Abstract
Crotalicidin (Ctn), a cathelicidin-related peptide from the venom of a South American rattlesnake, possesses potent antimicrobial, antitumor, and antifungal properties. Previously, we have shown that its C-terminal fragment, Ctn(15–34), retains the antimicrobial and antitumor activities but is less toxic to healthy cells and has improved serum stability. Here, we investigated the mechanisms of action of Ctn and Ctn(15–34) against Gram-negative bacteria. Both peptides were bactericidal, killing ∼90% of Escherichia coli and Pseudomonas aeruginosa cells within 90–120 and 5–30 min, respectively. Studies of ζ potential at the bacterial cell membrane suggested that both peptides accumulate at and neutralize negative charges on the bacterial surface. Flow cytometry experiments confirmed that both peptides permeabilize the bacterial cell membrane but suggested slightly different mechanisms of action. Ctn(15–34) permeabilized the membrane immediately upon addition to the cells, whereas Ctn had a lag phase before inducing membrane damage and exhibited more complex cell-killing activity, probably because of two different modes of membrane permeabilization. Using surface plasmon resonance and leakage assays with model vesicles, we confirmed that Ctn(15–34) binds to and disrupts lipid membranes and also observed that Ctn(15–34) has a preference for vesicles that mimic bacterial or tumor cell membranes. Atomic force microscopy visualized the effect of these peptides on bacterial cells, and confocal microscopy confirmed their localization on the bacterial surface. Our studies shed light onto the antimicrobial mechanisms of Ctn and Ctn(15–34), suggesting Ctn(15–34) as a promising lead for development as an antibacterial/antitumor agent.
Keywords: antimicrobial peptide (AMP), atomic force microscopy (AFM), surface plasmon resonance (SPR), confocal microscopy, Gram-negative bacteria, bacterial membrane disruption, bactericidal mechanism, time-resolved flow cytometry
Introduction
New antimicrobial drugs are urgently needed to address the growing challenge of bacterial resistance to existing antibiotics. Misuse of classical antibiotics has increased the number of superbugs and created a critical situation whereby previously controlled pathogens could in the future cause major morbidity or mortality (1, 2). This alarming growth of multidrug-resistant pathogens has prompted an intensive search for anti-infective drugs with novel mechanisms of action (3, 4). In particular, antimicrobial peptides (AMPs)9 have emerged as promising alternatives due to their broad-spectrum activity (including superbugs), selectivity, and mechanisms of action that potentially hinder the development of resistance (5).
AMPs are ancient weapons of the host defense machinery, present in all life domains (6). Although they can act in several possible ways to accomplish microbial cell death (e.g. membrane disruption, apoptosis induction, and internal target inhibition) (7, 8), an initial common step in the process is their recruitment onto the bacterial cell surface (9, 10). Accordingly, most AMPs display fairly conserved structural and physicochemical properties, such as positive net charge, high content of hydrophobic amino acid residues, or amphipathic structure, all favoring interaction with and insertion into membranes (11, 12). Cathelicidins are a large family of AMPs whose unifying feature is the presence of a conserved cathelin (cathepsin L inhibitor) domain at the N terminus. In contrast, their C-terminal domains contain a mature and active AMP and display high inter- and intraspecies diversity (13). Cathelicidins have been shown to be active against a broad range of targets, including bacteria, enveloped viruses, and fungi (14). In addition to causing direct pathogen killing, cathelicidins can modulate the immune response by assisting with pathogen clearance (15). Cathelicidins have been isolated from a wide range of organisms, including mammals (16–18), birds (19), fish (20), frogs (21), and marine (22) and terrestrial snakes (23–25).
We recently identified a new family of cathelicidin-like peptides named vipericidins in the venom glands of various South American pit viper snakes and experimentally validated them as AMPs (26). Crotalicidin (Ctn), the most active vipericidin, is a 34-residue helical peptide found in Crotalus durissus terrificus and was selected for further study. Ctn displays powerful antimicrobial, as well as antitumor and antifungal, activity. However, it is moderately hemolytic and unstable in serum (27, 28). To overcome these limitations, a rational dissection of the Ctn sequence was undertaken, aimed at identifying active motifs with enhanced properties. The search produced as main lead Ctn(15–34), a structurally disordered 20-mer that spans the C terminus and replicates the antibacterial and antitumor activities of the parental peptide but is less toxic toward healthy cells and significantly more stable in human serum (27). Investigating the mechanism of action on bacteria of both Ctn and Ctn(15–34) is of considerable interest, particularly given the potential therapeutic applications of the latter peptide.
In this study, the mode of action of Ctn and Ctn(15–34) toward two Gram-negative bacterial species was investigated in detail. We determined bactericidal concentrations and quantitative kinetics of cell death and measured peptide accumulation on bacterial cell surfaces. Combining flow cytometry and colony count procedures, we established that bacterial death is accomplished by membrane disruption. We also monitored peptide uptake and membrane permeabilization in real time to ascertain whether both effects are achieved simultaneously or peptide pre-accumulation precedes loss of viability. To visualize peptide localization and structural damage on bacteria surface, we carried out confocal and atomic force microscopy experiments. Furthermore, to gain insights into the cell selectivity and the ability to disrupt lipid bilayers of the fragment Ctn(15–34), we performed experiments with model vesicles mimicking healthy human and bacterial cell membranes. Altogether, our results provide strong evidence that Ctn and Ctn(15–34) act by inducing disruption of the bacterial cell; they also exemplify how a set of optimized methodologies can be combined to evaluate the action of AMPs at the membrane level.
Results
Synthetic peptides
The amino acid sequences in Table 1 were prepared in C-terminal amide form by Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid-phase synthesis and purified to >95% purity (see Fig. S1 for HPLC and MS data). For the N-terminal rhodamine B (RhB)-labeled peptides, the two peaks observed in the chromatograms of purified compounds (Fig. S1a) are due to RhB atropisomerism. The overall hydrophobicity of unlabeled and RhB-labeled peptides can be compared by the percentage of acetonitrile (ACN) at which they elute in HPLC (Table 1). Values for unlabeled Ctn and Ctn(15–34) were 30.9 and 30.3% ACN, respectively, whereas RhB-labeled versions eluted at 34.7 and 34.2% ACN, respectively, underlining the increase in hydrophobicity brought about by RhB labeling.
Table 1.
Peptides used in this study
| Peptide | Amino acid sequence | Theoretical massa | Experimental massb | Purityc | ACN gradientd | Retention time | ACNe |
|---|---|---|---|---|---|---|---|
| Da | Da | % | min | % | |||
| Ctnf | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF-amide | 4151.39 | 4152.45 | 98 | 15–40 | 9.556 | 30.9 |
| RhB-Ctn | RhB-KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF-amide | 4576.94 | 4575.97 | 99 | 20–40 | 10.997 | 34.7 |
| Ctn(15–34) | KKRLKKIFKKPMVIGVTIPF-amide | 2371.11 | 2371.23 | 99 | 10–40 | 10.131 | 30.3 |
| RhB-Ctn(15–34) | RhB-KKRLKKIFKKPMVIGVTIPF-amide | 2796.66 | 2795.90 | 98 | 20–40 | 10.670 | 34.2 |
a Theoretical molecular mass was calculated using GPMAW version 8.10.
b Experimental molecular mass was determined from the MS spectra shown in Fig. S1b.
c Peptide purity was estimated by peak integration of the analytical HPLC chromatograms shown in Fig. S1a.
d ACN gradient over 15 min used to run the analytical HPLC. Range indicates the initial and final ACN concentrations.
e Percentage of ACN corresponding to the eluted peptide, calculated from the gradient used and the retention time.
f Protein Data Bank accession code of Ctn: 2MWT.
Antibacterial and bactericidal activity
We evaluated the antimicrobial activity of Ctn and Ctn(15–34) against two Gram-negative bacterial strains: Escherichia coli ATCC 25922 (E. coli) and Pseudomonas aeruginosa ATCC 27853 (P. aeruginosa). As shown in Table 2, Ctn has the lowest minimal inhibitory concentration (MIC) for E. coli as well as for P. aeruginosa (0.78 and 1.56 μm, respectively). Ctn(15–34) also has a low MIC for E. coli (3.13 μm), but the concentration required to prevent P. aeruginosa visible growth is higher (12.5 μm). To determine whether the peptides were bactericidal or bacteriostatic, the minimal bactericidal concentrations (MBCs) were also determined (Table 2). The data show both Ctn and Ctn(15–34) to be bactericidal, as MBCs exceed MICs by <2-fold (29). The MIC/MBC determinations reveal that more peptide is required for bacterial death when higher bacterial inocula are used. This difference is significant for E. coli, for which up to 3 times more peptide is needed. These results support previous observations suggesting that the therapeutic effect of AMPs depends on the peptide/cell ratio rather than directly on peptide concentration (30, 31). In tune with this suggestion, and to avoid the results being influenced by the use of different peptide/cell ratios, peptide concentrations for mechanism-of-action studies were adjusted to the bacterial inoculum used for each specific experiment.
Table 2.
