Abstract
Elucidating the full repertoire of molecular mechanisms that promote stem cell maintenance requires sophisticated techniques for identifying and characterizing gene function in stem cells in their native environment. Ovarian germline stem cells in the fruit fly, Drosophila melanogaster, are an ideal model to study the complex molecular mechanisms driving stem cell function in vivo. A variety of new genetic tools make RNAi a useful complement to traditional genetic mutants for the investigation of the molecular mechanisms guiding ovarian germline stem cell function. Here, we provide a detailed guide for using targeted RNAi knockdown for the discovery of gene function in ovarian germline stem cells and their progeny.
Keywords: Germline, Germ cell, Stem cell, Drosophila, Ovary, Oogenesis
1 Introduction
Unlike most specialized cells, which divide to produce identical daughter cells, stem cells are endowed with the property of long-term self-renewal [1]. During cell division, a stem cell retains its unspecialized fate while producing a non-identical daughter destined for differentiation. Stem cells in the developing embryo and in some adult tissues allow for the continuous production of cells that can be instructed to differentiate into a variety of specialized fates. This unique ability, combined with recent advances in cell reprogramming and gene editing, has heightened clinical interest in the properties of stem cells [2]. Safe and effective use of stem cells and their progeny in regenerative therapies will require a detailed knowledge of the molecular factors that regulate stem cell function.
Maintenance of stem cell self-renewal is influenced by a variety of factors, including the cell’s chromatin state, metabolic status, and responsiveness to both the local microenvironment (the stem cell niche) and long-range physiological cues [3–6]. Identification and characterization of the molecular pathways that drive stem cell self-renewal thus require a model system wherein stem cells can be observed and manipulated within their native environment. While a variety of model systems have been employed to study stem cells, many of our fundamental models of stem cell function were developed as a result of experiments performed in vivo in the fruit fly, Drosophila melanogaster [7]. Like humans, Drosophila have a variety of tissue-resident stem cell populations that are critical for the continued production of specialized progeny [8–12]. The ease with which Drosophila are reared, the broad variety of genetic tools for cell-specific gene manipulation, and the high degree of conservation between many human and Drosophila cell signaling pathways make fruit flies a powerful model organism for biomedical research [13]. Drosophila are well-suited for rapid, large-scale genetic screens [14], and the availability of transgenic libraries, stock centers, and genome and bioinformatics tools [15–18] make them an efficient and cost-effective resource to study stem cell biology within the context of the whole organism.
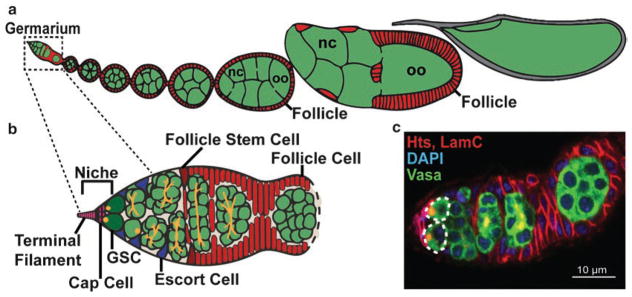
In particular, germline stem cells (GSCs) in the Drosophila ovary have provided fundamental insights into stem cell regulation. GSCs are a specialized population of stem cells that are maintained to replenish stocks of germ cells (oocytes), whose numbers are depleted by gamete production. GSCs reside in a somatic niche composed of cap cells and terminal filament cells, located at the anterior tip of each ovariole (the functional unit of the adult Drosophila ovary; Fig. 1a) in a specialized structure called the germarium (Fig. 1b) [12, 19]. GSCs divide asymmetrically to self-renew and produce a differentiating daughter cell (cystoblast), which undergoes four subsequent rounds of mitosis with incomplete cytokinesis (Fig. 1b, c). During this process, germ cells are specified into either oocyte or nurse cell fates, and oocytes will transition into meiosis. While all germ cells in the germarium contain endoplasmic reticulum-like organelles called fusomes (Fig. 1b, c), GSCs are readily identifiable by their anteriorly localized fusome juxtaposed to the somatic cap cells [20–23]. Additional somatic cells guide the development of the germ cells. Triangular escort cells extend long projections to move the dividing cysts laterally across the germarium [24]. Follicle cells (the daughters of a second population of stem cells, follicle stem cells) envelop each germline cyst, forming an epithelial monolayer that accompanies the developing germline cyst outside of the germarium [25–27]. The resulting follicle undergoes 14 distinct phases of development, culminating in a fertilizable oocyte [19].
Fig. 1.
Drosophila ovarian germline stem cells fuel oogenesis. (a, b) The Drosophila ovary is composed of 15–20 ovarioles (one is represented in a), consisting of a germarium (b) and a progressive series of follicles in different stages of development. The germarium houses two stem cell populations: germline stem cells (GSC; dark green) and follicle stem cells (FSC; dark red). GSCs reside in a somatic niche composed of terminal filament cells and cap cells (pink). Escort cells (blue) help guide developing germline cysts posteriorly, where they are encapsulated by follicle cells (red), forming a follicle (also called an egg chamber). Germ cells (green) differentiate into oocytes (oo) or nurse cells (nc). (c) Confocal micrograph of a germarium labeled with anti-Vasa (all germ cells; green), anti-Hts (fusomes and follicle cell membranes; red), anti-LaminC (nuclear envelope of cap cells; red), and DAPI (all nuclei). Dotted lines designate GSCs. Scale bar = 10 μm
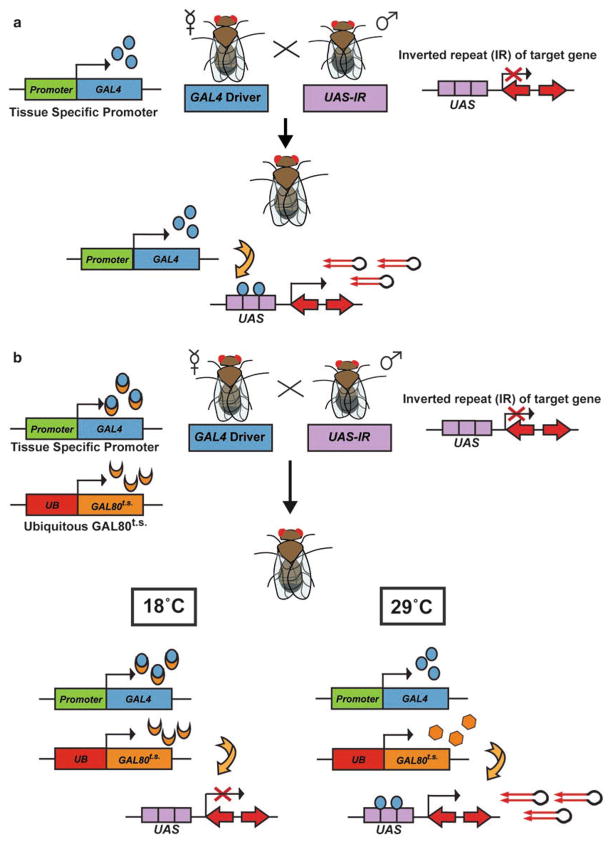
Targeted RNA interference, or RNAi, is a widely used experimental technique for gene silencing that has been successfully applied to identify novel genes that control stem cell fate and proliferation in Drosophila [28–32]. While the creation of genetic mutant alleles can be time-consuming and labor intensive, generation of RNAi against a target of interest is relatively easy and inexpensive [32–34]. Further, while complete loss-of-function genetic mutant alleles are useful tools, they may result in severe phenotypes that are difficult to interpret or developmentally lethal, precluding analysis of gene function in adult cells. In contrast, RNAi frequently reduces, but does not abolish, the targeted mRNA, resulting in hypomorphic phenotypes. By inducing RNAi using the UAS-GAL4 system, spatial control over gene knockdown in developing or adult fly tissues is readily accomplished (Fig. 2a). In this system, promoter or enhancer elements drive the expression of the yeast transcription factor Gal4 [35]. Once expressed, Gal4 binds a DNA response element, the upstream activating sequence (UAS), which precedes a gene fragment containing an inverted repeat with exact complementarity to a given gene of interest. Expression of the inverted repeat results in the formation of double-stranded RNA hairpins that trigger a sequence-specific RNAi silencing response in the cells in which it is expressed [36–39]. Thus, RNA hairpins are generated under the control of tissue- or cell-specific drivers, allowing for targeted knockdown of a gene of interest in specific cell population(s). RNAi experiments can be combined with additional genetic tools to restrict knockdown to specific temporal windows (Gal80t.s.; Fig. 2b) or to a clonal population of cells within a wild-type environment (mosaic analysis with a repressible cell marker; MARCM) [40–42]. RNAi in Drosophila ovarian GSCs thus presents an elegant experimental system to discover the molecular regulations that govern stem cell fate and function in vivo.
