Abstract
The type VI secretion system (T6SS) delivers toxic effectors between Gram-negative bacteria. Most antibacterial T6SS effectors are peptidoglycanases, nucleases, or lipases. In the current work, Tang et al. structurally and functionally characterize a novel family of NAD(P)+-hydrolyzing effectors (NADases), thus expanding the documented types of T6SS substrates. Bioinformatic identification of NADase family members putatively secreted by the bacteriolytic type VII secretion system (T7SS) of Gram-positive bacteria further points to NADases as a diverse and important class of effectors.
Keywords: protein secretion, toxicity, bacteria, Pseudomonas, NAD
Introduction
Bacterial species most often exist as part of a highly organized, dynamic community (1). How bacterial species can coexist with, or dominate, other organisms is a valuable area of research, as it not only provides fundamental knowledge of how diverse bacterial communities are established and maintained but also has clinical relevance for the treatment of pathogens, which need to compete with the host microbiota or other potential host pathogens to establish a niche. Bacteria employ a variety of secretory mechanisms to mediate interbacterial interactions. Contact-dependent antagonistic interactions among Gram-negative bacteria are mediated by the T6SS2 (2), whereas the T7SS is thought to be involved in those occurring among Gram-positive Firmicutes (3, 4). In either case, the breadth of molecules mediating interbacterial killing remains only partially characterized.
The T6SS structurally resembles an inverted bacteriophage tail anchored to the bacterial cell wall via a membrane-associated complex. The “tail” is composed of a sheath complex that encompasses a spiked tube. When the sheath contracts, it propels the spiked tube across both bacterial membranes and into adjacent cells (5). Secretion of effectors by this system requires association with one of the structural components of the spiked tube, hemolysin-coregulated protein (Hcp), valine–glycine repeat protein G (VgrG), or proline–alanine–alanine–arginine (PAAR) repeat proteins. Associations can be either physical, via protein–protein interactions, or genetic, as C-terminal extensions of structural T6SS components (5). Thus, proteins that possess a PAAR domain are putative T6SS effectors. Bacteria harboring T6SS effectors are protected from self-intoxication by immunity proteins, which specifically inactivate their cognate toxin. Proteomic, genetic, and bioinformatic studies have revealed that most antibacterial T6SS effectors are peptidoglycanases, lipases, or nucleases (6); however, a previous report identified the T6SS effector Tse6 of Pseudomonas aeruginosa as an NADase (7). Until now, it had been largely unknown whether NADase effectors are unique to P. aeruginosa or whether they represent a novel class of T6SS effectors.
In the current work, Tang et al. (8) searched for proteins containing PAAR domains in the genome of the plant commensal Pseudomonas protegens and identified two hypothetical T6SS effectors, RhsA (PFL_6096) and Tne2 (PFL_6209). RhsA shared homology with proteins of the rearrangement hotspot family of T6SS effectors, which commonly function as nucleases. In contrast, Tne2 lacked homology to any protein of known function. Bacterial competition assays showed that both RhsA and Tne2 provide P. protegens a T6SS-dependent fitness advantage over P. putida, leading the authors to investigate the specific function of Tne2. To this end, the 1.7-Å X-ray crystal structure of the novel T6SS effector in complex with its immunity protein Tni2 (PFL_6210) was determined. The structure of Tne2 showed substantial similarity to that of the previously identified T6SS NADase Tse6 (7), suggesting that Tne2 could function as an NADase. Indeed, the authors showed that Tne2 degrades NAD(P)+ in vitro by cleaving the nicotinamide group. Furthermore, Tne2 reduces cellular NAD+ levels when expressed heterologously in Escherichia coli, and this activity is abrogated by mutation of residues modeled as having direct contact with the NAD+ substrate. Altogether, the data indicate that the T6SS effector Tne2 of P. protegens is a bona fide NADase.
Despite the structural similarities between Tse6 and Tne2 (7, 8), these two effectors also present considerable differences, specifically in their respective activating loops and NAD+-binding pockets. Moreover, the cognate immunity protein for each effector, Tsi6 and Tni2, respectively, present prominent differences in their overall folds. Accordingly, Tsi6 neither interacts with Tne2 nor prevents Tne2-based toxicity. In light of these results, the authors proposed that Tse6 and Tne2 represent two distinct families of T6SS NADases. Because Tse6 was identified first, it was designated as a member of the type VI secretion NADase effector family 1 (Tne1), whereas Tne2 is the founding member of the Tne2 family. Bioinformatic analysis revealed more than 100 orthologous proteins and putative T6SS effectors for each family. Remarkably, nearly 50 additional Tne2 orthologs contain predicted LXG or WXG-100 domains at their N terminus. Because LXG and WXG-100 proteins are substrates of the T7SS (4), these results suggest that NADases may represent a prevalent class of effectors secreted by both Gram-positive and Gram-negative bacteria to mediate interbacterial interactions (Fig. 1).
Figure 1.
