manufacturer's protocol (Day 0). On Day 1, the media was discarded and the cells were washed with pre-warmed 1x PBS for two times. Fresh media was added to the cells immediately after. On Day 2, the first batch of lentivirus containing supernatant was collected. To eliminate cellular debris, the supernatant was centrifuged at 4°C, 2500 rpm in a tabletop centrifuge for 10 minutes and run through a 0.45 μM filter. The second batch of the lentiviral supernatant was collected on Day 3. The second batch supernatant was also purified by the centrifugation and the filtration. Two batches of the supernatant were combined, aliquoted, and stored at −80°C. shRNA were as follows:
| lacZ shRNA | 5′- GACTACACAAATCAGCGAT-3′ |
| 3′- CTGATGTGTTTAGTCGCTA-5′ | |
| Staufen1 shRNA 2 | 5′-GCAGTCCACCTACAGCTAT-3′ |
| 3′-CGTCAGGTGGATGTCGATA-5′ | |
| Staufen1 shRNA 4 | 5′-GCCAGAAAGGTTGGAGGTA-3′ |
| 3′-CGGTCTTTCCAACCTCCAT-5′ |
After culturing DRG neurons for 2 days at 37°C, lentivirus-containing supernatant was added to the neurons. The neurons were incubated at 37°C for overnight. The supernatant was discarded the next day, and an aliquot of the fresh Neurobasal media was added to the neurons. The cells were incubated at 37°C for additional four days.
Western blot
Primary antibodies used for western blot were: Staufen1 (1:500 dilution, Abcam), RhoA (1:1000 dilution, Santa Cruz), or GAPDH (1:1000 dilution, Abcam). Chemiluminescent signals were captured with the chemiluminescence dock (BioRad).
Generating RNA-Seq libraries
Staufen1 was depleted using shRNA delivered by lentivirus in DRG neurons. Total RNA samples were prepared with ReliaPrep RNA Cell Miniprep System (Promega) following the manufacturer's protocol. cDNA was synthesized using 10 ng of the RNA samples and SMARTer Ultra Low Input RNA Kit for Sequencing V3 (Clontech Laboratory). The concentration of the cDNA samples was measure with Quant-iT DNA Assay Kit, high sensitivity (Invitrogen). 10 ng of the cDNA samples was used to generate libraries using Nextera XT DNA Library Preparation Kit (Illumina) following the manufacturer's protocol. Sequences were analyzed using Flexbar, STAR, Samtools, Bedtools, HTseq, and DESeq2.
qRT-PCR
RNA samples were prepared using RNeasy Miniprep Kit (QIAGEN) following the manufacturer's protocol. cDNA samples were synthesized from the total RNA samples using SuperScript III (Invitrogen). The cDNA samples were then used to perform qRT-PCR using MasterCycler RealPlex2 (Eppendorf) and iQ SYBR Green Supermix (BioRad). The cDNA samples were probed for 18s rRNA, RPL19, β-actin, Rac1 and Nrp1 with the following primer sets that were designed with Primer3:
| 18s rRNA FW | 5′-AGGAATTGACGGAAGGGCACCA-3′ |
| 18s rRNA RV | 5′-TTATCGGAATTAACCAGACAA ATCG-3′ |
| RPL19 FW | 5′-AATGCCAACTCCCGTCAGCA-3′ |
| RPL19 RV | 5′-ACAGTCACAGGCTTGCGGATGA-3′ |
| β-actin FW | 5′-TGGTGGGAATGGGTCAGAAGGA-3′ |
| β-actin RV | 5′-TCAATGGGGTACTTCAGGGTCAGG-3′ |
| Rac1 FW | 5′-TGGGAGACGGAGCTGTTGGTAA-3′ |
| Rac1 RV | 5′-GGCCCAGATTCACTGGTTTTCC-3′ |
| Nrp1 FW | 5′-GGCCACAGAGAAGCCAACCATT-3′ |
| Nrp1 RV | 5′-TTGTGAGAGCCCCAGCCAAA-3′ |
Culturing DRG neurons in microfluidic chambers
A microfluidic chamber with the following design was used. The microfluidic chamber has two channels. We designated one channel as the soma channel and the other channel as the axon channel. The soma and axon channels are separated by 450 μm microgrooves. Each channel is connected to two reservoirs or compartments. The microfluidic chamber was placed onto a poly-D-lysine-coated glass bottom dish. All the channels and reservoirs were then coated with laminin. Approximately 2 × 105 neurons were introduced to one of the soma compartment. After letting the neurons attach for 10 minutes, 100 μl of Neurobasal media containing 25 ng/ml NGF, 20 μM 5-FU and 5 μM aphidicoline was added to the soma compartment. 200 μl of NBM containing 200 ng/ml NGF, 20 μM 5-FU and 5 μM aphidicoline was added to the axon compartment. Two days after plating the neurons, the media was changed to 100 μl of Neurobasal media with 5 ng/ml NGF, 20 μM 5-FU and 5 μM aphidicoline in the soma reservoirs. A fresh aliquot of 200 μl Neurobasal media with 200 ng/ml NGF, 20 μM 5-FU and 5 μM aphidicoline was added to the axon reservoirs after removing 2 day-old media.
