Abstract
Infections by ranaviruses such as Frog virus 3 (Fv3), are significantly contributing to worldwide amphibian population declines. Notably, amphibian macrophages (Mφs) are important to both the Fv3 infection strategies and the immune defense against this pathogen. However, the mechanisms underlying amphibian Mφ Fv3 susceptibility and resistance remain unknown. Mφ differentiation is mediated by signaling through the colony-stimulating factor-1 receptor (CSF-1R) which is now known to be bound not only by CSF-1, but also by the unrelated interleukin-34 (IL-34) cytokine. Pertinently, amphibian (Xenopus laevis) Mφs differentiated by CSF-1 and IL-34 are highly susceptible and resistant to Fv3, respectively. Accordingly, in the present work, we elucidate the facets of this Mφ Fv3 susceptibility and resistance. Because cellular resistance to viral replication is marked by expression of antiviral restriction factors, it was intuitive to find that IL-34-Mφs possess significantly greater mRNA levels of select restriction factor genes than CSF-1-Mφs. Xenopodinae amphibians have highly expanded repertoires of antiviral interferon (IFN) cytokine gene families, and our results indicated that in comparison with the X. laevis CSF-1-Mφs, the IL-34-Mφs express substantially greater transcripts of representative IFN genes, belonging to distinct gene family clades, as well as their cognate receptor genes. Finally, we demonstrate that IL-34-Mφ–conditioned supernatants confer IFN-mediated anti-Fv3 protection to the virally susceptible X. laevis kidney (A6) cell line. Together, this work underlines the differentiation pathways leading to Fv3-susceptible and -resistant amphibian Mφ populations and defines the molecular mechanisms responsible for these differences.
Keywords: antiviral agent, immunology, innate immunity, macrophage, Xenopus, : interleukin-34, Frog virus 3, Interleukin-34, antiviral immunity, colony-stimulating factor-1
Introduction
Amphibian population die-offs resulting from the Frog virus 3 (Fv3)3 ranavirus (family Iridoviridae) infections are significantly contributing to the worldwide amphibian declines (1, 2). Although much remains to be learned regarding the facets of amphibian immunity against these viral agents, macrophages (Mφs) are now known to be integral to both the Fv3 infection strategy and to the amphibian host immune responses against this pathogen (3, 4). Indeed, the capacity of the anuran (frogs and toads) amphibian Xenopus laevis to mount effective anti-Fv3 responses depends on appropriate Mφ differentiation (5, 6). As Mφ differentiation depends on the activation of the colony-stimulating factor-1 (CSF-1) receptor, it is compelling that in addition to CSF-1 (7–10), this receptor may be engaged by the unrelated interleukin-34 (IL-34) (11–14), presumably contributing to the functional heterogeneity seen across vertebrate Mφs. Our recent findings suggest that whereas the X. laevis Mφs generated by CSF-1 render animals more susceptible to Fv3, the IL-34-derived Mφ possess potent antiviral capacities and confer frog resistance to this viral pathogen (5, 6). However, the molecular mechanisms conferring CSF-1-Mφ susceptibility and IL-34-Mφ resistance to Fv3 remain to be fully defined.
Because antiviral interferon (IFN) cytokines represent a major pillar of vertebrate antiviral defenses (7, 9–11), it is particularly notable that amphibians hold a key stage in the evolution of these soluble effectors. Mammals, birds, and reptiles possess three types of IFNs, type I, II, and III IFNs (15), of which the type II IFNs mediate a variety of immunological roles, whereas the type I and type III IFNs predominantly participate in antiviral immunity (16). The type I IFNs of these higher vertebrates are encoded by intronless genes, whereas their type III IFNs (also known as IFNλs/IFNLs) are encoded by five exon/four intron gene transcripts (16). Conversely, teleost and cartilaginous fish do not appear to possess type III IFNs and encode type I IFNs with five-exon, four-intron organization (16). Intriguingly, Xenopodinae amphibians possess type I IFN genes with the five-exon, four-intron intron gene organization of the fish counterparts; intronless type I IFNs akin to those seen in higher vertebrates; and intronless and five exon/four intron-containing type III IFN genes (17, 18). Most notably, we recently demonstrated that X. laevis intron-containing type I and type III IFNs play important roles in anti-Fv3 responses (19–21).
Aside from antiviral IFN responses, the capacity of distinct cell types to support or minimize the replication of invading viral pathogens often correlates with their respective gene expression of antiviral restriction factors (24–27). Collectively, these cellular proteins present enormous structural and functional diversities and provide the first cellular lines of defense against viral infections by targeting and ablating distinct viral replication mechanisms (24–27). Whereas the expression of some of these restriction factors is enhanced by IFNs, others are constitutively expressed (24–27).
To delineate the mechanisms responsible for IL-34–derived Mφ antiviral resistance, and CSF-1-differentiated Mφ susceptibility to Fv3, we compared these cell populations for their gene expression of select intron-containing and intronless type I and type III IFNs as well as the respective IFN receptor genes. Additionally, we examined the restriction factor gene expression levels of these Mφ populations and compared the ability of Fv3 to replicate within CSF- and IL-34–derived Mφs. Moreover, we examined the ability of CSF-1-Mφ– and IL-34-Mφ–conditioned supernatants to restrict Fv3 growth in an otherwise highly Fv3-susceptible X. laevis kidney epithelial cell line (A6).
Results
IL-34-Mφs are more restrictive to Fv3 replication
We previously demonstrated that X. laevis Mφs differentiated with IL-34 are resistant to Fv3 infection, whereas CSF-1-derived Mφs are significantly more susceptible to this virus (5, 6). Accordingly, to examine whether CSF-1-Mφs support greater Fv3 replication than IL-34-Mφs, we differentiated IL-34- and CSF-1-Mφs from X. laevis bone marrow cells, challenged them with Fv3, and assessed the viral loads and expression of Fv3 82R (immediate early, IE), Fv3 95R (delayed early, E), and Fv3 93L (late, L) genes in these respective Mφ populations. As previously noted, CSF-1-Mφs possessed significantly greater Fv3 loads than detected in the IL-34-Mφs (Fig. 1A). Moreover, CSF-1-Mφs possessed substantially greater magnitudes of Fv3-82R, -95R, and -93L gene expression than detected in IL-34-Mφs (Fig. 1B). Of the examined Fv3 genes, 93L exhibited the greatest expression differences between CSF-1- and IL-34-Mφ (Fig. 1B).
Figure 1.