MIC and MBC of Ctn and Ctn(15–34) against two standard Gram-negative bacterial strains
5 × 105-cfu/ml or 107-cfu/ml bacterial suspensions were incubated with different concentrations of Ctn or Ctn(15–34). MIC was determined as the lowest peptide concentration that prevented visible bacterial growth. MBC was defined as the minimal peptide concentration where ≥99.9% bacteria death of the initial inoculum was observed.
| Peptide |
E. coli ATCC 25922 |
P. aeruginosa ATCC 27853 |
||
|---|---|---|---|---|
| 5 × 105 cfu/ml | 107 cfu/ml | 5 × 105 cfu/ml | 107 cfu/ml | |
| MIC (μm) | ||||
| Ctn | 0.78 | 6.25 | 1.56 | 3.13 |
| Ctn(15–34) | 3.13 | 50 | 12.5 | 25 |
| MBC (μm) | ||||
| Ctn | 1.56 | 6.25 | 3.13 | 3.13 |
| Ctn(15–34) | 6.25 | 50 | 25–50 | 25 |
Time-dependent bacterial death
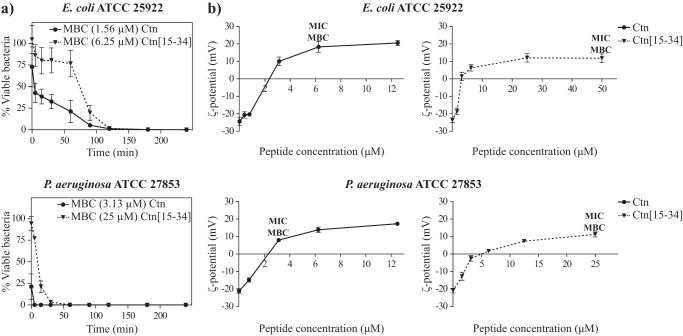
The killing kinetics of E. coli and P. aeruginosa by Ctn and Ctn(15–34) at their respective MBCs was evaluated using a time-kill assay. The results in Fig. 1a show that for both peptides, a bactericidal effect (99.9% reduction of bacterial viability) against E. coli is observed within 4 h. At 90 min, Ctn kills >90% of bacteria, whereas Ctn(15–34) needs 120 min for a similar effect. Ctn and Ctn(15–34) are faster at killing P. aeruginosa, requiring only 5 or 30 min for 90% cell death, respectively, with bactericidal effect observed at 15 and 90 min, respectively. This relatively fast effect against E. coli, even faster against P. aeruginosa, suggests that both peptides act by direct damage to the bacterial membrane rather than by inhibition of an internal target.
Figure 1.
Time-dependent bacterial death and bacterial surface neutralization induced by Ctn and Ctn(15–34). a, time-killing curves for E. coli and P. aeruginosa in the presence of Ctn and Ctn(15–34). 5 × 105 cfu/ml bacterial suspensions were incubated at 37 °C with Ctn (circles) or Ctn(15–34) (inverted triangles) at their MBC. At different time points, aliquots were collected, diluted, and spread onto trypticase soy agar plates. Bacterial colonies were counted after 24-h incubation at 37 °C, and viable bacteria are reported as a percentage of the control without peptide. Data correspond to mean ± S.D. (error bars) of three independent experiments. b, ζ potential measurements of E. coli and P. aeruginosa in the presence or absence of peptides. Ctn (circles) or Ctn(15–34) (inverted triangles) at different concentrations were added to 107 cfu/ml bacterial suspensions and allowed to equilibrate for 15 min at 25 °C, and 15 measurements were acquired. ζ potential (mV) was estimated using the Smoluchowski equation (55).
Membrane surface charge neutralization
To investigate whether surface charge can be neutralized by the accumulation of the peptides at the membrane of E. coli or P. aeruginosa, ζ potential measurements were carried out. The results show that, as expected (32, 33), in the absence of peptide, both bacteria possess a negative potential (Fig. 1b, ∼−24 mV for E. coli and ∼−21 mV for P. aeruginosa). In the presence of increasing peptide concentrations, a shift to positive ζ potential values is observed. In E. coli, complete neutralization of the surface charge occurs at 0.78–3.13 μm Ctn and 1.56–3.13 μm Ctn(15–34), whereas for P. aeruginosa, total surface charge neutralization is reached at 0.78–3.13 μm Ctn and 3.13–6.25 μm Ctn(15–34). For both bacteria, membrane neutralization occurs at concentrations below the MIC/MBC of either peptide.
Membrane permeabilization and viability correlation studies
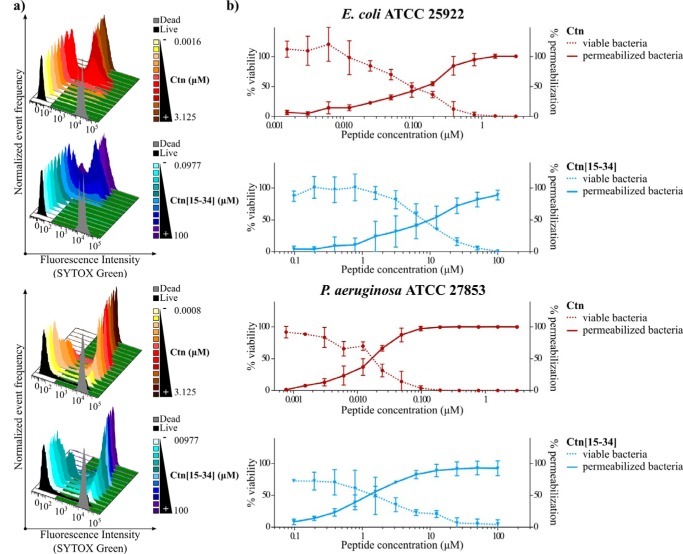
To clarify whether E. coli and P. aeruginosa death induced by Ctn and Ctn(15–34) is due to direct membrane disruption, correlation between membrane permeabilization and bacterial viability was studied. To this end, a flow cytometry assay (FCA) with SYTOX® Green to examine membrane permeabilization and a plate colony count to quantify viable cells were performed. As a cell-impermeable nucleic acid stain that only labels bacteria with compromised plasma membrane, SYTOX® Green is a useful tool in bacterial viability studies. As shown in Fig. 2, an increased percentage of bacterial cells with SYTOX® Green-permeable membrane is accompanied by a decrease in cell viability by the colony count method. FCA histograms on the SYTOX® Green channel (Fig. 2a) shift to higher fluorescence intensities indicative of E. coli and P. aeruginosa membrane disruption with increasing peptide concentration. Negative and positive controls (live and dead bacteria; black and gray histograms, respectively) were used to define gates and calculate permeabilization percentages, shown in Fig. 2b along with the viability percentages obtained in the colony count assay. A direct correlation between both sets of data (Fig. 2b) confirms that antibacterial activity of Ctn and Ctn(15–34) is mediated by membrane permeabilization.
Figure 2.
Bacterial cell viability and membrane permeabilization of E. coli and P. aeruginosa upon treatment with Ctn and Ctn(15–34). 5 × 105 cfu/ml bacterial suspensions were incubated at 37 °C with increasing peptide concentrations of Ctn (red) or Ctn(15–34) (blue). a, histograms on the SYTOX® Green channel detected by flow cytometry. Negative (live bacteria) and positive (dead bacteria) controls, used to define gates, are represented in black and gray, respectively. b, viable bacteria (dotted lines) and permeabilized bacteria (solid lines) are reported as a percentage of the control. Data correspond to mean ± S.D. (error bars) of three independent experiments.
To correlate the membrane permeabilization effect with peptide uptake (including both membrane-bound and intracellularly internalized), a parallel FCA experiment was performed with RhB-labeled versions of both peptides. From the SYTOX® Green and RhB channel readings (Fig. S2, a and b), the percentages of permeabilized and of peptide-internalizing cells, respectively, were determined (Fig. S2c). For both Ctn and Ctn(15–34), an increase in E. coli permeabilization coincides with increased peptide uptake, suggesting a direct correlation between the membrane damage observed and peptide entry/binding. The dose–response curve for RhB-Ctn (Fig. S2c, continuous red line) is similar to that for unlabeled Ctn (Fig. 2b, continuous red line), suggesting that RhB does not interfere with the peptide effect. In contrast, the profiles for RhB-labeled (Fig. S2c, continuous blue line) and unlabeled Ctn(15–34) (Fig. 2b, continuous blue line) are different. Whereas the latter induces 50% permeabilization in the 6.25–12.5 μm range, the percentage rises to 80% for the RhB-labeled analogue at similar concentrations. Moreover, at higher (25–100 μm) concentrations, a decrease in membrane permeabilization and uptake of RhB-Ctn(15–34) occurs, an effect not observed for unlabeled Ctn(15–34). This probably reflects a loss in integrity of the bacterial cell, no longer capable of retaining the labeled peptide within, and with its DNA (SYTOX® Green target) either damaged or leaking out. The observation of N-terminal RhB label interfering with the activity of Ctn(15–34) calls for caution in extrapolating results from labeled to unlabeled versions of this peptide.
