Abstract
Acid-sensing ion channels (ASICs) form both homotrimeric and heterotrimeric ion channels that are activated by extracellular protons and are involved in a wide range of physiological and pathophysiological processes, including pain and anxiety. ASIC proteins can form both homotrimeric and heterotrimeric ion channels. The ASIC3 subunit has been shown to be of particular importance in the peripheral nervous system with pharmacological and genetic manipulations demonstrating a role in pain. Naked mole-rats, despite having functional ASICs, are insensitive to acid as a noxious stimulus and show diminished avoidance of acidic fumes, ammonia, and carbon dioxide. Here we cloned naked mole-rat ASIC3 (nmrASIC3) and used a cell-surface biotinylation assay to demonstrate that it traffics to the plasma membrane, but using whole-cell patch clamp electrophysiology we observed that nmrASIC3 is insensitive to both protons and the non-proton ASIC3 agonist 2-guanidine-4-methylquinazoline. However, in line with previous reports of ASIC3 mRNA expression in dorsal root ganglia neurons, we found that the ASIC3 antagonist APETx2 reversibly inhibits ASIC-like currents in naked mole-rat dorsal root ganglia neurons. We further show that like the proton-insensitive ASIC2b and ASIC4, nmrASIC3 forms functional, proton-sensitive heteromers with other ASIC subunits. An amino acid alignment of ASIC3s between 9 relevant rodent species and human identified unique sequence differences that might underlie the proton insensitivity of nmrASIC3. However, introducing nmrASIC3 differences into rat ASIC3 (rASIC3) produced only minor differences in channel function, and replacing the nmrASIC3 sequence with that of rASIC3 did not produce a proton-sensitive ion channel. Our observation that nmrASIC3 forms nonfunctional homomers may reflect a further adaptation of the naked mole-rat to living in an environment with high-carbon dioxide levels.
Keywords: acid sensing ion channel (ASIC), ion channel, neuron, neurophysiology, pain
Introduction
Acid-sensing ion channels (ASICs)5 are part of the epithelial sodium channel/degenerin superfamily of ion channels and are implicated in a diverse range of physiological and pathophysiological processes, ranging from learning and memory to mechanosensation and pain (1). In mammals, there are 4 ASIC encoding genes, which generate 6 distinct ASIC subunits due to splice variants in the ACCN2 and ACCN1 genes producing a and b variants of the ASIC1 and ASIC2 subunits, respectively: ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3, and ASIC4. The crystal structure of ASIC1 demonstrated that ASICs form trimeric ion channels (2) and although evidence exists for the formation of ASIC/epithelial sodium channel heteromers (3, 4), it is largely thought that functional ASICs are the result of either homo- or heterotrimeric arrangement of ASIC subunits.
Unlike transient receptor potential vanilloid 1 (TRPV1) that produces a sustained inward current in response to extracellular protons (5), ASICs produce a transient inward current (6). However, being trimeric, the subunit configuration dictates the biophysical characteristics, such as the proton sensitivity for activation, the inactivation time constant, and the magnitude of the sustained current in subunit configurations where the current does not completely inactivate in the continued presence of agonist (7). Moreover, the sensitivity to different antagonists is also affected by subunit configuration. For example, the ASIC3 antagonist APETx2 inhibits ASIC3 homomers, as well as heteromers of ASIC3 with ASIC1a, ASIC1b, and ASIC2b, but does not inhibit ASIC2a + ASIC3 heteromers (8); APETx2 also relieves hyperalgesia in inflammatory pain models (9, 10). Furthermore, the non-proton ASIC3 agonist 2-guanidine-4-methylquinazoline (GMQ) causes ASIC3 activation at neutral pH (11), but also modulates the acid sensitivity of other ASIC subunits (12).
Of the 6 ASIC subunits, neither ASIC2b (13) nor ASIC4 (14, 15) form functional homomers, but they are able to form functional heteromers and modulate channel function (7, 13, 16), as can ASIC subunits that have been mutated to make them insensitive to protons as homomers (17, 18). The crystal structure of chicken ASIC1 (cASIC1) identified an acidic pocket region containing three carboxylate pairs (Asp238–Asp350, Glu239–Asp346, and Glu220–Asp408; cASIC1 numbering), which was suggested to be the primary site for proton sensing by ASICs (2). However, ASIC2a lacks Asp350 and is still functional, whereas ASIC2b also only lacks the Asp350 carboxylate of the acidic pocket and is not activated by protons (13, 17), results suggest that regions outside of the acidic pocket must be important for proton activation of ASICs. Indeed, we and others have identified a range of residues on ASIC1a and ASIC2a that when mutated alter proton sensitivity (17–21) and more recently we have shown that the first 87 amino acids of the extracellular domain of rat ASIC2a are required for its proton sensitivity (22).
Understanding the structure-function of ASIC3 is of particular interest because there is substantial evidence supporting involvement of ASIC3 in pain (9, 23–28), as well itch (29), mechanosensation (23, 30), and anxiety (31). Although ASIC3 expression was initially thought to be restricted to the peripheral nervous system (and hence its original name, dorsal root ganglia acid-sensing ion channel (DRASIC) (32)), we and others have demonstrated that ASIC3 is also expressed in both the spinal cord and numerous brain regions (33, 34), which makes it even more important to understand how ASIC3 functions because any potential drug that targets ASIC3 for the treatment of pain may also produce side effects with the central nervous system.
Here we investigated the function of ASIC3 cloned from the naked mole-rat, a species that we have previously shown to produce no behavioral response to acid as a noxious stimulus (35). This behavioral indifference to acid is not due to a lack of ASIC-like or TRPV1-like proton-gated currents in sensory neurons, but rather due to an amino acid variation in the voltage-gated Na+ channel NaV1.7 that confers enhanced acid block, such that acid acts like an anesthetic, rather than activator of naked mole-rat sensory neurons (36). This is likely a result of adaptation to living in a hypercapnic environment that may induce tissue acidosis (37) and indeed naked mole-rats also show reduced avoidance of ammonia and acid fumes (38, 39), as well as decreased aversion to CO2, absence of CO2-induced pulmonary edema, and enhanced ability to buffer against CO2-induced systemic acidosis (40). Although naked mole-rats can more efficiently buffer CO2, our previous data regarding the inability of acid to evoke action potentials in an ex vivo preparation (35) that arises from an amino acid variation in NaV1.7 (36) demonstrates that there are likely multiple adaptations to living in a hypercapnic environment. We recently compared ASIC expression between mice and naked mole-rats, finding that although ASIC4 is highly abundant in mouse tissues it is the most lowly expressed ASIC transcript in naked mole-rat tissues; the expression pattern of ASIC3 was similar between species (33). At a functional level, we have previously shown that naked mole-rat ASIC1a, ASIC1b, and TRPV1 are largely indistinguishable from the mouse orthologs (36, 41) and here we set out to explore the function of naked mole-rat ASIC3 considering its importance to a physiological and pathophysiological processes.