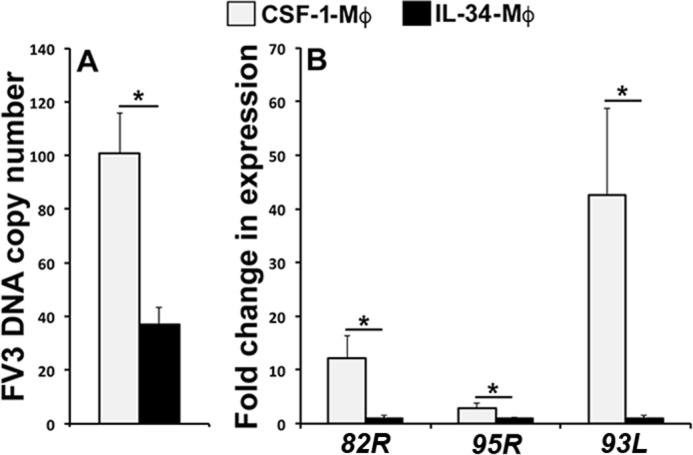
IL-34-Mφs are more restrictive to Fv3 replication than CSF-1-Mφs. X. laevis bone marrow cells were cultured in vitro with CSF-1 or IL-34 for 5 days, followed by Fv3 infection at 0.5 MOI for 24 h. A, CSF-1- and IL-34-Mφ Fv3 DNA loads, determined by absolute qPCR (n = 5). B, FV3-challenged CSF-1- and IL-34-Mφ gene expression of viral immediate early (82R), delayed early (95R), and late (93L) genes, quantified relative to the gapdh endogenous control (n = 5). Results are means ± S.E. (error bars); *, statistically significant difference (p < 0.05).
IL-34-Mφs exhibit greater restriction factor gene expression than CSF-1-Mφs
Because our results indicated that IL-34-Mφs are more restrictive to Fv3 infection while the expression of appropriate restriction factors is crucial to adequate cellular arrest of viral replication (24–27), we next examined whether IL-34-Mφ resistance to Fv3 could be attributed to more robust expression of such restriction factors. In comparison with CSF-1-Mφs, IL-34-Mφs exhibited a significantly greater expression levels of the apobec2 (apolipoprotein B mRNA-editing enzyme-2) and trim28 (tripartite motif–containing 28) restriction factors (Fig. 2A), which are known to suppress viral replication (22, 23). Moreover, Fv3 is a dsDNA virus, and our result showed that IL-34-Mφs also possessed more robust gene expression of samhd1 (SAM domain and HD domain-containing protein 1) and ctcf (CCCTC-binding factor) restriction factors, both of which are known to specifically target dsDNA viruses (24–27) (Fig. 2A). By contrast, CSF-1- and IL-34-Mφs had comparable mRNA levels of cyclophilin B (cypb) and rad21 (Fig. 2A).
Figure 2.
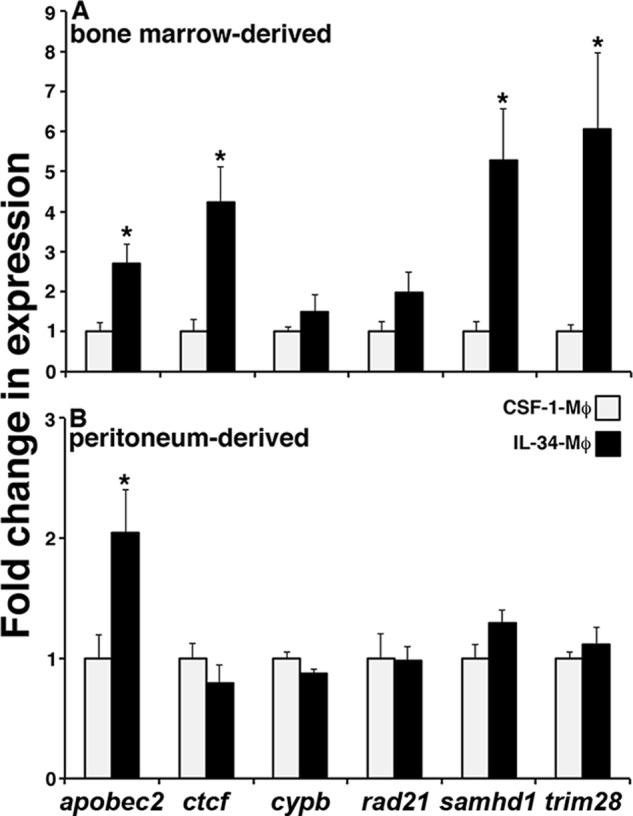
A, bone marrow–derived IL-34-Mφs exhibit greater restriction factor gene expression than CSF-1-Mφs. B, peritonea-derived IL-34-Mφs exhibit greater expression of apobec2 than CSF-1-Mφs. X. laevis bone marrow and peritonea-derived CSF-1- and IL-34-Mφ cultures were assessed by qPCR for gene expression of viral restriction factors, with all expression quantified relative to the gapdh endogenous control (n = 5). Results are means ± S.E. (error bars); *, statistically significant difference from CSF-1-Mφs (p < 0.05).
We showed previously that, like other vertebrate CSF-1 receptor ligands (28, 29), the frog CSF-1 and IL-34 chemoattract Mφ precursors and induce their functional differentiation (5, 6). In this respect, intraperitoneal injection of frogs with either recombinant CSF-1 or IL-34 results in peritoneal accumulation of Mφs that morphologically (85–90%) represent the respective CSF-1- or IL-34-Mφ populations (5, 6). Moreover, and akin to their bone marrow-derived counterparts, the peritonea-derived IL-34- and CSF-1-Mφs are respectively highly resistant and susceptible to Fv3 (5, 6). To examine the extent to which restriction factor expression correlates with the Fv3 susceptibility of these Mφ populations, we examined the expression of the same panel of restriction factor–encoding genes as above, in peritonea-derived CSF-1- and IL-34-Mφs (Fig. 2B). Of the examined genes and in comparison with peritonea-derived CSF-1-Mφs, the peritoneal IL-34-Mφs only expressed greater levels of apobec2 (Fig. 2B).