Time-resolved peptide uptake and bacterial death
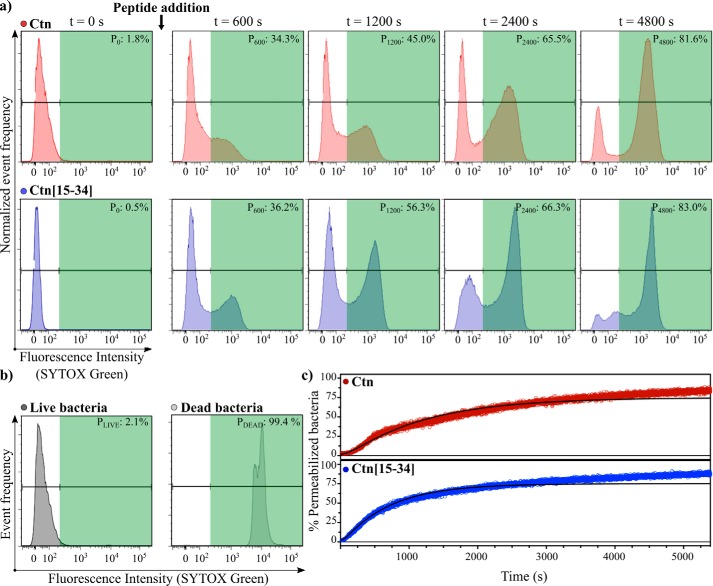
Changes in membrane permeabilization and peptide uptake were monitored by a time-resolved flow cytometry assay, which allows a more accurate study of the kinetics of peptide effect, compared with kinetic studies based on end-point sampling. We studied the membrane permeabilization of E. coli using SYTOX® Green dye right after Ctn or Ctn(15–34) addition, monitoring the changes from negative to positive gates for 90 min. Fig. 3 (a and b) shows time-course results and negative and positive controls (for the whole acquisition, see Movies S1a and S1b for Ctn and Ctn(15–34), respectively). As detailed under “Experimental procedures,” data from the FCA histograms were used to generate the kinetic curves on Fig. 3c. Data were fitted using the two-state kinetic model described previously for other AMPs (34), assuming that peptide-bacteria interaction consists of an initial binding step, followed by permeabilization of the membrane.
Figure 3.
Time-resolved bacterial membrane permeabilization. Ctn (red) or Ctn(15–34) (blue) at its MBC was added to 107 cfu/ml E. coli suspensions. Changes on the SYTOX® Green channel were monitorized during 90 min immediately after the peptide addition. a, flow cytometry histograms on the SYTOX® Green channel at different time points in the presence of Ctn and Ctn(15–34). Pn is the percentage of permeabilization at each time interval (n) (in s). b, flow cytometry histograms on the SYTOX® Green channel for the negative (live bacteria) and positive (dead bacteria) controls, used to define gates. c, percentages of permeabilized bacteria over time after the addition of the peptides. Data were normalized by controls, corrected using the calibration curve shown in Fig. S8a, and fitted with the two-state model (34) as described under “Experimental procedures.” The kinetic curve for the untreated control shown in Fig. S8b demonstrates that bacteria remain alive during the whole acquisition, and the changes observed in c are due to the peptide activity.
Although both peptides induce permeabilization of ∼90% of bacteria after 90 min (Fig. 3c), the process differs between peptides at the early stages; based on the tendency of the experimental data, Ctn(15–34) seems to start killing bacteria right after its addition, and the permeabilization takes place in a single process. In contrast, Ctn shows an initial lag phase. Fitting of the kinetic data to a two-state model results in statistically poor residuals for some time intervals (Fig. S3), suggesting that for both peptides, permeabilization is more complex than as assumed by the model. An additional slow event leading to permeabilization is suggested because the data deviate from the fit at longer times. In contrast, the fit is quite acceptable at shorter times. k0, the membrane attachment rate constant, is higher for Ctn(15–34) than for Ctn (2 × 10−2 s−1 versus 1 × 10−3 s−1) as well as the permeabilization rate constant (1 × 10−3 s−1 versus 9 × 10−4 s−1). Cooperativity is not observed in either case (f ∼ 0), contrary to other AMPs (34).
To determine whether permeabilization is concomitant with internalization, kinetic studies with RhB-labeled peptides were run. Correlograms (Fig. S4a) at different time points show the changes recorded on the SYTOX® Green and RhB channels (see Movies S2a and S2b for the entire acquisition of RhB-Ctn and RhB-Ctn(15–34), respectively). Kinetic curves (Fig. S4c) show both peptides becoming gradually bound to and/or internalized into the cells until an equilibrium is reached, but the uptake of RhB-Ctn(15–34) is faster than that of RhB-Ctn. Interestingly, RhB-Ctn(15–34) uptake precedes bacterial membrane damage, suggesting that peptide accumulation up to an effective threshold is necessary to trigger membrane permeabilization. In contrast, RhB-Ctn uptake shortly precedes membrane permeabilization, confirming that membrane becomes permeabilized as Ctn molecules are recruited to the surface.
Peptide distribution in bacteria
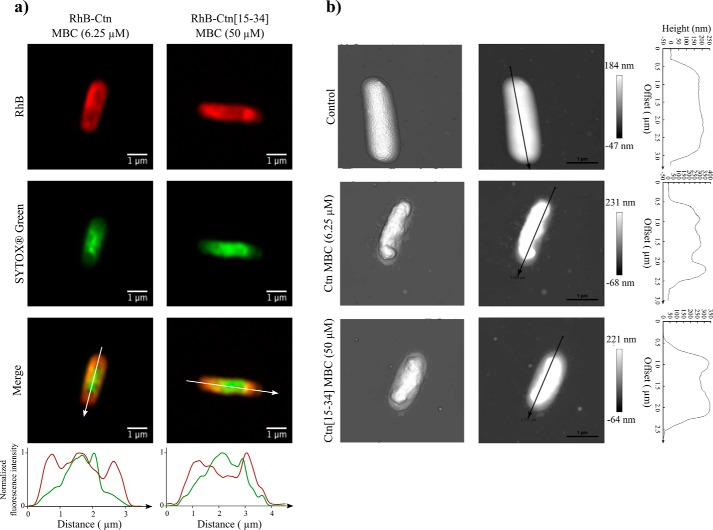
To investigate how peptides are localized within the bacteria, confocal microscopy was done on E. coli after incubation with RhB-labeled Ctn and Ctn(15–34) for 90 min. Images were also acquired using the SYTOX® Green dye, to correlate peptide location with bacterial membrane permeabilization. Single-cell images acquired using the superresolution Airyscan mode are shown in Fig. 4a. Both peptides are preferentially localized around the bacteria, probably on the membrane. In some images, it was also possible to observe both peptides apparently co-localizing with SYTOX® Green (as shown in the fluorescence distribution plot; Fig. 4a). This suggests partial internalization and binding to DNA by the peptide, but the finding is not general for all bacteria. Images acquired in the normal confocal mode (Fig. S5) show untreated controls and bacteria treated with non-labeled peptides.
Figure 4.
Peptide localization and structural changes on E. coli cell surface. a, confocal point scanning microscopy images of E. coli upon treatment with RhB-Ctn and RhB-Ctn(15–34). Peptides were added to 107 cfu/ml E. coli suspensions at their MBC. The fluorescence emission intensity of RhB (red signal) allows detection of the peptides' distribution. Fluorescence emission intensity of SYTOX® Green (green signal) allows detection of bacterial membrane permeabilization. Normalized fluorescence intensity (for red and green signals) along the white line indicated in the merged superposition image is also shown. b, atomic force microscopy images of untreated E. coli suspension at 107 cfu/ml or after incubation with Ctn and Ctn(15–34) at their MBC. Three-dimensional projections (left) and height images (middle) of E. coli cells and the profile (right) of the cross-line on height images are shown.
Imaging peptide effect on bacterial cells
To visualize structural changes on E. coli induced by Ctn and Ctn(15–34), individual bacteria were imaged by AFM after incubation with peptides at their MBC. As shown in Fig. 4b, both peptides induce major morphological alterations. Compared with control, the typical shape of the bacteria is lost, and substantial shrinkage, particularly at the central region, is observed. Additionally, treatment with either peptide also induces leakage of cytoplasmic content, as shown by images of bacterial surroundings. Cross-line profiles on Fig. 4b allow to visualize the location of three (for Ctn) or two (for Ctn(15–34)) protuberances in the septal poles of the bacteria and the aforementioned shrinkage at the middle.
Ability of Ctn(15–34) to target the bacterial surface
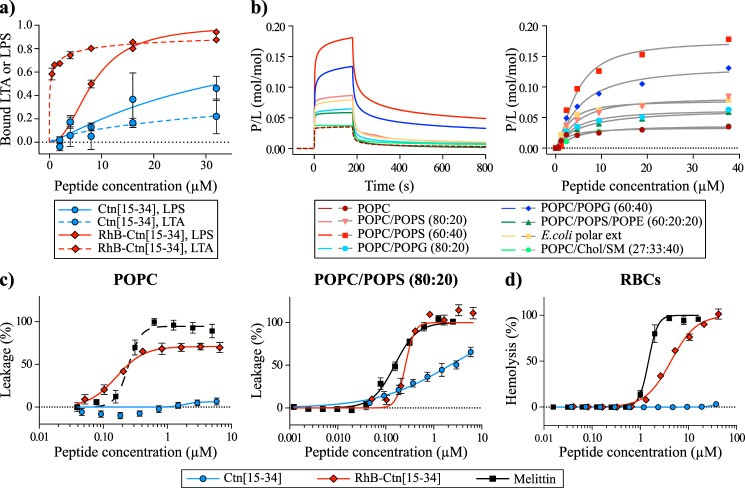
Gram-positive bacteria have lipoteichoic acid (LTA) at the membrane surface, whereas Gram-negative bacteria have lipopolysaccharide (LPS) (35). Both LTA and LPS are negatively charged and likely to be involved in the first contact of peptides with bacterial cells. To examine whether Ctn(15–34) and its labeled analogue RhB-Ctn(15–34) are able to bind to these molecules, we have used a Limulus amebocyte lysate (LAL) assay, which detects the presence of non-neutralized LTA and LPS (36). Results in Fig. 5a show that RhB-Ctn(15–34) neutralizes both LPS and LTA, whereas Ctn(15–34) is less efficient in doing it, especially for LTA.