Results
Staufen1 is detected near Upf1 in the DRG growth cones
Staufen1 has previously been shown to be present in axons due to its associated with axonally trafficked mRNA transport granules.6,20 Since Staufen1 can participate in SMD in a Upf1-dependent manner,11,14 we considered the possibility that axonal Staufen1 could have an additional function in controlling mRNA stability via SMD.
Upf1 is expressed in axons and particularly prominent in growth cones, which also are enriched in ribosomes and mRNA7. We reasoned that if Staufen1 interacts with Upf1 to promote Staufen-mediated mRNA decay, then Staufen1 should be localized adjacent to Upf1 in axons.
To test this, we examined localization of Staufen1 and Upf1 by immunofluorescence in the axons and the growth cones of embryonic day (E) 13, day in vitro (DIV) 4 mouse dorsal root ganglia (DRG) neurons. The axons and growth cones were counterstained using an antibody against β-tubulin and fluorescently tagged phalloidin, respectively. We readily detected punctate Staufen1 labeling along microtubules in the axons (Fig. 1A), as has been reported previously.20,25
Figure 1.
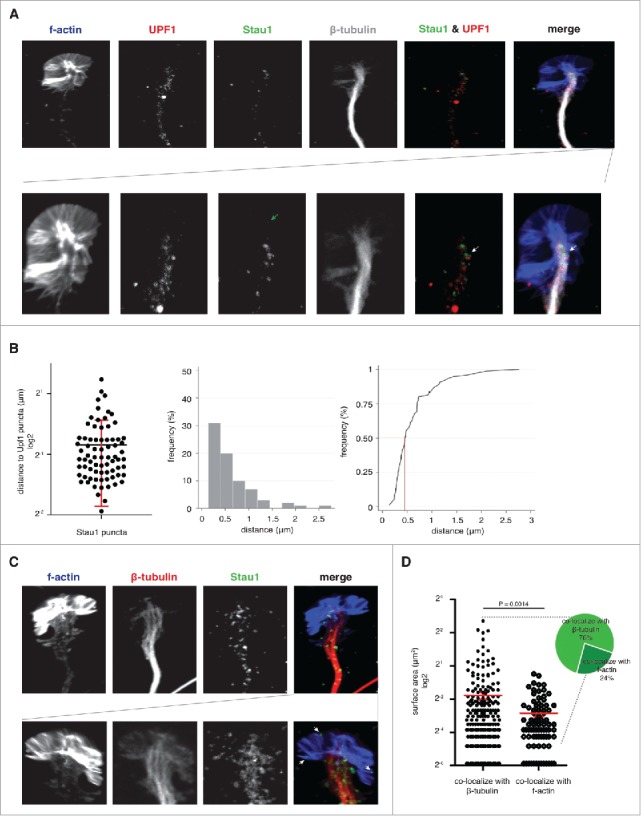
Staufen1 is detected near UPF1 in the mouse DRG axons and growth cones. A. Staufen1 is localized near UPF1 in the DRG growth cone. E14 DRG neurons were cultured for 4 DIV prior to staining to detect F-actin, Staufen1, UPF-1, and β-tubulin. The confocal images illustrate Staufen1 and UPF1's location within a DRG growth cone. Below is a magnified image of the growth cone to help visualize Staufen1 and UPF1 puncta. White arrows point to Staufen1 near UPF1 with a partial overlap. B. Half of Staufen1 puncta are located < 0.5 μm away from UPF1. The distance between Staufen1 and UPF1 was measured with ImageJ (n = 75) and plotted (left image) and represented as a histogram (middle image). A frequency distribution (right image) analysis shows that the majority of Staufen1 puncta are within 0.5 µm of a UPF1 puncta. The proximity of Staufen1 and UPF1 may be consistent with their interaction in SMD. C. Staufen1 particles show different sizes in axons compared to growth cones. Staufen1 and β-tubulin were detected by immunofluorescence and F-actin was detected using a CytoPainter F-actin staining kit (Abcam). Staufen1 puncta can be seen in axons in addition to growth cones. In many cases, Staufen1 puncta in growth cones were smaller than puncta in axons. White arrows point to examples of Staufen1 located at the distal ends of the growth cones. D. Average size of Staufen1-containing granules. Each dot represents a granule. Each granule was measured using ImageJ (n = 334 granules). The plot shows that the average size of the granules differs depending on their association with different cytoskeleton. Specifically, the granules at the distal ends are smaller than the granules close to β-tubulin.
In addition to axonal localization, we also noticed that Staufen1 and Upf1 were localized closely within growth cones (0.62 +/− 0.47 μm away from Upf1 puncta on average, n = 75, Fig. 1B). In several cases, partial Staufen1 and Upf1 overlap was detected (Fig. 1A). This raises the possibility that Stuafen1 and Upf1 could potentially interact in order to mediate SMD in axons and growth cones.
Staufen1 forms larger puncta in axons than growth cones
Although Staufen1 is normally considered a part of transport granules that are associated with microtubules and microtubule-based transport, we also noticed that Staufen1 puncta were also in growth cones (Fig. 1C). Often, Staufen1 was seen in the periphery of growth cones, which are sites where ribosomes and proteins synthesis has been detected.3 These sites are not normally associated with microtubules or microtubule-localized granules, suggesting that these are not likely to reflect Staufen1 granules that are being transported along microtubules. Thus, this pool of Stauen1 may have a different function than microtubule-proximal Staufen1.