Fig. 2.
Targeted RNAi using the UAS-GAL4 system. (a) Virgin female tissue-specific Gal4 driver flies are crossed to male upstream activating sequence (UAS)-RNAi responder flies (UAS-IR), resulting in hairpin expression and RNAi-mediated gene knockdown in specific cells in all progeny. (b) For temporal control over Gal4 expression, Gal4 driver flies also carry a transgene in which the Gal4 inhibitor, Gal80ts, is ubiquitously expressed. In the resulting progeny, Gal4 expression is temperature-dependent. At 18 °C, Gal80ts binds Gal4, suppressing hairpin production. At 29 °C, the conformation of the Gal80ts protein changes, such that it can no longer bind Gal4, making Gal4 available to initiate transcription of UAS-RNAi
In this protocol, we provide an experimental strategy utilizing targeted RNAi to knock down gene function in Drosophila ovarian GSCs. This protocol can be adapted to analyze the function of any number of genes in GSCs and their progeny and is particularly useful for gene discovery. In general, the phases of the protocol include (1) selecting or creating an appropriate UAS-RNAi transgenic line against a gene of interest, (2) choosing a germline-specific Gal4 driver, (3) Drosophila husbandry to generate transgenic flies, (4) dissecting adult ovaries, (5) immunostaining, (6) image acquisition via confocal microscopy, and (7) image analysis. This basic protocol also provides a platform for further analysis of stem cell function, including gene expression, cell cycle progression, and known markers of GSC self-renewal and daughter cell differentiation. Taken together with other recently published methods [43–49], this protocol highlights the power of the Drosophila ovarian GSC as a model for the study of stem cell function in its native environment.
2 Materials
2.1 Drosophila Strains and Culture
Drosophila strains (see Note 1), including a germline-compatible UAS-RNAi responder strain and a germline-specific Gal4 strain.
Standard fly culture medium (cornmeal/molasses/yeast/agar) in bottles and vials (see Note 2).
Yeast paste: 1–1 mixture of dry active yeast (see Note 3) and distilled water, mixed to the consistency of smooth peanut butter. Store covered with Parafilm at 4 °C to prevent drying.
Dissecting stereomicroscope with dual goose-neck illumination, 0.6–4× magnification range, and a flat black dissection base, equipped with a fly pad and air needle to deliver CO2 (Drosophila anesthetic; see Note 4).
2.2 Ovary Dissection and Immunostaining
1.5 mL microfuge tubes, pre-coated (see Note 5).
15-mL polypropylene centrifuge tubes.
50-mL polypropylene centrifuge tube.
Glass or plexiglass dissection dish.
Kimwipes.
Glass Pasteur pipettes and bulbs.
Two pairs of sharpened forceps (INOX, Dumont #5, Biologie point).
Two 27 × 1¼ gauge needles with 1 mL syringes.
Cold room-safe orbital nutator (also called a GyroMixer).
Grace’s Insect Medium without additives (Lonza; see Note 6).
Phosphate-buffered saline (PBS).
Wash solution: 0.1% Triton-X-100 in PBS.
Fixative: 5.3% formaldehyde in Grace’s media (see Note 7). Prepare fresh prior to dissection. For each sample, add 300 μL 16% formaldehyde (Ted Pella; see Note 8) and 600 μL Grace’s media. Keep on ice.
Blocking Solution: 5% bovine serum albumin (BSA), 5% normal goat serum, 0.1% Triton-X-100 in PBS (see Note 9). Prepare using sterile technique, and store at 4 °C. Discard if cloudy.
Primary Antibodies: In nearly all initial experiments, we use two primary antibodies that allow for identification of germ-line and somatic cells in the ovary (see Fig. 3c, d). Mouse anti-Hts (1B1-s; Developmental Studies Hybridoma Bank) is diluted in blocking solution to a final concentration of 1:10 and labels both the germline-specific fusome and follicle cell plasma membranes. Mouse anti-LaminC (LC28.26-s; Developmental Studies Hybridoma Bank) is diluted in blocking solution to a final concentration of 1:100 and strongly labels the nuclear envelope of cap cells (see Notes 10 and 11).
Secondary Antibodies: Species-matched secondary antibodies tagged with a fluorophore of interest. For the detection of anti-Hts and anti-LaminC (see Fig. 3c, d), we use Alexa Fluor goat anti-mouse-568 (Life Technologies; see Note 12), diluted to a final concentration of 1:200 in blocking solution.
4′,6-Diamidino-2-phenylindole (DAPI): For working concentration, prepare a 1:500 dilution of a 5.0 mg/mL DAPI stock solution (see Note 13) in 0.1% Triton-X-100 in PBS.
Mounting Media: 20 mg/mL n-propyl gallate in glycerol (see Note 14). In a 50 mL conical tube, combine 1.0 g n-propyl gallate (Sigma) with 5 mL PBS; vortex to mix. Add 45 mL 100% glycerol. Cover the conical tube in foil and rotate on a nutator at room temperature overnight. Media should be clear but very viscous. Store protected from light at 4 °C. A working stock can be poured into an opaque dropper bottle for ease of application.