Figure 5.
Interaction of Ctn(15–34) and RhB-Ctn(15–34) with LPS, LTA, model membranes, and RBCs. a, binding of Ctn(15–34) and of RhB-Ctn(15–34) to LTA and LPS as examined using the LAL assay. The concentration of RhB-Ctn(15–34) to neutralize 50% of LTA and LPS is 0.22 and 8.26 μm, respectively. Ctn(15–34) requires >32 μm to neutralize both LTA and LPS. b, binding of Ctn(15–34) to model membranes followed by surface plasmon resonance. Peptide samples were injected over lipid bilayer deposited onto an L1 chip surface; sensorgrams (left) obtained upon injection of 38 μm peptide over a particular lipid bilayer for 180 s (association phase) and the dissociation followed for 600 s (dissociation phase). Shown is the amount of Ctn(15–34) (right) obtained at the end of association phase (t = 170 s) upon injection of various peptide concentrations. The signal of sensorgrams and dose–response curves were normalized to peptide/lipid ratios (P/L, mol/mol) by converting response units into mol of peptide and normalized for the amount of lipid deposited onto the lipid surface (1 response unit = 1 pg/mm2 of peptide or lipid). c, membrane disruption induced by Ctn(15–34) and RhB-Ctn(15–34). The percentage of membrane leakage was determined by CF dequenching. LUVs composed of POPC or POPC/POPS (80:20) with total lipid concentration of 5 μm were incubated with various concentrations of peptide. Melittin, a membrane-disrupting peptide, was included as a positive control. d, toxicity against RBCs induced by Ctn(15–34) and RhB-Ctn(15–34). A suspension of RBCs (0.25%, v/v) was incubated with various concentrations of peptides. Percentage of hemolysis was detected by absorbance of hemoglobin released into the supernatant. Melittin was included as a control peptide with hemolytic properties. Error bars, S.D.
Interaction of Ctn(15–34) with model membranes
The ability of Ctn(15–34) to bind model membranes composed of different lipid mixtures was examined by surface plasmon resonance (SPR). Previous studies suggested a preferential binding of Ctn(15–34) to bacteria and cancer cells rather than healthy eukaryotic cells (27). In view of this, a number of phospholipid compositions mimicking various types of cell membranes were assayed. Vesicles composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), a zwitterionic phospholipid forming neutral, fluid lipid bilayers at 25 °C, were compared with membranes made of POPC/sphingomyelin (SM)/cholesterol (Chol) (POPC/SM/Chol; 27:33:40 molar ratio), which forms bilayers with the liquid-ordered phase. POPC mimics the neutral fluid portions, whereas POPC/Chol/SM simulates the more-ordered raftlike portions of the outer leaflet in healthy human cells (37, 38). The inner leaflet of healthy human cell plasma membranes possesses zwitterionic phosphatidylethanolamine (PE)-, anionic phosphatidylserine (PS)-, and phosphatidylcholine (PC)-phospholipids (37, 38); thus, the mixture POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS)/1-palmitoyl-2-oleoyl-sn-glycero-phospho-l-ethanolamine (POPE) (60:20:20) has been included to mimic the inner leaflet of healthy cell membranes. Tumor cells possess larger proportions of negatively charged phospholipids exposed to the outer leaflet than healthy cells; therefore, the potential effect of negatively charged phospholipids was further examined by testing the affinity to membranes composed of POPC/POPS in 80:20 or 60:40 proportions. Finally, bacterial cell membranes have high negative charge and large proportions of phosphatidylgylcerol (PG)-phospholipids; to mimic them, vesicles composed of POPC/POPG (80:20 or 60:40) or of an E. coli polar extract that contains a mixture of PE-phospholipids, PG-phospholipids, and cardiolipin were prepared.
Sensorgrams obtained with zwitterionic lipid systems (i.e. POPC and POPC/Chol/SM)) (Fig. 5b, left) showed rapid dissociation, suggesting that Ctn(15–34) is rapidly removed from neutral membranes, and dose–response curves (Fig. 5b, right) showed that Ctn(15–34) has a weak overall affinity for these lipid membranes. No significant differences were detected between fluid or liquid-ordered phases, suggesting an overall weak affinity for neutral membranes. In contrast, Ctn(15–34) has higher affinity for vesicles made of anionic phospholipids, as suggested by dose-response curves and sensorgrams obtained with POPC/POPS and POPC/POPG (see Fig. 5b and Table 3). Improved affinity is observed with higher content of anionic lipids (e.g. compare POPC with POPC/POPS (80:20) and with POPC/POPS (60:40)). The addition of PE-phospholipids to anionic membranes did not improve Ctn(15–34) binding, as shown with POPC/POPS (80:20) versus POPC/POPE/POPS (60:20:20). Although an improvement in affinity was observed for all anionic membranes, Ctn(15–34) has better affinity for POPS than for POPG, as demonstrated by comparing its binding with POPC/POPG (60:40) and POPC/POPS (60:40) (Fig. 5b).
Table 3.
Affinity of Ctn(15–34) for different lipid systems as followed by surface plasmon resonance
P/Lmax (mol/mol) is the maximum binding of Ctn(15–34) for a given lipid system. P/Lmax was calculated by fitting the dose–response curves shown in Fig. 5b (right) with one-site specific binding with the Hill slope equation.
| Lipid system | P/Lmax |
|---|---|
| × 10−1 mol/mol | |
| POPC | 0.33 ± 0.05 |
| POPC/Chol/SM (27:33:40) | 0.37 ± 0.02 |
| E. coli polar extract | 0.77 ± 0.04 |
| POPC/POPS (80:20) | 0.83 ± 0.11 |
| POPC/POPS (60:20) | 1.76 ± 0.15 |
| POPC/POPS/POPE (60:20:20) | 0.65 ± 0.09 |
| POPC/POPG (80:20) | 0.63 ± 0.07 |
| POPC/POPG (60:40) | 1.35 ± 0.15 |
To investigate whether N-terminal RhB derivatization alters the binding behavior of Ctn(15–34), the labeled and unlabeled versions were compared for binding to POPC and POPC/POPS (80:20). Similar to the case for bacterial cells (Fig. 2 and Fig. 2), the RhB version shows higher affinity for both neutral and anionic membranes (Fig. S6).
Membrane-disrupting properties of Ctn(15–34)
To examine whether peptides disturb pure lipid bilayers, vesicles loaded with carboxyfluorescein (CF) were incubated with various concentrations of Ctn(15–34) or RhB-Ctn(15–34). Fig. 5c shows that Ctn(15–34) does not induce dye leakage from neutral POPC vesicles, yet it disrupts negatively charged POPC/POPS (80:20) vesicles in a dose-dependent manner (LC50 2.17 ± 0.36 μm). In agreement with an overall increase in affinity for the membrane, RhB-Ctn(15–34) disrupts vesicles, including zwitterionic membranes, with much higher efficiency than its unlabeled version (LC50 0.24 ± 0.02 μm with POPC and 0.26 ± 0.02 μm with POPC/POPS (80:20)) (see Fig. 5c). Both peptides have a similar effect on POPC/POPCS (80:20) vesicles at 5, 10, and 15 min (Fig. S7), suggesting that disruption takes place within the first 5 min.
Toxicity to human red blood cells
The hemolytic properties of Ctn(15–34) and RhB-Ctn(15–34) were tested in human red blood cells (RBCs). Ctn(15–34) is not hemolytic up to 64 μm when tested against 0.25% (v/v) of RBCs (Fig. 5d). The lack of toxicity of Ctn(15–34) against RBCs is in agreement with its low affinity and inability to disturb neutral model membranes. On the other hand, derivatization of the peptide with RhB makes the peptide toxic, as shown with RhB-Ctn(15–34) (HC50 4.33 ± 0.18 μm) and is in agreement with this peptide possessing high affinity and the ability to disrupt lipid membranes.
Discussion
The aim of this work was to gain insights into the antimicrobial mechanisms of Ctn, an AMP derived from the venom gland of the rattlesnake Crotalus durissus terrificus, and of its fragment Ctn(15–34). Previous studies demonstrated that Ctn is toxic against bacteria, fungi, tumor, and healthy cells (26, 28). In contrast, Ctn(15–34) kills Gram-negative bacteria, fungi, and tumor cells but not healthy eukaryotic cells. Moreover, Ctn(15–34) has remarkable stability in human serum and is 40% smaller than Ctn (20 versus 34 amino acid residues) (27). For this reason, Ctn(15–34) is regarded as a promising anti-infective lead. However, its mechanism of action had not been examined until now.