In many cases, we observed Staufen1 puncta that were larger in axons along microtubules than in growth cones. To quantify these differences, we measured Staufen1 puncta cross-sectional area from 334 puncta using ImageJ. We divided the Staufen1 puncta into two groups depending on their localization to axons or growth cones based on their co-localization with β-tubulin or F-actin, respectively. The average size of the Staufen1 puncta associated with β-tubulin was significantly larger than the puncta associated with F-actin (288 +/− 44 nm2 vs. 138 +/− 16 nm2). Taken together, this demonstrates that the average size of Staufen1 puncta depends on its localization to axons or growth cones. The different sizes of Staufen1 puncta raise the possibility that Staufen1 could have distinct roles in axons and growth cones.
Staufen1 regulates DRG growth cone morphology
We next asked if Staufen1 has a transport-independent function in growth cones. To examine the role of Staufen1, we first generated lentiviral Staufen1-knockdown constructs. We cultured DRG neurons and infected them with lentivirus expressing a GFP marker and shRNA against lacZ (control) or Staufen1. Lentiviral expression of either of two shRNA (shRNA2 and shRNA4) resulted in substantial reduction in the levels of Staufen1 protein after four days (Fig. 2A).
Figure 2.
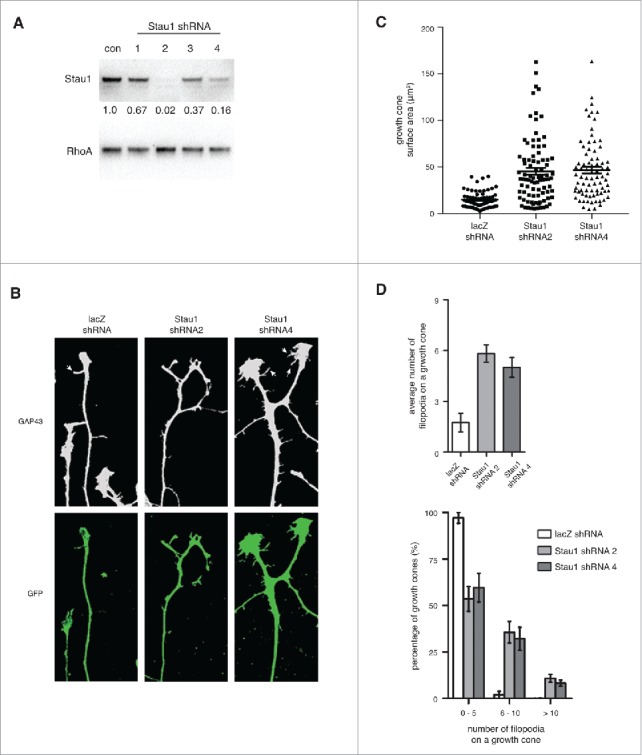
Depleting Staufen1 causes abnormal DRG growth cone morphologies. A. Staufen1 was depleted in neurons using a lentivirus-mediated shRNA expression. Lysates were prepared from the control and Staufen1-depleted neurons and Staufen1 was detected by immunoblotting. About 98% and 80% of endogenous Staufen1 was depleted by shRNA2 and shRNA4, respectively. B. Growth cones show changes in morphology following Staufen1 depletion. DRG neurons were cultured for two days. Then, shRNA against lacZ (control) and Staufen1 was delivered to the neurons by lentivirus infection. After removing the lentivirus, the neurons were cultured for additional four days. The neurons were fixed, permeabilized, and stained for Gap43 (1:1000, Abcam) and GFP (1:1000, Abcam). White arrows point to filopodia on a growth cones. C. Growth cones are enlarged when Staufen1 is depleted from neurons. A dot plot compares the averages size of the growth cones between the control and Staufen1-depleted neurons. Each dot represents the size of a growth cone (lacZ shRNA: circle, n = 67, Staufen1 shRNA 2: square, n = 88, Staufen1 shRNA 4: triangle, n = 78). The average sizes of the growth cones were 15.1 μm, 45.4 μm, and 46.7 μm for the control and Staufen1 depletion by shRNA2 and shRNA4, respectively. D. The average number of filopodia per growth cone significantly increases in the Staufen1-depleted compare to the control neurons. A bar graph shows the average number of filopodia per growth cone in the control (2 per growth cone, n = 59) and Staufen1-depleted neurons (shRNA 2: six per growth cone, n = 68, shRNA 4: five per growth cone, n = 63). A histogram shows a distribution of the growth cones from a low to high number of filopodia per growth cone. This indicates that Staufen1 is required for proper development of the growth cones.
We next examined the effect of Staufen1-depletion on growth cone morphology. DRG neurons expressing Staufen1 shRNA were stained for GAP43 to detect growth cones and for GFP to visualize axons of lentivirus-infected neurons. Staufen1-depleted growth cones appeared strikingly different from control (lacZ shRNA) growth cones (Fig. 2B). The Staufen1-depleted growth cones were enlarged and the mean number of filopodia per growth cone was increased (Fig. 2B and Supplemental Figure 1B). To quantify this difference, we measured the size of growth cones from control (n = 67) and Staufen1-depleted neurons (n = 88 for shRNA2 and n = 78 for shRNA4). The Staufen1-depleted growth cones were approximately three-fold larger (∼45.41 +/− 3.74 μm2 for shRNA2 and ∼46.67 +/− 3.65 μm2 for shRNA4) than the control growth cones (∼15.1 +/− 1.08 μm2, Fig. 2C).