IL-34-Mφs exhibit greater gene expression of intron-containing and intronless type I and type III IFNs than CSF-1-Mφs
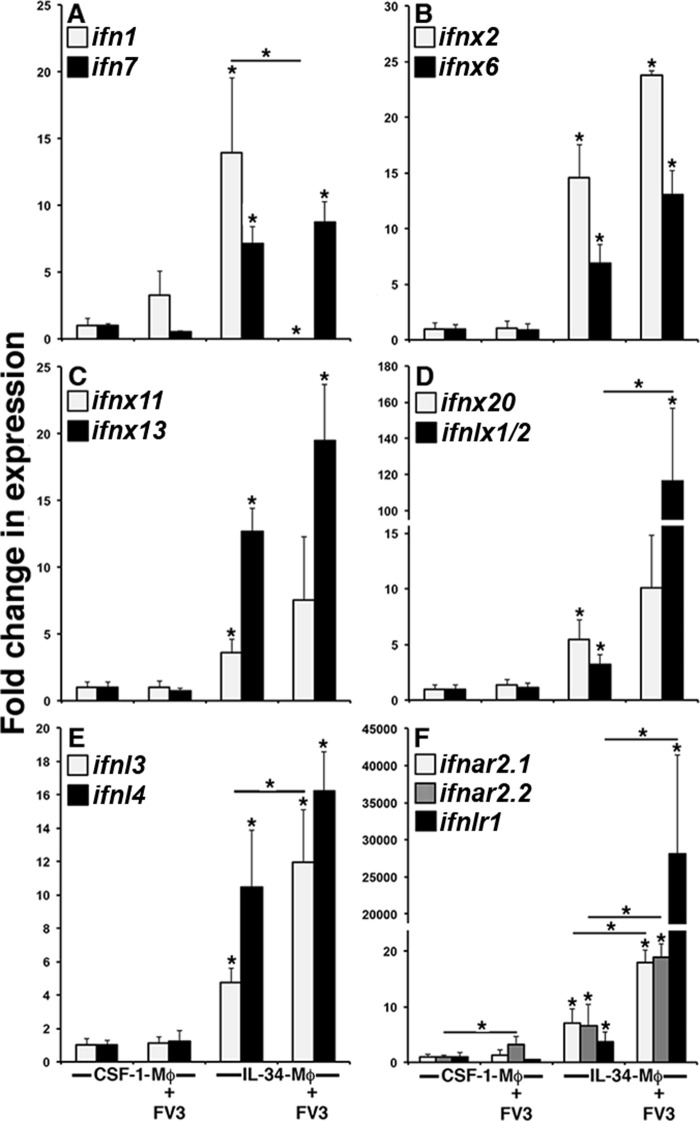
Xenopodinae encode highly expanded repertoires of phylogenetically distinct intron containing and intronless type I and type III IFNs (ifn, ifnx, ifnl, and ifnlx, respectively (17, 18)). Accordingly, to define the possible mechanisms conferring the disparities in IL-34- and CSF-1-Mφ-Fv3 resistance, we generated CSF-1- and IL-34-Mφs, challenged them with Fv3, and examined their gene expression of type I and III IFNs belonging to each of the distinct clades, described in recent reports (17, 18). In comparison with the CSF-1-Mφ cultures, IL-34-Mφs exhibited significantly greater gene expression of intron-containing type I IFNs (ifn1 and ifn7; Fig. 3A), intronless type I IFNs (ifnx2, ifnx6, ifnx11, ifnx13, and ifnx20; Fig. 3, B–D), and intron-containing and intronless type III IFNs (ifnlx1/2, ifnl3, and ifnl4, respectively; Fig. 3, D and E). Virally challenging CSF-1-Mφ did not result in significant changes to their gene expression of any of the examined IFNs (Fig. 3, A–E). Fv3 challenge completely abrogated the gene expression of IL-34-Mφ ifn1 (Fig. 3A) and had no significant affect on ifn7, ifnx2, ifnx6, ifnx11, ifnx13, ifnx20, or ifnl4 gene expression (Fig. 3, B–E). By contrast, Fv3-challenged IL-34-Mφs possessed significantly greater transcript levels of ifnl3 and ifnlx1/2 (Fig. 3, D and E, respectively).
Figure 3.
Gene expression analysis of intron-containing and intronless type I and type III IFNs (IFN, IFNX, IFNL, and IFNLX, respectively) and their receptors in CSF-1- and IL-34-Mφs. X. laevis bone marrow–derived CSF-1- and IL-34-Mφs were challenged with Fv3 and examined for gene expression of ifn1 and ifn7 (A); ifnx2 and ifnx6 (B); ifnx11 and ifnx13 (C); ifnx20 and ifnlx1/2 (D); ifnl3 and ifnl4 (E); and ifnar2.1, ifnar2.2, and ifnlr1 (F). All expression was quantified relative to the gapdh endogenous control (n = 5). Results are means ± S.E. (error bars). *, statistical difference from respective CSF-1-Mφs; * (above horizontal lines), statistical difference between the treatment groups denoted by the respective lines (p < 0.05).
IL-34-Mφs express greater levels of type I and type III IFN receptors than CSF-1-Mφs
We previously demonstrated that IL-34-Mφs are more responsive to type I IFN stimulation and inhibit Fv3 replication with greater efficiency than CSF-1-Mφs (3). The type I IFN receptor complex comprises the ligand-binding IFNAR2 chain and the signal-propagating IFNAR1 chain (12). Conversely, type III IFN receptor complex consists of a ligand-binding IFNLR1 chain and signal-propagating interleukin-10 receptor-2 (IL-10R2) chain (13, 14). Because IL-34-Mφ expressed greater levels of all examined IFN genes compared with CSF-1-Mφ (Fig. 3, A–E), we next examined whether these cells also possessed greater gene expression of type I and III IFN ligand binding chains (ifnar2 and ifnlr1, respectively; Fig. 2F). Notably, X. laevis possess two isoforms of the ifnar2 (ifnar2.1 and ifnar2.2), both of which were significantly more expressed in IL-34-Mφ than in CSF-1-Mφ (Fig. 3F). Following Fv3 challenge, IL-34-Mφs displayed a robust and significant up-regulation of their ifnar2.1, IFNAR2.2, and IFNLR1 gene expression, whereas CSF-1-Mφs exhibited a modest but significant expression increase in only the ifnar2.2 gene (Fig. 3F).