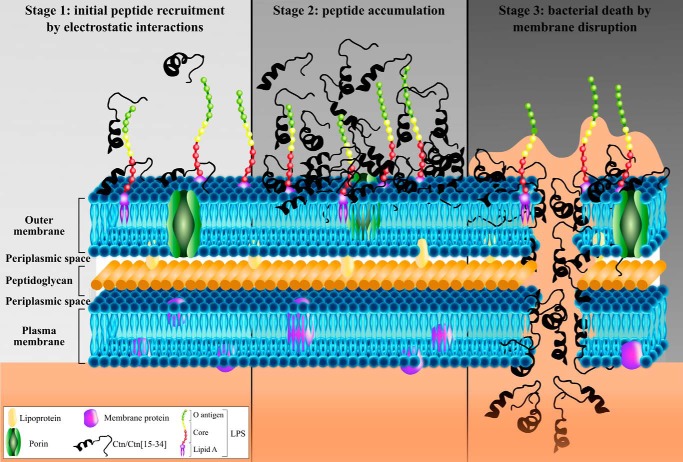
To investigate the mode of action, we compared the antimicrobial potency and ability to disrupt membranes of Ctn and Ctn(15–34), using Gram-negative E. coli and P. aeruginosa as model organisms. We confirmed that both peptides are bactericidal at low micromolar concentrations (Table 1) and demonstrated that bacterial death is accomplished remarkably fast (Fig. 1a), particularly against P. aeruginosa. A fast induction of bacterial death is normally associated with loss of physical integrity of the membrane (39), as described previously for AMPs such as melittin (40), C16G2 (41), vAMP 059, and vCPP 2319 (42). The current results confirm a mode of action related to membrane disruption and suggest different stages in the permeabilization process (Fig. 6): (Stage 1) initial peptide recruitment; (Stage 2) peptide accumulation; and (Stage 3) cell death by membrane disruption.
Figure 6.
Mechanism of action proposed for Ctn and Ctn(15–34). Both Ctn and Ctn(15–34) present a membrane-disruptive effect comprising three main steps: (Stage 1) initial peptide recruitment due to electrostatic attractions between cationic residues of peptide and anionic molecules in the bacterial outer membrane; (Stage 2) accumulation of peptide on the surface of the membrane up to an effective threshold concentration; and (Stage 3) bacterial cell death by membrane disruption.
Stage 1: Initial peptide recruitment
There is wide agreement that AMPs are usually drawn to microbial surfaces by electrostatic attraction between cationic (Lys, Arg, and His) residues in the peptide and anionic components in the bacterial surface, such as LPS and LTA on Gram-negative or Gram-positive surfaces, respectively, and/or anionic phospholipids (PG and cardiolipin) in the plasma membrane (7). In the present case, the high cationicity of both Ctn and Ctn(15–34) (net charge +16 and +8, respectively) and their preferential action on Gram-negative bacteria suggest that both peptides might bind LPS, as proposed for other snake-derived cathelicidins, such as Pb-CATH (43) and HC-CATH (22). As shown in Fig. 5a, Ctn(15–34) is not highly efficient at neutralizing both LPS and LTA, but even so, the binding is higher for LPS than for LTA, in concordance with its preference for Gram-negative bacteria. The fact that both peptides show a preference for cardiolipin (see below in AFM results) also reinforces an electrostatically driven initial recruitment step.
Stage 2: Peptide accumulation
Build-up of Ctn and Ctn(15–34) onto the bacterial surface was confirmed by ζ potential studies, because the negatively charged surface of both E. coli and P. aeruginosa was neutralized by increasing peptide concentrations, which caused a shift to positive ζ potential values (Fig. 1b). In all cases, neutralization occurred at concentrations lower than MIC or MBC (Fig. 1b), suggesting that surface charge equalization is not enough and that more peptide is necessary to induce bacterial death, which takes place after a threshold concentration of Ctn and Ctn(15–34) on the bacteria surface has been reached and peptides can begin to cross/enter the membrane.
Stage 3: Cell death by membrane disruption
Bacterial death results from membrane disruption and consequent loss of functionality. FCA data in Fig. 2 and Fig. S2 show a direct correlation between peptide uptake, membrane permeabilization, and viability loss. A mechanism dependent on the ability to target the membrane was also confirmed by imaging techniques. AFM studies showed important disturbances in the E. coli surface upon incubation with Ctn and Ctn(15–34) (Fig. 4b). The leakage of cytoplasmic content observed after peptide treatment is in agreement with a lytic mechanism of action, and the shrinkage observed in the middle region of the bacteria could be due to preferential accumulation of peptides in cardiolipin-rich domains, namely in the apical and septal area of the cell (44), as described previously for other AMPs (32). Confocal microscopy experiments found RhB peptides in the periphery of E. coli bacteria (Fig. 4a), probably located on the membrane, in tune with the membrane-disruption hypothesis, although they were also found internalized inside cells in some cases. Thus, we propose that both peptides are initially recruited and accumulate around the surface until a threshold concentration is reached, after which they begin to enter/cross the lipid bilayer, causing cell death.
The membrane-lytic mechanism proposed here for Ctn and Ctn(15–34) is in agreement with results described for other snake-derived, evolutionarily related cathelicidins (e.g. cathelicidin-BF (Cbf), a 30-residue AMP from the elapid snake Bungarus fascatus, also described as membrane-disruptive against bacteria) (45, 46). Analogs Cbf-K16 and Cbf-A7A13, able to kill multiresistant E. coli in a few hours, can also penetrate bacteria cell membrane as well as bind to DNA (47), suggesting that these peptides are not simply retained on the lipid bilayer. The N-terminal fragment, BF-15, and its analog ZY13, both active against resistant Candida albicans and less hemolytic than Cbf, also act in a membrane-disruptive manner (48). The aforementioned Pb-CATH and HC-CATH also possess bacterial membrane lytic properties (22, 43), and OH-CATH and NA-CATH, two cathelicidins found in cobra species, possess 79% identity with Ctn and are also membrane-active (24, 49).
Despite evidence suggesting a common membrane disruption mechanism for snake cathelicidins, how this process unfolds over time is not well studied, in part due to the limited information available by standard methods. To shed light on this issue, we employed time-resolved flow cytometry (34, 50), a high-resolution technique that allows monitoring peptide uptake and membrane permeabilization over time. This is particularly relevant to membrane-lytic peptides, for which binding and accumulation on the bacterial surface are early-onset events not easily monitored by end-point experiments. For instance, FCA kinetic curves (Fig. 3) reveal differences undetectable with the colony count method. Ctn(15–34) acts faster than Ctn due to faster binding and membrane permeabilization (Fig. 3). The slower stages in the action of Ctn cause a lag in its kinetic curve, although no cooperativity is detected, which demonstrates that common-sense reasoning that assigns lag phase directly to positive cooperativity (34, 51, 52) might be an oversimplification.
The mechanism of action of both Ctn and Ctn (15–34) includes steps whose kinetic curves deviate from the two-state model. Interestingly, the related NA-CATH cathelicidin seems to switch its mechanism of action from membrane disruption to pore-based lysis in a biphasic model, mimicking the E. coli membrane with an impact on kinetics similar to that described for Ctn (49). Given the high structural similarities between both peptides (an N-terminal α-helical segment followed by a disordered tail) (27, 53), it is plausible that Ctn may act similarly, and such behavior would explain the complex cell death kinetic pattern here reported. This dual behavior might be tentatively related to the composite structure (α-helix plus random tail) of both NA-CATH and Ctn, each moiety of the peptide embedding into the membrane in a different fashion, as suggested for NA-CATH (49). The fact that removal of the N-terminal helix of Ctn to give Ctn(15–34) results in a simplified killing mechanism corroborates our hypothesis.
RhB-labeled versions of Ctn and Ctn(15–34) show different uptake kinetics relative to unlabeled peptides (Fig. S4). RhB-Ctn binds slower than RhB-Ctn(15–34) (Fig. S4), but membrane permeabilization is much faster once the peptides are bound. Use of RhB-labeled Ctn(15–34) (Fig. S4) made possible the monitoring of peptide attachment to bacteria simultaneously with membrane permeabilization. RhB fluorophore emission confirms that RhB-Ctn(15–34) binds faster to membranes than RhB-Ctn, in agreement with the two-state model analyses for both peptides (k0 = 7 × 10−4 s−1 versus k0 = 2 × 10−2 s−1). In contrast, upon membrane binding, membrane permeabilization is faster for RhB-Ctn (k2 = 1 × 10−2 s−1 versus k2 = 1 × 10−3 s−1). For both labeled peptides, f ∼ 0, which shows that no cooperativity exists. In general, kinetic and membrane-affinity results for RhB-Ctn(15–34) appear to be affected by RhB labeling (Fig. 5 (a, c, and d) and Figs. S2 and S6); hence, conclusions cannot be extrapolated to the unlabeled version.
The previously described putative selectivity of Ctn(15–34) for anionic domains (27) was corroborated by experiments with model membranes showing a preference for anionic, particularly PS-rich vesicles (Fig. 5b). LUV leakage (Fig. 5c) and RBC hemolysis (Fig. 5d) studies show that Ctn(15–34) preferentially interacts and disrupts vesicles mimicking bacterial and tumor rather than healthy eukaryotic cells. These results are in agreement with data from other snake cathelicidins (e.g. OH-CATH), which suggest a crucial implication of the four N-terminal amino acid residues in hemolytic activity (24).
In conclusion, by deploying a set of high-resolution methodologies, we have achieved accurate spatiotemporal characterization of events involved in the bactericidal action of Ctn and Ctn(15–34) on E. coli and P. aeruginosa. The study sheds light on the mechanism whereby the peptides accumulate and disrupt bacterial and model membranes and reveals slight differences in their behavior, including different kinetics of access to bacterial surfaces and different toxicity. By dissecting Ctn into its Ctn(15–34) fragment, we have produced a smaller, simpler, and more cost-efficient AMP displaying selectivity for bacterial-like, PS-rich membranes and a simplified mechanism of action. Taken together, our findings augur a promising role for Ctn(15–34) as an anti-infective lead and, in a broader sense, provide valuable insights into AMP lethal mechanisms that should be useful in the design and development of antimicrobials against resistant bacteria.