Growth cones of Staufen1-depleted neurons also exhibited increased numbers of filopodia. Control growth cones had 2.08 filopodia per growth cone on average (n = 59, Fig. 2D) while Staufen1-depleted growth cones had approximately 4.85 to 5.99 filopodia per growth cone (n = 69 for shRNA2, n = 63 for shRNA4).
Next, we analyzed the number of filopodia per growth cone across a population of growth cones. We found that 97% of control growth cones had 0–5 filopodia and 1.9% had 6–10 filopodia. However, about 53–60% of Staufen1-depleted growth cones had 0–5 filopodia, 32–35% had 6–10 filopodia, and 8–11% had more than 10 filopodia (Fig. 2D). These data indicate that Staufen1 depletion results in a marked increase in the fraction of growth cones that have increased numbers of filopodia.
Taken together, these data show that Staufen1 is required for proper growth cone size and morphology and regulates the number of filopodia in growth cones.
Depleting Staufen1 alters the transcriptome in DRG neurons
We next wanted to identify targets of Staufen-mediated mRNA decay in DRG neurons. To identify Staufen1 targets, we monitored mRNA expression levels in control and Staufen1-depleted DRG neurons. cDNA libraries were prepared from poly(A) RNA for next-generation sequencing in three biological replicates. All three replicates were highly correlated with high R2 values ranging between 0.762 and 0.990 (Supplemental Figure 2B).
We first analyzed the sequencing data for differential gene expression in control and Staufen1-depleted neurons (Supplemental Figure 2A). The genomic data was presented using MA plots.26 The MA plots show that many transcripts exhibit significantly altered expression (1810 and 4233 genes for shRNA2 and shRNA4, Fig. 3A) when Staufen1 was depleted. Since Staufen1 depletion is expected to prevent SMD, SMD targets would show increased expression in Staufen1-depleted cells. However, of these expression changes, less than half were upregulated after Staufen1 depletion (729 for shRNA2 and 1962 for shRNA4 respectively). Notably, the effect of Staufen1-depletion is not primarily an increase in transcript expression as would be expected for a protein involved in RNA decay. This is consistent with the emerging view that Staufen1 might function in multiple RNA pathways in cells.15,16
Figure 3.
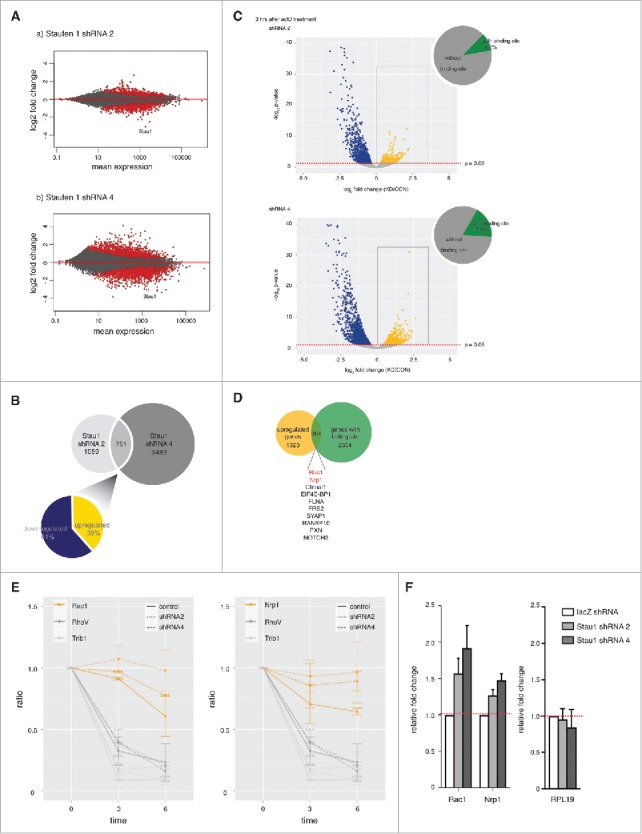
Depleting Staufen1 changes transcriptome patterns in DRG neurons and prolongs Staufen1 targets' RNA decay rate in DRG neurons. A. Staufen1 depletion causes significant changes in transcript abundance. Staufen1 was depleted in DRG neurons by infecting with lentivirus expressing Staufen1-specific shRNA. The neurons were incubated for four additional days after removing the lentivirus. Total RNA samples and RNA levels were measured by RNA-Seq. MA plots were generated using DESeq2. Red dots represent transcripts with a statistically significant change in abundance. B. Venn diagram of transcripts with altered expression level in different Staufen1-depletion experiments. Staufen1 depletion by shRNA2 showed a change in 1,810 transcripts, while Staufen1 depletion by shRNA4 showed a change in 4,233 transcripts. 751 transcripts were changed in both of these datasets. The pie chart shows the percentage of the 751 transcripts that are upregulated (61%, yellow) and downregulated (39%, dark blue). C. An actinomycin D time course to measure transcript decay rates reveals a subset of transcripts whose decay rate is dependent on Staufen1. In these experiments, we measured how transcript levels reduced at different time points after actinomycin D (ActD, 0.1 ng/ml) treatment for 0, 3, or 6 hours. RNA-Seq was performed to measure transcript abundance. The Staufen1-depleted neurons were compared to the control neurons at 3 hours after the ActD treatment. These results were presented in a volcano plots, which shows that there are a significant number of transcripts that show increased or decreased degradation rates upon Staufen1 depletion. Blue dots represent downregulated transcripts when Staufen1 was depleted. These transcripts appear to have degraded more quickly when Staufen1 was depleted. Yellow dots represent upregulated transcripts when Staufen1 was depleted. These transcripts exhibited greater stability when Staufen1 was depleted and may therefore represent SMD targets. These results identify potential SMD targets in neurons and suggest that some of the transcripts that show increased abundance in Staufen1-depleted cells show this increase because they have decreased decay rates after Staufen1 depletion. D. Identification of putative SMD targets. Shown is a Venn diagram comparing the upregulated transcripts (i.e., increased stability upon Staufen1 depletion) from each time point (yellow) and transcripts with known Staufen1-binding sites (green). 