Abstract
The bacterial type VI secretion system (T6SS) mediates antagonistic cell–cell interactions between competing Gram-negative bacteria. In plant-beneficial bacteria, this pathway has been shown to suppress the growth of bacterial pathogens; however, the identification and mode of action of T6SS effector proteins that mediate this protective effect remain poorly defined. Here, we identify two previously uncharacterized effectors required for interbacterial antagonism by the plant commensal bacterium Pseudomonas protegens. Consistent with the established effector-immunity paradigm for antibacterial T6SS substrates, the toxic activities of these effectors are neutralized by adjacently encoded cognate immunity determinants. Although one of these effectors, RhsA, belongs to the family of DNase enzymes, the activity of the other was not apparent from its sequence. To determine the mechanism of toxicity of this latter effector, we determined its 1.3 Å crystal structure in complex with its immunity protein and found that it resembles NAD(P)+-degrading enzymes. In line with this structural similarity, biochemical characterization of this effector, termed Tne2 (Type VI secretion NADase effector family 2), demonstrates that it possesses potent NAD(P)+ hydrolase activity. Tne2 is the founding member of a widespread family of interbacterial NADases predicted to transit not only the Gram-negative T6SS but also the Gram-positive type VII secretion system, a pathway recently implicated in interbacterial competition among Firmicutes. Together, this work identifies new T6SS effectors employed by a plant commensal bacterium to antagonize its competitors and broadly implicates NAD(P)+-hydrolyzing enzymes as substrates of interbacterial conflict pathways found in diverse bacterial phyla.
Keywords: microbiology, bacteria, protein secretion, protein structure, protein–protein interaction, type VI secretion, interbacterial antagonism, Pseudomonas protegens, NADase effectors
Introduction
Bacteria use secreted proteins to interact with their environment. Among the protein secretion systems that facilitate these interactions, the predominant function of the type VI secretion system (T6SS)3 is to mediate antagonistic interactions between contacting Gram-negative bacteria (1). The T6SS is a membrane-embedded protein complex that resembles the tail component of contractile bacteriophage (2). Through cycles of extension and contraction, this pathway delivers effector proteins from the cytoplasm of a donor bacterium directly into the periplasm or cytoplasm of a recipient bacterium (3, 4). Used by both pathogenic and commensal bacteria of eukaryotes, the T6SS mediates a complex interplay of bacterial cell–cell interactions between beneficial and disease-causing bacteria (5–9).
The antibacterial properties of the T6SS are due to the toxic activities of secreted effector proteins. These effectors degrade conserved molecules required for cell viability, including cell wall peptidoglycan, membrane phospholipids, and chromosomal DNA (10). Intracellular intoxication and intoxication of sister cells are prevented by the existence of cognate immunity proteins, which directly bind to and inhibit the activity of their associated effector (11). Effectors transit the T6SS through physical association with one of two families of secreted structural T6SS apparatus proteins. Large, multidomain effectors interact with members of the valine-glycine repeat protein G (VgrG) family of proteins, whereas small, single-domain effectors associate with the lumen of ring-shaped haemolysin-coregulated protein (Hcp) (12–14). The former of these two effector groups often contains conserved N-terminal adaptor domains that facilitate their physical interaction with VgrG (15), the best characterized of which is the Pro-Ala-Ala-Arg (PAAR) domain (16). The cone-shaped PAAR domain possesses a flat hydrophobic surface at its base that strongly associates with a highly complementary surface at the C terminus of VgrG proteins (17).
Pseudomonas fluorescens and other closely related plant-associated bacteria are well known for their ability to both promote plant growth and suppress pathogens (18). Pathogen suppression by these organisms is accomplished, in part, by the production of diffusible small molecules that inhibit the growth of many disease-causing fungi (19). More recently, the T6SS has been demonstrated to play a role in suppressing the growth of bacterial pathogens. For example, the T6SS of P. fluorescens MFE01 can protect potato tubers from blackleg disease caused by Pectobacterium atrosepticum (20). Similarly, the K1-T6SS from Pseudomonas putida KT2440 allows it to outcompete Xanthomonas campestris during co-infection of plant leaves (21). However, the contribution of individual effector proteins in interbacterial competition was not assessed in either study.
Despite the increasing number of characterized T6SS effectors in both Hcp- and VgrG-linked groups, their contribution to interbacterial killing by plant-protective Pseudomonas spp. has yet to be examined. Pseudomonas protegens Pf-5 (formerly known as P. fluorescens Pf-5) is a well-characterized plant commensal bacterium that is of significant agricultural interest due to its biocontrol properties (22). In previous work, we showed that this bacterium possesses a potent antibacterial T6SS (23). Furthermore, three Hcp-linked effectors have been identified by informatic and proteomic approaches (12, 24). The contribution of two of these effectors, Tae3 and Tse4, in interbacterial competition has not been examined, although the third, Tge2, has been shown to confer a T6SS-dependent fitness advantage to P. protegens when co-cultured with competitor bacteria on low-salt media (23). Tge2 is thought to hydrolyze the glycan strands of cell wall peptidoglycan, potentially explaining its efficacy when cells are grown under hypoosmotic conditions; however, the contribution of Tge2 relative to the overall fitness advantage conferred by the T6SS of this organism is minimal. These findings suggest that either Tae2 and/or Tse4 are responsible for much of the T6SS-dependent growth inhibition or that there exist additional uncharacterized effectors in this organism.
In this work, we sought to identify T6SS effectors responsible for interbacterial killing by P. protegens Pf-5. This led us to characterize two PAAR domain-containing effectors, which we subsequently show play a substantial role in interbacterial competition by this organism. Structural determination of one of these effectors revealed its mode of action as an NAD(P)+-hydrolyzing (NADase) enzyme, a biochemical activity not frequently associated with T6SS effectors. To further explore the potential diversity of NADase effectors, we next used iterative homology search algorithms and identified two phylogenetically distinct families of T6SS NADases. One of these families also contains a significant number of proteins associated with the bacterial type VII secretion system (T7SS), a pathway recently implicated in interbacterial antagonism between Gram-positive bacteria (25, 26). Taken together, our findings reveal a broad distribution of interbacterial NADase enzymes and provide insight into how a plant commensal bacterium inhibits the growth of its bacterial competitors.
Results
Identification of novel T6SS effector-immunity pairs in P. protegens
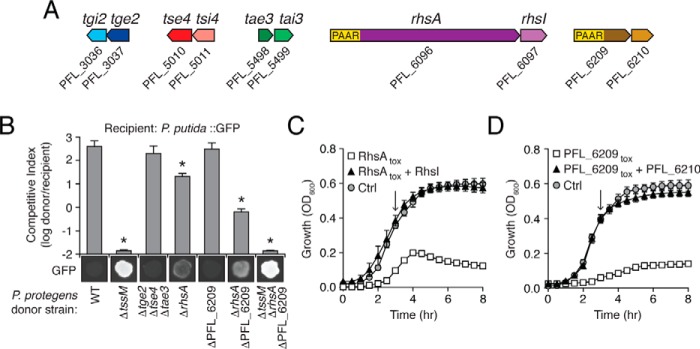
The recent finding that PAAR domains function as interaction partners of VgrG proteins has enabled the facile identification of PAAR domain-containing T6SS effectors in diverse bacteria (17). We searched the genome of P. protegens Pf-5 for PAAR-encoding genes and identified two hypothetical T6SS effectors, PFL_6096 and PFL_6209 (Fig. 1A). The former of these genes, which we named rhsA, belongs to the rearrangement hotspot (Rhs) family of T6SS effectors. Proteins encoded by rhs genes typically contain an N-terminal PAAR domain, a large internal segment composed of multiple tyrosine-aspartate (YD) repeats, and a polymorphic C-terminal toxin domain (27). The second effector, PFL_6209, also contains a predicted PAAR domain at its N terminus; however, it lacks identifiable YD repeats, and its C-terminal domain does not bear homology to characterized proteins.