IL-34-Mφ–conditioned supernatants protect frog kidney epithelial cells from Fv3 infection
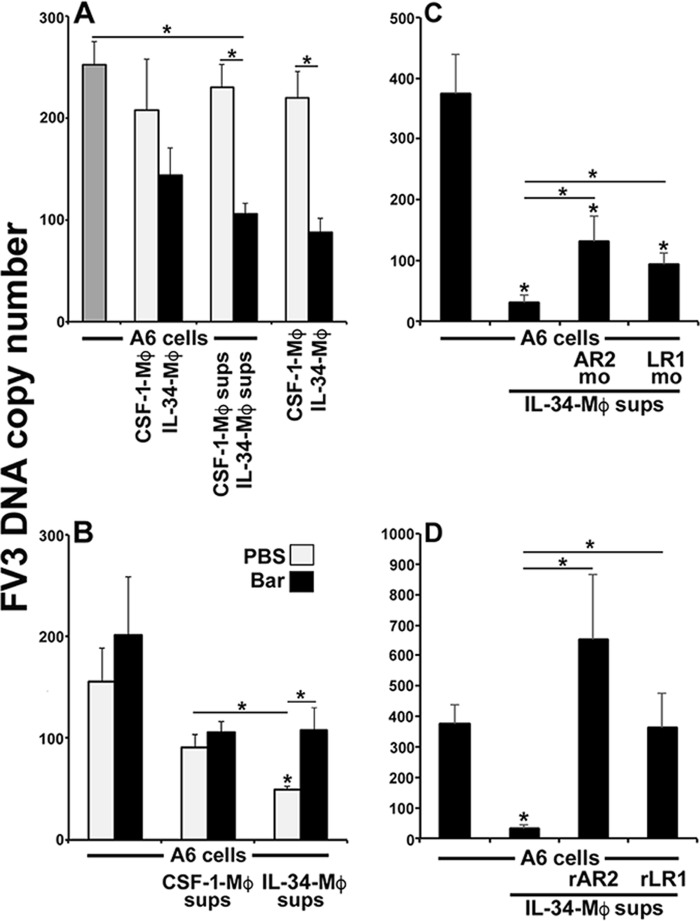
The kidneys of anuran amphibians represent a central target for Fv3 replication (30). Because our results indicate that IL-34-Mφs express greater levels of antiviral IFNs (Fig. 3, A–E), we next examined whether co-incubation of the X. laevis kidney-derived, Fv3-susceptible epithelial cell line (A6) with IL-34-Mφs or with the supernatants conditioned by these cells (and presumably containing IFNs) would grant A6 cells protection from Fv3. To this end, A6 cells were cultured with CSF-1-Mφs, IL-34-Mφs, or medium conditioned by these respective Mφ populations and infected with Fv3 for 24 h, and Fv3 DNA loads were assessed (Fig. 4A). IL-34-Mφs (cultured without A6 cells) exhibited substantially reduced Fv3 loads compared with those seen in the CSF-1-Mφ cultures (Fig. 4A), confirming that IL-34-Mφs are more resistant to Fv3 than CSF-1-Mφs. Fv3-infected A6 cells co-cultured with IL-34-Mφs exhibited modestly (not significantly) reduced Fv3 loads, compared with A6 cultured alone (Fig. 4A). Most notably, A6 cells cultured with IL-34 Mφ-derived supernatants possessed significantly reduced Fv3 loads compared with those seen in control Fv3-infected A6 cultures (Fig. 4A).
Figure 4.
Supernatants from IL-34 Mφ cultures confer anti-Fv3 protection to the frog A6 kidney epithelial cell line. A, A6 cells, cultured for 3 h with CSF-1- or IL-34-Mφs or supernatants (sups) obtained from these respective Mφ cultures were challenged with Fv3 (0.5 MOI) for 24 h, and Fv3 DNA viral loads were examined by absolute qPCR (n = 5). B, A6 cells were incubated for 3 h with medium conditioned by CSF-1- or IL-34-Mφ cultures, with or without the Jak1/Tyk2 inhibitor (Bar or PBS, respectively), and infected with Fv3 (0.5 MOI) for 24 h, and Fv3 DNA viral loads were examined by absolute qPCR (n = 5). C, A6 cells were transfected with antisense mo against ifnar2.1/2.2 or ifnlr1, incubated for 3 h with medium conditioned by IL-34-Mφs, and infected with Fv3 (0.5 MOI) for 24 h before Fv3 DNA viral load analysis (n = 5). D, A6 cells were co-incubated with recombinant extracellular domains of IFNAR2.1 (rAR2) or IFNLR1 (rLR1) and medium conditioned by IL-34-Mφs for 3 h and infected with Fv3 (0.5 MOI) for 24 h before Fv3 DNA viral load analysis (n = 5). Results are means ± S.E. (error bars). * (above horizontal lines), statistical difference between the treatment groups denoted by the respective lines (p < 0.05).
IL-34-Mφ–conditioned supernatants contain IFNs
The type I and type III IFNs of higher vertebrates signal through distinct receptor complexes but employ the same downstream signaling pathways, including the Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2) (30–32). Concomitantly, we recently showed that the antiviral effects of both X. laevis type I and type III IFNs could be pharmacologically inhibited with a Jak1/Tyk2 inhibitor, baricitinib (Bar) (21). To test our hypothesis that the anti-Fv3 A6 cell protection conferred by IL-34-Mφ–conditioned medium is due to the presence of IFNs within these supernatants, we cultured A6 cells with CSF-1-Mφ– and IL-34-Mφ–conditioned media, with or without Bar (Fig. 4B). Bar inhibition of A6 cell IFN signaling significantly abolished the antiviral protection conferred by IL-34-Mφ supernatants (Fig. 4B), supporting the notion that the anti-Fv3 protection mediated by IL-34-Mφ–derived supernatants is (at least in part) due to the presence of IFNs within these supernatants.
To confirm that the anti-Fv3 protection elicited by IL-34-Mφ supernatants was mediated by antiviral IFNs, we examined the capacity of these supernatants to protect A6 cultures subsequent to their antisense morpholino (mo) knockdown of type I or type III IFN receptor chains (both isoforms of ifnar2.1/2.2 and ifnlr1; Fig. 4C), which are responsible for binding of the respective ligands (33, 34). Pretreating A6 cells with either ifnar2-mo or ifnlr1-mo significantly (but not entirely) reduced the anti-Fv3 protection offered by the IL-34-Mφs (Fig. 4C).
As a parallel approach to confirming the antiviral IFN content of IL-34-Mφ supernatants, we produced the extracellular domains of IFNAR2 (isoform 1) and IFNLR1 in recombinant form (rAR2 and rLR1, respectively) and added these to the A6 cells cultured with IL-34-Mφ supernatants to bind up any type I or III IFNs, thereby ablating any protection offered by these cytokines (Fig. 4D). The addition of either rAR2 or rLR1 resulted in complete abolishment of the anti-FV3 protection offered by the IL-34-Mφ supernatants to A6 cultures (Fig. 4D). Culturing A6 cells with either rAR2 or rLR1 alone had no effect on their Fv3 susceptibility (data not shown).
CSF-1- and IL-34-Mφs may be counterpolarized
To discern whether CSF-1 and IL-34 could be functionally counterpolarized with IL-34 and CSF-1, respectively, CSF-1- and IL-34-Mφs were differentiated for 5 days and counterstimulated for 24 h with IL-34 or CSF-1, respectively, and their IFN ligand and receptor gene expression was examined (Fig. 5, A and B). Akin to our earlier observations (Fig. 3, A–E), IL-34-Mφ exhibited consistently greater expression of all examined IFNs (ifn7, ifnx13, ifnlx1/2, and ifnl4) as compared with CSF-1-Mφ (Fig. 5A). Notably, CSF-1-Mφ stimulated with IL-34 as well as IL-34-Mφs treated with CSF-1 possessed ifn7, ifnx13, ifnlx1/2, and IFNL4 transcript levels comparable with IL-34-Mφs (Fig. 5A). Both CSF-1-Mφs stimulated with IL-34 and IL-34-Mφs treated with CSF-1 had significantly greater ifnar2.1 (but not ifnar2.2) gene expression levels than those observed in IL-34- or CSF-1-Mφs (Fig. 5B). By contrast, whereas the CSF-1-Mφ stimulated with IL-34 had ifnlr1 transcript levels similar to those detected in IL-34-Mφs, IL-34-Mφs stimulated with CSF-1 had drastically decreased expression levels of this gene compared with IL-34-Mφs (Fig. 5B).