Experimental procedures
Peptides
Peptides in Table 1 were assembled as C-terminal carboxamides on Rink amide resin as described previously (27). For N-terminal RhB coupling, the peptide-resin was N-deprotected with piperidine/N,N-dimethylformamide (20:80, v/v) and treated with a 5-fold molar excess of RhB/N,N′-diisopropylcarbodiimide (1:1 molar ratio) in dichloromethane, followed by extensive dichloromethane and N,N-dimethylformamide washes. Analytical reversed-phase HPLC, preparative HPLC, and LC-MS analyses were as reported previously (27). Peptide stock solutions were prepared in sterile Milli Q water and stored at −20 °C.
Antibacterial and bactericidal assays
The antimicrobial activity of Ctn and Ctn(15–34) was studied against Gram-negative reference strains E. coli ATCC 25922 and P. aeruginosa ATCC 27853. Minimal inhibitory and bactericidal concentrations (MIC and MBC) were determined by the broth microdilution method as described in document M26-A of the Clinical and Laboratory Standards Institute (54). Briefly, bacterial suspensions were prepared by the direct colony suspension method into BBLTM Mueller Hinton II Broth (cation-adjusted) medium (MHBII) (BD Biosciences) and incubated for 18 h at 37 °C with 2-fold serial peptide dilutions on 96-well microtiter polystyrene plates (Corning Inc.). Final bacterial concentration was 5 × 105 cfu/ml or 107 cfu/ml, and peptide concentrations were in the 0.1–100 μm range. After incubation, MIC values were determined as the lowest peptide concentration preventing visible bacterial growth. MBC values were subsequently determined as described previously (42). All assays were performed in triplicate.
Time-kill curves
The time-dependent bactericidal effect of Ctn and Ctn(15–34) were assessed against both E. coli and P. aeruginosa. Bacterial suspensions prepared on MHBII at 5 × 105 cfu/ml were treated with the peptides at a final concentration corresponding to their MBC and incubated at 37 °C and 200 rpm. As control, bacterial suspensions without peptide were prepared under the same conditions. Aliquots of untreated and peptide-treated bacterial suspensions were withdrawn at different time points (t = 5, 15, 30, 60, 90, 120, 180, and 240 min), serially diluted, plated onto trypticase soy agar plates, and incubated at 37 °C. After 24 h, the number of bacterial colonies was counted, and viable bacteria (in cfu/ml) at each t were calculated as percentage of the untreated control at t0. The assay was performed in triplicate.
ζ potential
For the ζ potential studies, E. coli or P. aeruginosa suspensions were prepared in HEPES buffer (10 mm HEPES, 150 mm NaCl, pH 7.4) at 107 cfu/ml, and Ctn or Ctn(15–34) was added at different concentrations. Treated and untreated suspensions were injected into zeta cells and allowed to equilibrate at 25 °C for 15 min, and measurements were acquired on a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK). ζ potential was calculated from the mean of 15 measurements, using the Smoluchowski equation as described previously (55). Experiments were performed in triplicate.
Viability and membrane permeabilization correlation studies
To determine bacterial viability and membrane permeabilization, bacterial suspensions at 108 cfu/ml in MHBII were centrifuged 10 min at 4000 × g and diluted to 5 × 105 cfu/ml in HEPES buffer. Incubation with 2-fold serial peptide dilutions was done at 37 °C and 200 rpm, for 90 min for E. coli and 60 min for P. aeruginosa. Untreated bacteria were used as negative control. To estimate the percentage of permeabilized bacteria and percentage of viable bacteria, each sample was respectively evaluated by FCA and colony count methods using protocols described previously (42).
Membrane permeabilization induced by RhB-labeled peptides
Samples for FCA were prepared as described above. Data were acquired on a BD FACSAria III cell sorter (BD Biosciences) using blue (488-nm) and yellow-green (561-nm) lasers and emission filters BP530/30 (LP505) and BP586/15 to detect SYTOX® Green and RhB, respectively. 5000 events corresponding to bacterial population as previously defined by forward scatter and side scatter were recorded. Experiments were performed in triplicate.
Peptide uptake and bacterial death kinetics
Peptide uptake and bacterial permeabilization were monitored in real time as described elsewhere (34). Briefly, a 108 cfu/ml bacterial inoculum was prepared in MHBII, centrifuged for 10 min at 4000 × g, and diluted to 107 cfu/ml on HEPES buffer. Dead bacteria were prepared by incubation at 37 °C with 70% (v/v) isopropyl alcohol for 60 min. SYTOX® Green was added to the bacterial suspension at 0.05 μm final concentration and incubated for at least 10 min. Ctn, RhB-Ctn, Ctn(15–34), or RhB-Ctn(15–34) was added to a final concentration equal to its MBC. Measurements were performed on a Fortessa X-20 instrument (BD Biosciences) using blue (488 nm) and yellow-green (561 nm) lasers and emission filters BP530/30 (LP505) and BP586/15 to detect SYTOX® Green and RhB, respectively. A total of 10,000 events were recorded for controls. For kinetic experiments, acquisition was done for 90 min immediately after the peptide addition. A control of untreated E. coli was also recorded for 90 min to ensure that bacteria remained alive during the total acquisition (see Fig. S8b). The bacterial population was selected according to the forward scatter/side scatter and the positive/negative SYTOX® Green gates (SG+/SG−), and positive/negative RhB gates (RhB+/RhB−) were established using controls. For kinetic analysis, events (#) on the SG+ and SG− gates were clustered for time intervals (n) equal to 1 s, and the number of events for each interval (#n) was counted. The ratios of permeabilized bacteria (P) and peptide uptake (Y) (bound to the membrane or internalized) for each n were calculated as follows.
| (1) |
| (2) |
Estimated percentages of permeabilized bacteria and peptide uptake for each n were calculated by the following equations.
| (3) |
| (4) |
The percentage of permeabilized bacteria was corrected using the equation derived from the calibration curve shown on Fig. S8a, obtained by combining known percentages of dead and live bacteria.
Data were fitted to the two-state model described previously (34), using the following equation,
| (5) |
where k′1 = k0xf; y0 = initial % permeabilized bacteria; Bl = final % permeabilized bacteria; k2 = permeabilization kinetic constant; k0 = membrane attachment constant; and f = cooperativity binding factor.
Confocal microscopy
An E. coli suspension was prepared at 108 cfu/ml in MHBII, centrifuged for 10 min at 4000 × g, and diluted to 107 cfu/ml on HEPES buffer. RhB-labeled or unlabeled peptides were added to the bacterial suspensions at a final concentration equal to their MBC, and the samples were incubated at 37 °C and 200 rpm for 90 min. Untreated bacteria were used as a control. After incubation, samples were placed on ice, and SYTOX® Green was added at 5 μm final concentration. Samples were placed on an ibiTreat-coated 8-well μ-slide (Ibidi, Munich, Germany) before imaging. Acquisition was made on a confocal point-scanning Zeiss LSM 880 microscope (Carl Zeiss, Jena, Germany) equipped with an alpha Plan-Apochromat ×100 oil immersion objective (1.46 numerical aperture). The 488-nm line from an argon laser and the DPSS 561-20 laser were used to excite SYTOX® Green and RhB, respectively. In the normal confocal mode, ×0.6 zoom images were recorded at 1024 × 1024 resolution. In the Airyscan superresolution mode, 5.5× zoom was applied to select one single bacteria cell, and the optimal resolution was selected. ZEN software was used for image acquisition and Airyscan image processing. Fiji software was used for background subtraction and brightness and contrast adjust (identically for compared image sets). At least 12 total images were acquired in three independent replicates.
Atomic force microscopy
Morphological changes on E. coli cells induced by Ctn and Ctn(15–34) were visualized by AFM imaging. 108 cfu/ml suspensions were prepared on MHBII, centrifuged for 10 min at 4000 × g, and diluted to 107 cfu/ml in HEPES buffer. Bacterial suspensions in the absence or the presence of Ctn or Ctn(15–34) at their MBC were incubated at 37 °C and 200 rpm for 90 min. Untreated bacteria were used as a control. Slide preparation, image acquisition, and processing were as described previously (32).
Binding affinity for LTA and LPS
Neutralization of LTA and LPS by Ctn(15–34) or RhB-Ctn(15–34) was examined using the end-point chromogenic LAL assay kit (QLC-1000, Lonza) following the same protocol as before (36).
Liposome preparation
POPC; POPS; POPG; POPE; E. coli polar lipid extract containing PE-phospholipids, PG-phospholipids, and cardiolipin in the proportion 67:23.2:9.8 (w/w/w); and octadecanoyl SM extracted from porcine brain were purchased from Avanti Polar Lipids (Alabaster, AL). Lipids were solubilized in spectroscopy-grade chloroform and mixed as required to prepare defined lipid compositions. LUVs (100-nm diameter) or SUVs (50-nm diameter) were prepared in HEPES buffer by extrusion, as described previously (56). LUVs were used in fluorescence spectroscopy assays, whereas SUVs were used for SPR studies to facilitate bilayer deposition on the chip surface.
Vesicle leakage assay
Content leakage from POPC or POPC/POPS (80:20 molar ratio) vesicles was quantified by CF fluorescence dequenching using LUVs loaded with 50 mm CF as described previously (57). Briefly, 2-fold dilutions of peptides (starting with 10 μm) were incubated for 20 min with LUVs (5 μm of lipid concentration) in HEPES buffer. Fluorescence emission intensity (excitation at 494 nm and emission at 515 nm) was measured in an M1000 microplate reader (Tecan, Männedorf, Switzerland), and the leakage percentage was calculated as described previously (57).