Figure 1.
Novel T6SS effectors in P. protegens provide a competitive growth advantage during interspecies competition. A, schematic of genes encoding effector-immunity pairs in P. protegens Pf-5. Locus tag numbers are indicated below each gene. For each effector-immunity bicistron, shading is used to indicate the effector (dark) and its cognate immunity (light). B, growth experiments measuring the fitness of the indicated P. protegens donor strains in co-culture with a GFP-labeled P. putida recipient. For each competition, donor and recipient were mixed and incubated on solid medium for 8 h at 30 °C. Initial and final populations were enumerated on non-selective media (P. protegens) or selective media (P. putida). The competitive index was determined by comparing final and initial ratios for each competition. Asterisks indicate outcomes statistically different from the wildtype competition (n = 3, p < 0.05). Fluorescence images are a qualitative indication of the remaining P. putida at the end of each competition. C and D, growth in liquid media of E. coli cells expressing RhsAtox or RhsAtox and RhsI (C) or PFL_6209tox or PFL_6209tox and PFL_6210 (D). In both experiments, a vector control was included. Protein expression was induced at the indicated time point (arrow).
To assess the contribution of RhsA and PFL_6209 to interbacterial fitness under cell–contact-promoting conditions, various P. protegens donor strains were co-cultured with P. putida, a bacterium that also inhabits soil environments. P. putida was chosen as the competitor because we previously determined that the T6SS of P. protegens confers a significant fitness advantage over this organism (23). We first compared the competitive fitness of wildtype P. protegens to that of a P. protegens strain lacking the three previously identified Hcp-associated effectors Tae3, Tge2, and Tse4. Under our experimental conditions, a P. protegens strain lacking these three effectors outcompeted P. putida to the same extent as the wildtype strain (Fig. 1B).
We next assessed the competitive index of P. protegens strains bearing inactivating mutations in either rhsA or PFL_6209. Although the fitness of a P. protegens strain lacking PFL_6209 was indistinguishable from wildtype, a ΔrhsA mutant exhibited a moderate reduction in its ability to compete with P. putida. Additionally, a ΔPFL_6209 ΔrhsA double mutant displayed a reduction in competitive index that far exceeded that of the ΔrhsA donor strain. Importantly, this reduction in competitive fitness was not additive with a strain lacking T6SS apparatus function, indicating an epistatic relationship between these genes. In total, these data indicate that rhsA and PFL_6210 play a significant role in interspecies competition between P. protegens and P. putida.
Multidomain T6SS effectors typically harbor domains with toxin activity at their C terminus (28). In line with this precedent, we found that expression of the C-terminal domain of RhsA (RhsAtox) significantly inhibited the growth of both E. coli and P. putida (Fig. 1C and Fig. S1). This growth inhibition was abrogated upon co-expression with PFL_6097, indicating that the protein product of this gene (henceforth referred to as rhsI) confers immunity to RhsAtox-mediated toxicity. The C-terminal toxin domains of several previously characterized Rhs proteins possess DNase activity (29). Similarly, the C terminus of RhsA belongs to a superfamily of restriction endonucleases possessing a conserved aspartic acid dyad (NCBI conserved domain accession number cl00516) (Fig. S2A). Site-specific mutation of one of these residues, Asp-1399, abrogated growth inhibition suggesting that this effector also exerts toxicity by hydrolyzing DNA (Fig. S2B).
In contrast to the toxin domain of RhsA, the C terminus of PFL_6209 does not share homology with proteins of known function. Nonetheless, we found that heterologous expression of the C-terminal domain of PFL_6209 (PFL_6209tox) inhibits both E. coli and P. putida growth, demonstrating that like RhsA, the C terminus of PFL_6209 also encodes an antibacterial toxin (Fig. 1D and Fig. S1). Additionally, co-expression of the downstream gene, PFL_6210, confers immunity to the activity of this toxin. Taken together, our data suggest that P. protegens employs a nucleic acid-degrading effector and an effector of unknown activity to gain a fitness advantage over competitor bacteria.
Structure of the PFL_6209tox–PFL_6210 effector–immunity complex
We next sought to determine the molecular basis for the antibacterial activity exhibited by PFL_6209. To this end, we first determined the 1.7-Å co-crystal structure of PFL_6209tox in complex with PFL_6210 using selenium-incorporated protein and the selenium single wavelength anomalous dispersion technique (30). The PFL_6209tox–PFL_6210 complex crystallized in the monoclinic space group C2 with two copies each of PFL_6209tox and PFL_6210 in the asymmetric unit. The electron density allowed for both copies of the toxin-immunity complex to be built in their entirety with the exception of the N-terminal hexahistidine tag and several vector-encoded amino acids that were fused to PFL_6209tox to facilitate purification of the complex. The final model was refined to an Rwork of 18.1% and Rfree of 21.6% (Table 1).
Table 1.
X-ray data collection and refinement statistics
| Tne2tox –Tni2 (selenomethionine) | |
|---|---|
| Data collection | |
| Beamline | 21-ID-F |
| Wavelength (Å) | 0.979 |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 176.7, 40.6, 95.9 |
| α, β, γ (°) | 90.0, 97.2, 90.0 |
| Resolution (Å) | 68.94–1.71 (1.77–1.71)a |
| Total no. of reflections | 145,128 |
| Total no. of unique reflections | 72,673 |
| Rmerge (%)b | 5.4 (64.6)a |
| I/σI | 13.8 (1.2)a |
| Completeness (%) | 98.7 (89.4)a |
| Redundancy | 2.0 (2.0)a |
| Refinement | |
| Rwork/Rfree (%)c | 18.1/21.6 |
| No. of atoms | |
| Protein | 4236 |
| Water | 574 |
| Average B-factors (Å2) | |
| Protein | 31.6 |
| Water | 40.6 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.89 |
| Ramachandran plot (%)d | |
| Total favored | 98.1 |
| Total allowed | 100.0 |
| Coordinate error (Å)e | 0.23 |
a Values in parentheses correspond to the highest resolution shell.
b Rmerge = Σ Σ |I(k) − 〈I〉|/Σ I(k), where I(k) and 〈I〉 represent the diffraction intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
c Rwork = Σ ‖Fobs| − k|Fcalc‖/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
d Data were calculated using MOLPROBITY (49).
e Maximum-likelihood based coordinate error was determined by PHENIX (50).