Results
Naked mole-rat ASIC3 is insensitive to protons
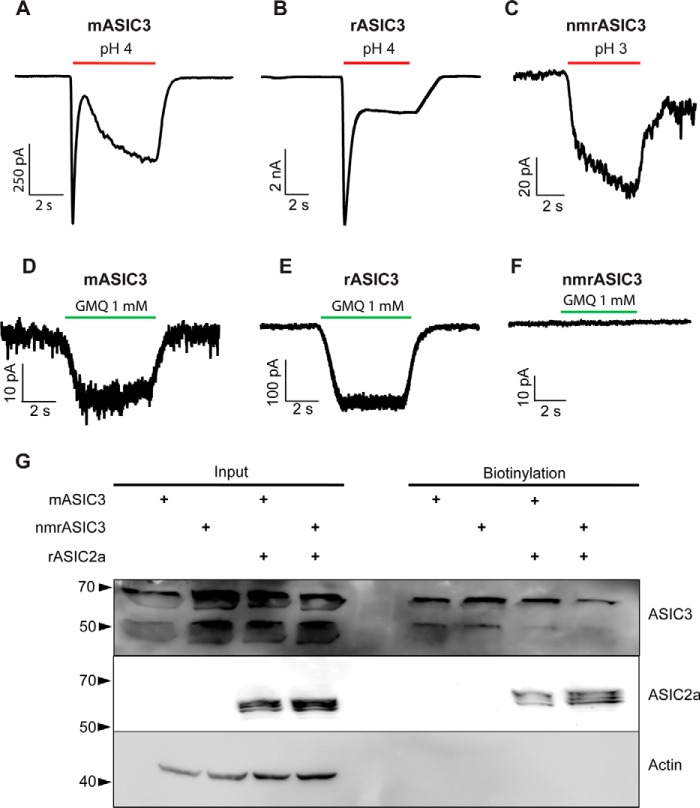
Primers for cloning mouse ASIC3 (mASIC3) and naked mole-rat ASIC3 (nmrASIC3) were designed based upon the published genome sequences and constructs were made using pIRES2-EGFP or pTarget vectors; rat ASIC3 (rASIC3 in pTracer) was a kind gift from G. Lewin. All constructs were expressed in Chinese hamster ovary (CHO) cells that lack endogenous ASICs (17), and whole-cell patch clamp electrophysiology was used to measure responses to a pH 4.0 stimulus from a starting pH of pH 7.4. Although mASIC3 and rASIC3 robustly responded to protons with a stereotypical transient ASIC-like current (mASIC3: 69 ± 13 pA/pF, n = 26, Fig. 1A, and rASIC3: 627 ± 92 pA/pF, n = 18, Fig. 1B), nmrASIC3 failed to respond with an ASIC-like response to protons, even using a pH 3.0 stimulus, but rather produced a very small, non-inactivating response, similar to that which we have observed previously in non-transfected CHO cells (17) and in CHO cells transfected with the proton-insensitive ASIC2b (22) (pH 4.0, n = 24 and pH 3.0, n = 13, 7 separate transfections, Fig. 1C); a summary of all data are given in Table 1. We also investigated whether the non-proton ASIC3 agonist GMQ could activate nmrASIC3, but whereas we observed GMQ-mediated inward currents in cells expressing mASIC3 (Fig. 1D, 14.5 ± 3.0 pA/pF, 6 of 18 pH-sensitive cells responded) and rASIC3 (Fig. 1E, 180.6 ± 55.1 pA/pF, n = 8, all pH-sensitive cells responded), nmrASIC3 failed to respond (Fig. 1F, n = 11 cells from 4 transfections). One possibility is that nmrASIC3 is retained in the endoplasmic reticulum, as has been proposed for the proton-insensitive mASIC2b, which only gets to the plasma membrane when coexpressed with the proton-sensitive ASIC2a (42). However, using a biotinylation assay to determine plasma membrane expression of mASIC3 and nmrASIC3, transfected either alone, or with rASIC2a, we observed that nmrASIC3 traffics to the plasma membrane, regardless of whether it is transfected alone or with rASIC2a, just like mASIC3 (Fig. 1G, a second experiment showed the same plasma membrane trafficking of nmrASIC3). Therefore, the insensitivity of nmrASIC3 to protons and GMQ cannot be explained by a lack of membrane expression.
Figure 1.
Representative traces of currents recorded during stimulation with low pH and GMQ. Currents recorded from CHO cells expressing mASIC3 (A), rASIC3 (B), or nmrASIC3 (C) stimulated with either pH 4.0 or 3.0 solution, and stimulated with 1 mm GMQ (D–F). G, Western blot of whole cell lysates (input) and biotinylated surface fraction from cells transfected with mASIC3, nmrASIC3, or co-transfected with m/nmrASIC3 and rASIC2a and stained with anti-ASIC3 antibody, anti-ASIC2 antibody, or anti-β-Actin antibody.
Table 1.
Summary data of peak current density, Isus/Ipeak, EC50, and Hill coefficient for all constructs tested
Numbers in parentheses refer to number of cells tested, details of statistical comparisons are not included for clarity, refer to graphs.
| Peak current density at pH 4.0 | Isus/Ipeak at pH 4.0 | Inactivation time constant at pH 4.0 | pH EC50 | pH Hill coefficient | Peak current density GMQ (1 mm) | |
|---|---|---|---|---|---|---|
| pA/pF | % | ms | pA/pF | |||
| nmrASIC1b | 20 ± 3 (21) | 12 ± 2 (17) | 118 ± 7 (21) | 6.02 ± 0.02 (8) | 1.42 ± 0.1 | NDa |
| nmrASIC3 + nmrASIC1b | 67 ± 13 (18) | 34 ± 5 (18) | 125 ± 7 (17) | 5.58 ± 0.07 (12) | 0.89 ± 0.09 | ND |
| nmrASIC3 + rASIC2a | 455 ± 217 (10) | 19 ± 4 (6) | 1637 ± 62 | 4.39 ± 0.17 (11) | 1.17 ± 0.16 | ND |
| mASIC3 | 69 ± 13 (26) | 48 ± 4 (23) | 202 ± 12 (23) | 6.01 ± 0.14 (9) | 0.68 ± 0.1 | 14 ± 3 (6) |
| mASIC3-ATG | 109 ± 51 (8) | ND | 352 ± 31 (9) | 6.02 ± 0.14 (10) | 0.65 ± 0.07 | ND |
| mASIC3 + nmrASIC1b | 193 ± 48 (14) | 31 ± 4 (15) | 93 ± 5 (14) | 5.66 ± 0.08 (8) | 0.96 ± 0.09 | ND |
| mASIC3 + rASIC2a | 129 ± 23 (13) | 141 ± 25 (13) | 80 ± 10 (4) | 4.45 ± 0.15 (15) | 0.72 ± 0.06 | ND |
| rASIC3 | 627 ± 92 (18) | 35 ± 5 (18) | 387 ± 52 (12) | 6.39 ± 0.17 (10) | 0.91 ± 0.18 | 181 ± 55 (8) |
| rASIC3-A62E | 705 ± 219 (7) | 43 ± 31 (7) | 343 ± 47 (13) | 5.91 ± 0.20 (6) | 0.76 ± 0.07 | ND |
| rASIC3-A62E/R102H | 673 ± 213 (8) | 26 ± 6 (8) | 191 ± 14 (8) | 6.15 ± 0.13 (7) | 0.72 ± 0.06 | ND |
| rASIC2a | 421 ± 58 (50) | 29 ± 6 (8) | 1220 ± 108 (17) | 4.44 ± 0.08 (14) | 1.62 ± 0.51 | ND |
a ND, not determined.
nmrASIC3 is functional in dorsal root ganglion neurons
We have previously demonstrated that naked mole-rat dorsal root ganglion (DRG) neurons produce ASIC-like currents in response to acid (36) and that these neurons express nmrASIC3 mRNA (33, 36) and thus we used APETx2, an inhibitor of most ASIC3-containing ASIC trimers, to determine whether nmrASIC3 contributes to acid-sensitivity of these neurons. A pH 5.0 stimulus evoked two types of inward currents in naked mole-rat DRG neurons: rapidly-inactivating, ASIC-like currents and sustained, TRPV1-like currents (Fig. 2, A and B). Exposure to 2 μm APETx2 for 30 s caused a significant decrease in the amplitude of the ASIC-like responses evoked by a second pH 5.0 stimulus (61 ± 16 pA/pF versus 33 ± 10 pA/pF, n = 8, p ≤ 0.05, Fig. 2, A and C), but had no effect upon TRPV1-like responses 5 ± 2 versus 4 ± 1 pA/pF, n = 14, Fig. 2, B and C); the inhibition of ASIC-like responses was reversible (47 ± 11 pA/pF, p ≤ 0.05). Considering that APETx2 is selective for ASIC3 homomers and most ASIC3 heteromers (8), and that nmrASIC3 does not form proton-sensitive homomers (Fig. 1), these results suggest that nmrASIC3 can form proton-sensitive heteromers with other ASIC subunits, much like the other proton-insensitive ASIC subunits ASIC2b and ASIC4 (13, 16).