264 upregulated transcripts have Staufen1-binding sites. This identifies 264 transcripts that both bind Staufen1 and show increased stability upon Staufen1 depletion and may represent SMD targets. E. Count plots illustrate RNA reads at different time points relative to 0 hour for selected transcripts. Yellow lines represent Rac1 and Nrp1 (controls). Grey lines represent RhoV and Trib1. As shown in the plots, control transcripts RhoV and Trib1 with known half-life have a decay rate that was comparable to control when Staufen1 was depleted. However, for Rac1 and Nrp1, the decay rate was markedly reduced when Staufen1 was depleted. F. Confirmation by qRT-PCR that Rac1 and Nrp1 are upregulated when Staufen1 is depleted. cDNA samples were probed for RPL19, Rac1 and Nrp1 by qRT-PCR. The control transcript RPL19 was unaffected while predicted SMD targets Rac1 and Nrp1 show increased expression upon depletion of Staufen1.
Nevertheless, we next sought to identify mRNAs that could be Staufen-mediated mRNA decay targets. We first identified the transcripts whose levels were changed in both Staufen1 knockdown experiments, i.e., neurons expressing Staufen1 shRNA2 and in neurons expressing Staufen1 shRNA4. This resulted in 751 genes (Fig. 3B). Of these 751 genes, 39% were upregulated and 61% were downregulated (Fig. 3B). These upregulated genes were considered potential targets of Staufen-mediated mRNA decay in neurons in the subsequent experiments.
A subset of transcripts show enhanced stability in Staufen1-deficient neurons
We next wanted to determine if any of the upregulated transcripts in Staufen1-deficient cells might be increased due to impaired SMD. Since SMD reduces the half-life of mRNA, we asked if any of these transcripts show increased half-lives when Staufen1 is depleted.
To identify genes that show an increase in their half-life, we again depleted Staufen1 in DRG neurons and generated NGS libraries as described earlier but in the presence of actinomycin D (ActD), at three different time points (0, 3, and 6 hours after the ActD treatment). We prepared two replicates per condition per time point. The replicates were highly correlated, R2 value ranging between 0.76 and 0.95 (Supplemental Figure 3A). Volcano plots were prepared comparing the expression level of each transcript in the control and Staufen1-depleted neurons at 3 hours (Fig. 3C).
We reasoned that the expression level for a SMD target would be higher in the Staufen1-depleted neurons compared to control knockdown neurons at every time point after actinomycin D treatment. Thus, we specifically were looking for transcripts that appeared to be upregulated after actinomycin D treatment in Staufen1-depleted cells. These transcripts are indicated as yellow dots (352 for shRNA2 and 631 for shRNA4). In contrast, the blue dots indicate transcripts with downregulated RNA abundance in the Staufen1-depleted cells (1031 for shRNA2 and 1606 for shRNA4). These transcripts appear to be degrading faster after Staufen1 depletion and therefore are unlikely to be SMD targets.
We also compared the control and Staufen1-depleted neurons at 6 hours and observed a similar result (Supplemental Figure 3B).
The volcano plots (Fig. 3C and Supplemental Figure 4) clearly demonstrate that there was a small number of genes that showed delayed degradation in Staufen1-depleted neurons compared to control. This suggests that Staufen1 is required for proper decay of select transcripts in DRG neurons.
Rac1 and Nrp1 are SMD targets
We next sought to validate individual transcripts that are predicted to be SMD targets. To generate a high-confidence list of potential SMD targets, we combined the upregulated gene lists for shRNA2 and shRNA4 and then overlapped the upregulated gene lists from Fig. 3C to a list of genes with known Staufen1-binding sites.15,16 We found 264 genes were upregulated in both datasets and contained Staufen1-binding sites (Fig. 3D).
Since our goal was to eventually test whether any of these putative neuronal SMD targets are also targeted by SMD within axons, we wanted to identify transcripts that are also present in axons of embryonic DRG neurons. Thus, we compared the 264 genes to a set of DRG axonal transcripts. These transcripts were identified based on RNA-Seq analysis of axons obtained from DRG neurons cultured in microfluidic chambers. We found that about 20% of the 264 transcripts were also abundantly expressed in the DRG axons (top 10% by expression, Supplemental Figure 4B). This analysis resulted in a set of transcripts that could potentially be under Staufen1 control in axons.