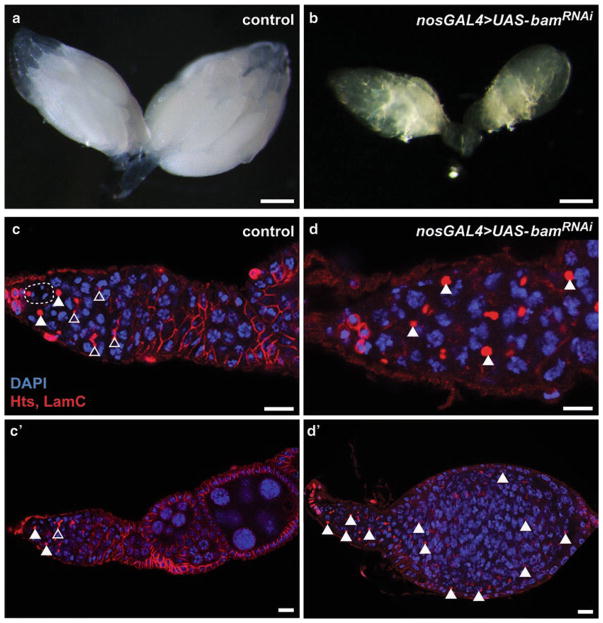
Fig. 3.
Germline-targeted RNAi knockdown of bam results in small ovaries filled with undifferentiated germ cells. (a, b) Bright-field images of intact control (a) and germline-specific bam knockdown (b) ovaries 5 days after eclosion. (c, d) Confocal micrographs of representative ovarioles labeled with anti-Hts (fusomes and follicle cell membranes; red), anti-LaminC (nuclear envelope of cap cells; red), and DAPI (all nuclei). While nos-Gal4 control germaria (c) contain GSCs (dashed line), cystoblasts (filled arrowheads), and differentiating germ cells (recognized by branched fusomes; open arrowheads), bam knockdown germaria have accumulated GSC- and cystoblast-like cells throughout the germarium, at the expense of differentiated germ cells. Further, bam knockdown results in a block to oogenesis, as evidenced by the lack of progressively developed follicles (d’) typically found in control ovarioles (c’). Scale bar = 200 μm (a, b), 10 μm (c, d)
2.3 Sample Preparation and Confocal Microscopy
Two needle holders with sharpened tungsten needles; one should have an “L”-shaped bend at the tip (see Note 15).
Glass microscope slides and coverslips (1 μm thickness, 22 × 22 mm).
Steel weight, measuring approximately 250 g.
Fingernail polish (Sally Hansen Tough as Nails, any color).
Laser scanning confocal microscope, inverted, equipped with 63× oil immersion lens (n.a. = 1.4) and 1.5–3× optical zoom (see Note 16).
Confocal image acquisition and analysis software (see Note 17).
3 Methods
3.1 Drosophila Strains and Culture
3.1.1 Selecting a Gene-Specific UAS-RNAi Responder Transgene
Although a wide variety of target-specific UAS-RNAi transgenes are available in Drosophila [32], care must be taken to ensure successful knockdown of your gene of interest in female GSCs. A variety of commonly used Drosophila genetic tools, including the UASt responder lines that promote strong expression in somatic cells, do not work effectively in the female germline [39, 40, 50, 51]. To circumvent this problem, Ni and colleagues developed two RNAi vectors (Valium20 and Valium22) specifically designed for optimal expression of RNAi hairpins in the germline [50]. Valium20 promotes strong knockdown in both the ovarian germ-line and soma; in contrast, the Valium22 vector, based on the germline-compatible UASp vector [51], promotes strong expression in the germline, but only moderate to weak expression in the soma [50, 52]. A large collection of target-specific Valium20- and Valium22-based UAS-RNAi transgenic lines (henceforth referred to as “UAS-RNAi responders”) are now available from the Transgenic RNAi Project at Harvard Medical School (TRiP; www.flyrnai.org; see Note 18) via the Bloomington Drosophila Stock Center (see Note 1; Table 1). Another large collection of UAS-RNAi responders are available from the Vienna Drosophila Stock Center (VDRC). The VDRC maintains two libraries of UAS-RNAi responders that differ primarily in the insertion site of the UAS-RNAi into the Drosophila genome [36]. Although neither of these libraries were created in germline-optimized vectors, reliable expression of gene-specific RNAi hairpins has been achieved in the germline via co-expression of Dicer2 (UAS-Dcr2) [29, 30].
Table 1.
Resources for RNAi-mediated gene knockdown in Drosophila ovarian GSCs and somatic support cells
| Transgenes | Source | Adult ovarian expression pattern | References | |
|---|---|---|---|---|
| Gal4 drivers | ||||
|
| ||||
| Germline | nos-Gal4::VP16 (III)a | BLMb #4937 | Moderate expression in GSCs, strong expression in germline cysts | [51, 55] |
| nos-NGT40; UAS-Dcr2 | BLM #25751 | Very weak expression in GSCs | [29, 30] | |
| MTD-Gal4 | BLM #31777 | Moderate expression in GSCs, very strong expression in germline cysts | [54] | |
| ET-Flpx2Gal4–168 | Drosophila community | Germline clones | [41] | |
|
| ||||
| Niche | bab1-Gal4 | BLM #6802; Drosophila community | Cap cells and escort cells | [61, 62] |
|
| ||||
| c587-Gal4 | Drosophila community | Escort cells, follicle stem cells, prefollicle cells | [69] | |
|
| ||||
| Tj-Gal4 | KYOc #104055 | Terminal filament & cap cells | [70] | |
|
| ||||
| ET-Flpx2Gal4-688A | Drosophila community | Escort cell clones | [41] | |
|
| ||||
| 13C06-Gal4 | BLM #47860 | Escort cells, follicle stem cells, and prefollicle cells | [71] | |
|
| ||||
| ET-Flpx2Gal4-398A | Drosophila community | Escort cells, follicle stem cells, and prefollicle cell clones | [41] | |
|
| ||||
| UAS-RNAi responders | ||||
|
| ||||
| Long hairpins | VDRC KK | VDRCd | Germline expression with UAS-Dcr2 | [36]
|
| VDRC GD | VDRC | Germline expression with UAS-Dcr2 | [36] | |
|
| ||||
| Short hairpins | TRiPe Valium10 | BLM | Expression limited to somatic cells | [39] |
| TRiP Valium20 | BLM | Expression in somatic and germline cells | [50] | |
| TRiP Valium22 | BLM | Strong expression in germline cells, weaker in somatic cells | [50] | |
Note that a second insertion is available on the X chromosome; however, its expression is weaker than the insertion on the III
Bloomington Drosophila Stock Center at Indiana University, Bloomington, IN, USA
Kyoto Stock Center at the Kyoto Institute of Technology, Kyoto, Japan
Vienna Drosophila Resource Center, Vienna, Austria
Transgenic RNAi Resource Project at Harvard Medical School, Boston, MA, USA
While an ever-increasing number of germline-compatible UAS-RNAi responders are being made public (see FlyBase, www.flybase.org, for a current listing), these tools may not yet be available for some genes of interest. In this case, it is possible to create a Valium20- or Valium22-based UAS-RNAi transgene using standard molecular cloning techniques and have this transgene commercially injected into embryos for the establishment of transgenic lines (see Note 19). While creation of customized UAS-RNAi lines does require time to establish new transgenic lines, it is a viable and cost-effective option for genes with few existing tools. Detailed protocols, cloning vectors, and primer sequences are available from the TRiP (www.flyrnai.org) [50].