Figure 2.
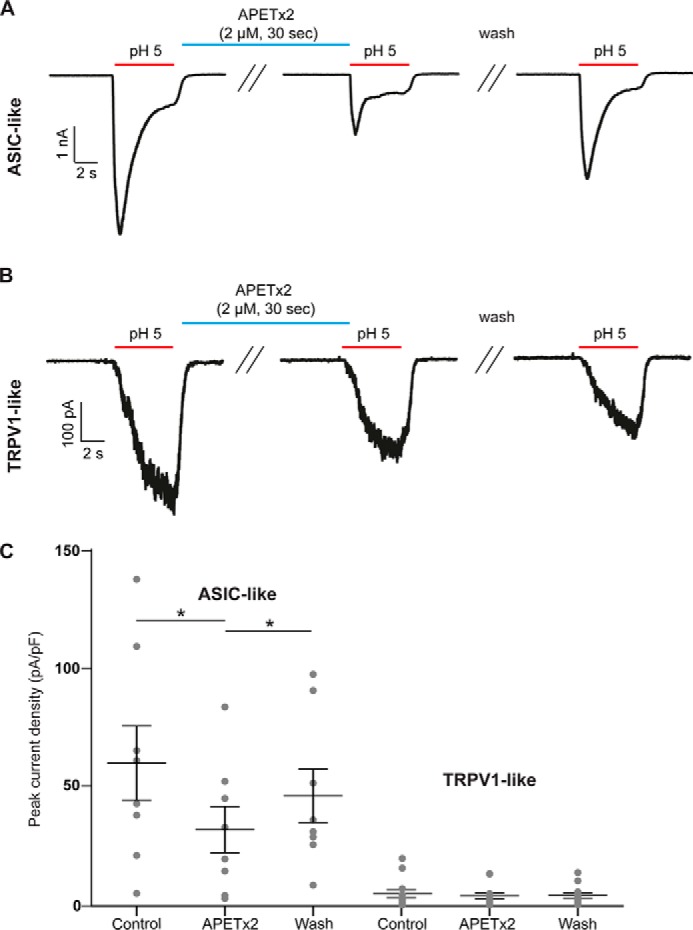
APETx2 blocks transient currents in naked mole-rat DRG neurons. Representative ASIC-like (A) or TRPV1-like (B) current traces recorded from DRG neurons stimulated with pH 5.0 before APETx2 application, immediately after application of 2 μm APETx2 for 30 s, and after 30 s wash at pH 7.4. C, quantification of results showing that ASIC-like transient currents were significantly and reversibly inhibited by APETx2, whereas TRPV1-like sustained currents were not affected. Bars represent mean ± S.E. Data were analyzed by paired t test. **, p < 0.01.
nmrASIC3 forms functional ASIC heteromers with other ASIC subunits
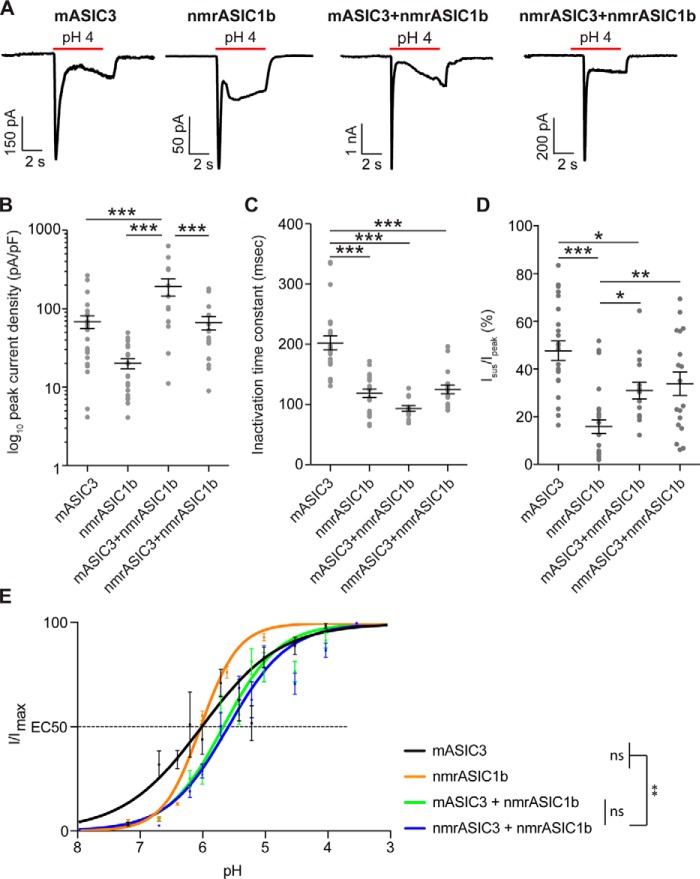
To determine whether nmrASIC3 can form proton-sensitive heteromers with other ASIC subunits as DRG neuron data would suggest, we cotransfected either nmrASIC3 or mASIC3 with nmrASIC1b and compared the properties of currents recorded from these cotransfected cells with those only transfected with either mASIC3 or nmrASIC1b. Using a pH 4.0 stimulus, currents recorded from cells transfected with mASIC3 had a peak current density of 69 ± 13 pA/pF (n = 26), an inactivation time constant of 202 ± 12 ms (n = 23) and Isus/Ipeak, the sustained current as a percentage of the peak current, was 48 ± 4% (n = 23, Fig. 3, A–D). Currents recorded from cells transfected with nmrASIC1b had a peak current density of 20 ± 3 pA/pF, an inactivation time constant of 118 ± 7 ms (n = 21), and Isus/Ipeak was 12 ± 2% (n = 17, Fig. 3, A–D). In cells cotransfected with nmrASIC1b and nmrASIC3, currents were recorded in all instances suggesting that nmrASIC3 does not have a dominant-negative effect. Properties of nmrASIC3 + nmrASIC1b currents were as follows: peak current density, 67 ± 13 pA/pF (n = 18), inactivation time constant, 125 ± 7 ms (n = 17), and Isus/Ipeak was 34 ± 5% (n = 18, Fig. 3, A–D). Similar currents were observed in cells expressing mASIC3 + nmrASIC1b: peak current density, 193 ± 48 pA/pF (n = 14, p ≤ 0.001 versus nmrASIC3 + nmrASIC1b), inactivation time constant, 93 ± 5 ms (n = 14), and Isus/Ipeak 31 ± 4% (n = 15, Fig. 3, A–D). The inactivation time constant of mASIC3 was significantly slower than the inactivation time constants of nmrASIC1b and both mASIC3 + nmrASIC1b and nmrASIC3 + nmrASIC1b (p ≤ 0.001, Fig. 3C). Importantly, for both nmrASIC3 + nmrASIC1b- and mASIC3 + nmrASIC1b-mediated currents, the Isus/Ipeak was significantly greater than that of nmrASIC1b homomers (p ≤ 0.01 and p ≤ 0.05, respectively, Fig. 3D; the Isus/Ipeak mASIC3 + nmrASIC1b was significantly less than that of mASIC3 homomers, p ≤ 0.05, respectively, but for nmrASIC3 + nmrASIC1b there was no significant difference compared with mASIC3 homomers Fig. 3D). These results suggest that both nmrASIC3 and mASIC3 form heteromers with nmrASIC1b to produce currents with a substantial sustained component. Although the large sustained component measured in cells expressing mASIC3 and nmrASIC1b could be the result of measuring a mixture of mASIC3 homomeric currents (large Isus/Ipeak) and nmrASIC1b homomers (small Isus/Ipeak) this cannot explain the large Isus/Ipeak measured in cells expressing nmrASIC3 and nmrASIC1b because nmrASIC3 does not form proton-sensitive homomers and thus it is likely that ASIC3 and ASIC1b form heteromers that have a substantial Isus/Ipeak as has been shown by others for rASIC3 + rASIC1b (7). A second piece of evidence suggesting that nmrASIC3 can form functional ASIC heteromers is that pH-response curves show that the effective concentration 50 (EC50) for mASIC3 + nmrASIC1b heteromers was not significantly different from that of nmrASIC3 + nmrASIC1b (pH 5.66 ± 0.08, n = 8, versus pH 5.58 ± 0.07, n = 12, p = 0.91), however, heteromers of nmrASIC3 + nmrASIC1b were significantly different from the EC50 of either mASIC3 or nmrASIC1b homomers (mASIC3, pH 6.01 ± 0.14, n = 9 and nmrASIC1b, pH 6.03 ± 0.02, n = 9, p ≤ 0.01 Fig. 3E), but not mASIC3 + nmrASIC1b (p = 0.053 and 0.05, respectively).