PFL_6209tox resembles an interbacterial NADase and depletes cellular NAD+
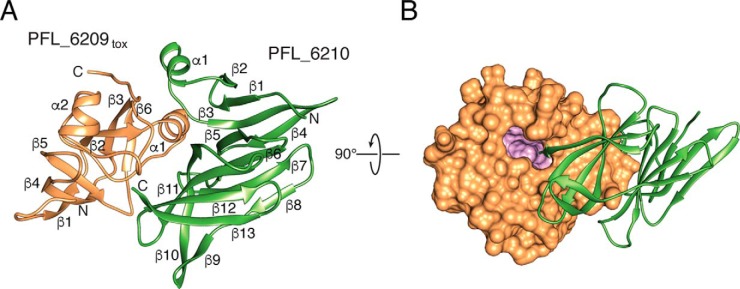
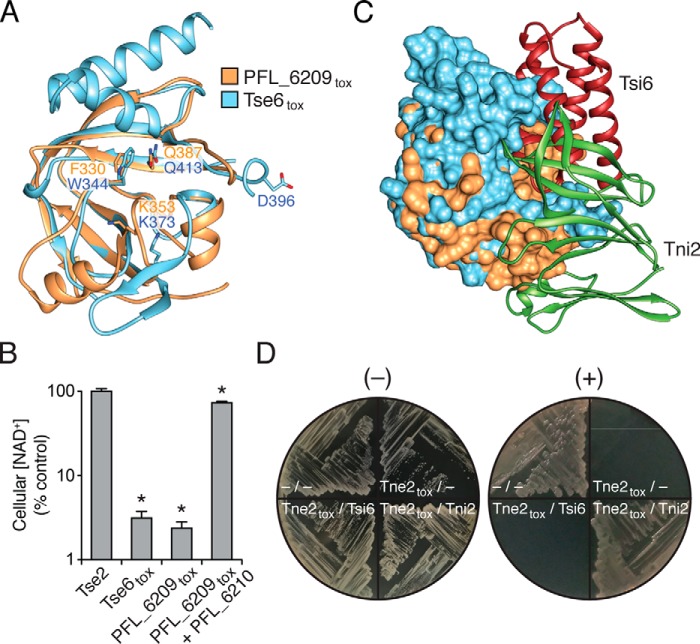
PFL_6209tox adopts a mixed α/β fold consisting of two α-helices and six β-strands (Fig. 2A). The toxin exhibits an overall globular form with the most notable surface feature being a deep cavity that extends ∼12 Å into the core of the protein. This cavity, which we hypothesize includes the active site of PFL_6209tox, is partially occluded by the C terminus of PFL_6210 (Fig. 2B). A search of the PDB using the DALI server identified the toxin domain of the Pseudomonas aeruginosa T6SS effector Tse6 (Tse6tox) as the top structural homolog of PFL_6209tox with a Z-score of 10.0. Despite having only 17% sequence identity between them, the toxin domains of PFL_6210 and Tse6 superimpose with a root-mean-square deviation (r.m.s.d.) of 2.2 Å over 97 equivalent Cα positions (Fig. 3A).
Figure 2.
Overall structure of PFL_6209tox–PFL_6210 complex. A, ribbon representation of PFL_6209tox (orange) in complex with PFL_6210 (green). Secondary structure elements and the location of the N and C termini of each protein are indicated. B, space-filling representation PFL_6209tox in complex with a ribbon representation of PFL_6210 rotated 90° with respect to A. Residues lining the cavity that extends into the core of PFL_6209tox are colored pink.
Figure 3.
PFL_6209tox resembles the NADase domain of the T6SS effector Tse6. A, structural alignment of PFL_6209tox with the NADase domain of Tse6. Predicted NAD+-binding residues Trp-344, Lys-373, and Gln-413 of Tse6tox superpose with Phe-330, Lys-353, and Gln-413 of PFL_6209tox. Asp-396, which is required for optimal NAD+ hydrolysis by Tse6tox, does not overlay with an equivalent residue in PFL_6209tox. B, relative NAD+ levels in E. coli cells expressing a bacteriostatic toxin that does not possess NADase activity (Tse2), Tse6tox, PFL_6209tox, or PFL_6209tox–PFL_6210 complex (hereafter referred to as Tne2tox and Tni2, respectively). NAD+ levels were assayed 60 min after induction of expression and compared with E. coli cells harboring a vector control. Error bars represent ± S.D. (n = 3). Asterisks indicate statistically significant differences in NAD+ levels compared with vector control. C, structural superposition of Tne2tox and Tse6tox in complex with their respective immunity determinants, Tni2 and Tsi6. The structurally aligned toxin domains are depicted as surface representations and are colored as in A, and the structurally distinct immunity proteins are shown as ribbon representations. D, Tsi6 does not confer immunity to Tne2tox-mediated toxicity. Growth of E. coli cells co-expressing the indicated genes under non-inducing (−) or inducing (+) conditions. Dashes represent vector controls.
Tse6 is a cytotoxic effector delivered to target bacteria by the Hcp secretion island I-encoded T6SS (H1-T6SS) of P. aeruginosa (12). It exerts toxicity by depleting cells of the essential dinucleotides NAD+ and its phosphorylated derivative NADP+ (28). Although its mechanism of catalysis is not known, it possesses several conserved NAD+-binding residues found in bacterial mono-ADP-ribosylating toxins (31). Two of these residues in Tse6, Trp-344 and Gln-413, superimpose with Phe-330 and Gln-387, respectively, of PFL_6209tox (Fig. 3A). Furthermore, Tse6 and PFL_6209 also share a conserved lysine residue near this predicted NAD+-binding site. Despite these similarities, PFL_6209 lacks an invariant aspartate residue (Asp-396) found in Tse6 and homologous toxins that is required for optimal NAD+ hydrolysis. This aspartate residue is located within a highly flexible region of Tse6tox termed the “activation loop” that forms a lid over the NAD+-binding site in the absence of Tsi6. In contrast, the equivalent region in PFL_6209tox does not contain any conserved residues and is significantly shorter. In summary, PFL_6209tox possesses a putative NAD+-binding pocket with both similarities and differences to those of Tse6 and mART toxins.
Given its overall structural similarity to Tse6tox, we next employed a coupled enzyme assay to assess NAD+ levels in E. coli cells undergoing intoxication by PFL_6209tox. As controls, the amount of NAD+ in cells expressing a bacteriostatic toxin that does not possess NADase activity (Tse2) and in cells expressing Tse6tox was examined (28, 32). Consistent with it possessing NADase activity, a marked reduction in NAD+ levels in cells expressing PFL_6209tox was observed (Fig. 3B). Furthermore, NAD+ concentrations were restored to near control levels by co-expressing PFL_6210. These observations suggest that PFL_6209 is a NADase enzyme and thus we henceforth refer to this effector as Type VI secretion NADase effector family 2 (Tne2). Given that PFL_6210 confers immunity to Tne2, we refer to this protein as Type VI secretion NADase immunity family 2 (Tni2).
Interbacterial NADase effectors are inhibited by diverse immunity proteins
Tni2 is composed of three β-sheets each formed by four antiparallel β-strands (Fig. 2A). Additionally, a single α-helix located between the β2 and β3 strands of the first β-sheet wraps around the α1 helix of Tne2tox and comprises a significant proportion of the 1129Å2 of total buried surface area between the toxin-immunity complex. Interface analysis by the PDBePISA server estimates that the free energy of complex formation between Tne2tox and Tni2 is −12.4 kcal/mol and that their association is mediated by numerous hydrogen-bonding interactions (33). Consistent with this observation, isothermal titration calorimetry indicates that complex formation between Tne2tox and Tni2 is exothermic and that the proteins interact with a dissociation constant of 8.1 nm (Fig. S3A). A comparison of the Tni2 structure to known structures in the PDB identified several β-propeller motif-containing proteins as having the highest structural similarity. These proteins include the substrate-gating domain of the trilobed protease from Pryococcus furiosus (Z-score of 8.4, 5.0-Å r.m.s.d. over 114 equivalent Cα positions), a putative TolB homolog from Agrobacterium tumefaciens (Z-score of 8.3, r.m.s.d. of 3.8Å over 112 equivalent Cα positions) and the substrate-gating domain of oligopeptidase B from Leishmania major (Z-score of 8.2, r.m.s.d. of 3.6Å over 112 equivalent Cα positions (34, 35). β-Propeller motifs are similarly composed of four-stranded, antiparallel, twisted β-sheets; however, they differ from Tni2 in that their β-sheets are radially arranged around a central pore.