Three common issues arise when using targeted UAS-RNAi approaches. First, the strength of knockdown of a gene of interest can vary between UAS-RNAi transgenes. This can be assessed by measuring RNA or protein levels using a variety of techniques but is perhaps best visualized in vivo by immunofluorescence against the protein of interest if suitable reagents are available. Second, a given RNAi hairpin may target multiple genes, resulting in phenotypes not due to loss of the gene of interest (“off-target effects”). This is particularly true for the VDRC KK library [53]. To minimize the chances of off-target effects, choose more than one hairpin, targeted at different regions of the gene of interest, for RNAi analysis. The most effective control is to design hairpins with high sequence similarity to the experimental hairpin, but with several mismatched bases, thus precluding the induction of RNAi [33, 34]. Lastly, for genes that encode multiple isoforms, hairpins could target one, many, or all isoforms of the mRNA, depending on the targeted region. It is important to recognize the location of the hairpin sequence with regard to the predicted transcript, as this could impact the interpretation of results.
3.1.2 Selecting a Germline-Specific Gal4 Driver Transgene
As in the case of the UAS-RNAi responder, a germline-compatible Gal4 driver must also be selected to achieve gene-specific knockdown in GSCs. As mentioned above, ubiquitous expression of a UAS-RNAi responder could cause developmental lethality, thus precluding analysis of adult GSC phenotypes. Further, some drivers that are considered to be “ubiquitous” (such as the hsGal4 used in many somatic experiments) do not work well in the germline [35, 40, 51]. Thus, a germline-specific Gal4 driver is required to restrict UAS-RNAi responder expression to GSCs. Unfortunately, very few germline-specific drivers exist, and even fewer drive expression robustly in GSCs; however, a few lines can be used to successfully knock down gene expression in GSCs and their daughters without directly modifying the surrounding somatic cells (Table 1).
Perhaps the most frequently used germline drivers in the Drosophila ovary are the nos-Gal4::VP16 and the “maternal triple” MTD-Gal4 [43, 51, 54, 55]. Two important caveats should be noted, however, when employing the nos-Gal4::VP16 driver in GSC experiments. First, nos-Gal4::VP16 expression is not limited to adult GSCs [51, 55]. This driver is expressed in the developing primordial germ cells, larval/pupal germ cell precursors, adult GSCs, and nearly all GSC progeny. It is therefore difficult to conclusively demonstrate that a given gene of interest functions solely in adult GSCs (i.e., GSC function versus GSC establishment). (Unfortunately, the Gal80t.s. system cannot help in this regard, as Gal4::VP16 lacks the Gal80-binding site [56].) Second, while nos-Gal4::VP16 is expressed in adult GSCs, it is expressed at much higher levels in differentiating daughter cells (particularly 2-, 4-, 8-, and 16-cell cysts) [51, 55]. Thus, any knockdown experiments that result in phenotypes impacting germline differentiation must be carefully interpreted. For example, nos-Gal4::VP16-mediated knockdown of the differentiation factor bag of marbles (bam) using targeted RNAi results in a block in germline differentiation, leading to the accumulation of single germ cells and 2-cell germ cysts at the expense of 4-, 8-, and 16-cell cysts (Fig. 3). As an isolated experiment, these results could indicate that bam is required for the differentiation of 2-cell cysts; however, analysis of Bam protein expression and a bam genetic null allele clearly demonstrates that bam is required for the initial differentiation of the immediate GSC daughter cell (the cystoblast) [57–60]. Similar considerations should be made when using MTD-Gal4, a compound driver consisting of three independent Gal4 insertions (nos-Gal4::VP16 plus pCOG-Gal4::VP16 and nos.NGT40-Gal4) [43, 51, 54]. Intriguingly, neither pCOG-Gal4::VP16 (also called otu-Gal4::VP16) nor nos.NGT40-Gal4 are highly expressed in adult GSCs (E. Ables unpublished and Ref. [51]); however, several research groups have obtained GSC-related phenotypes when using this driver (in combination with UAS-Dcr2) in large-scale screens [29, 30], indicating that low levels of transgene expression are sufficient to induce phenotypes in some situations. Finally, a newly characterized Gal4 driver, ET-Flpx2Gal4-398A, is also available for clonal expression of UAS-RNAi responders in the germ-line [41], but has not yet been widely utilized.
A variety of somatic Gal4 drivers are also available that can be used to identify microenvironmental factors that influence GSC function (Table 1). Most of these somatic Gal4 drivers robustly promote the expression of a wide variety of UAS-RNAi transgenes (except the Valium22 series). Researchers should note, however, that many of these drivers are expressed in overlapping populations of somatic cells and may also be expressed outside of the ovary. For example, bab1-Gal4 is frequently used to drive UAS-RNAi expression in the somatic cap cells of the ovary, but it also shows strong expression in the somatic polar and stalk cells present in more developed stages of oogenesis [61, 62]. Researchers should test driver expression in multiple tissues to preclude non-autonomous effects prior to any knockdown experiment and check the expression pattern of the driver over the course of the experiment to ensure the pattern does not change due to the genetic manipulation or nutrient status of the fly.
3.1.3 Drosophila Husbandry
Maintain fly cultures using standard procedure (see Note 20).
Culture at least two bottles each of the Gal4 driver line and an isotype control (typically yw; see Note 21) and one bottle of the UAS-RNAi responder line (see Note 22). Collect virgin females from the Gal4 driver line and the isotype control.
Set control (Gal4 driver with yw males; UAS-RNAi responder with yw female virgins) and experimental (Gal4 driver with UAS-RNAi responder) crosses (Fig. 2a) in vials at a density of five pairs of flies per vial (see Note 23). Crosses should be set in replicates. Addition of dry yeast to the culture medium provides optimal conditions to increase egg laying. Maintain vials at 25 °C, transferring adults to new vials every 3 days. New flies should emerge within 10 days of this date (at this temperature). For experiments using a Gal80ts repressor (Fig. 2b), crosses should be maintained at 18 °C, and new flies will emerge within 18–20 days.
Allows crosses to eclose for 1–2 days. Collect the female flies you will dissect and an equal number of accompanying males (typically 12–15 pairs of flies per vial), and add them to fresh vials supplemented with wet yeast paste (see Note 24).
Transfer adults into freshly yeasted vials daily, and maintain at a constant temperature (typically 25 °C or 29 °C; see Note 25) until the day of dissection. For experiments using a Gal80ts repressor, flies should be maintained at 29 °C. The age of the flies at dissection (determined by the number of days post-eclosion) is determined by the experimenter. For the analysis of GSC self-renewal, it is appropriate to collect flies at a series of timepoints (i.e., 0 days, 4 days, 8 days, and 12 days after eclosion) to chart GSC maintenance over time. The timing of dissection post-eclosion will depend on the number of lines being screened and the exact phenotypes expected/discovered and should be determined experimentally.
3.2 Ovary Dissection and Immunostaining (Fig. 4)
Fig. 4.