Figure 5.
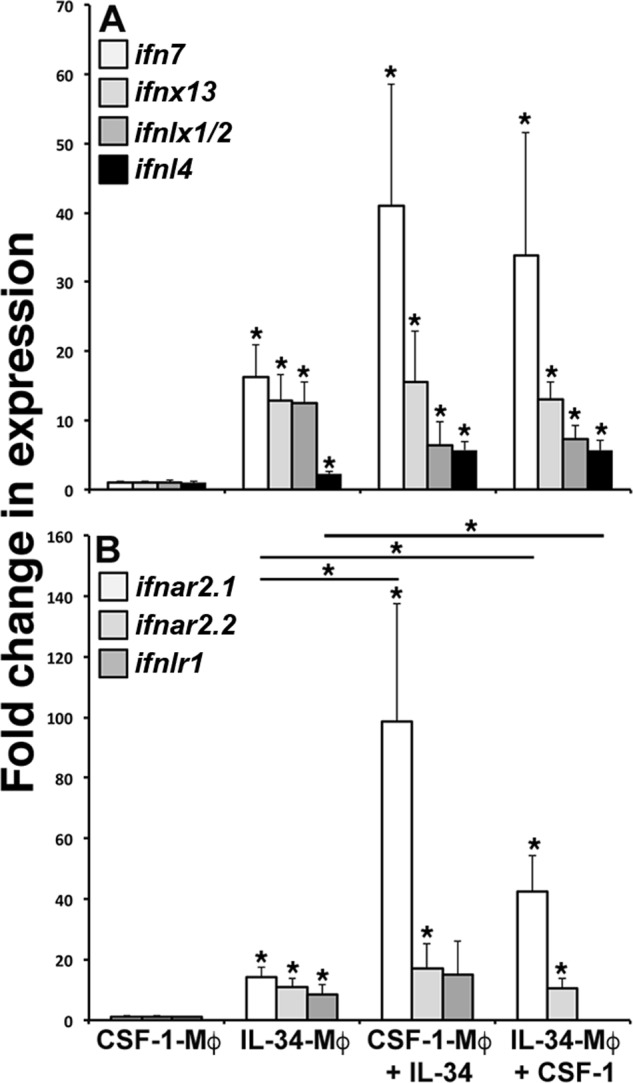
CSF-1-Mφs may be counterpolarized with IL-34. CSF-1- and IL-34-Mφs were cultured in vitro for 5 days and stimulated with reciprocal cytokines (IL-34 and CSF-1, respectively) for 24 h. Mφ cultures were examined for gene expression of ifn7, ifnx13, ifnlx1/2, and ifnl4 (A) and ifnar2.1, ifnar2.2, and ifnlr1 (B). All expression was quantified relative to the gapdh endogenous control (n = 5). Results are means ± S.E. (error bars). *, statistical difference from respective CSF-1-Mφs; * (above horizontal lines), statistical difference between the treatment groups denoted by the respective lines (p < 0.05).
Discussion
Mφs are integral to the physiology and immune defenses of all vertebrate species. Perhaps thus, many pathogens have coevolved to evade Mφ immunity and utilize these cells toward their infection strategies. Ranaviruses such as Fv3 represent ecologically and commercially relevant viral pathogens that have espoused this strategy to thrive within their cold-blooded vertebrate hosts (3). Fv3 infects, persists, and disseminates within amphibian Mφs (3, 35, 36). Whereas Mφs differentiated by CSF-1 are highly permissive to this Fv3, IL-34-Mφs not only appear to be resistant to this virus, but may also represent a crucial effector subset in the defense against this pathogen (5, 6). Presently, we show that the means of these cells' resistance to Fv3 infection and replication reflect their heightened expression of antiviral restriction factors, whereas their protective properties are attributed at least in part to their production of antiviral interferon cytokines.
Because the peritonea-derived, Fv3-resistant IL-34-Mφs only exhibit greater expression of apobec2 than peritoneal CSF-1-Mφs, this restriction factor may be particularly important to anti-Fv3 immunity. However, we emphasize that the differentiation of immune cells into distinct effectors results in lineage/subset-specific activation of unique and often non-overlapping gene regulatory networks (37, 38). In turn, it is this coordinated activation of specific repertoires of genes that grants those specialized cell types their respective functionally distinct capacities (37, 38). We postulate that the functional differences between CSF-1- and IL-34-Mφs reflect their unique Mφ subset “programing.” In turn, we propose that Mφs differentiated by IL-34 are polarized toward a broad antiviral state through their expression of coordinated networks of genes related to antiviral defenses. Moreover, we suspect that the repertoires of these expressed genes far exceed those examined here, and the resistance of IL-34-Mφs to Fv3 is conferred by potentially redundant or otherwise complementary mechanisms. This is supported by our observations that, while being highly resistant to Fv3 (6), frog peritonea-derived IL-34-Mφs only express one of a subset of the (examined) restriction factors expressed by the bone marrow IL-34-Mφs, as compared with the respective CSF-1-Mφs. Moreover, we have previously shown that an elevated respiratory burst capacity of IL-34-Mφs may also contribute to their Fv3 resistance (6). Frogs presumably possess multiple distinct subsets of IL-34-Mφs, all of which are presumably resistant to Fv3, but as the result of at least partially non-overlapping antiviral mechanisms. The antiviral efficacies of these frog IL-34-Mφ subsets may reflect their heightened production of and autocrine stimulation by IFNs, their expression of antiviral genes (such as restriction factors, IFN-stimulated genes), and their robust respiratory burst responses and more than likely result from other immunological parameters that are not considered here.
Many aspects of Mφ biology are interdependent on the activation of the colony-stimulating factor-1 receptor (39, 40). Considering the array of functionally disparate Mφ subsets described across vertebrates, it is intuitive that some of this heterogeneity would stem from differential ligation of this receptor. CSF-1 was known to add to this Mφ functional diversity (41), and the more recently discovered IL-34 cytokine appears to likewise aggrandize the Mφ functional spectrum. The roles of IL-34 in vertebrate immunity appear to be complex and multifaceted, as this cytokine has been linked to the development of Langerhans cells (42, 43), osteoclasts (44, 45), B cell–stimulating myeloid cells (46), and microglia (42). Moreover, IL-34 is produced by fibroblasts and synoviocytes of patients with rheumatoid arthritis (47), and its absence is correlated with reduced synaptic loss and less severe associated memory impairment in West Nile virus–infected mice (48), suggesting that IL-34-Mφs have pro-inflammatory roles. The IL-34 produced during inflammatory responses has also been proposed to have anti-inflammatory roles (49). This is supported by the recent observation that IL-34 is produced by T regulatory cells and contributes to transplant tolerance (50). Our work using the amphibian X. laevis-Fv3 infection model indicates that the frog IL-34 is crucial to the development of an antiviral Mφ subset, and the absence of proper IL-34 production by amphibian tadpoles may be an important underlying cause of their greater susceptibility to this pathogen (5). It will be interesting to learn whether the antiviral properties of IL-34–derived Mφs are conserved across vertebrate species or represent an amphibian phenomenon.