Tse6 is also encoded adjacent to an immunity gene, termed tsi6, whose protein product inhibits its NADase activity. In contrast to Tni2, the Tsi6 immunity protein adopts an all-α-helical fold that binds to a surface of Tse6tox that has minimal overlap with the equivalent Tni2-binding site on Tne2tox despite the overall similarity of the toxins (Fig. 3C). In accordance with the high level of interaction specificity observed between T6SS effector-immunity pairs, Tsi6 does not interact with Tne2tox, and co-expression of Tsi6 in E. coli cells expressing Tne2tox did not abrogate Tne2tox-based toxicity (Fig. 3D and Fig. S3B). Given the stark differences between their overall folds, we speculate that the inhibitory activity of Tni2 and Tsi6 toward their respective NADase toxins arose through convergent evolution.
Tne2 possesses an NAD+-binding pocket and degrades NAD(P)+ in vitro
We next sought to identify Tne2tox residues involved in NAD+ hydrolysis. Given that the solely characterized catalytic residue of Tse6, Asp-396, is absent in Tne2tox, little insight into its mode of NAD+ binding and catalysis could be ascertained from comparison with this enzyme. Therefore, we performed in silico ligand docking studies to identify potential NAD+-binding residues. The highest scoring solution from this approach placed the nicotinamide moiety of NAD+ deep into the Tne2tox cavity that is partially occluded by Tni2 (Fig. 4A). In this position, NAD+ binding is predicted to be highly favorable with a calculated free energy of binding of −12.1 kcal/mol. Lending further support to our docking results is the observation that the adenosine group of NAD+, which lies outside the Tne2tox cavity, is predicted to sterically clash with Tni2 (Fig. S4, A and B). Thus, even though Tni2 does not completely occlude the predicted NAD+-binding pocket, it appears to interact with the same surface of Tne2tox as the adenosine moiety of NAD+ potentially explaining its effectiveness as a Tne2-specific immunity determinant.
Figure 4.
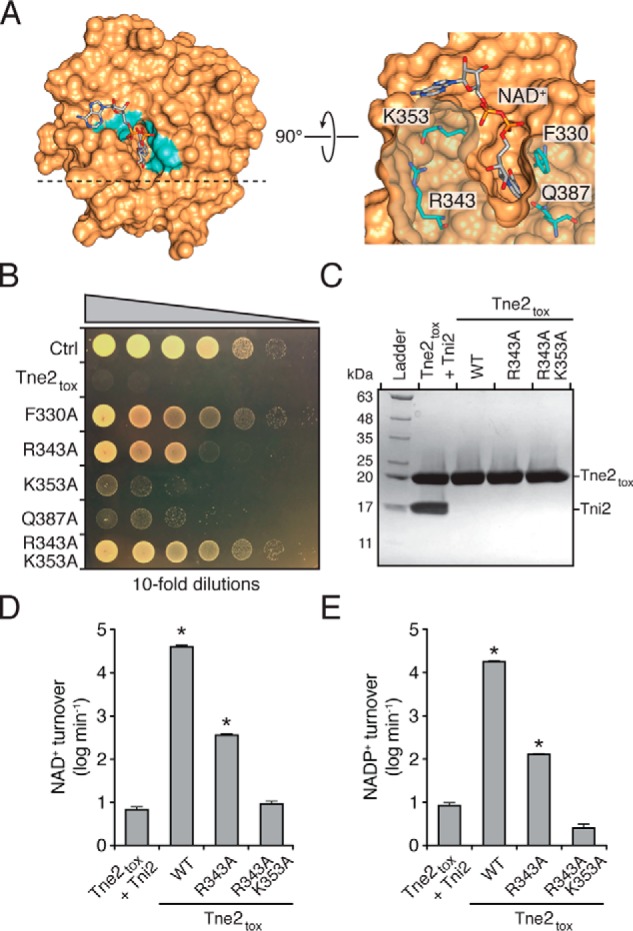
Identification of residues required for optimal NAD(P)+ hydrolysis by Tne2tox. A, molecular docking places NAD+ in the predicted active-site pocket of Tne2tox. Phe-330, Arg-343, Lys-353, and Gln-413, which are conserved among Tne2 orthologous proteins and line the NAD+-binding pocket, are indicated. B, growth of E. coli cells expressing a vector control (Ctrl), Tne2tox, or the indicated site-specific variants of Tne2tox. C, SDS-PAGE analysis of the Tne2tox–Tni2 complex and refolded Tne2tox and the indicated site-specific mutants thereof. Tne2tox and its variants possess an N-terminal hexahistidine tag, whereas Tni2 is untagged. Proteins were visualized using Coomassie Blue staining. D and E, hydrolysis rates of NAD+ (D) and NADP+ (E) by the purified proteins indicated in C. Error bars represent ± S.D. (n = 3). Asterisks indicate NAD(P)+ hydrolysis rates that are significantly higher than the Tni2-inhibited control.
To pinpoint functionally important amino acids that compose the NAD+-binding pocket of Tne2tox, we searched for conserved residues in this region of the toxin. Notably, the four most conserved residues within Tne2tox, Phe-330, Arg-343, Lys-353, and Gln-387, all localize to this region of the protein (Fig. 4A and Fig. S4C). Phe-330 and Gln-387 are predicted to interact with the ribose moiety and the hydroxyl group in the 2-position of nicotinamide riboside, respectively, whereas Lys-353 is predicted to interact with the adenosine-linked phosphate group of the dinucleotide substrate. In contrast, Arg-343 is not predicted to interact with NAD+; rather, its guanidinium moiety forms a hydrogen bond with the carbonyl group of the Lys-353 peptide bond. In this location, Arg-343 positions the loop region between α2 and β3 of Tne2tox, which is required for the formation of the NAD+-binding cavity. Mutation of this residue would be predicted to cause a substantial widening of this cavity resulting in the movement of a significant number of predicted NAD+-binding residues away from the substrate-binding site. To test the functional importance of these four residues, we expressed variants of Tne2tox bearing alanine substitutions in each of these positions in E. coli. This analysis showed that mutation of Lys-353 or Gln-387 results in a modest decrease in growth inhibition, whereas the alanine substation of Phe-330 or Arg-343 almost completely abrogated Tne2tox toxicity. We additionally generated an R343A/K353A double mutant, and the growth of this variant was indistinguishable from E. coli harboring a vector control (Fig. 4B).