Interactions with lipid bilayers
Peptide-membrane interactions were monitored by SPR at 25 °C with a L1 biosensor chip in a Biacore 3000 instrument (GE Healthcare Australia, Parramatta, Australia). SUVs composed of POPC, POPC/POPE (80:20 molar ratio), POPC/POPG (80:20), POPC/POPG (60:40), POPC/Chol/SM (27:33:40), POPC/POPS (80:20), POPC/POPS (60:40), POPC/POPS/POPE (60:20:20), or E. coli polar extract were prepared using HEPES buffer and deposited onto a L1 chip by injecting SUV suspensions with 0.5 mm lipid at a flow rate of 2 μl/min for 40 min. Peptides were injected over the lipid bilayers at a flow rate of 5 μl/min for 180 s (association phase), and dissociation was monitored for 600 s (58, 59). HEPES was used as running buffer, and the chip was regenerated as before (60). All solutions were freshly prepared and filtered using a 0.22-μm pore size filter. The response units were normalized to peptide/lipid ratio as described previously (58).
Hemolytic studies
Fresh human blood was collected from three healthy donors following protocols approved by the Human Research Ethics Unit at the University of Queensland. RBCs were isolated by centrifugation (1500 × g, 1 min), washed three times, and resuspended in PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4) at 0.25% (v/v). RBC suspensions were incubated with increasing concentrations of Ctn(15–34) or with RhB-Ctn(15–34) for 1 h at 37 °C, and the percentage of hemolysis was quantified by the 405-nm absorbance of released hemoglobin. Samples incubated with PBS or with Triton X-100 0.1% (v/v) were used to define 0 and 100% of hemolysis, respectively, as described previously (61). Melittin, a hemolytic peptide, was included as a control.
Author contributions
C. P.-P., S. A. D., M. M. D., J. M. F., D. G., M. A. R. B. C., S. T. H., A. S. V., and D. A. designed the experiments. C. P.-P., S. A. D., M. M. D., D. G., A. S. V., A. H. B., S. T. H., and D. A. ran experiments. C. P.-P., S. A. D., M. M. D., J. M. F., A. S. V., S. T. H., and D. A. analyzed the data. C. P.-P., A. S. V., S. T. H., and D. A. wrote the manuscript with contributions from all other authors.
Supplementary Material
Acknowledgment
We thank Aina Ollé for help with script design.
This work was supported by Spanish Ministry of Economy and Competitiveness (MINECO) Grants SAF2011-24899 and AGL2014-52395-C2, by Fundação para a Ciência e a Tecnologia (FCT, Portugal) Grants PTDC/QEQ-MED/4412/2014, and by EU Marie Skłodowska-Curie Research and Innovation Staff Exchange (RISE) program Grant 644167, 2015–2019. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Movies S1a, S1b, S2a, and S2b.
- AMP
- antimicrobial peptide
- ACN
- acetonitrile
- AFM
- atomic force microscopy
- CF
- 5(6)-carboxyfluorescein
- Chol
- cholesterol
- Ctn
- crotalicidin
- FCA
- flow cytometry assay
- LPS
- lipopolysaccharide
- LTA
- lipoteichoic acid
- LUV
- large unilamellar vesicle
- MBC
- minimal bactericidal concentration
- MHBII
- Mueller Hinton broth cation-adjusted
- MIC
- minimal inhibitory concentration
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-phospho-l-ethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-rac-glycerol
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- PS
- phosphatidylserine
- RBC
- red blood cells
- RhB
- rhodamine B
- SM
- sphingomyelin
- SPR
- surface plasmon resonance
- SUV
- small unilamellar vesicle
- LAL
- Limulus amebocyte lysate
- Cbf
- cathelicidin-BF
- CATH
- cathelicidin
- cfu
- colony-forming units.
References
- 1. Thabit A. K., Crandon J. L., and Nicolau D. P. (2015) Antimicrobial resistance: impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin. Pharmacother. 16, 159–177 10.1517/14656566.2015.993381 [DOI] [PubMed] [Google Scholar]
- 2. Michael C. A., Dominey-Howes D., and Labbate M. (2014) The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health 2, 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Högberg L. D., Heddini A., and Cars O. (2010) The global need for effective antibiotics: challenges and recent advances. Trends Pharmacol. Sci. 31, 509–515 10.1016/j.tips.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 4. Wright G. D., and Sutherland A. D. (2007) New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 13, 260–267 10.1016/j.molmed.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 5. Rios A. C., Moutinho C. G., Pinto F. C., Del Fiol F. S., Jozala A., Chaud M. V., Vila M. M., Teixeira J. A., and Balcão V. M. (2016) Alternatives to overcoming bacterial resistances: state-of-the-art. Microbiol. Res. 191, 51–80 10.1016/j.micres.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 6. Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 7. Yeaman M. R., and Yount N. Y. (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- 8. Hancock R. E., and Sahl H. G. (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 10.1038/nbt1267 [DOI] [PubMed] [Google Scholar]
- 9. Sani M. A., and Separovic F. (2016) How membrane-active peptides get into lipid membranes. Acc Chem. Res. 49, 1130–1138 10.1021/acs.accounts.6b00074 [DOI] [PubMed] [Google Scholar]
- 10. Li J., Koh J. J., Liu S., Lakshminarayanan R., Verma C. S., and Beuerman R. W. (2017) Membrane active antimicrobial peptides: translating mechanistic insights to design. Front. Neurosci. 11, 73 10.3389/fnins.2017.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marín-Medina N., Ramírez D. A., Trier S., and Leidy C. (2016) Mechanical properties that influence antimicrobial peptide activity in lipid membranes. Appl. Microbiol. Biotechnol. 100, 10251–10263 10.1007/s00253-016-7975-9 [DOI] [PubMed] [Google Scholar]
- 12. Chen L., Zhang Q., Yuan X., Cao Y., Yuan Y., Yin H., Ding X., Zhu Z., and Luo S. Z. (2017) How charge distribution influences the function of membrane-active peptides: lytic or cell-penetrating? Int. J. Biochem. Cell Biol. 83, 71–75 10.1016/j.biocel.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 13. Tomasinsig L., and Zanetti M. (2005) The cathelicidins: structure, function and evolution. Curr. Protein Pept. Sci. 6, 23–34 10.2174/1389203053027520 [DOI] [PubMed] [Google Scholar]
- 14. Kościuczuk E. M., Lisowski P., Jarczak J., Strzałkowska N., Jóźwik A., Horbańczuk J., Krzyżewski J., Zwierzchowski L., and Bagnicka E. (2012) Cathelicidins: family of antimicrobial peptides: a review. Mol. Biol. Rep. 39, 10957–10970 10.1007/s11033-012-1997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong J. H., Ye X. J., and Ng T. B. (2013) Cathelicidins: peptides with antimicrobial, immunomodulatory, anti-inflammatory, angiogenic, anticancer and procancer activities. Curr. Protein Pept. Sci. 14, 504–514 10.2174/13892037113149990067 [DOI] [PubMed] [Google Scholar]
- 16. Dürr U. H., Sudheendra U. S., and Ramamoorthy A. (2006) LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 1758, 1408–1425 10.1016/j.bbamem.2006.03.030 [DOI] [PubMed] [Google Scholar]
- 17. Termén S., Tollin M., Olsson B., Svenberg T., Agerberth B., and Gudmundsson G. H. (2003) Phylogeny, processing and expression of the rat cathelicidin rCRAMP: a model for innate antimicrobial peptides. Cell Mol. Life Sci. 60, 536–549 10.1007/s000180300045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J., Wong E. S., Whitley J. C., Li J., Stringer J. M., Short K. R., Renfree M. B., Belov K., and Cocks B. G. (2011) Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS One 6, e24030 10.1371/journal.pone.0024030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao Y., Cai Y., Bommineni Y. R., Fernando S. C., Prakash O., Gilliland S. E., and Zhang G. (2006) Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281, 2858–2867 10.1074/jbc.M507180200 [DOI] [PubMed] [Google Scholar]
- 20. Uzzell T., Stolzenberg E. D., Shinnar A. E., and Zasloff M. (2003) Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 24, 1655–1667 10.1016/j.peptides.2003.08.024 [DOI] [PubMed] [Google Scholar]
- 21. Mu L., Zhou L., Yang J., Zhuang L., Tang J., Liu T., Wu J., and Yang H. (2017) The first identified cathelicidin from tree frogs possesses anti-inflammatory and partial LPS neutralization activities. Amino Acids 49, 1571–1585 10.1007/s00726-017-2449-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei L., Gao J., Zhang S., Wu S., Xie Z., Ling G., Kuang Y. Q., Yang Y., Yu H., and Wang Y. (2015) Identification and characterization of the first cathelicidin from sea snakes with potent antimicrobial and anti-inflammatory activity and special mechanism. J. Biol. Chem. 290, 16633–16652 10.1074/jbc.M115.642645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao H., Gan T. X., Liu X. D., Jin Y., Lee W. H., Shen J. H., and Zhang Y. (2008) Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 29, 1685–1691 10.1016/j.peptides.2008.06.008 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Zhao H., Yu G. Y., Liu X. D., Shen J. H., Lee W. H., and Zhang Y. (2010) Structure-function relationship of king cobra cathelicidin. Peptides 31, 1488–1493 10.1016/j.peptides.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 25. Blower R. J., Barksdale S. M., and van Hoek M. L. (2015) Snake cathelicidin NA-CATH and smaller helical antimicrobial peptides are effective against Burkholderia thailandensis. PLoS Negl. Trop. Dis. 9, e0003862 10.1371/journal.pntd.0003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Falcao C. B., de La Torre B. G., Pérez-Peinado C., Barron A. E., Andreu D., and Rádis-Baptista G. (2014) Vipericidins: a novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 46, 2561–2571 10.1007/s00726-014-1801-4 [DOI] [PubMed] [Google Scholar]