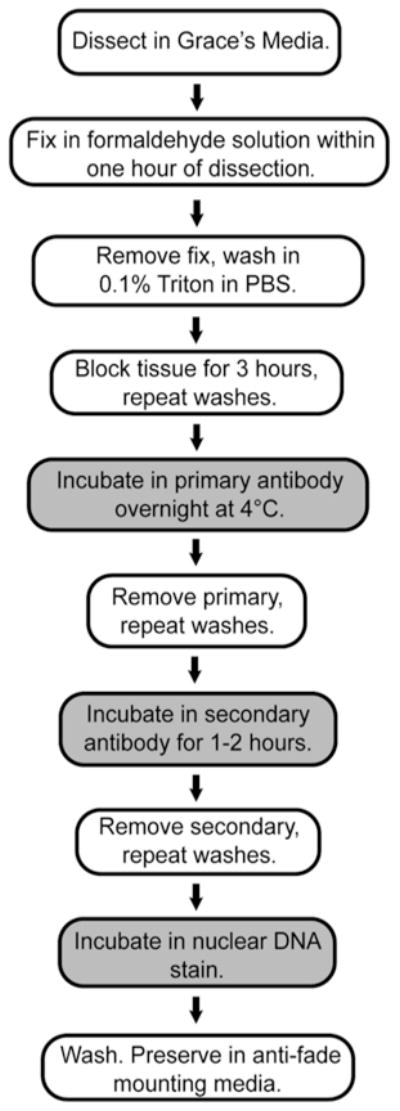
Experimental work-flow for ovary immunostaining. Preparation of Drosophila ovaries for confocal imaging is outlined, highlighting the addition of primary and secondary antibodies and nuclear counterstain
Use CO2 to anesthetize flies. Remove males and discard. Dissect ovaries from female flies in cold Grace’s medium in a glass or plexiglass dissecting dish. To dissect ovaries (see Note 26), grasp the anterior end of the female’s abdomen with forceps in the left hand. Using forceps in the right hand, pinch the last stripe in the female’s abdomen and pull away from the body. The forceps will grab not only the cuticle but also the urogenital system (frequently including the ovaries). Repeat dissection for all of the females in that vial. Collect dissected ovaries in cold Grace’s medium, and quickly break open the outer muscle sheath surrounding the ovarioles to separate individual ovarioles. Using a glass Pasteur pipet, remove BSA from a labeled, pre-coated microfuge tube and discard. Using the same pipet (which is now also coated with BSA), move the dissected ovaries in Grace’s medium to the microfuge tube. Place the tube on ice.
Repeat dissection with additional vials of flies. Proceed with fixation (below) within an hour of dissection of the first fly.
Allow ovaries to collect in the bottom of the microfuge tube by gravity. Remove as much Grace’s media as possible (see Note 27), and add 1.0 mL of Fixative. Invert the microfuge tube several times to ensure all ovaries are suspended, and rotate on a nutator for 13 min at room temperature.
Remove fixative to an appropriate waste container, and quickly add 1.0 mL of wash solution. Invert the tube several times, and remove the wash to the waste container. Add fresh wash solution, invert the tube to suspend the ovaries, and rotate on a nutator for 15 min.
Discard the solution, and wash ovaries two more times in 1.0 mL of wash solution for greater than 10 min.
Discard the solution, and add 1.0 mL of blocking solution. Rotate ovaries for 3 h on nutator at room temperature.
Discard the solution, and add 400 μL of primary antibody mixture (see Notes 28 and 29). Incubate ovaries overnight or over two nights at 4 °C on a nutator.
Remove antibody (see Note 30) and wash ovaries four times in 1.0 mL wash solution for at least 30 min each on a nutator.
Discard the last wash, and add 500 μL of secondary antibody mixture. Incubate ovaries for 1–2 h at room temperature on a nutator. From this step forward, all samples should be protected from light to minimize loss of fluorescence.
Discard antibody and wash ovaries four times in 1.0 mL wash solution for at least 30 min each on a nutator.
Discard the last wash, and add 500 μL of DAPI. Incubate ovaries for 15 min on a nutator.
Discard DAPI, and wash ovaries two times in 1.0 mL wash solution for 10 min each on a nutator.
Discard the last wash, removing as much liquid as possible. Add four drops of mounting media directly on top of the ovaries. Samples can be stored upright at 4 °C for up to a month.
3.3 Sample Preparation and Confocal Imaging
Prepare samples on glass slides for imaging on the day of or day before imaging (see Note 31).
Using a glass Pasteur pipet, move ovaries and mounting media onto a slide. Remove excess mounting media. Isolate (tease) individual ovarioles apart. For optimal imaging of GSCs, remove all large follicles (see Note 32). Spread ovarioles out across the slide, making sure there are no large clumps.
Once all large follicles are removed and ovarioles are sufficiently spread away from each other in the center of the slide, add 1–2 small drops of mounting media back to slide. Drop a glass coverslip onto the center of the slide, and allow capillary action to spread the mounting media and ovarioles across the coverslip. If any air remains between the coverslip and the slide, use a Pasteur pipet to slowly add a little more mounting media at the edges of the coverslip.
Place Kimwipes and a steel weight on top of the coverslip/slide to flatten the ovarioles. After 10–15 min, remove the weight and seal the coverslip to the slide using fingernail polish. Allow slides to dry before imaging.
Use an inverted confocal microscope with a 63× oil immersion lens and an optical zoom (1.5–3×) to image samples. Images are collected as confocal z-stacks (1 μm optical sections). Each germarium on the slide should be imaged for analysis (typically 75–100 germaria per slide).
3.4 Phenotypic Analysis, Quantification, and Next Steps
RNAi-mediated knockdown of genes that function in the Drosophila germline may result in a variety of phenotypes. Loss of expression of genes that function in an early step of germline establishment (i.e., during development) or germline differentiation may result in a complete block to oogenesis. Knockdown of genes that maintain the GSC fate may result in a complete loss of GSCs (and, thus, all daughter germ cells), resulting in rudimentary ovaries devoid of germ cells. Loss of expression of genes that function in early cyst growth (independent of GSC function or germ-line differentiation) may result in ovaries lacking more developed follicles. Although each of these possible scenarios results in a similar “small ovary” phenotype, knockdown of some genes may result in subtle phenotypes that do not alter overall ovarian morphology. It is therefore important to perform a thorough characterization of ovariole structure and cellular composition as a starting point to more detailed molecular analyses.
Following the immunofluorescent detection of anti-Hts and anti-LaminC, early germ cells can be recognized by the presence of the Hts-positive fusome (see Fig. 3c, c’). GSCs are unambiguously identified via their anteriorly localized fusome adjacent to the cap cells (which express high levels of LaminC). While the fusome is typically round in shape, changes in fusome morphology (elongation and fragmentation) correlate with the different phases of the GSC cell cycle and can be used as an indicator of cell cycle progression [20, 23]. When the GSC divides to form a cystoblast, a new fusome forms, located in the posterior of the cystoblast but with a similar round shape as the GSC fusome (arrowheads, Fig. 3c). As the cystoblast continues to divide with incomplete cytokinesis to form multicellular germline cysts, the fusome branches such that a single branch endpoint is located in each cell cytoplasm (open arrowheads, Fig. 3c). Initial quantification of germ cell number at each stage of differentiation (GSCs, cystoblasts, 2-cell, 4-cell, 8-cell cysts) can thus provide a snapshot of GSC maintenance and the ability of the daughter germ cells to divide and differentiate.