Our results clearly indicate that IL-34-Mφs are significantly more resistant to Fv3 infection and viral replication than CSF-1-Mφs. We propose that a significant source of this resistance is the IL-34-Mφ expression of antiviral restriction factors. Notably, cellular resistance to viral replication is marked by expression of antiviral restriction factors (24–27), such as those up-regulated by IL-34-Mφs and including apobec (51) and trim (52) family members, samhd1 (53), and ctcf (54). Each of these gene products represents a barrier at a distinct step in viral replication, so the relative magnitudes of CSF-1- and IL-34-Mφ expression of these genes probably contribute to their respective susceptibility and resistance to Fv3.
It is noteworthy that whereas CSF-1-Mφs supported greater expression of Fv3 IE, E, and L genes, these cells had particularly high expression of the Fv3 93L late gene compared with IL-34-Mφs. Although the functional roles of the 93L gene product await elucidation, it is tempting to speculate that the resulting protein may have an important role in immune evasion and/or viral dissemination. It will be interesting to learn with further studies whether CSF-1-Mφ susceptibility to Fv3 is somehow related to the elevated 93L gene expression within these cells.
Our past work indicates that administration of IL-34 to tadpoles and adult frogs confers significant anti-Fv3 protection and reduces animal viral loads (5, 6), whereas our present findings indicate that IL-34-Mφ–conditioned supernatants possess factor(s) that render the frog A6 kidney epithelial line considerably more resistant to Fv3. Concomitantly, type I and type III IFNs are key mediators of vertebrate antiviral defenses (7, 9–11), whereas amphibians encode highly expanded repertoires of phylogenetically distinct type I and type III IFNs (17, 18). In this respect, it is particularly compelling that IL-34-Mφs express significantly greater levels of type I and III IFN genes belonging to these diverse clades as well as their cognate receptors. Moreover, it appears that the A6 cell anti-Fv3 protection offered by IL-34–conditioned medium depends on IFN signaling, supporting our hypothesis that this Mφ population offers antiviral protection by production of these antiviral cytokines. It will be interesting to learn the roles of the respective IFNs in amphibian antiviral immunity and the in vivo contexts in which IL-34-Mφs produce these mediators.
The lack of X. laevis CSF-1- and IL-34-Mφ–specific markers has precluded the investigation of the roles of these cells in vivo. However, it is notable that IL-34 gene expression is higher than that of CSF-1 in the frog spleen tissues (5), suggesting that IL-34 may be more prominently involved in the frog innate and possibly adaptive immune defenses. The tadpoles of anuran amphibians are more susceptible to and die from Fv3, whereas adult frogs also suffer from productive infections but are usually able to clear these pathogens (19, 55–58). The tadpole and adult frog kidneys are a central site of Fv3 replication (35, 36), and during FV3 infections, adult frogs up-regulate their kidney expression of both csf-1 and il-34 genes, whereas Fv3-challenged tadpoles increase their kidney csf-1 but not il-34 gene expression (5). Thus, the adult frogs presumably produce both the Fv3-susceptible CSF-1-Mφs and the antiviral IL-34-Mφ in response to viral challenge, whereas the failure of tadpoles to mount an IL-34 response and hence generate IL-34-Mφs may contribute to their Fv3 susceptibility.
Whereas Mφs represent the foundation of vertebrate immunity, the mechanisms controlling the ontogeny of the many functionally disparate Mφ subsets seen across vertebrata await elucidation. Presently, we demonstrate that CSF-1- and IL-34-Mφ growth factors contribute to this functional spectrum by producing virally susceptible and resistant effector subsets. We believe that further insight into the functional dichotomies of CSF-1- and IL-34-Mφs will not only permit greater understanding of the facets of amphibian susceptibility and resistance to ranaviruses, but will also grant a more comprehensive view of vertebrate Mφ-pathogen interactions.
Experimental procedures
Animals
Out-bred adult X. laevis were purchased from Xenopus1 and housed and handled under strict laboratory and institutional animal care and use committee regulations.
Recombinant cytokines and cytokine receptors, A6 cell and Mφ culture media, and conditions
The production of recombinant X. laevis cytokines, isolation and culture of bone marrow Mφs, and maintenance of A6 cells were performed as described previously (6, 20, 59). The generation of recombinant extracellular domains of IFNAR2 and IFNLR1 was achieved using the same methodology described previously (6, 20, 59). Bone-marrow IL-34- and CSF-1-Mφ were generated by incubating 104 X. laevis bone marrow cells with 250 ng/ml recombinant CSF-1 or IL-34 in individual wells of 96-well plates for 5 days, with a second respective cytokine treatment added on the third day of culture. During reciprocal differentiation experiments, Mφs were differentiated with CSF-1 or IL-34 as above and on the fifth day of culture, washed, and incubated with 250 ng/ml of the reciprocal cytokine for an additional 24 h. For A6 cell protection experiments, 104 A6 cells were incubated with CSF-1 or IL-34 Mφs or supernatants derived from these cultures for 3 h before a 24-h Fv3 challenge (multiplicity of infection (MOI) = 0.5). Peritonea-derived IL-34- and CSF-1-Mφs were generated as described previously (6).
Morpholino targeting of IFNAR1 and IFNLR2
Translation-blocking mo, specific for the X. laevis ifnar2 (mo 5′-CAGCAGAAGACACAGCCCTGCCATG-3′; targets both isoforms 2.1 and 2.2) and ifnlr1 (5′-GCCCAGGTGAACCAGACAGACATAT-3′) were designed by and purchased from GeneTools, LLC and introduced into A6 cells using the Endo-Porter PEG reagent according to the manufacturer's directions (GeneTools, LLC).
Quantitative gene expression analysis
RNA and DNA isolation, cDNA synthesis, Mφ and Fv3 gene expression, and Fv3 DNA load analyses were performed as described previously. All primers were validated before use, and all primer sequences are listed in Table S1.
Statistical analysis
All data were examined using analysis of variance and Student's t test (Prism, GraphPad Software, La Jolla, CA); p < 0.05.
Author contributions
A.Y., M.P., and L.G. designed and performed all of the experiments and analyzed the results. A.Y. and L.G. wrote the manuscript and prepared the figures.
Supplementary Material
This work was supported by start-up funding from George Washington University (to L. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
- Fv3
- frog virus 3
- CSF-1
- colony-stimulating factor-1
- IFN
- interferon
- IL
- interleukin
- Mφ
- macrophage
- Bar
- baricitinib
- mo
- morpholino
- MOI
- multiplicity of infection
- qPCR
- quantitative PCR.