Figure 3.
Characterization of CHO cells co-expressing nmrASIC1b and mASIC3 or nmrASIC3. A, currents recorded from CHO cells expressing mASIC3, nmrASIC1b, mASIC3 + nmrASIC1b, or nmrASIC3 + nmrASIC1b. B–D, quantification of log10 peak current density (B), inactivation time constant (C), and Isus/Ipeak (D). Bars represent mean ± S.E. E, pH-response curves of mASIC3, nmrASIC1b, mASIC3 + nmrASIC1b, and nmrASIC3 + nmrASIC1b. Data were analyzed by ANOVA with Tukey's multiple comparison test. ***, p < 0.001; **, p < 0.01 comparing all conditions.
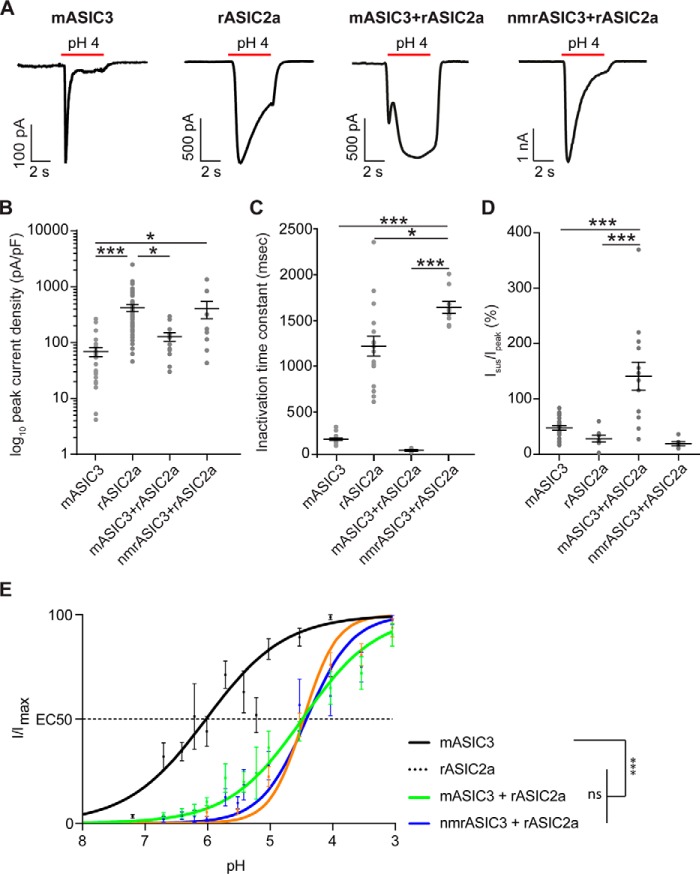
We also investigated the ability of mASIC3 and nmrASIC3 to form heteromers with rASIC2a and found that as shown previously (7, 43) coexpression of mASIC3 and rASIC2a resulted in currents with an Isus/Ipeak that was significantly greater than that produced by either mASIC3 or rASIC2a homomers (mASIC3 + rASIC2a: 141 ± 25, n = 13 versus mASIC3: 48 ± 4%, n = 23, p < 0.001 and rASIC2a 29 ± 6%, n = 8, p < 0.001, Fig. 4, A and D). By contrast, cells expressing nmrASIC3 and rASIC2a produced currents that did not produce a large Isus/Ipeak (Fig. 4A and D) and appeared largely indistinguishable from rASIC2a homomers; the lack of a large sustained current in cells expressing nmrASIC3 + rASIC2a in response to protons is not necessarily a sign that heteromers are not formed because rASIC2a + rASIC3 heteromers have been reported to have an Isus/Ipeak of ∼30% (28), not to dissimilar to the 19 ± 4% observed here. Moreover, a small, but significant difference in the inactivation time constant was observed suggesting that heteromers may be formed: nmrASIC3 + rASIC2a heteromers inactivated significantly more slowly than mASIC3 + rASIC2a heteromers and homomers of either rASIC2a or mASIC3 (nmrASIC3 + rASIC2a, 1637 ± 62 ms, n = 9 versus mASIC3 + rASIC2a, 80 ± 10 ms, n = 4, p ≤ 0.001, versus rASIC2a, 1220 ± 108 ms, n = 17, p < 0.05, and versus mASIC3, 202 ± 12 ms, n = 23, p ≤ 0.001, Fig. 4C). Currents mediated by nmrASIC3 + rASIC2a were of significantly greater magnitude than those mediated by mASIC3 (nmrASIC3 + rASIC2a 455 ± 217 pA/pF, n = 10, versus 69 ± 13 pA/pF, n = 26, p ≤ 0.05), whereas those mediated by rASIC2a were significantly greater than those mediated by mASIC3 + rASIC2a and mASIC3 (421 ± 58 pA/pF, n = 50, versus mASIC3 + rASIC2a, 129 ± 23 pA/pF, n = 13, p ≤ 0.05, and 69 ± 13 pA/pF, n = 26, p ≤ 0.001, respectively, Fig. 4B). Examination of pH-response curves showed that both mASIC3 + rASIC2a and nmrASIC3 + rASIC2a produced currents that were significantly less sensitive to protons than mASIC3, but not significantly different from each other (EC50s: mASIC3 + rASIC2a, pH 4.45 ± 0.15, n = 15, versus nmrASIC3 + rASIC2a, pH 4.39 ± 0.17, n = 13, p = 0.98, and versus mASIC3, pH 6.01 ± 0.12, n = 10, p ≤ 0.0001, Fig. 4E), and these were also indistinguishable from that of rASIC2a (EC50: rASIC2a, pH 4.44 ± 0.05, n = 14, p = 0.99 and p = 0.99, respectively). In summary, based upon biophysical characterization, nmrASIC3 forms functional heteromers with nmrASIC1b (Fig. 3), but the evidence is less clear for heteromeric formation with rASIC2a (Fig. 4) although these two subunits are both present at the plasma membrane when cotransfected (Fig. 1G) and the fact that ASIC-like currents are sensitive to inhibition by APETx2 in DRG neurons from naked mole-rats (Fig. 2) strongly supports the premise that although nmrASIC3 produces proton-insensitive homomers it can form functional heteromers in vivo.