Alterations in GSC or cystoblast number are the initial phenotypic indicators of a change in GSC function. Reduction or complete loss of GSCs (quantified as the average number of GSCs per germarium) is indicative of a failure to maintain the GSC population. Additional analyses can then be performed to elucidate the cause of GSC loss. For example, apoptosis assays (such as immunofluorescence for TUNEL or cleaved caspase) can indicate whether GSCs are lost due to cell death, and molecular markers for each of the differentiated stages can be assessed to determine whether GSCs are lost due to premature differentiation. Such markers may include phosphorylated Mothers Against Decapentaplegic (pMad, highly expressed in GSCs), Bam (expressed in cystoblasts and 2- and 4-cell cysts), Ataxin-2-binding protein 1 (A2BP1, expressed in 4- and 8-cell cysts) [63], and C-3-G and Orb (expressed in oocytes). Expansion of the number of GSCs and/or cystoblast-like cells per germarium may indicate an increased rate of GSC proliferation or a failure of the immediate daughter cell to initiate differentiation. To evaluate differences in GSC proliferation, the S-phase indicator 5-ethynyl-2′-deoxyuridine (EdU) or the M-phase indicator phospho-histone H3 can be added to the analysis. These markers are also useful in examining changes in cystoblast differentiation and proliferation. Immunofluorescence analysis in targeted RNAi knockdown ovaries can be complemented by molecular analysis of the ovary, including gene expression (RT-PCR, RNA-seq, or in situ hybridization) or protein analysis (Western blotting). All phenotypes derived from targeted RNAi should be validated using other gene loss-of-function models, including the phenotypic assessment of genetic mutants by mosaic clonal analysis [45, 48].
Acknowledgments
Many thanks to Todd Nystul, Daniela Drummond-Barbosa, Acaimo González-Reyes, the Bloomington Stock Center, the Kyoto Stock Center, the Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies. Thanks also to the TRiP at Harvard Medical School for pVALIUM vectors, valuable protocols, bioinformatics resources, and advice on building our own UAS-RNAi responders. This work was supported by supported by the East Carolina University Thomas Harriot College of Arts and Sciences and Division of Research and Graduate Studies, and the March of Dimes Foundation (5-FY14-62).
Footnotes
Many of these lines can be obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu), the Vienna Drosophila Resource Center (http://stockcenter.vdrc.at), or the Kyoto Drosophila Stock Center (http://www.dgrc.kit.ac.jp).
We recommend Nutri-Fly MF, mixed using the manufacturer’s instructions and supplemented with Tegosept to prevent mold growth. Nutri-Fly MF is a molassess-based media formulation available from Genesee Scientific (https://geneseesci.com) that is easily mixed using an immersion blender and convection hot plate. Because it can be made in relatively small batches, it is a good option for small Drosophila labs. Fly culture medium is best when used fresh (within 3–4 weeks of pouring). We use 6 oz. square bottom polypropylene bottles and narrow polystyrene vials, both sealed with Flugs.
We use Fleischmann’s instant dry yeast, available in 1 lb. packages from Sam’s club.
Drosophila husbandry and anesthetic supplies can be purchased from Genesee Scientific.
Tubes are pre-coated in a 3% bovine serum albumin (BSA) solution [diluted from a 30% solution (Sigma) in distilled water] to prevent the ovaries from sticking to the sides of the tubes. Add 250 μL of 3% BSA into microfuge tubes. Place tubes on a nutator at room temperature for 1 h to ensure complete coverage of inside of tube. Store tubes at 4 °C.
We make 50 mL sterile aliquots of Grace’s media and store at 4 °C until ready for dissection. A fresh batch should be used if the aliquot appears cloudy.
Optimal fixative concentration and fixation time may vary between primary antibodies and should be experimentally determined. We have found that the concentration and incubation time given here are optimal for the primary and secondary antibodies used in this protocol.
Formaldehyde rapidly degrades when in contact with air. For best results, we order 10 mL ampules of ultrapure 16% formaldehyde, transfer the liquid to a 15 mL conical tube, and store for use no longer than 1 week at 4 °C.
Ideal blocking solutions help to reduce background staining and may vary with primary and secondary antibodies. It is important to choose serum for blocking solution that matches the host species of the secondary antibodies used in the immunostaining reaction.
We have also found that some concentrated aliquots of LC28.26 will also label the nuclear envelopes of GSCs, germ cells, and, occasionally, follicle cells (faintly).
This protocol can be adapted to stain with other molecular markers, including apoptosis assays (TUNEL) and proliferation markers (EdU) or other primary antibodies. Simply adjust the antibody concentration (if necessary) and select the appropriate secondary antibody in order to maintain specificity and avoid cross reactivity.
Upon arrival, we add an equal volume of 100% glycerol to each commercial secondary antibody, mix well, and store at −20 °C in 100 μL aliquots.
Prepare a 5 mg/mL DAPI (Life Technologies, Cat #D1306) stock solution according to manufacturer’s instructions in deionized water.
While a variety of commercially available glycerol-based anti-fade mounting media are available, we prefer to make our own solution. We have found that it is less expensive and preserves fluorescence intensity equally (if not better) than commercially available brands. We have previously used mounting media directly supplemented with DAPI but have found that these formulations do not penetrate well into the ovary, resulting in uneven nuclear staining.
Different members of our lab separate ovarioles (“teasing”) using different methods. Having one needle with an “L” or “shepherd’s crook” shape helps some of our lab members pull the largest follicles off the ovarioles in a left-right motion (think of a cane pulling a person off a stage in old movies).
We recommend the Zeiss LSM700 system, particularly for small labs with routine use.
We use Zeiss Black image acquisition software, and Zeiss Blue image analysis software. Many common analyses can also be conducted in ImageJ, which is freely available (http://imagej.nih.gov/ij/).
Information on the specific vector used to generate a UAS-RNAi of interest can be found in FlyBase by clicking on the “inserted element” link, followed by the “associated sequence features” link. In general, UAS-RNAi lines beginning with “JF” or “HMS” were created in non-germline-compatible vectors (Valium1 or Valium10), while “HMJ,” “HMC,” “GL,” or “GLC” were created in the Valium20 or Valium22 vectors.
A wide variety of companies now provide transgenic injection services for Drosophila, including BestGene (www.thebestgene.com), Rainbow Transgenic Flies, Inc. (www.rainbowgene.com), and Genetic Services, Inc. (www.geneticservices.com).
Methods for Drosophila culture maintenance and husbandry have been described elsewhere.
y1w1 (abbreviated yw in this protocol) is frequently used as an inbred isotype control because of its prevalent use in generating transgenic flies. These are available from the Bloomington Stock Center (BLM #1495).
Two control crosses are appropriate to ensure that any experimental phenotypes are the result of knockdown of the gene of interest. Since the UAS-RNAi should not be expressed in the absence of Gal4, cross UAS-RNAi responder males with yw female virgins. Likewise, the Gal4 should not cause ovarian defects due to genomic insertion site or a background mutation. Thus, cross Gal4 driver female virgins with yw males.