To rule out the possibility that the differences we observed in E. coli growth are due to variable toxin expression levels, we next examined NAD+ hydrolysis rates using purified toxin. To accomplish this, we co-expressed wildtype Tne2tox, Tne2toxR343A, and Tne2toxR343A/K353A as binary complexes with Tni2 to prevent toxicity during overexpression in E. coli. Purified toxin-immunity complexes were then subjected to denaturing conditions to separate immunity, and the toxins were refolded using renaturing buffer (Fig. 4C). We then measured the rates of NAD+ turnover using an end-point assay that detects a base-catalyzed fluorescent breakdown product of NAD+ and found that wildtype Tne2tox catalyzes an NAD+ hydrolysis rate of ∼40,000 min−1 (Fig. 4D). Mirroring our E. coli toxicity data, Tne2toxR343A exhibited significantly reduced NADase activity (∼400 min−1), whereas the activity of Tne2toxR343A/K353A was further reduced to the level of a Tni2-inhibited control (∼10 min−1). A similar trend was observed when we examined NADP+ hydrolysis rates; however, Tne2tox is ∼2-fold less efficient at hydrolyzing the phosphorylated form of the dinucleotide (Fig. 4E). Analysis of the reaction products by LC-MS indicates that the enzyme severs the nicotinamide moiety from ADP-ribose (Fig. S5). In total, our biochemical data support our molecular docking studies and indicate that NAD(P)+ hydrolysis by Tne2tox requires a constellation of conserved charged and polar residues that line the substrate-binding pocket of the toxin.
Tne2 is the founding member of a widespread NADase effector family
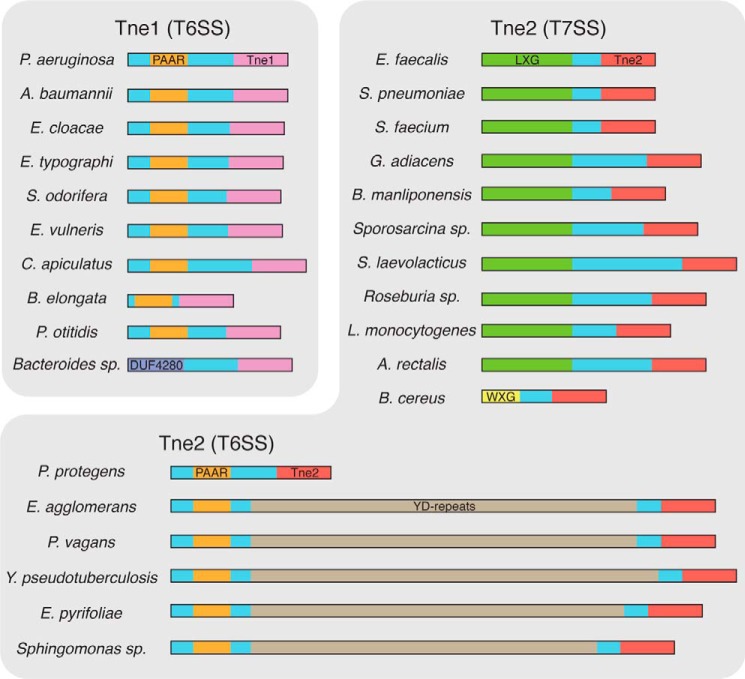
Our finding that Tne2 is a NAD(P)+-hydrolyzing effector involved in interbacterial antagonism prompted us to examine the prevalence of T6SS effectors harboring this biochemical activity. Because Tse6 was the first identified interbacterial NADase, we initiated our search by examining the phylogenetic distribution of its toxin domain (Tse6tox). Using the iterative homology search algorithm jackHAMMR to query the UniProtKB database, 127 Tse6tox orthologous proteins were identified. Because NAD+ hydrolase activity has been associated with host cell targeting toxins that do not utilize the T6SS for export, we refined our list by excluding proteins that do not possess N-terminal trafficking domains associated with the T6SS or are not encoded adjacent to T6SS structural genes. Within our list of candidate NADase effectors, 37 possess PAAR domains, three harbor a domain belonging to the DUF4280 family of proteins, and an additional 64 are encoded in proximity to T6SS structural genes (Table S3 and Fig. S6). PAAR domains are typically associated with proteobacterial T6SSs (T6SSi), whereas the DUF4280 domains, which are predicted to adopt a similar cone-shaped three-dimensional structure, are linked to the T6SSiii found in the phylum Bacteroidetes (17, 36). In keeping with our nomenclature for Tne2, we propose to name members of this family of NADase effectors Tne1.
We next examined the abundance and distribution of Tne2tox orthologous proteins and identified 937 sequences. Of these, only 175 possess trafficking domains or are adjacent to structural genes implicating their transit through the T6SS (Table S4 and Fig. S7). However, in contrast to the Tne1 family, which is restricted to Gram-negative bacteria, we found that ∼200 Tne2 orthologous proteins are found in the Gram-positive phylum Firmicutes. Within this subset of proteins, we noted that 43 contain predicted LXG or WXG-100 domains at their N termini. LXG and WXG-100 proteins are substrates of the T7SS, and we and others have recently implicated this pathway in interbacterial antagonism among Gram-positive Firmicutes (25, 26). Thus, it appears that NAD(P)+-hydrolyzing enzymes mediate bacterial competition via multiple specialized protein secretion systems found in diverse bacterial phyla (Fig. 5).
Figure 5.
Overview of the Tne families. Shown are predicted domain structures of representative genera identified in the Tne1 and Tne2 families of NADase enzymes. Depicted domains include Tne1 toxins (pink), Tne2 toxins (red), PAAR domains (orange), DUF4280 (blue), YD-repeats (tan), LXG domains (green) and WXG-100 domains (WXG, yellow). Tne2 family members are subdivided into those predicted to transit the T6SS (bottom) or the T7SS (right). Accession numbers for all Tne1 and Tne2 family members are available in supplemental Tables S3 and S4, respectively.
Discussion
In this study, we sought to identify T6SS effectors required for interbacterial antagonism by the plant commensal bacterium P. protegens. This observation led us to identify and characterize RhsA and Tne2 as new substrates of the T6SS in this organism. Although RhsA belongs to a superfamily of endonucleases, the activity of Tne2 was not apparent from its sequence. Our structural and biochemical characterization of this protein revealed that it possesses potent NAD(P)+ hydrolase activity and that it is the founding member of a widespread NADase effector family.
Previously, three T6SS effectors had been identified in P. protegens; however, we have found that these effectors do not significantly contribute to co-culture fitness on rich media. This observation, taken together with our prior work demonstrating that Tge2 confers a co-culture fitness advantage on low-salt media, suggests that different T6SS effectors have variable efficacy depending on environmental conditions (23). Tge2 and Tae3 degrade peptidoglycan, and thus their effectiveness under low salinity is consistent with the requirement of the cell wall to withstand turgor pressure (24). The mode of action of Tse4 has not been determined, but it is conceivable that the efficacy of this effector is also influenced by the extracellular milieu. In contrast, RhsA and Tne2 degrade cytoplasmic molecules essential to all cellular life, and therefore, their ability to inhibit the growth of bacterial competitors may not be as dependent on the conditions of the external environment. It is worth noting that Tne2 only contributes to co-culture fitness in a strain lacking rhsA. This phenotype could be due to effector synergy as the predicted consequence of RhsA intoxication would be DNA damage, the repair of which would require NAD+-dependent DNA ligases. Alternatively, deletion of both tne2 and rhsA may prevent the export of additional yet unidentified effectors. Future work will be aimed at distinguishing between these two possibilities.