Figure 4.
Characterization of CHO cells co-expressing rASIC2a and mASIC3 or nmrASIC3. A, currents recorded from CHO cells expressing mASIC3, rASIC2a, mASIC3 + rASIC2a, or nmrASIC3 + rASIC2a. B–D, quantification of log10 peak current density (B), inactivation time constant (C), and sustained current in proportion to peak current (D). Bars represent mean ± S.E. E, pH-response curves of mASIC3, rASIC2a, mASIC3 + rASIC2a, and nmrASIC3 + rASIC2a. Data were analyzed by ANOVA with Tukey's multiple comparison test. ***, p < 0.001 comparing all conditions.
Amino acid variations specific to nmrASIC3 do not account for homomeric proton-insensitivity
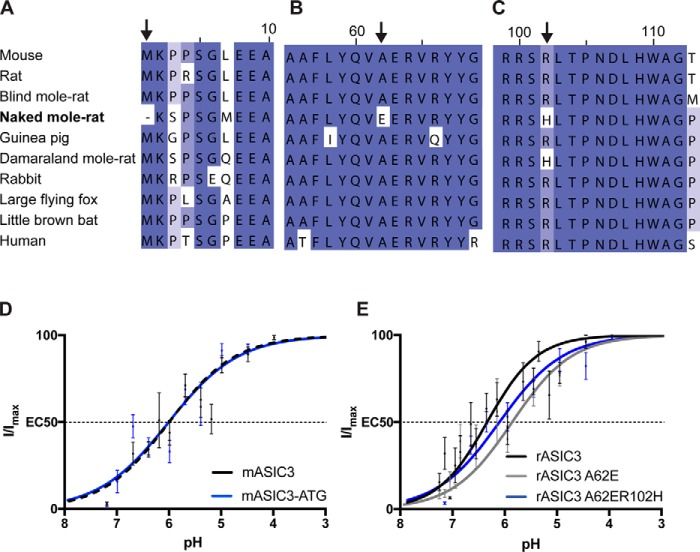
To determine the molecular basis for proton-insensitivity of nmrASIC3, we aligned the nmrASIC3 amino acid sequence with that of 9 other species (Fig. 5, A–C). We found only three instances where naked mole-rat residues differed in a region otherwise conserved in all species. Importantly, we included guinea pig, a rodent more closely related to naked mole-rat than mouse or rat, and guinea pig sensory neurons are activated by the non-proton agonist of ASIC3, GMQ demonstrating the functionality of ASIC3 in this species (27). We first identified that nmrASIC3 is missing the first methionine of the protein sequence, having instead a methionine at position 7 (Fig. 5A). We thus created a version of mASIC3 lacking the initial ATG and further mutated position 7 leucine for methionine (L7M), termed mASIC3-ATG, and an nmrASIC3 with an added methionine at position 1 and an M7L mutation termed nmrASIC3 + ATG. mASIC3-ATG responded to low pH and the EC50 was 6.02 ± 0.14 (n = 10), not significantly different than that of wildtype mASIC3 (p = 0.97, Fig. 5D). By contrast, nmrASIC3 + ATG, like wildtype nmrASIC3, did not respond to acid. Variation in the initial 7 amino acids of nmrASIC3 cannot therefore explain its proton insensitivity.
Figure 5.
Multiple sequence alignment of rodent and human ASIC3 protein including species closely related to naked mole-rat or living in similarly hypoxic/hypercapnic habitats. A, amino acids 1–10, methionine is missing in the naked mole-rat (arrow). B, amino acids 55–69, change of alanine 62 to glutamate in naked mole-rat (arrow). C, amino acids 99–112, change of arginine to histidine in naked mole-rat and Damaraland mole-rat (arrow). D, pH-response curves of mASIC3 and mASIC3-ATG. E, pH-response curves of rASIC3, rASIC3-A62E, rASIC3-A62E/R102H. Data were analyzed by a t test.
A second difference exclusive to nmrASIC3 in the comparison made was replacement of alanine at position 62 with glutamate (A62E, Fig. 5B), a residue likely within transmembrane domain 1, but close to the start of the extracellular domain (2). Because rASIC3 produced larger currents than mASIC3, we used this construct from this point onwards to determine whether mutations altered pH sensitivity. A pH 4.0 stimulus produced inward currents of a similar amplitude in cells expressing either rASIC3A62E or rASIC3 (rASIC3A62E: 705 ± 219 pA/pF, n = 7, and rASIC3: 499 ± 145, n = 10), but currents inactivated significantly more rapidly (rASIC3A62E: 343 ± 47 ms, n = 13 versus rASIC3: 580 ± 33 ms, n = 5, p ≤ 0.001). The pH-response curve was slightly, but nonsignificantly, shifted to the right (rASIC3A62E: pH 5.91 ± 0.2, n = 6, and rASIC3: 6.39 ± 0.17, n = 10, p = 0.09, Fig. 5E).
A third amino acid variation was identified at residue 102, a conserved arginine being replaced in both the naked mole-rat and Damaraland mole-rat with histidine (Fig. 5C) and thus rASIC3-A62E was further mutated to produce rASIC3-A62E/R102H. The pH-response curve for the double mutant rASIC3-A62E/R102H was not significantly different than that of rASIC3 (pH 6.15 ± 0.13, n = 7, p = 0.33, Fig. 5E). As for rASIC3-A62E, currents mediated by rASIC3-A62E/R102H inactivated significantly faster than wildtype rASIC3 (rASIC3-A62E/R102H, 191 ± 14 ms, n = 8, p < 0.001). Thus, it would appear that neither Ala62 nor Arg102 are of crucial importance in proton activation of ASIC3 and indeed when expressed in CHO cells neither nmrASIC3-E62A nor nmrASIC3-H102R resulted in the rescuing of nmrASIC3 proton-sensitivity.
Discussion
Sensitivity to acid as a noxious stimulus is largely conserved throughout the animal kingdom (44), but the naked mole-rat is behavioral insensitive to acid (35) due to a variation in NaV1.7, which results in acid anesthetizing, rather than activating, naked mole-rat sensory neurons (36). Moreover, naked mole-rats show behavioral indifference to both ammonia and acid fumes (38, 39), as well as CO2 (40) at levels producing avoidance in mice. All of these findings demonstrate likely adaptations to having evolved in a subterranean, hypoxic/hypercapnic environment (45, 46). Here we undertook to investigate the properties of nmrASIC3 because evidence supports a role for ASIC3 in a wide variety of situations, including: pain (9, 23–26), as well itch (29), mechanosensation (23, 30), and anxiety (31). Although nmrASIC3 displays a similar expression profile to mASIC3 (33), we show here that it is insensitive to both protons and the non-proton agonist GMQ when expressed in CHO cells, even though it traffics to the plasma membrane. Considering the 82.7% identity with mASIC3 and 81.6% rASIC3 at the amino acid level, including 95% similarity of the EC domain (EMBOSS matcher algorithm) (47), these findings were unexpected. Much like ASIC2b and ASIC4, which are also insensitive to protons (13, 16), we have produced pharmacological evidence that nmrASIC3 contributes to heteromeric ASIC formation in DRG neurons, such that the ASIC3 subunit containing antagonist APETx2 reversibly inhibits ASIC-like currents, but has no effect on TRPV1-like currents, recorded from naked mole-rat DRG neurons. It has been well characterized that although homomeric ASIC currents can occur in both peripheral and central neurons (9, 48), it is perhaps more common for ASICs to form heteromers (9, 49) and variations of the ASIC subunit configuration has a significant effect upon the sensitivity of channels to protons, their inactivation time constants, and sensitivity to different pharmacological agents (7, 8, 50, 51). As well as the reversible inhibition of proton-gated currents in DRG neurons by APETx2, in a heterologous expression system we observed that nmrASIC3 appears to form heteromers with both ASIC1b and ASIC2a, which would suggest that in vivo it acts to modulate ASIC currents, just like ASIC2b and ASIC4.