Experimental crosses and their controls should always have the same diet, ambient temperature, and be age-matched.
Oogenesis is exquisitely tuned to the female fly’s diet [64]. To ensure that any phenotypes identified are the result of gene knockdown and not effects of suboptimal diet, flies must be carefully cultured. Feeding flies daily with fresh yeast paste is necessary to maintain the optimal conditions for egg laying. Despite the waste of vials, it is also critical to discard vials with old, dried yeast paste: flies will reduce egg production in the absence of fresh yeast.
As with any UAS/Gal4 experiment, the strength of RNAi knockdown can be temperature-dependent, even in the absence of the Gal80 system. The nos-Gal4::VP16 driver is particularly sensitive to temperature; weak phenotypes may be enhanced by maintaining flies at 29 °C rather than 25 °C.
A variety of reviews have recently published excellent descriptions of ovary dissection techniques [65–68]; we therefore refer readers there for additional photographs of ovary dissection and separating ovarioles. In particular, we have found that a YouTube video from Scott Ferguson (https://www.youtube.com/watch?v=T94be2i5qb4) is helpful for first-time ovary dissectors.
At each liquid exchange throughout the rest of the protocol, allow a few minutes for ovaries to fall to the bottom of the microfuge tube by gravity before removing the solution. With the exception of the fixative and primary antibody solutions, we use a vacuum trap flask equipped with a thin pipet tip to remove liquid from the ovaries. We typically leave about 50–100 μL of solution on dissected ovaries at any given time in this protocol. This helps to avoid accidental suction of the ovaries into waste and prevents the samples from drying.
Although this protocol details immunostaining for two primary antibodies raised in the same species (and thus detected using a single secondary antibody), we routinely use multiple antibodies to localize distinct proteins or cell labels in the ovary. In theory, immunostaining with multiple antibodies of different species should work well by combining all primary antibodies together at this step. We achieve vastly decreased background staining, however, by incubating primary antibodies in series over several nights, separated by extensive wash steps (four times 30 min each). In effect, primary antibodies are layered on top of the sample. In contrast, secondary antibodies against different species are typically grouped together into a single solution.
If two antibodies are raised in the same species, but it is necessary to image them separately, apply one antibody followed by washes, and then the appropriate secondary, as written. Then wash ovaries five times for 30–45 min each in wash solution, re-block for 1 h, and apply the second primary antibody. Following incubation, proceed with wash and second secondary (with a different fluorophore) as written.
The mixture of anti-Hts and anti-LamC at the concentrations given can be collected, stored at 4 °C, and reused for one additional experiment.
Samples on slides will dry out over time, decreasing fluorescence intensity. Since image acquisition is frequently the bottleneck to these experiments, we store all samples in mounting media until the day before imaging.
Follicles can be staged based on a variety of morphological features [19]. Stage 10 follicles are easily identified under a dissecting stereomicroscope, because the size of the oocyte approximates one-half the size of the whole follicle. We remove all follicles larger than a Stage 10 for optimal imaging of GSCs.
References
- 1.Daley GQ. Stem cells and the evolving notion of cellular identity. Philos Trans R Soc Lond Ser B Biol Sci. 2015;370(1680):20140376. doi: 10.1098/rstb.2014.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanherkar RR, Bhatia-Dey N, Makarev E, Csoka AB. Cellular reprogramming for understanding and treating human disease. Front Cell Dev Biol. 2014;2:67. doi: 10.3389/fcell.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ables ET, Laws KM, Drummond-Barbosa D. Control of adult stem cells in vivo by a dynamic physiological environment: diet-dependent systemic factors in Drosophila and beyond. Wiley Interdiscip Rev Dev Biol. 2012;1(5):657–674. doi: 10.1002/wdev.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess RJ, Agathocleous M, Morrison SJ. Metabolic regulation of stem cell function. J Intern Med. 2014;276(1):12–24. doi: 10.1111/joim.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iglesias-Bartolome R, Gutkind JS. Signaling circuitries controlling stem cell fate: to be or not to be. Curr Opin Cell Biol. 2011;23(6):716–723. doi: 10.1016/j.ceb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakada D, Levi BP, Morrison SJ. Integrating physiological regulation with stem cell and tissue homeostasis. Neuron. 2011;70(4):703–718. doi: 10.1016/j.neuron.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21(1):159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Cuevas M, Matunis EL. The stem cell niche: lessons from the drosophila testis. Development. 2011;138(14):2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homem CC, Repic M, Knoblich JA. Proliferation control in neural stem and progenitor cells. Nat Rev Neurosci. 2015;16(11):647–659. doi: 10.1038/nrn4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Edgar BA. Intestinal stem cell function in Drosophila and mice. Curr Opin Genet Dev. 2012;22(4):354–360. doi: 10.1016/j.gde.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahai-Hernandez P, Castanieto A, Nystul TG. Drosophila models of epithelial stem cells and their niches. Wiley Interdiscip Rev Dev Biol. 2012;1(3):447–457. doi: 10.1002/wdev.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie T. Control of germline stem cell self-renewal and differentiation in the Drosophila ovary: concerted actions of niche signals and intrinsic factors. Wiley Interdiscip Rev Dev Biol. 2013;2(2):261–273. doi: 10.1002/wdev.60. [DOI] [PubMed] [Google Scholar]
- 13.Wangler MF, Yamamoto S, Bellen HJ. Fruit flies in biomedical research. Genetics. 2015;199(3):639–653. doi: 10.1534/genetics.114.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 15.dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM. FlyBase: introduction of the Drosophila melanogaster release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43(Database issue):D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohr SE, Hu Y, Kim K, Housden BE, Perrimon N. Resources for functional genomics studies in Drosophila melanogaster. Genetics. 2014;197(1):1–18. doi: 10.1534/genetics.113.154344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim HS, Miller A, Housden A, Foos M, Randkelv S, Kelley C, Namgyal P, Villalta C, Liu LP, Jiang X, Huan-Huan Q, Xia W, Fujiyama A, Toyoda A, Ayers K, Blum A, Czech B, Neumuller R, Yan D, Cavallaro A, Hibbard K, Hall D, Cooley L, Hannon GJ, Lehmann R, Parks A, Mohr SE, Ueda R, Kondo S, Ni JQ, Perrimon N. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venken KJ, Sarrion-Perdigones A, Vandeventer PJ, Abel NS, Christiansen AE, Hoffman KL. Genome engineering: Drosophila melanogaster and beyond. Wiley Interdiscip Rev Dev Biol. 2015 doi: 10.1002/wdev.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spradling A. Developmental genetics of oogenesis. In: Bate M, editor. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Plainview, NY: 1993. pp. 1–70. [Google Scholar]