We next selected Rac1 and Nrp1 for further analysis. We calculated their relative RNA decay rate in DRG neurons by comparing their normalized counts at 3- and 6- to 0- hour time point after actinomycin D treatment. The relative RNA decay rate of control transcripts RhoV and Trib1 (grey lines), did not change between the control (solid line) and Staufen1-depleted neurons (dotted lines, Fig. 3E). However for Rac1 and Nrp1 (yellow lines), Staufen1-depleted neurons (dotted lines) showed an increase in the relative RNA decay rate compared to the control neurons (solid line). Notably, Rac1 exhibited statistically significant prolonged decay at the 3- and 6-hour time points while Nrp1 displayed statistically significant prolonged RNA decay at the 6-hour time points.
As an additional test, we asked if Rac1 and Nrp1 are upregulated when Staufen1 is depleted. To test this, we measured their relative RNA amount in Staufen1-depleted neurons using qRT-PCR (Fig. 3F). The relative RNA amount for Rac1 and Nrp1 was increased about 1.2 (Nrp1) to 2 (Rac1) fold upon Staufen1 depletion. Together, these data are consistent with the idea that Rac1 and Nrp1 are SMD targets.
The SMD function of Staufen1 controls levels of Rac1 in axons
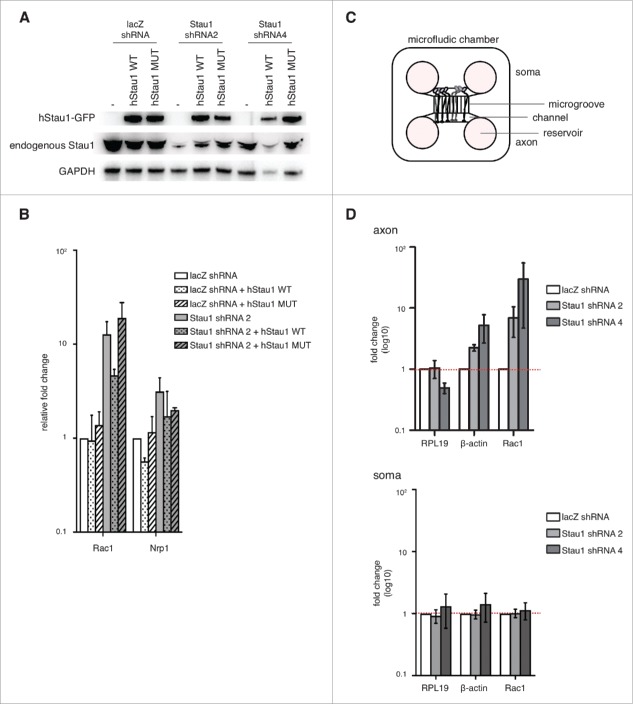
We found that Staufen1 is required for proper Rac1 and Nrp1 decay in DRG neurons by depleting endogenous Staufen1. However Staufen1 also has transport/granule roles. Hence, it is not clear if the effects of Staufen1 depletion RNA levels reflect a loss of its SMD function or a loss of its transport function. To address the question, we used a Staufen1 quadruple mutant (A375E, R376A, L472S, S473E) that can transport mRNA but does not mediate SMD.27 We depleted endogenous Staufen1 and rescued with either wild-type or mutant Staufen1 in DRG neurons (Fig. 4A). We then collected total RNA for qRT-PCR as described earlier. When we rescued with wild-type Staufen1, we observed Rac1 levels amount comparable to the control neurons. In contrast, when we rescued with Staufen1 that cannot mediate SMD, the RNA levels remained above the control. This indicated that the loss of Staufen1 decay function accounts for the increased Rac1 levels in Staufen1-deficient DRG neurons. This further suggested that the Staufen1 decay function influences the expression levels of specific transcripts in DRG neurons.
Figure 4.