With regard to understanding the basis of nmrASIC3 proton-insensitivity, manipulation of the few amino acid variations that we identified from a multiple sequence alignment neither abolished rASIC3 proton-sensitivity, nor rescued nmrASIC3 proton-sensitivity and thus it remains unclear why nmrASIC3 fails to respond to protons. Of the 3 carboxylate pairs contained within the acidic pocket identified from the crystal structure of cASIC1 that were proposed to be critical for proton activation of ASICs (Asp238–Asp350, Glu239–Asp346, and Glu220–Asp408; cASIC1a numbering) (2), Asp238 is replaced by glutamate and Asp346 is actually replaced by a serine in mASIC3 and rASIC3. Lacking the full set of carboxylates in the proton-sensitive mASIC3 and rASIC3 confirms earlier results from ourselves and others highlighting that although the acidic pocket is involved in proton activation of ASICs, it alone is not responsible for proton activation of ASICs (17–22). In nmrASIC3, Glu220 becomes Asp210, Glu239 becomes Asp229, and Asp346 is actually retained Asp352, thus nmrASIC3 actually has a full set of carboxylates in the acidic pocket, and although two glutamates are replaced by aspartates, which would have a different pKa, they are still protonatable residues. A series of mutations have been made in rASIC3, two of which were similar to those in this study, E63A and R102A, and although EC50 values for these mutants are not reported, both responded to protons and E63A was observed to slow down the rate of inactivation (52), whereas here we observed that the inactivation time constant was more rapid in rASIC3-E62A. However, Cushman et al. (52) stimulated using pH 6.0 from a starting point of pH 8.0, whereas here we stimulated using pH 4.0 from a starting point of pH 7.4, which may explain the difference observed. Taken together, from the results presented here it remains unclear why nmrASIC3 is proton-insensitive; there are further nmrASIC3 amino acid variations that remain to be tested, which may yet account for the proton-insensitivity observed, although these are far less species specific than those tested here.
Considering the varied physiology with which ASIC3 is concerned, the fact that nmrASIC3 forms non-functional homomers may be a further adaptation to living in a hypercapnic environment. Future experiments determining the role of ASIC3 in different brain regions of the naked mole-rat will be required to understand just how the proton insensitivity of nmrASIC3 influences brain function.
Experimental procedures
Animals
All experiments were conducted in accordance with the United Kingdom Animal (Scientific Procedures) Act 1986 Amendment Regulations 2012 under a Project License (70/7705) granted to E. St. J. S. by the Home Office; the University of Cambridge Animal Welfare Ethical Review Body also approved procedures. Young adult naked mole-rats were used in this study: 2 males and 1 female aged between 3.5 and 4.5 years. Animals were maintained in a custom-made caging system with conventional mouse/rat cages connected by different lengths of tunnel. Bedding and nesting material were provided along with a running wheel. The room was warmed to 28 °C, with a heat cable to provide extra warmth running under 2–3 cages, and red lighting (08:00–16:00) was used.
Chinese hamster ovary cell culture and transfection
CHO cells (Sigma) were grown using standard procedures in the following medium: Ham's F-12 Nutrient Mixture (Life Technologies), 10% fetal bovine serum (Sigma), 1% penicillin/streptomycin, 100 units/ml (Life Technologies). 24 h before transfecting cells, 35-mm dishes (Fisher) were coated with 100 μg/ml of poly-l-lysine (Sigma) and cells from a 70–80% confluent flask were trypsinized, resuspended in 5 ml of CHO medium, and a volume was taken to seed cells at a 1:10 dilution, 2 ml/dish. For transfections, an EGFP expression vector was used to enable identification of transfected cells and DNA was transfected at a ratio of 10:1, ASICx:GFP, or 5:5:1 in c-transfection experiments, using 0.9 μg of ASICx DNA and 0.09 μg of EGFP DNA (2 μg of DNA was used for nmrASIC3); the transfection reagent Lipofectamine LTX (Life Technologies) was used according to the manufacturer's protocol.
Cloning and mutagenesis
mASIC3 and nmrASIC3 was amplified from mouse and naked mole-rat whole brain cDNA, respectively, using forward (fw, mASIC3, atgaaacctccctcaggactgga; nmrASIC3, aagagccctcgggatggagga) and reverse primers (rv, mASIC3, ctagagccttgtcacgaggtaaca; nmrASIC3, ctagaatcactagtttgcccgggat), cloned into pIRES or pTarget expression plasmid and confirmed by sequencing. Rat ASIC2a cDNA in a pCI expression plasmid and nmrASIC1b in pEGFP-N3 have been previously described (22, 36). Mutations were inserted with the FastCloning method (53) using primer pairs specific to the construct (mASIC3-ATG, fw, aaacctccctcaggatggagga, rv, cattcctgagggaggttttaccgtcgactgcagaattcga; nmrASIC3 + ATG, fw, AAGAGCCCCTCGGGGCTGGAGGAGGCTCGGAGAA; rv, CCGAGGGGCTCTTCATGCTAGCGGAT; rASIC3-A62E, fw, tctaccaggtggaggagcgggttcg, rv, cctggtagaggaaggccgccagcga; rASIC3-A62E/R102H, fw, cccactgcgccgctcaca, tk;2rv, gtgaggtgtgagcggcgca; nmrASIC3-E62A, fw, ctaccaggtggctgagcgggtacgcta, rv, acctggtagaggaaggctgccagcga; nmrASIC3-H102R, fw, cgctcacgcctcactcccaacga, rv, cgtgagcggcgcagcgggttgatgtt).
Biotinylation
CHO cells were transfected using polyethylenimine (PEI). The cells cultured in 75 cm2 flasks were ∼75% confluent. For transfection, to 1 ml of serum-free DMEM, 25 μg of total plasmid DNA encoding mASIC3, nmrASIC3, or rASIC2a was mixed with 15 μl of 7.5 mm polyethylenimine. When a combination of plasmids was to be transfected, the concentration of plasmids was split equally. This transfection mixture was incubated for 10 min at room temperature and added drop-by-drop to the flask that was replaced with fresh growth media prior to addition of the transfection mixture. For isolation of cell-surface biotinylated proteins, 48-h post-transfection, the growth medium was removed from cells. Ice-cold HEPES buffer saline (HBS) (140 mm NaCl, 1.5 mm Na2HPO4·2H2O, 50 mm HEPES, pH 7.05) containing 0.2 mg/ml of biotin-sulfo-NHS (Thermo Fisher Scientific, catalog number 21331) was added to cells and incubation was carried out for 60 min on ice. Subsequently, biotin containing HBS was removed and cells were washed at least 3 times with 15 ml of Tris-buffered-saline (25 mm Tris-HCl, 150 mm NaCl, 10 mm EDTA, pH 7.4). Cells were collected in the same buffer, pelleted at 1,000 × g for 5 min at 4 ºC. The pellet was solubilized in solubilization buffer (25 mm Tris-HCl, 150 mm NaCl, 10 mm EDTA, 1% Triton X-100, and 1 mg/ml of protease inhibitor (Roche Applied Science)) for 60 min at 4 °C with continuous mixing. This lysis mixture was centrifuged at 50,000 × g for 60 min at 4 °C and the supernatant was incubated with 50 μl of monomeric avidin-coated agarose beads (Thermo Fisher Scientific) for 2 h at 4 °C with continuous mixing. The protein–bead complexes were collected by centrifugation at 20,800 × g for 10 min, washed with solubilization buffer at least 3 times with a mixing time of 5 min between washes. The protein was eluted from the beads using 50 μl of Laemmli buffer for immunoblotting. For electrophoresis, 20 μl of the protein/Laemmli buffer mixture was loaded in the lanes of 10% acrylamide and SDS-PAGE was carried out. Proteins were then transferred onto a polyvinylidene difluoride membrane, blocked with 5% milk/TBS/Tween 20 solution for 60 min at room temperature, probed with primary antibody at 4 °C overnight (anti-ASIC2, Abcam, ab77384 (1:250), and anti-ASIC3, Boster, PA1938 (1:250)), washed with 5% milk/TBS/Tween 20 solution and incubated with secondary antibody (anti-mouse horseradish peroxidase, used at 1:1000, Thermo Fisher Scientific, catalog number 31430; anti-rabbit horseradish peroxidase, used at 1:1000, Bio-Rad, catalog number 1706515) for 2 h at room temperature. Blots were washed in distilled water and then developed with West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Protein samples from beads were checked for any “biotin-permeabilization” by probing for actin (negative control, A2228, 1:500, Sigma).