- 20.de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125(15):2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- 21.Ables ET, Drummond-Barbosa D. The steroid hormone ecdysone functions with intrinsic chromatin remodeling factors to control female germline stem cells in Drosophila. Cell Stem Cell. 2010;7(5):581–592. doi: 10.1016/j.stem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu HJ, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci U S A. 2009;106(4):1117–1121. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313(2):700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris LX, Spradling AC. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138(11):2207–2215. doi: 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232(3):559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 26.Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121(11):3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- 27.Nystul T, Spradling A. An epithelial niche in the Drosophila ovary undergoes long-range stem cell replacement. Cell Stem Cell. 2007;1(3):277–285. doi: 10.1016/j.stem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Neumuller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, Knoblich JA. Genome-wide analysis of self-renewal in Drosophila neural stem cells by trans-genic RNAi. Cell Stem Cell. 2011;8(5):580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan D, Neumuller RA, Buckner M, Ayers K, Li H, Hu Y, Yang-Zhou D, Pan L, Wang X, Kelley C, Vinayagam A, Binari R, Randklev S, Perkins LA, Xie T, Cooley L, Perrimon N. A regulatory network of Drosophila germline stem cell self-renewal. Dev Cell. 2014;28(4):459–473. doi: 10.1016/j.devcel.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira FK, Sanchez CG, Hurd TR, Seifert JR, Czech B, Preall JB, Hannon GJ, Lehmann R. ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat Cell Biol. 2015;17(5):689–696. doi: 10.1038/ncb3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng X, Han L, Singh SR, Liu H, Neumuller RA, Yan D, Hu Y, Liu Y, Liu W, Lin X, Hou SX. Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 2015;10(7):1226–1238. doi: 10.1016/j.celrep.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2(8):a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr SE. RNAi screening in Drosophila cells and in vivo. Methods. 2014;68(1):82–88. doi: 10.1016/j.ymeth.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014;15(9):591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 36.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 37.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18(8):896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 38.Lam G, Thummel CS. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr Biol. 2000;10(16):957–963. doi: 10.1016/s0960-9822(00)00631-x. [DOI] [PubMed] [Google Scholar]
- 39.Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34(1–2):1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 41.Huang P, Sahai-Hernandez P, Bohm RA, Welch WP, Zhang B, Nystul T. Enhancer-trap flippase lines for clonal analysis in the Drosophila ovary. G3 (Bethesda) 2014;4(9):1693–1699. doi: 10.1534/g3.114.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1(6):2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- 43.Hudson AM, Cooley L. Methods for studying oogenesis. Methods. 2014;68(1):207–217. doi: 10.1016/j.ymeth.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konig A, Shcherbata HR. Visualization of adult stem cells within their niches using the Drosophila germline as a model system. Methods Mol Biol. 2013;1035:25–33. doi: 10.1007/978-1-62703-508-8_3. [DOI] [PubMed] [Google Scholar]
- 45.Laws KM, Drummond-Barbosa D. Genetic mosaic analysis of stem cell lineages in the Drosophila ovary. Methods Mol Biol. 2015;1328:57–72. doi: 10.1007/978-1-4939-2851-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim RS, Osato M, Kai T. Isolation of undifferentiated female Germline cells from adult Drosophila ovaries. Curr Protoc Stem Cell Biol. 2015;34:2E 3 1–2E 3 15. doi: 10.1002/9780470151808.sc02e03s34. [DOI] [PubMed] [Google Scholar]
- 47.Luo L, Chai PC, Cai Y. Immunostaining of germline stem cells and the niche in Drosophila ovaries. Methods Mol Biol. 2013;1035:1–7. doi: 10.1007/978-1-62703-508-8_1. [DOI] [PubMed] [Google Scholar]
- 48.Rubin T, Huynh JR. Mosaic analysis in the Drosophila melanogas-ter ovary. Methods Mol Biol. 2015;1328:29–55. doi: 10.1007/978-1-4939-2851-4_3. [DOI] [PubMed] [Google Scholar]
- 49.Singh SR, Liu Y, Kango-Singh M, Nevo E. Genetic, immunofluorescence labeling, and in situ hybridization techniques in identification of stem cells in male and female germ-line niches. Methods Mol Biol. 2013;1035:9–23. doi: 10.1007/978-1-62703-508-8_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8(5):405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78(1–2):113–118. doi: 10.1016/s0925-4773(98)00157-9. http://dx.doi.org/10.1016/S0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 52.Ables ET, Bois KE, Garcia CA, Drummond-Barbosa D. Ecdysone response gene E78 controls ovarian germline stem cell niche formation and follicle survival in Drosophila. Dev Biol. 2015;400(1):33–42. doi: 10.1016/j.ydbio.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11(3):222–223. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- 54.Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134(4):703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 55.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8(4):243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 56.Faucherre A, Lopez-Schier H. Delaying Gal4-driven gene expression in the zebrafish with morpholinos and Gal80. PLoS One. 2011;6(1):e16587. doi: 10.1371/journal.pone.0016587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13(20):1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 58.McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121(9):2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- 59.McKearin DM, Spradling AC. Bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4(12B):2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 60.Ohlstein B, McKearin D. Ectopic expression of the Drosophila bam protein eliminates oogenic germline stem cells. Development. 1997;124(18):3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- 61.Bolivar J, Pearson J, Lopez-Onieva L, Gonzalez-Reyes A. Genetic dissection of a stem cell niche: the case of the Drosophila ovary. Dev Dyn. 2006;235(11):2969–2979. doi: 10.1002/dvdy.20967. [DOI] [PubMed] [Google Scholar]
- 62.Cabrera GR, Godt D, Fang PY, Couderc JL, Laski FA. Expression pattern of Gal4 enhancer trap insertions into the bric a brac locus generated by P element replacement. Genesis. 2002;34(1–2):62–65. doi: 10.1002/gene.10115. [DOI] [PubMed] [Google Scholar]
- 63.Tastan OY, Maines JZ, Li Y, McKearin DM, Buszczak M. Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development. 2010;137(19):3167–3176. doi: 10.1242/dev.050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231(1):265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- 65.Groen CM, Tootle TL. Visualization of actin cytoskeletal dynamics in fixed and live Drosophila egg chambers. Methods Mol Biol. 2015;1328:113–124. doi: 10.1007/978-1-4939-2851-4_8. [DOI] [PubMed] [Google Scholar]
- 66.Legent K, Tissot N, Guichet A. Visualizing microtubule networks during Drosophila oogenesis using fixed and live imaging. Methods Mol Biol. 2015;1328:99–112. doi: 10.1007/978-1-4939-2851-4_7. [DOI] [PubMed] [Google Scholar]
- 67.Manning L, Starz-Gaiano M. Culturing Drosophila egg chambers and investigating developmental processes through live imaging. Methods Mol Biol. 2015;1328:73–88. doi: 10.1007/978-1-4939-2851-4_5. [DOI] [PubMed] [Google Scholar]
- 68.Thompson L, Randolph K, Norvell A. Basic techniques in Drosophila ovary preparation. Methods Mol Biol. 2015;1328:21–28. doi: 10.1007/978-1-4939-2851-4_2. [DOI] [PubMed] [Google Scholar]
- 69.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, Xie T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131(6):1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 70.Tanentzapf G, Devenport D, Godt D, Brown NH. Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol. 2007;9(12):1413–1418. doi: 10.1038/ncb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sahai-Hernandez P, Nystul TG. A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development. 2013;140(22):4490–4498. doi: 10.1242/dev.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]