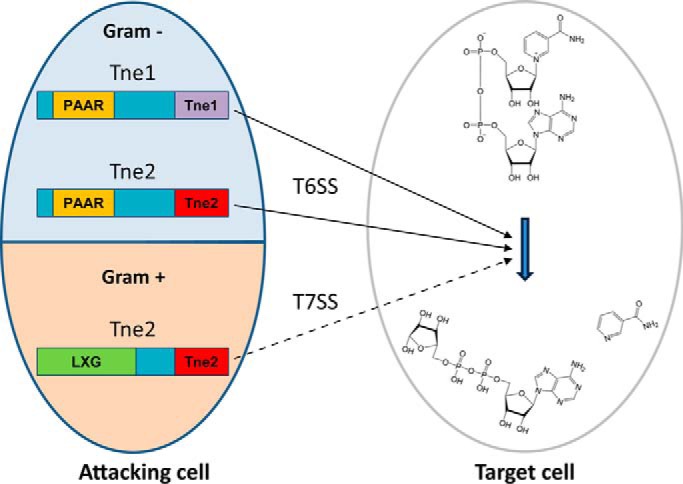
NADase effector families Tne1 and Tne2 mediate interbacterial antagonism. The T6SS of Gram-negative bacteria (solid black arrows) secretes toxic NADase effectors from both Tne families into adjacent bacteria (7, 8). Similarly, Gram-positive bacteria encode Tne2 orthologs possessing domains that suggest they are secreted by the T7SS (dashed black line); however, this remains to be experimentally shown (see text for details).
The current study thus characterizes a novel class of T6SS effectors that has, until now, been largely underappreciated. Structural comparisons between Tne2 and the only other NADase T6SS effector characterized to date, Tse6 (7), enabled the authors to define the two effectors as founding members of two novel families of NADases, Tne2 and Tne1, respectively. Furthermore, the finding that a subset of Tne2 orthologs possesses domains associated with T7SS effectors indicates that the T6SS and T7SS may translocate similar toxins to vastly different bacterial cell types. However, it must be noted that the putative T7SS NADases have yet to be validated.
The authors also found that the previously identified T6SS effectors of P. protegens, namely Tge2, Tae3, and Tse4, appear not to be involved in bacterial antagonism under the experimental condition tested. The finding of T6SS effectors with no observable antibacterial activity has also been documented for Acinetobacter baumannii (9, 10). It is possible that the role of these effectors is dependent on environmental conditions, as was determined for Tge2 (8), or that they have eukaryotic targets. Furthermore, these effectors may play other roles (e.g. nutrient acquisition) or may represent novel classes of uncharacterized effectors. Altogether, the findings of this study provide a wealth of information for future investigation and greatly improve our understanding of the plethora of molecules that mediate interbacterial interactions. Furthermore, the finding of this novel effector class broadens the possible targets for innovative antimicrobials. Molecules targeting bacterial dinucleotide metabolism could constitute the basis for novel antibacterial strategies.
Acknowledgment
Work in the Feldman lab is supported by National Institutes of Health Grant 1R01AI125363-01.
The authors declare that they have no conflicts of interests with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- T6SS
- type VI secretion system
- T7SS
- type VII secretion system.
References
- 1. Hibbing M. E., Fuqua C., Parsek M. R., and Peterson S. B. (2010) Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., and Mekalanos J. J. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cao Z., Casabona M. G., Kneuper H., Chalmers J. D., and Palmer T. (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2, 16183 10.1038/nmicrobiol.2016.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whitney J. C., Peterson S. B., Kim J., Pazos M., Verster A. J., Radey M. C., Kulasekara H. D., Ching M. Q., Bullen N. P., Bryant D., Goo Y. A., Surette M. G., Borenstein E., Vollmer W., and Mougous J. D. (2017) A broadly distributed toxin family mediates contact-dependent antagonism between Gram-positive bacteria. eLife 6, e26938 10.7554/eLife.26938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cianfanelli F. R., Monlezun L., and Coulthurst S. J. (2016) Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 6. Lien Y.-W., and Lai E.-M. (2017) Type VI secretion effectors: Methodologies and biology. Front Cell Infect Microbiol. 7, 254 10.3389/fcimb.2017.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitney J. C., Quentin D., Sawai S., LeRoux M., Harding B. N., Ledvina H. E., Tran B. Q., Robinson H., Goo Y. A., Goodlett D. R., Raunser S., and Mougous J. D. (2015) An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619 10.1016/j.cell.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang J. Y., Bullen N. P., Ahmad S., and Whitney J. C. (2017) Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J. Biol. Chem. 293, 1504–1514 10.1074/jbc.RA117.000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weber B. S., Hennon S. W., Wright M. S., Scott N. E., de Berardinis V., Foster L. J., Ayala J. A., Adams M. D., and Feldman M. F. (2016) Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. MBio 7, e01253–16 10.1128/mBio.01253-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lazzaro M., Feldman M. F., and García Véscovi E. (2017) A transcriptional regulatory mechanism finely tunes the firing of type VI secretion system in response to bacterial enemies. MBio 8, e00559–17 10.1128/mBio.00559-17 [DOI] [PMC free article] [PubMed] [Google Scholar]