Electrophysiology
DRG neurons were cultured as described previously (36, 41). Whole-cell patch clamp recordings from CHO cells were performed at room temperature 24 h after transfection and recordings from DRG neurons 24 h after culturing. For all ASIC experiments, the intracellular solution contained 110 mm KCl, 10 mm NaCl, 1 mm MgCl2, 1 mm EGTA, 10 mm HEPES, 2 mm Na2ATP, 0.5 mm Na2GTP in MilliQ water; pH was set to pH 7.3 by adding KOH, and the osmolality was adjusted to 310–315 mosmol with sucrose. The extracellular solution contained 140 mm NaCl, 4 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm HEPES (solutions > pH 6) or MES (solutions < pH 6), 4 mm glucose in MilliQ water; osmolality was adjusted to 300–310 mosmol with sucrose and pH was adjusted with NaOH and HCl; unless stated otherwise, e.g. for test pH solutions, the extracellular solution was pH 7.4. Patch pipettes were pulled from glass capillaries (Hilgenberg) using a model P-97, Flaming/Brown puller (Sutter Instruments) and had a resistance of 4–10 megohms. Data were acquired using an EPC10 amplifier (HEKA) and Patchmaster software (HEKA). 2 μm APETx2 were added to the pH 7.4 solution for DRG neuron experiments and solutions containing the synthetic ASIC3 agonist GMQ (Sigma) in pH 7.4 were diluted from a stock solution of 50 mm dissolved in DMSO. For measurement of current amplitude and inactivation time constant, a protocol of 5 s of pH 7.4 followed by a 5-s stimulus with pH 3 or 4, then a return to pH 7.4 solution for 5 s was used; the holding potential was −60 mV for both DRG neurons and CHO cells. For pH-response curves, a 2.5-s stimuli between pH 3 and 6 (ASIC2) or 1-s stimuli between pH 4 and 7.2 (ASIC1, ASIC3) were applied in random order with 30 s of bath solution between stimuli to minimize desensitization, although this is not a prominent feature of ASIC1b, ASIC2a, or ASIC3 (22, 54).
Data analysis
Statistical analysis was performed in Prism (GraphPad), which was also used to plot data. Peak current density was analyzed by measuring the size of the peak current compared with the baseline current (average current measured over 4 s prior to stimulation). The absolute current size was then divided by capacitance of the cell to result in normalized peak current density (pA/pF). Peak current density data were transformed using yi = log10(xi). The inactivation time constant, τ, was measured using a built-in function of Fitmaster. Statistical analysis was performed in GraphPad Prism using repeated measures analysis of variance (ANOVA) with Tukey's multiple comparisons for DRG neuron data or ordinary one-way ANOVA and Tukey's multiple comparisons, comparing data from each construct with every other construct of the same experiment for transformed peak current density data and inactivation data of CHO cell experiments. Results are expressed as mean ± S.E., unless otherwise stated; this might, however, not necessarily represent the statistical differences for peak current density data, which were transformed as above. Sustained current to transient current ratio was calculated by measuring the size of the sustained current at the end of the stimulus compared with the baseline current and dividing this value by the peak current (Isus/Ipeak × 100). For pH-response curves, all measurements were transformed to percent of the maximum peak current (I/Imax × 100) for each cell. The EC50 and Hill coefficient were determined by plotting individual pH-response curves for every cell using GraphPad Prism. Outliers were identified using Prism's ROUT method with Q = 1% and those cells were eliminated from the dataset. The mean EC50 and Hill coefficient for each condition were then used to calculate an average curve using the Hill equation, which was plotted along with the mean ± S.E. of measured values. Figures were made using Adobe Illustrator.
Multiple sequence alignment
ASIC3 protein sequences were obtained from the NCBI genome database or ENSEMBL (mouse NM_183000, naked mole-rat PREHGLG00000022115, rat NM_173135.1, guinea pig ENSCPOT00000020999, blind mole-rat XM_008847215.1, Damaraland mole-rat XM_010637570.1, rabbit ENSRNOG00000058308, human ENSG00000213199, large flying fox ENSPVAG00000007091, little brown bat ENSMLUG00000028175). Naked mole-rat sequences that were not previously annotated were identified using NCBI's basic local alignment search tool (BLAST) online. Sequences were aligned using MAFFT version 7 using default settings with an unalignment factor of 0.8. Sequences were visualized and manipulated in Jalview. Alignments are shaded by BLOSUM62 score, which indicates the likelihood that two amino acids are aligned because they are homologous (55). Dark blue indicates that the residues agree with the consensus sequence (>80% homology), medium blue that they have a positive BLOSUM62 score (corresponding to >62% likelihood of homology), light blue represents >40% homology and white residues are not homologous (<40% homology).
Author contributions
L. N. S. and E. St. J. S. conceived and designed the study. L. N. S. created constructs, conducted electrophysiology experiments, and analyzed the data. S. S. conducted biotinylation experiments. G. C. conducted some CHO cell electrophysiology experiments and analyzed the data. E. St. J. S. conducted DRG neuron electrophysiology experiments and analyzed the data. All authors contributed to writing the manuscript.
Acknowledgments
All authors thank members of the Smith lab for assistance, especially J. Bartlett, J. Raby, and L. Rockall, and are grateful to the support of animal technicians in particular H. R. Forest and A. J. Robinson. We are also grateful for discussion of the project with Prof. G Lewin and Dr. D. Omerbašić.
The authors declare that they have no conflicts of interest with the contents of this article.
- ASIC
- acid-sensing ion channels
- TRPV1
- transient receptor potential vanilloid 1
- GMQ
- 2-guanidine-4-methylquinazoline
- DRG
- dorsal root ganglion
- CHO
- Chinese hamster ovary
- EGFP
- enhanced green fluorescent protein
- ANOVA
- analysis of variance
- TBS
- Tris-buffered saline.