Staufen1 is required for local RNA decay in the DRG neurons. A. Expression of wild-type and mutant Staufen1. E14 DRG neurons were depleted of Staufen1 using lentivirus expressing Staufen1-specific shRNA. Staufen1 depletion was rescued by expression wild-type Staufen1 (WT) or a Staufen1 mutant (MUT) that can transport mRNA but cannot mediate SMD. Lysates were immunoblotted to examine Staufen1 and GAPDH (load control) expression levels. As shown in the immunoblots, ectopic Staufen1 WT and MUT were successfully expressed in the neurons. B. An SMD-deficient Staufen1 is unable to rescue Staufen1 depletion. Staufen1 depletion causes increased expression of Rac1 and Nrp1. We rescued Staufen1 depletion with wild-type Staufen1 and a mutant that lacks SMD activity.27 Total RNA was extracted and18S rRNA, RPL19, Rac1 and Nrp1 were detected by qRT-PCR. As expected, Rac1 is upregulated when Staufen1 is depleted. When Staufen1 WT is used to rescue the knockdown, Rac1 levels are reduced towards the level in control neurons. In contrast, when Staufen1 MUT is expressed, the Rac1 amount remained similar to the Staufen1 depletion. This indicates that Staufen1 MUT cannot reverse Rac1 upregulation caused by Staufen1 depletion. This suggests that Staufen1's SMD is required for its ability to regulate Rac1 expression. C. Schematic of microfluidic chambers using for isolating axons. The schematic diagram shows the cell body (left) and axonal (right) compartments separated by 450 µm-long microgrooves. The microgrooves are small enough to allow axons to travel through, but prevent cell bodies from appearing in the axonal compartment. Approximately 20,000–30,000 neurons are plated in the cell body compartment, with axons reaching the axonal compartment by DIV2. D. Rac1 and β-actin are SMD targets in axons. Axonal transcript levels were detected by qRT-PCR after isolation of axons prepared from neurons cultured in microfluidic chambers. Three to four days after plating neurons in microfluidic chambers, lentivirus was added to the neurons to deplete Staufen1. Total RNA was extracted from the axons and the soma. qRT-PCR was performed on axonal and soma cDNA samples to probe β-actin and Rac1 (white: control, light grey: shRNA2, dark grey: shRNA4). Axonal β-actin and Rac1 show an increase in transcript levels compared to the control neurons when Staufen1 is depleted. In contrast, somatic β-actin and Rac1 transcript levels do not increase compared to the control neurons when Staufen1 is depleted. This indicates that Staufen1 is required to properly regulated β-actin and Rac1 in the DRG axons. This further suggests that SMD functions locally in the axons.
Staufen1 influences Rac1 levels in axons
After identifying SMD targets in whole DRG neuronal cells, we then asked if SMD occurs within the axonal compartment of DRG neurons. To selectively isolate axonal mRNA for quantitative analysis of specific transcripts, we used a microfluidic chamber to separate the axons from the soma.23,28,29 (Fig. 4C) We focused on Rac1 due to its role in axonal morphology and F-actin dynamics, which we previously found were affected in Staufen1-depleted neurons (see Fig. 1). For these experiments, we cultured E14 DRG neurons in microfluidic chambers and depleted Staufen1 transcripts throughout the neuron by applying Staufen1-specific shRNA to the somatic compartment. After 4 DIV, we collected RNA from the axons separately from the soma to analyze using qRT-PCR. Rac1 increased in the axons while the control transcript RPL19, remained unchanged when Staufen1 was depleted (Fig. 4D).
Strikingly, another RNA with known Staufen1-binding sites β-actin, also increased in the axons. In contrast, β-actin did not change compared to the control in the soma (Fig. 4D). Taken together, this suggested that SMD occurs in the axons for Rac1 and β-actin but not in the soma.
Discussion
Staufen1 is a double-stranded RNA binding protein that is well known to be present in neurons in both axons and dendrites. Because Staufen1 is a major component of RNA transport granules, its neuronal function has been primarily linked to mRNA transport. In this study, we show that Staufen1 has an additional function involving RNA degradation of a subset of transcripts in DRG neurons. Furthermore, we show that Staufen1 is present in axons and growth cones along with Upf1, and can function locally within axons where its targets include Rac1 and β-actin. These studies demonstrate that local RNA degradation can be mediated by a local SMD pathway.
The concept that Staufen1 may regulate mRNA levels in growth cones is supported by Staufen1 knockdown studies. The morphological effects of Staufen1 knockdown on growth cones is compatible with the idea that Staufen1 can influence local translation. A major function of local translation is to affect the axonal and growth cone cytoskeleton.29-33 Thus, the altered morphologic features suggest altered mRNA and translation regulation in growth cones. Notably, our studies suggest that β-actin and Rac1 are Staufen1 targets in axons. β-actin is the major component of the growth cone. Additionally, Rac1 has previously been shown to influence morphology of growth cones.34-36 Thus, the morphologic effects of Staufen1 depletion may reflect elevated levels of Rac1 and β-actin protein levels.
We demonstrated that Staufen1 readily degraded β-actin and Rac1 via SMD in the DRG axons. However, β-actin was not regulated by Staufen1 in the soma based on soma-specific isolation using a microfluidic chamber (see Fig. 4D). Furthermore, when we generated RNA-Seq libraries using the DRG whole cell RNA, β-actin did not show any statistically significant change (data not shown). It only became evident that β-actin is a Staufen1 target when we analyzed DRG axonal RNA using qRT-PCR. Even though β-actin is upregulated in axons upon Staufen1 depletion from neurons, this upregulation is masked by the lack of increase of the abundant levels of β-actin RNA in the cell body. It will be interesting to identify other Staufen1 targets in the axons and elucidate the extent of axonal SMD in determining the axonal transcriptome.
The altered morphology of Staufen1-labeled puncta also suggests that Staufen1 may have a specialized role in growth cones. We found that Staufen1 is present in different sized particles in growth cones compared to axons. The smaller size of Staufen1-puncta in growth cones and their localization away from microtubules suggest that these puncta have non-transport roles. Thus, Staufen1 in growth cones may function in a growth cone SMD pathway.
Although our studies show that Staufen1 can affect mRNA stability in growth cones, we cannot exclude the possibility that the effects of Staufen1 on growth cone morphology are mediated by additional pathways such as direct control of translation by Staufen1 as was described in non-neuronal cells.15
Although Staufen1 has been shown to mediate RNA decay in various studies, SMD was not readily detected in other studies.15,16 This raises the possibility that the levels of SMD may be dependent on the cell type. However, even in neurons, SMD does not appear to be the major function of Staufen1 since more transcripts were downregulated than upregulated in Staufen1-deficient neurons. Nevertheless, SMD may still function in specific subcellular compartments, such as growth cones. The presence of both Staufen1 and Upf1 in a specific compartment may facilitate SMD in growth cones.