The lack of sequence identity between Tne2tox and characterized NADase toxins demonstrates that proteins possessing this enzymatic activity can vary significantly in sequence while retaining the ability to hydrolyze NAD+. This sequence variation is likely a result of a molecular “arms race” resulting from effector co-evolution with cognate immunity arising from selective pressure exerted by T6SS-dependent interbacterial antagonism. We found that Tsi6 (Tni1) and Tni2 differ in both sequence and structure, bind to non-overlapping sites on their associated toxin, and do not cross-neutralize non-cognate NADase effectors. Given these remarkable differences, we speculate that each Tne-Tni pair evolved independently from one another, and bacteria possessing a NADase effector-immunity pair from one family would not be resistant to intoxication by species harboring an effector-immunity pair from the other family. In this way, bacterial species harboring compatible immunity could co-exist in a community while at the same time exclude bacteria with incompatible immunity.
The Tne families identified herein greatly expand the diversity of known interbacterial NADase enzymes. Previously, Tse6 (Tne1) was the sole example of a T6SS NADase effector (28). We show here that the toxin domain of this effector belongs to a family of T6SS NADases found in the Gram-negative phyla Proteobacteria and Bacteroidetes. Several Tne2 family members are also predicted substrates of the T6SS; however, we also identified a significant number of homologs in this family that are predicted substrates of the T7SS found in Gram-positive organisms. These genes encode N-terminal domains belonging to the WXG-100 and LXG families of proteins. WXG-100 domains are established signal sequences for the T7SS and LXG domains, given their predicted structural similarity to WXG-100 domains, and likely also fulfill this role (37, 38). Our recent characterization of T7SS-exported LXG substrates from Streptococcus intermedius B196 identified the TelB protein as an interbacterial NADase secreted by this organism (25). TelB bears no homology to the Tne1 or Tne2 families of NADases; rather, it belongs to a previously defined family of host cell-targeting NADases. The founding member of this family (formerly termed DUF4237) is tuberculosis necrotizing toxin (TNT) from Mycobacterium tuberculosis (39). Unlike the Tne families, TNT family members are broadly distributed in fungi in addition to many species of pathogenic bacteria. TNT does not require the T7SS for export and facilitates its own entry into host cells. We speculate that host cell-targeting NADases evolved from ancestral interbacterial predecessors. Furthermore, their ability to intoxicate eukaryotes arose through the acquisition of trafficking domains that alter their targeting specificity to different cell types. In this way, the rapid evolution of antibacterial effectors arising from interbacterial antagonism may play a significant role in the ongoing evolution of virulence.
Experimental procedures
Bacterial strains, plasmids, and growth conditions
All P. protegens strains generated were derived from the sequenced strain Pf-5 (22). The P. putida strain used in this study was derived from the sequenced strain KT2440 (40). P. protegens mutants were generated by allelic exchange as described previously (23). E. coli strains XL1-Blue, BL21(DE3) CodonPlus, and SM10 were used for plasmid maintenance, gene expression, and conjugative transfer, respectively. A detailed list of strains and plasmids used in this study can be found in Tables S1 and S2. P. protegens and P. putida strains were grown at 30 °C in LB medium supplemented with 30 μg ml−1 gentamicin when needed. E. coli strains were grown in LB at 37 °C supplemented with 50 μg ml−1 kanamycin, 150 μg ml−1 carbenicillin, 200 μg ml−1 trimethoprim, 0.1% w/v l-rhamnose, and 0.5 mm IPTG as required.
Bacterial competition assays
Interspecies competition assays between P. protegens and P. putida were performed as described previously with a few minor modifications. Briefly, overnight cultures of P. protegens donor strains and the P. putida recipient strain were mixed in a 10:1 ratio (determined by A600), and 5 μl of the mixture was spotted onto a 0.2-μm nitrocellulose membrane overlaid on a 3% LB agar plate. Competitive indices were calculated by enumerating donor/recipient colony-forming units after 8 h of growth at 30 °C. All competitive indices were adjusted by the donor/recipient ratio in the initial inoculum. The P. putida recipient strain expresses a gfp construct from the attTn7 site to enable its differentiation from unlabeled P. protegens donor via selection on gentamicin or GFP fluorescence.
Growth curves and toxicity assays
For growth-curve experiments, stationary phase E. coli BL21 (DE3) CodonPlus cells harboring plasmids expressing the indicated genes were back-diluted 200-fold into LB broth containing the appropriate antibiotics. Cultures were grown at 37 °C with shaking in 96-well plates, and A600 readings were taken every 30 min for 3 h using a Synergy 4 Microplate Reader (BioTek Instruments). Expression of toxin and immunity genes was induced by the addition of 0.1% (w/v) l-rhamnose and 200 μm IPTG, respectively, followed by an additional 5-h incubation under the same conditions.
For toxicity assays, E. coli BL21 (DE3) CodonPlus cells harboring the indicated plasmids were either struck out on LB agar containing the appropriate antibiotics, 0.1% (w/v) l-rhamnose, and 150 μm IPTG (for Tsi6/Tni2 rescue experiment) or diluted 1:106, and each 10-fold dilution was spotted onto 3% LB agar plates containing 200 μg ml−1 trimethoprim and 0.1% (w/v) l-rhamnose (for Tne2 point mutants).
Protein expression and purification
Protein expression constructs were transformed into E. coli BL21 (DE3) CodonPlus cells, and growth in LB broth was supplemented with the appropriate antibiotics. Cells were grown at 37 °C with shaking to an A600 prior to induction with 1 mm IPTG. Post-induction, cells were grown for an additional 5 h at 30 °C before being pelleted by centrifugation and flash-frozen. For the Tne2tox–Tni2 complex used in crystallization, cells expressing this complex were resuspended in Buffer A (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10 mm imidazole) prior to lysis by sonication. Cell lysates were then cleared by centrifugation, loaded onto a gravity flow column containing Ni2+-nitrilotriacetic acid (Ni-NTA)-conjugated agarose, and washed with 50 ml of Buffer A. Bound complex was then eluted with Buffer A supplemented with 400 mm imidazole. The Tne2tox–Tni2 complex was then further purified by size-exclusion chromatography using a 16/600 HiLoadS200 run in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl. Proteins used for binding studies and enzyme assay were purified using the same chromatographic procedures except that the buffer used was Buffer B (10 mm sodium/potassium phosphate, pH 7.4, 137 mm NaCl, 2.7 mm KCl) with added imidazole when needed. For experiments requiring purified Tne2tox or point mutants thereof, a denaturation and refolding protocol was employed. Briefly, the Ni-NTA-agarose-bound Tne2tox–Tni2 complex was denatured with 50 ml of Buffer B containing 8 m urea to remove untagged Tni2. Tne2tox was then refolded in 50 ml of Buffer B prior to elution from the column. Refolded proteins were further purified by size-exclusion chromatography run in Buffer B.
Crystallization and structure determination
Purified selenomethionine-incorporated Tne2tox–Tni2 complex was concentrated to 20 mg ml−1 by spin filtration and screened for crystallization conditions using the hanging drop vapor diffusion technique and commercially available crystallization suites (MCSG1–4, Anatrace). After 1 week, polycrystalline clusters were obtained in 30% (v/v) PEG 400, 100 mm MgCl2, 100 mm MES, pH 6.5. The clusters were used to generate a seed stock that was used for subsequent streak seeding into an optimization screen that systematically varied PEG 400 concentration and MES pH. This approach yielded singular diffraction quality crystals in a condition containing 34% (v/v) PEG 400, 100 mm MgCl2, 100 mm MES-NaOH, pH 7.0. Crystals were flash-frozen without any added cryoprotectant, and diffraction data were collected on beamline 21-ID-F at the Advanced Photon Source. X-ray data were processed with HKL-2000, and experimental phases were obtained using the Phenix AutoSol, and an initial model was generated using Phenix AutoBuild (41, 42). Minor model adjustments were made manually using COOT between iterative rounds of refinement using Phenix. refine (43, 44). The final model of Tne2tox–Tni2 was refined to an Rwork/Rfree of 18.1 and 21.6%, respectively (Table 1).