- 27. Falcao C. B., Pérez-Peinado C., de la Torre B. G., Mayol X., Zamora-Carreras H., Jiménez M. Á., Rádis-Baptista G., and Andreu D. (2015) Structural dissection of crotalicidin, a rattlesnake venom cathelicidin, retrieves a fragment with antimicrobial and antitumor activity. J. Med. Chem. 58, 8553–8563 10.1021/acs.jmedchem.5b01142 [DOI] [PubMed] [Google Scholar]
- 28. Cavalcante C. S., Falcão C. B., Fontenelle R. O., Andreu D., and Rádis-Baptista G. (2017) Anti-fungal activity of Ctn[15–34], the C-terminal peptide fragment of crotalicidin, a rattlesnake venom gland cathelicidin. J. Antibiot. 70, 231–237 [DOI] [PubMed] [Google Scholar]
- 29. Levison M. E., and Levison J. H. (2009) Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect. Dis. Clin. North Am. 23, 791–815, vii 10.1016/j.idc.2009.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roversi D., Luca V., Aureli S., Park Y., Mangoni M. L., and Stella L. (2014) How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem. Biol. 9, 2003–2007 10.1021/cb500426r [DOI] [PubMed] [Google Scholar]
- 31. Hällbrink M., Oehlke J., Papsdorf G., and Bienert M. (2004) Uptake of cell-penetrating peptides is dependent on peptide-to-cell ratio rather than on peptide concentration. Biochim. Biophys. Acta 1667, 222–228 10.1016/j.bbamem.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 32. Alves C. S., Melo M. N., Franquelim H. G., Ferre R., Planas M., Feliu L., Bardají E., Kowalczyk W., Andreu D., Santos N. C., Fernandes M. X., and Castanho M. A. (2010) Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 285, 27536–27544 10.1074/jbc.M110.130955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shephard J., McQuillan A. J., and Bremer P. J. (2008) Mechanisms of Cation Exchange by Pseudomonas aeruginosa PAO1 and PAO1 wbpL, a Strain with a truncated lipopolysaccharide. Appl. Environ. Microbiol. 74, 6980–6986 10.1128/AEM.01117-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freire J. M., Gaspar D., de la Torre B. G., Veiga A. S., Andreu D., and Castanho M. A. (2015) Monitoring antibacterial permeabilization in real time using time-resolved flow cytometry. Biochim. Biophys. Acta 1848, 554–560 10.1016/j.bbamem.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 35. Silhavy T. J., Kahne D., and Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Torcato I. M., Huang Y. H., Franquelim H. G., Gaspar D., Craik D. J., Castanho M. A., and Troeira Henriques S. (2013) Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim. Biophys. Acta 1828, 944–955 10.1016/j.bbamem.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 37. Daleke D. L. (2008) Regulation of phospholipid asymmetry in the erythrocyte membrane. Curr. Opin. Hematol. 15, 191–195 10.1097/MOH.0b013e3282f97af7 [DOI] [PubMed] [Google Scholar]
- 38. van Meer G., Voelker D. R., and Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahlapuu M., Håkansson J., Ringstad L., and Björn C. (2016) Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell Infect. Microbiol. 6, 194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Faust J. E., Yang P. Y., and Huang H. W. (2017) Action of antimicrobial peptides on bacterial and lipid membranes: a direct comparison. Biophys. J. 112, 1663–1672 10.1016/j.bpj.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaplan C. W., Sim J. H., Shah K. R., Kolesnikova-Kaplan A., Shi W., and Eckert R. (2011) Selective membrane disruption: mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob. Agents Chemother. 55, 3446–3452 10.1128/AAC.00342-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dias S. A., Freire J. M., Pérez-Peinado C., Domingues M. M., Gaspar D., Vale N., Gomes P., Andreu D., Henriques S. T., Castanho M. A. R. B., and Veiga A. S. (2017) New potent membrane-targeting antibacterial peptides from viral capsid proteins. Front. Microbiol. 8, 775 10.3389/fmicb.2017.00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim D., Soundrarajan N., Lee J., Cho H. S., Choi M., Cha S. Y., Ahn B., Jeon H., Le M. T., Song H., Kim J. H., and Park C. (2017) Genomewide analysis of the antimicrobial peptides in Python bivittatus and characterization of cathelicidins with potent antimicrobial activity and low cytotoxicity. Antimicrob. Agents Chemother. 61, e00530–17 10.1128/AAC.00530-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mileykovskaya E., and Dowhan W. (2000) Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182, 1172–1175 10.1128/JB.182.4.1172-1175.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou H., Dou J., Wang J., Chen L., Wang H., Zhou W., Li Y., and Zhou C. (2011) The antibacterial activity of BF-30 in vitro and in infected burned rats is through interference with cytoplasmic membrane integrity. Peptides 32, 1131–1138 10.1016/j.peptides.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 46. Wang Y., Zhang Z., Chen L., Guang H., Li Z., Yang H., Li J., You D., Yu H., and Lai R. (2011) Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS One 6, e22120 10.1371/journal.pone.0022120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hao Q., Wang H., Wang J., Dou J., Zhang M., Zhou W., and Zhou C. (2013) Effective antimicrobial activity of Cbf-K16 and Cbf-A7 A13 against NDM-1-carrying Escherichia coli by DNA binding after penetrating the cytoplasmic membrane in vitro. J. Pept. Sci. 19, 173–180 10.1002/psc.2488 [DOI] [PubMed] [Google Scholar]
- 48. Jin L., Bai X., Luan N., Yao H., Zhang Z., Liu W., Chen Y., Yan X., Rong M., Lai R., and Lu Q. (2016) A designed tryptophan- and lysine/arginine-rich antimicrobial peptide with therapeutic potential for clinical antibiotic-resistant Candida albicans vaginitis. J. Med. Chem. 59, 1791–1799 10.1021/acs.jmedchem.5b01264 [DOI] [PubMed] [Google Scholar]
- 49. Samuel R., and Gillmor S. (2016) Membrane phase characteristics control NA-CATH activity. Biochim. Biophys. Acta 1858, 1974–1982 10.1016/j.bbamem.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 50. Freire J. M., Veiga A. S., Rego de Figueiredo I., de la Torre B. G., Santos N. C., Andreu D., Da Poian A. T., and Castanho M. A. (2014) Nucleic acid delivery by cell penetrating peptides derived from dengue virus capsid protein: design and mechanism of action. FEBS J. 281, 191–215 10.1111/febs.12587 [DOI] [PubMed] [Google Scholar]
- 51. Koshland D. E. Jr., Némethy G., and Filmer D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 10.1021/bi00865a047 [DOI] [PubMed] [Google Scholar]
- 52. Coutinho A., and Prieto M. (2003) Cooperative partition model of nystatin interaction with phospholipid vesicles. Biophys. J. 84, 3061–3078 10.1016/S0006-3495(03)70032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du H., Samuel R. L., Massiah M. A., and Gillmor S. D. (2015) The structure and behavior of the NA-CATH antimicrobial peptide with liposomes. Biochim. Biophys. Acta 1848, 2394–2405 10.1016/j.bbamem.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 54. Barry A. L., Craig W. A., Nadler H., Reller L. B., Sanders C. C., and Swenson J. M. (1999) Methods for determining bactericidal activity of antimicrobial agents; approved guideline. NCCLS document M26-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 55. Domingues M. M., Santiago P. S., Castanho M. A., and Santos N. C. (2008) What can light scattering spectroscopy do for membrane-active peptide studies? J. Pept. Sci. 14, 394–400 10.1002/psc.1007 [DOI] [PubMed] [Google Scholar]
- 56. Henriques S. T., Pattenden L. K., Aguilar M. I., and Castanho M. A. (2009) The toxicity of prion protein fragment PrP(106–126) is not mediated by membrane permeabilization as shown by a M112W substitution. Biochemistry 48, 4198–4208 10.1021/bi900009d [DOI] [PubMed] [Google Scholar]
- 57. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., and Craik D. J. (2009) The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J. Biol. Chem. 284, 20699–20707 10.1074/jbc.M109.003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henriques S. T., Huang Y. H., Castanho M. A., Bagatolli L. A., Sonza S., Tachedjian G., Daly N. L., and Craik D. J. (2012) Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 287, 33629–33643 10.1074/jbc.M112.372011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., and Craik D. J. (2011) Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J. Biol. Chem. 286, 24231–24241 10.1074/jbc.M111.253393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Henriques S. T., Pattenden L. K., Aguilar M. I., and Castanho M. A. (2008) PrP(106–126) does not interact with membranes under physiological conditions. Biophys. J. 95, 1877–1889 10.1529/biophysj.108.131458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang Y. H., Colgrave M. L., Clark R. J., Kotze A. C., and Craik D. J. (2010) Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J. Biol. Chem. 285, 10797–10805 10.1074/jbc.M109.089854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.