Notably, Staufen1 is also present in dendrites where it may also influence mRNA levels and the local protein synthesis capacity at synapses. Future studies may reveal the role of Staufen1 in local degradation in dendrites as well as the signaling pathways that regulate the activation of SMD in neurons.
Supplementary Material
Disclosure of potential conflicts of interest
The authors have no conflicts of interest.
Acknowledgments
We thank members of the Jaffrey laboratory for helpful comments and suggestions. We thank L. Maquat for the mutant Staufen1 plasmid and M. Kiebler for Staufen1 antibodies for immunofluorescence.
Funding
This work was supported by NIH grant R01 NS056306 (S.R.J.) and the Clinical Translational Science Center (CTSC) (J.Y.K.).
Accession Codes Sequencing data have been deposited in the GEO database.
References
- 1.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001. December 20;32(6):1013–26. doi: 10.1016/S0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 2.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012. May;13(5):308–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006. October;9(10):1247–56. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J Cell Biol. 1970. January;44(1):62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003. April 15;23(8):3251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006. September 21;51(6):685–90. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013. June 6;153(6):1252–65. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci. 2014. March 26;34(13):4481–93. doi: 10.1523/JNEUROSCI.4944-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007. July 13;130(1):179–91. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ. Introns in UTRs: Why we should stop ignoring them. Bioessays. 2012. December;34(12):1025–34. doi: 10.1002/bies.201200073. [DOI] [PubMed] [Google Scholar]
- 11.Kim YK, Furic L, DesGroseillers L, Maquat LE. Mammalian Staufen1 Recruits Upf1 to Specific mRNA 3′UTRs so as to Elicit mRNA Decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 12.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013. Jul-Aug;4(4):423–35. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamachi N, Tani H, Akimitsu N. Up-frameshift protein 1 (UPF1): Multitalented entertainer in RNA decay. Drug Discov Ther. 2012. April;6(2):55–61. [PubMed] [Google Scholar]
- 14.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007. June 06;26(11):2670–81. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol. 2014. January;21(1):26–35. doi: 10.1038/nsmb.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015. March 26;519(7544):491–4. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. JNeurosci. 1999. 1999/01/01/;19(1):288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kîhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Molecular Biology of the Cell. 1999 1999/09//;10(9):2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001. November 8;32(3):463–75. doi: 10.1016/S0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 20.Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, Goetze B, Lupas A, St Johnston D, Kiebler M. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J Neurosci. 2003. July 02;23(13):5778–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, et al.. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci U S A. 2008. October 21;105(42):16374–9. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deglincerti A, Liu Y, Colak D, Hengst U, Xu G, Jaffrey SR. Coupled local translation and degradation regulate growth cone collapse. Nat Commun. 2015;6:6888. doi: 10.1038/ncomms7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker BA, Hengst U, Kim HJ, Jeon NL, Schmidt EF, Heintz N, Milner TA, Jaffrey SR. Reprogramming axonal behavior by axon-specific viral transduction. Gene Ther. 2012. September;19(9):947–55. doi: 10.1038/gt.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker BA, Ji SJ, Jaffrey SR. Intra-axonal translation of RhoA promotes axon growth inhibition by CSPG. J Neurosci. 2012. October 10;32(41):14442–7. doi: 10.1523/JNEUROSCI.0176-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price TJ, Flores CM, Cervero F, Hargreaves KM. The RNA binding and transport proteins staufen and fragile X mental retardation protein are expressed by rat primary afferent neurons and localize to peripheral and central axons. Neuroscience. 2006. September 15;141(4):2107–16. doi: 10.1016/j.neuroscience.2006.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002. February 15;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol. 2013. April;20(4):515–24. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji SJ, Jaffrey SR. Intra-axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron. 2012. April 12;74(1):95–107. doi: 10.1016/j.neuron.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009. August;11(8):1024–30. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32(6):1013–26. doi: 10.1016/S0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 31.Campbell DS, Regan AG, Lopez JS, Tannahill D, Harris WA, Holt CE. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. JNeurosci. 2001. 2001/11/01/;21(21):8538–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005. August 18;436(7053):1020–4. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006. October;9(10):1265–73. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Saint-Cyr-Proulx E, Aktories K, Lamarche-Vane N. Rac1 and Cdc42 but not RhoA or Rho kinase activities are required for neurite outgrowth induced by the Netrin-1 receptor DCC (deleted in colorectal cancer) in N1E-115 neuroblastoma cells. Journal of Biological Chemistry. 2002;277(17):15207–14. doi: 10.1074/jbc.M109913200. [DOI] [PubMed] [Google Scholar]
- 35.Fukata M, Nakagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Current Opinion in Cell Biology. 2003;15(5):590–7. doi: 10.1016/S0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 36.Thies E, Davenport RW. Independent roles of Rho-GTPases in growth cone and axonal behavior. Journal of Neurobiology. 2003;54(2):358–69. doi: 10.1002/neu.10135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.