References
- 1. Carey C., Cohen N., and Rollins-Smith L. (1999) Amphibian declines: an immunological perspective. Dev. Comp. Immunol. 23, 459–472 10.1016/S0145-305X(99)00028-2 [DOI] [PubMed] [Google Scholar]
- 2. Daszak P., Berger L., Cunningham A. A., Hyatt A. D., Green D. E., and Speare R. (1999) Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5, 735–748 10.3201/eid0506.990601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grayfer L., Andino Fde J., Chen G., Chinchar G. V., and Robert J. (2012) Immune evasion strategies of ranaviruses and innate immune responses to these emerging pathogens. Viruses 4, 1075–1092 10.3390/v4071075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grayfer L., and Robert J. (2016) Amphibian macrophage development and antiviral defenses. Dev. Comp. Immunol. 58, 60–67 10.1016/j.dci.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grayfer L., and Robert J. (2014) Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. J. Leukoc. Biol. 96, 1143–1153 10.1189/jlb.4A0614-295R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grayfer L., and Robert J. (2015) Distinct functional roles of amphibian (Xenopus laevis) colony-stimulating factor-1- and interleukin-34-derived macrophages. J. Leukoc. Biol. 98, 641–649 10.1189/jlb.4AB0315-117RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang T., Hanington P. C., Belosevic M., and Secombes C. J. (2008) Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J. Immunol. 181, 3310–3322 10.4049/jimmunol.181.5.3310 [DOI] [PubMed] [Google Scholar]
- 8. Pixley F. J., and Stanley E. R. (2004) CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14, 628–638 10.1016/j.tcb.2004.09.016 [DOI] [PubMed] [Google Scholar]
- 9. Hanington P. C., Wang T., Secombes C. J., and Belosevic M. (2007) Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J. Biol. Chem. 282, 31865–31872 10.1074/jbc.M706278200 [DOI] [PubMed] [Google Scholar]
- 10. Garceau V., Smith J., Paton I. R., Davey M., Fares M. A., Sester D. P., Burt D. W., and Hume D. A. (2010) Pivotal advance: avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J. Leukoc. Biol. 87, 753–764 10.1189/jlb.0909624 [DOI] [PubMed] [Google Scholar]
- 11. Belosevic M., Hanington P. C., and Barreda D. R. (2006) Development of goldfish macrophages in vitro. Fish Shellfish Immunol. 20, 152–171 10.1016/j.fsi.2004.10.010 [DOI] [PubMed] [Google Scholar]
- 12. Droin N., and Solary E. (2010) Editorial: CSF1R, CSF-1, and IL-34, a “menage a trois” conserved across vertebrates. J. Leukoc. Biol. 87, 745–747 10.1189/jlb.1209780 [DOI] [PubMed] [Google Scholar]
- 13. Lin H., Lee E., Hestir K., Leo C., Huang M., Bosch E., Halenbeck R., Wu G., Zhou A., Behrens D., Hollenbaugh D., Linnemann T., Qin M., Wong J., Chu K., et al. (2008) Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 320, 807–811 10.1126/science.1154370 [DOI] [PubMed] [Google Scholar]
- 14. Wei S., Nandi S., Chitu V., Yeung Y. G., Yu W., Huang M., Williams L. T., Lin H., and Stanley E. R. (2010) Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 88, 495–505 10.1189/jlb.1209822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sadler A. J., and Williams B. R. (2008) Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou J., and Secombes C. J. (2011) Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 35, 1376–1387 10.1016/j.dci.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 17. Sang Y., Liu Q., Lee J., Ma W., McVey D. S., and Blecha F. (2016) Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity. Sci. Rep. 6, 29072 10.1038/srep29072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gan Z., Chen S. N., Huang B., Hou J., and Nie P. (2017) Intronless and intron-containing type I IFN genes coexist in amphibian Xenopus tropicalis: insights into the origin and evolution of type I IFNs in vertebrates. Dev. Comp. Immunol. 67, 166–176 10.1016/j.dci.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 19. Grayfer L., De Jesús Andino F., and Robert J. (2014) The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. J. Virol. 88, 5766–5777 10.1128/JVI.00223-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grayfer L., De Jesús Andino F., and Robert J. (2015) Prominent amphibian (Xenopus laevis) tadpole type III interferon response to the frog virus 3 ranavirus. J. Virol. 89, 5072–5082 10.1128/JVI.00051-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wendel E. S., Yaparla A., Koubourli D. V., and Grayfer L. (2017) Amphibian (Xenopus laevis) tadpoles and adult frogs mount distinct interferon responses to the Frog Virus 3 ranavirus. Virology 503, 12–20 10.1016/j.virol.2017.01.001 [DOI] [PubMed] [Google Scholar]
- 22. Shi K., Carpenter M. A., Banerjee S., Shaban N. M., Kurahashi K., Salamango D. J., McCann J. L., Starrett G. J., Duffy J. V., Demir Ö., Amaro R. E., Harki D. A., Harris R. S., and Aihara H. (2017) Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 24, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolf D., Hug K., and Goff S. P. (2008) TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 12521–12526 10.1073/pnas.0805540105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Badia R., Angulo G., Riveira-Muñoz E., Pujantell M., Puig T., Ramirez C., Torres-Torronteras J., Marti R., Pauls E., Clotet B., Ballana E., and Esté J. A. (2016) Inhibition of herpes simplex virus type 1 by the CDK6 inhibitor PD-0332991 (palbociclib) through the control of SAMHD1. J. Antimicrob. Chemother. 71, 387–394 10.1093/jac/dkv363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ballana E., and Esté J. A. (2015) SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol. 23, 680–692 10.1016/j.tim.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 26. Lang F., Li X., Vladimirova O., Hu B., Chen G., Xiao Y., Singh V., Lu D., Li L., Han H., Wickramasinghe J. M., Smith S. T., Zheng C., Li Q., Lieberman P. M., et al. (2017) CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci. Rep. 7, 39861 10.1038/srep39861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li D. J., Verma D., Mosbruger T., and Swaminathan S. (2014) CTCF and Rad21 act as host cell restriction factors for Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog. 10, e1003880 10.1371/journal.ppat.1003880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dwyer A. R., Mouchemore K. A., Steer J. H., Sunderland A. J., Sampaio N. G., Greenland E. L., Joyce D. A., and Pixley F. J. (2016) Src family kinase expression and subcellular localization in macrophages: implications for their role in CSF-1-induced macrophage migration. J. Leukoc. Biol. 100, 163–175 10.1189/jlb.2A0815-344RR [DOI] [PubMed] [Google Scholar]