Determination of cellular NAD+ levels
The determination of relative cellular NAD+ levels was performed as reported previously (28). Briefly, overnight cultures of E. coli strains harboring the appropriate expression plasmids and a vector control were back-diluted 200-fold into LB broth and grown at 37 °C with shaking to mid-log phase prior to induction of protein expression with 0.1% (w/v) l-rhamnose and 150 μm IPTG. One-hour post-induction, cultures were diluted to A600 = 0.5, and 500 μl of cells were collected by microcentrifugation. Cells were then lysed in 0.2 m NaOH, 1% (w/v) cetyltrimethylammonium bromide followed by treatment with 0.4 m HCl at 60 °C for 15 min. After neutralization with 0.5 m Tris base, samples were then mixed with an equal volume of NAD/NADH-Glo detection reagent (Promega) prepared immediately before use as per the instructions of the manufacturer. Luciferin bioluminescence was measured continuously for 2 h using a Synergy H1 plate reader. The slope of the luciferin signal from the linear range of the assay was used to determine relative NAD+ concentration compared with a vector control strain.
Isothermal titration calorimetry
Purified Tni2, Tsi6, and Tne2tox were dialyzed into PBS buffer, and each solution was degassed prior to experimentation. ITC measurements were performed using a Nano ITC (TA Instruments). Titrations were carried out with 130 μm Tni2 or Tsi6 in the syringe and 13 μm Tne2tox in the cell. Each titration experiment consisted of 20 2.5-μl injections with 300-s intervals between each injection. The ITC data were analyzed using NanoAnalyze 2.1 (TA Instruments) and fit using a single-site binding model.
Substrate docking
Molecular docking of NAD+ onto the Tne2tox structure was performed using AutoDock Vina with the AutoDock Tools GUI (45, 46). A search volume that included the entire surface of Tne2tox was queried, and potential NAD+-binding sites were ranked based on predicted free energy of binding. The most thermodynamically favorable ligand-bound structure was used as a guide for subsequent mutational analyses of Tne2tox.
NAD(P)+ hydrolysis assay
Hydrolysis rates of NAD(P)+ and NAD(P)+ by Tne2tox were determined using a base-catalyzed fluorescence end-point assay (47). 100-μl reactions in PBS buffer contained either 0.2 mm NAD+ or 0.15 mm NADP+ and the following concentrations of each enzyme: Tne2tox (25 pm); Tne2tox–Tni2 (100 nm); Tne2toxR343A (2 nm); and Tne2toxR343A/K353A (100 nm). Each enzyme concentration was chosen to ensure <10% of the total substrate was consumed. Reactions were incubated at 37 °C for 30 min and subsequently terminated by the addition of 50 μl of 6 m NaOH. Following a 30-min incubation at room temperature in the dark, the base-catalyzed fluorescent breakdown product of the remaining NAD+ and NADP+ was measured using a Synergy 4 Microplate Reader (BioTek Instruments) at excitation and emission wavelengths of 360 and 420 nm, respectively. The amount of NAD+ and NADP+ hydrolyzed in each reaction was determined by interpolation from a standard curve.
Mass spectrometry
1 mm NAD+ was incubated for 30 min at room temperature in the presence or absence of 100 nm Tne2tox. Samples were then passed through a spin filtration unit (10-kDa cutoff, Millipore), and the reaction products were analyzed in an LCMS system including an Agilent 1200 series HPLC system coupled with a Bruker MicroTOF II mass spectrometer. For each run, 10 μl of sample was injected onto an Eclipse XDB-C18 (2.1 × 100 mm, 3.5 μm) reverse phase C18 column at a flow rate of 0.15 ml min−1, using a mobile phase consisting of 5 mm ammonium formate (A) and methanol (B). Reaction products were eluted using a linear gradient that began at 95% (A) and ended at 10 min with 70% (B). The mobile phase was then held at 70% (B) for 5 min. Finally, the mobile phase was returned to 95% (A) at 15.1 min, and the column was reequilibrated with 95% (A) for 4 min. The flow rate was increased to 0.40 ml min−1 during this washing and reequilibration phase and then returned to the initial flow rate of 0.15 ml at 19.1 min. NAD+ and ADP-ribose were ionized by electrospray ion source, using N2 for nebulization (pressure of 2.0 Bar) and drying (flow of 6 liters min−1, temperature of 200 °C). Capillary voltage was 4500 V, end plate offset −500 V, hexapole radio frequency 450.0 Vpp, energy transfer time of 49.0 μs, and pre-pulse storage of 10.0 μs. Mass spectrometry data were collected in positive electrospray ionization mode. Data were acquired in centroid mode spanning an m/z range of 100–800. The resulting mass spectra were analyzed using the Hystar 3.2 software package.
Informatic identification of NADase families
Protein sequences for the toxin domains of Tse6 (Tne1, residues 282–430) and Tne2 (residues 260–408) were subjected to an iterative homology search using the HMMER algorithm (jackHMMER) (48). In both cases, the homology searches converged, resulting in 127 and 937 sequences for the Tne1 and Tne2 groups, respectively. Sequences were then filtered for those containing trafficking domains associated with T6SSs (PAAR) or T7SSs (LXG and WXG100).
Author contributions
J. Y. T., N. P. B., S. A., and J. C. W. data curation; J. Y. T., N. P. B., S. A., and J. C. W. formal analysis; J. Y. T., N. P. B., S. A., and J. C. W. investigation; J. C. W. conceptualization; J. C. W. supervision; J. C. W. funding acquisition; J. C. W. writing-original draft; J. C. W. project administration; J. C. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Matthew Walker for assistance with ITC experiments; Nicola Henriquez for mass spectrometry assistance; Peter Stogios for initial screening of protein crystals; Zdzislaw Wawrzak for assistance with X-ray data collection and structure determination; and Brian Coombes for helpful comments on the manuscript. Equipment was provided by the Michael DeGroote Institute for Infectious Disease Research.
This work was supported by start-up funds from McMaster University and Discovery Grant RGPIN-2017-05350 from the Natural Sciences and Engineering Research Council of Canada (to J. C. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S7, Tables S1–S4, and supporting Refs. 1–9.
The atomic coordinates and structure factors (code 6B12) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T6SS
- type VI secretion system
- T7SS
- type VII secretion system
- ITC
- isothermal titration calorimetry
- Ni-NTA
- Ni2+-nitrilotriacetic acid
- r.m.s.d.
- root-mean-square deviation
- TNT
- tuberculosis necrotizing toxin
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- PDB
- Protein Data Bank.
References
- 1. Russell A. B., Peterson S. B., and Mougous J. D. (2014) Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basler M., Pilhofer M., Henderson G. P., Jensen G. J., and Mekalanos J. J. (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ho B. T., Fu Y., Dong T. G., and Mekalanos J. J. (2017) Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 114, 9427–9432 10.1073/pnas.1711219114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vettiger A., and Basler M. (2016) Type VI secretion system substrates are transferred and reused among sister cells. Cell 167, 99–110 10.1016/j.cell.2016.08.023 [DOI] [PubMed] [Google Scholar]
- 5. Hecht A. L., Casterline B. W., Earley Z. M., Goo Y. A., Goodlett D. R., and Bubeck Wardenburg J. (2016) Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 17, 1281–1291 10.15252/embr.201642282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson M. C., Vonaesch P., Saffarian A., Marteyn B. S., and Sansonetti P. J. (2017) Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21, 769–776 10.1016/j.chom.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 7. Sana T. G., Flaugnatti N., Lugo K. A., Lam L. H., Jacobson A., Baylot V., Durand E., Journet L., Cascales E., and Monack D. M. (2016) Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A. 113, E5044–E5051 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y., Waldor M. K., and Mekalanos J. J. (2013) Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663 10.1016/j.chom.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wexler A. G., Bao Y., Whitney J. C., Bobay L. M., Xavier J. B., Schofield W. B., Barry N. A., Russell A. B., Tran B. Q., Goo Y. A., Goodlett D. R., Ochman H., Mougous J. D., and Goodman A. L. (2016) Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 113, 3639–3644 10.1073/pnas.1525637113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durand E., Cambillau C., Cascales E., and Journet L. (2014) VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507 10.1016/j.tim.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 11. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., and Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitney J. C., Beck C. M., Goo Y. A., Russell A. B., Harding B. N., De Leon J. A., Cunningham D. A., Tran B. Q., Low D. A., Goodlett D. R., Hayes C. S., and Mougous J. D. (2014) Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542 10.1111/mmi.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hachani A., Allsopp L. P., Oduko Y., and Filloux A. (2014) The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289, 17872–17884 10.1074/jbc.M114.563429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Silverman J. M., Agnello D. M., Zheng H., Andrews B. T., Li M., Catalano C. E., Gonen T., and Mougous J. D. (2013) Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 51, 584–593 10.1016/j.molcel.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bondage D. D., Lin J. S., Ma L. S., Kuo C. H., and Lai E. M. (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940 10.1073/pnas.1600428113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cianfanelli F. R., Alcoforado Diniz J., Guo M., De Cesare V., Trost M., and Coulthurst S. J. (2016) VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12, e1005735 10.1371/journal.ppat.1005735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shneider M. M., Buth S. A., Ho B. T., Basler M., Mekalanos J. J., and Leiman P. G. (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weller D. M. (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256 10.1094/PHYTO-97-2-0250 [DOI] [PubMed] [Google Scholar]
- 19. Haas D., and Défago G. (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- 20. Decoin V., Barbey C., Bergeau D., Latour X., Feuilloley M. G., Orange N., and Merieau A. (2014) A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS One 9, e89411 10.1371/journal.pone.0089411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernal P., Allsopp L. P., Filloux A., and Llamas M. A. (2017) The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987 10.1038/ismej.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paulsen I. T., Press C. M., Ravel J., Kobayashi D. Y., Myers G. S., Mavrodi D. V., DeBoy R. T., Seshadri R., Ren Q., Madupu R., Dodson R. J., Durkin A. S., Brinkac L. M., Daugherty S. C., Sullivan S. A., et al. (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23, 873–878 10.1038/nbt1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitney J. C., Chou S., Russell A. B., Biboy J., Gardiner T. E., Ferrin M. A., Brittnacher M., Vollmer W., and Mougous J. D. (2013) Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J. Biol. Chem. 288, 26616–26624 10.1074/jbc.M113.488320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Russell A. B., Singh P., Brittnacher M., Bui N. K., Hood R. D., Carl M. A., Agnello D. M., Schwarz S., Goodlett D. R., Vollmer W., and Mougous J. D. (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549 10.1016/j.chom.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitney J. C., Peterson S. B., Kim J., Pazos M., Verster A. J., Radey M. C., Kulasekara H. D., Ching M. Q., Bullen N. P., Bryant D., Goo Y. A., Surette M. G., Borenstein E., Vollmer W., and Mougous J. D. (2017) A broadly distributed toxin family mediates contact-dependent antagonism between Gram-positive bacteria. eLife 6, e26938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao Z., Casabona M. G., Kneuper H., Chalmers J. D., and Palmer T. (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2, 16183 10.1038/nmicrobiol.2016.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma J., Sun M., Dong W., Pan Z., Lu C., and Yao H. (2017) PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 19, 345–360 10.1111/1462-2920.13621 [DOI] [PubMed] [Google Scholar]
- 28. Whitney J. C., Quentin D., Sawai S., LeRoux M., Harding B. N., Ledvina H. E., Tran B. Q., Robinson H., Goo Y. A., Goodlett D. R., Raunser S., and Mougous J. D. (2015) An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619 10.1016/j.cell.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koskiniemi S., Lamoureux J. G., Nikolakakis K. C., t'Kint de Roodenbeke C., Kaplan M. D., Low D. A., and Hayes C. S. (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 110, 7032–7037 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rice L. M., Earnest T. N., and Brunger A. T. (2000) Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D Biol. Crystallogr. 56, 1413–1420 10.1107/S0907444900010039 [DOI] [PubMed] [Google Scholar]
- 31. Fieldhouse R. J., and Merrill A. R. (2008) Needle in the haystack: structure-based toxin discovery. Trends Biochem. Sci. 33, 546–556 10.1016/j.tibs.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 32. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., and Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 34. Bosch J., Tamura T., Tamura N., Baumeister W., and Essen L. O. (2007) The β-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology. Acta Crystallogr. D Biol. Crystallogr. 63, 179–187 10.1107/S0907444906045471 [DOI] [PubMed] [Google Scholar]
- 35. McLuskey K., Paterson N. G., Bland N. D., Isaacs N. W., and Mottram J. C. (2010) Crystal structure of Leishmania major oligopeptidase B gives insight into the enzymatic properties of a trypanosomatid virulence factor. J. Biol. Chem. 285, 39249–39259 10.1074/jbc.M110.156679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell A. B., Wexler A. G., Harding B. N., Whitney J. C., Bohn A. J., Goo Y. A., Tran B. Q., Barry N. A., Zheng H., Peterson S. B., Chou S., Gonen T., Goodlett D. R., Goodman A. L., and Mougous J. D. (2014) A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 10.1016/j.chom.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ates L. S., Houben E. N., and Bitter W. (2016) Type VII secretion: a highly versatile secretion system. Microbiol. Spectr. 4 10.1128/microbiolspec.VMBF-0011-2015 [DOI] [PubMed] [Google Scholar]
- 38. Zhang D., de Souza R. F., Anantharaman V., Iyer L. M., and Aravind L. (2012) Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J., Siroy A., Lokareddy R. K., Speer A., Doornbos K. S., Cingolani G., and Niederweis M. (2015) The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol. 22, 672–678 10.1038/nsmb.3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nelson K. E., Weinel C., Paulsen I. T., Dodson R. J., Hilbert H., Martins dos Santos V. A., Fouts D. E., Gill S. R., Pop M., Holmes M., Brinkac L., Beanan M., DeBoy R. T., Daugherty S., Kolonay J., et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808 10.1046/j.1462-2920.2002.00366.x [DOI] [PubMed] [Google Scholar]
- 41. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 10.1107/S0907444909012098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson S. L., and Morrison D. L. (1970) The alkaline reaction of nicotinamide adenine dinucleotide, a new transient intermediate. J. Biol. Chem. 245, 4519–4524 [PubMed] [Google Scholar]
- 48. Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.