- 29. Grayfer L., Hanington P. C., and Belosevic M. (2009) Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol. 26, 406–413 10.1016/j.fsi.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 30. Donnelly R. P., and Kotenko S. V. (2010) Interferon-λ: a new addition to an old family. J. Interferon Cytokine Res. 30, 555–564 10.1089/jir.2010.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durbin R. K., Kotenko S. V., and Durbin J. E. (2013) Interferon induction and function at the mucosal surface. Immunol. Rev. 255, 25–39 10.1111/imr.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kotenko S. V. (2011) IFN-λs. Curr. Opin. Immunol. 23, 583–590 10.1016/j.coi.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uzé G., and Monneron D. (2007) IL-28 and IL-29: newcomers to the interferon family. Biochimie 89, 729–734 10.1016/j.biochi.2007.01.008 [DOI] [PubMed] [Google Scholar]
- 34. Uzé G., Schreiber G., Piehler J., and Pellegrini S. (2007) The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 316, 71–95 [DOI] [PubMed] [Google Scholar]
- 35. Morales H. D., and Robert J. (2007) Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J. Virol. 81, 2240–2248 10.1128/JVI.01104-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robert J., Abramowitz L., Gantress J., and Morales H. D. (2007) Xenopus laevis: a possible vector of Ranavirus infection? J. Wildl. Dis. 43, 645–652 10.7589/0090-3558-43.4.645 [DOI] [PubMed] [Google Scholar]
- 37. Laslo P., Pongubala J. M., Lancki D. W., and Singh H. (2008) Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin. Immunol. 20, 228–235 10.1016/j.smim.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 38. Singh H., Khan A. A., and Dinner A. R. (2014) Gene regulatory networks in the immune system. Trends Immunol. 35, 211–218 10.1016/j.it.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 39. Lichanska A. M., Browne C. M., Henkel G. W., Murphy K. M., Ostrowski M. C., McKercher S. R., Maki R. A., and Hume D. A. (1999) Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1. Blood 94, 127–138 [PubMed] [Google Scholar]
- 40. Guilbert L. J., and Stanley E. R. (1980) Specific interaction of murine colony-stimulating factor with mononuclear phagocytic cells. J. Cell Biol. 85, 153–159 10.1083/jcb.85.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamilton T. A., Zhao C., Pavicic P. G. Jr., and Datta S. (2014) Myeloid colony-stimulating factors as regulators of macrophage polarization. Front. Immunol. 5, 554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greter M., Lelios I., Pelczar P., Hoeffel G., Price J., Leboeuf M., Kündig T. M., Frei K., Ginhoux F., Merad M., and Becher B. (2012) Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060 10.1016/j.immuni.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., and Colonna M. (2014) Interkeukin-34, a cytokine crucial for the differentiation and maintenance of tissue resident macrophages and Langerhans cells. Eur. J. Immunol. 44, 1575–1581 10.1002/eji.201344365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baud'huin M., Renault R., Charrier C., Riet A., Moreau A., Brion R., Gouin F., Duplomb L., and Heymann D. (2010) Interleukin-34 is expressed by giant cell tumours of bone and plays a key role in RANKL-induced osteoclastogenesis. J. Pathol. 221, 77–86 10.1002/path.2684 [DOI] [PubMed] [Google Scholar]
- 45. Chen Z., Buki K., Vääräniemi J., Gu G., and Väänänen H. K. (2011) The critical role of IL-34 in osteoclastogenesis. PLoS One 6, e18689 10.1371/journal.pone.0018689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamane F., Nishikawa Y., Matsui K., Asakura M., Iwasaki E., Watanabe K., Tanimoto H., Sano H., Fujiwara Y., Stanley E. R., Kanayama N., Mabbott N. A., Magari M., and Ohmori H. (2014) CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34. J. Leukoc. Biol. 95, 19–31 10.1189/jlb.0613311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chemel M., Le Goff B., Brion R., Cozic C., Berreur M., Amiaud J., Bougras G., Touchais S., Blanchard F., Heymann M. F., Berthelot J. M., Verrecchia F., and Heymann D. (2012) Interleukin 34 expression is associated with synovitis severity in rheumatoid arthritis patients. Ann. Rheum. Dis. 71, 150–154 10.1136/annrheumdis-2011-200096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vasek M. J., Garber C., Dorsey D., Durrant D. M., Bollman B., Soung A., Yu J., Perez-Torres C., Frouin A., Wilton D. K., Funk K., DeMasters B. K., Jiang X., Bowen J. R., Mennerick S., Robinson J. K., Garbow J. R., Tyler K. L., Suthar M. S., Schmidt R. E., Stevens B., and Klein R. S. (2016) A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature 534, 538–543 10.1038/nature18283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guillonneau C., Bézie S., and Anegon I. (2017) Immunoregulatory properties of the cytokine IL-34. Cell Mol Life Sci. 74, 2569–2586 10.1007/s00018-017-2482-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bézie S., Picarda E., Ossart J., Tesson L., Usal C., Renaudin K., Anegon I., and Guillonneau C. (2015) IL-34 is a Treg-specific cytokine and mediates transplant tolerance. J. Clin. Invest. 125, 3952–3964 10.1172/JCI81227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trang K., Raposo R. A., Lowe M. M., Krow-Lucal E. R., Yonemoto W., Cabido V. D., SenGupta D., and McCune J. M. (2017) Relative mRNA expression levels of restriction factors and antiviral genes in fetal and adult human monocytes and monocyte-derived macrophages. Viral Immunol. 30, 142–148 10.1089/vim.2016.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nisole S., Stoye J. P., and Saïb A. (2005) TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3, 799–808 10.1038/nrmicro1248 [DOI] [PubMed] [Google Scholar]
- 53. Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W., Hofmann H., Kim B., Lindemann D., and Landau N. R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 10.1186/1742-4690-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pentland I., and Parish J. L. (2015) Targeting CTCF to control virus gene expression: a common theme amongst diverse DNA viruses. Viruses 7, 3574–3585 10.3390/v7072791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bayley A. E., Hill B. J., and Feist S. W. (2013) Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Dis. Aquat. Organ 103, 171–183 10.3354/dao02574 [DOI] [PubMed] [Google Scholar]
- 56. Hoverman J. T., Gray M. J., and Miller D. L. (2010) Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis. Aquat. Organ. 89, 97–107 10.3354/dao02200 [DOI] [PubMed] [Google Scholar]
- 57. Landsberg J. H., Kiryu Y., Tabuchi M., Waltzek T. B., Enge K. M., Reintjes-Tolen S., Preston A., and Pessier A. P. (2013) Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, U.S.A. Dis. Aquat. Organ. 105, 89–99 10.3354/dao02625 [DOI] [PubMed] [Google Scholar]
- 58. Reeve B. C., Crespi E. J., Whipps C. M., and Brunner J. L. (2013) Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica). Ecohealth 10, 190–200 10.1007/s10393-013-0834-6 [DOI] [PubMed] [Google Scholar]
- 59. Yaparla A., Wendel E. S., and Grayfer L. (2016) The unique myelopoiesis strategy of the amphibian Xenopus laevis. Dev. Comp. Immunol. 63, 136–143 10.1016/j.dci.2016.05.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.