References
- 1. Boscardin E., Alijevic O., Hummler E., Frateschi S., and Kellenberger S. (2016) The function and regulation of acid-sensing ion channels (ASICs) and the epithelial Na+ channel (ENaC): IUPHAR review 19: ASIC and ENaC nomenclature. Br. J. Pharmacol. 173, 2671–2701 10.1111/bph.13533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jasti J., Furukawa H., Gonzales E. B., and Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 10.1038/nature06163 [DOI] [PubMed] [Google Scholar]
- 3. Jeggle P., Smith E. S. J., Stewart A. P., Haerteis S., Korbmacher C., and Edwardson J. M. (2015) Atomic force microscopy imaging reveals the formation of ASIC/ENaC cross-clade ion channels. Biochem. Biophys. Res. Commun. 464, 38–44 10.1016/j.bbrc.2015.05.091 [DOI] [PubMed] [Google Scholar]
- 4. Kapoor N., Lee W., Clark E., Bartoszewski R., McNicholas C. M., Latham C. B., Bebok Z., Parpura V., Fuller C. M., Palmer C. A., and Benos D. J. (2011) Interaction of ASIC1 and ENaC subunits in human glioma cells and rat astrocytes. Am. J. Physiol. Cell Physiol. 300, C1246–C1259 10.1152/ajpcell.00199.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tominaga M., Caterina M. J., Malmberg A. B., Rosen T. A., Gilbert H., Skinner K., Raumann B. E., Basbaum A. I., and Julius D. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543 10.1016/S0896-6273(00)80564-4 [DOI] [PubMed] [Google Scholar]
- 6. Gründer S., and Pusch M. (2015) Biophysical properties of acid-sensing ion channels (ASICs). Neuropharmacology 94, 9–18 10.1016/j.neuropharm.2014.12.016 [DOI] [PubMed] [Google Scholar]
- 7. Hesselager M., Timmermann D. B., and Ahring P. K. (2004) pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J. Biol. Chem. 279, 11006–11015 10.1074/jbc.M313507200 [DOI] [PubMed] [Google Scholar]
- 8. Diochot S., Baron A., Rash L. D., Deval E., Escoubas P., Scarzello S., Salinas M., and Lazdunski M. (2004) A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 23, 1516–1525 10.1038/sj.emboj.7600177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., and Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 10.1038/emboj.2008.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J. Y., Saez N. J., Cristofori-Armstrong B., Anangi R., King G. F., Smith M. T., and Rash L. D. (2017) Inhibition of acid-sensing ion channels by diminazene and APETx2 evoke partial and highly variable antihyperalgesia in a rat model of inflammatory pain: ASICs in inflammatory pain. Br. J. Pharmacol. 10.1111/bph.14089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu Y., Chen Z., Li W.-G., Cao H., Feng E.-G., Yu F., Liu H., Jiang H., and Xu T.-L. (2010) A nonproton ligand sensor in the acid-sensing ion channel. Neuron 68, 61–72 10.1016/j.neuron.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 12. Alijevic O., and Kellenberger S. (2012) Subtype-specific modulation of acid-sensing ion channel (ASIC) function by 2-guanidine-4-methylquinazoline. J. Biol. Chem. 287, 36059–36070 10.1074/jbc.M112.360487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lingueglia E., de Weille J. R., Bassilana F., Heurteaux C., Sakai H., Waldmann R., and Lazdunski M. (1997) A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J. Biol. Chem. 272, 29778–29783 10.1074/jbc.272.47.29778 [DOI] [PubMed] [Google Scholar]
- 14. Akopian A. N., Chen C. C., Ding Y., Cesare P., and Wood J. N. (2000) A new member of the acid-sensing ion channel family. Neuroreport 11, 2217–2222 10.1097/00001756-200007140-00031 [DOI] [PubMed] [Google Scholar]
- 15. Grunder S., Geissler H. S., Bassler E. L., and Ruppersberg J. P. (2000) A new member of acid-sensing ion channels from pituitary gland. Neuroreport 11, 1607–1611 10.1097/00001756-200006050-00003 [DOI] [PubMed] [Google Scholar]
- 16. Donier E., Rugiero F., Jacob C., and Wood J. N. (2008) Regulation of ASIC activity by ASIC4–new insights into ASIC channel function revealed by a yeast two-hybrid assay. Eur. J. Neurosci. 28, 74–86 10.1111/j.1460-9568.2008.06282.x [DOI] [PubMed] [Google Scholar]
- 17. Smith E. S., Zhang X., Cadiou H., and McNaughton P. A. (2007) Proton binding sites involved in the activation of acid-sensing ion channel ASIC2a. Neurosci. Lett. 426, 12–17 10.1016/j.neulet.2007.07.047 [DOI] [PubMed] [Google Scholar]
- 18. Paukert M., Chen X., Polleichtner G., Schindelin H., and Gründer S. (2008) Candidate amino acids involved in H+ gating of acid-sensing ion channel 1a. J. Biol. Chem. 283, 572–581 10.1074/jbc.M706811200 [DOI] [PubMed] [Google Scholar]
- 19. Li T., Yang Y., and Canessa C. M. (2009) Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J. Biol. Chem. 284, 4689–4694 10.1074/jbc.M805302200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liechti L. A., Bernèche S., Bargeton B., Iwaszkiewicz J., Roy S., Michielin O., and Kellenberger S. (2010) A combined computational and functional approach identifies new residues involved in pH-dependent gating of ASIC1a. J. Biol. Chem. 285, 16315–16329 10.1074/jbc.M109.092015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Della Vecchia M. C., Rued A. C., and Carattino M. D. (2013) Gating transitions in the palm domain of ASIC1a. J. Biol. Chem. 288, 5487–5495 10.1074/jbc.M112.441964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schuhmacher L.-N., Srivats S., and Smith E. S. (2015) Structural domains underlying the activation of acid-sensing ion channel 2a. Mol. Pharmacol. 87, 561–571 10.1124/mol.114.096909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Price M. P., McIlwrath S. L., Xie J., Cheng C., Qiao J., Tarr D. E., Sluka K. A., Brennan T. J., Lewin G. R., and Welsh M. J. (2001) The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron 32, 1071–1083 10.1016/S0896-6273(01)00547-5 [DOI] [PubMed] [Google Scholar]
- 24. Izumi M., Ikeuchi M., Ji Q., and Tani T. (2012) Local ASIC3 modulates pain and disease progression in a rat model of osteoarthritis. J. Biomed. Sci. 19, 77 10.1186/1423-0127-19-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ikeuchi M., Kolker S. J., Burnes L. A., Walder R. Y., and Sluka K. A. (2008) Role of ASIC3 in the primary and secondary hyperalgesia produced by joint inflammation in mice. Pain. 137, 662–669 10.1016/j.pain.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hiasa M., Okui T., Allette Y. M., Ripsch M. S., Sun-Wada G.-H., Wakabayashi H., Roodman G. D., White F. A., and Yoneda T. (2017) Bone pain induced by multiple myeloma is reduced by targeting V-ATPase and ASIC3. Cancer Res. 77, 1283–1295 10.1158/0008-5472.CAN-15-3545,10.1158/1538-7445.AM2017-1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Callejo G., Castellanos A., Castany M., Gual A., Luna C., Acosta M. C., Gallar J., Giblin J. P., and Gasull X. (2015) Acid-sensing ion channels detect moderate acidifications to induce ocular pain. Pain 156, 483–495 10.1097/01.j.pain.0000460335.49525.17 [DOI] [PubMed] [Google Scholar]
- 28. Reimers C., Lee C.-H., Kalbacher H., Tian Y., Hung C.-H., Schmidt A., Prokop L., Kauferstein S., Mebs D., Chen C.-C., and Gründer S. (2017) Identification of a cono-RFamide from the venom of Conus textile that targets ASIC3 and enhances muscle pain. Proc. Natl. Acad. Sci. U.S.A. 114, E3507–E3515 10.1073/pnas.1616232114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng Z., Li W.-G., Huang C., Jiang Y.-M., Wang X., Zhu M. X., Cheng X., and Xu T.-L. (2015) ASIC3 mediates itch sensation in response to coincident stimulation by acid and nonproton ligand. Cell Rep. 13, 387–398 10.1016/j.celrep.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 30. Jones R. C. 3rd, Xu L., and Gebhart G. F. (2005) The mechanosensitivity of mouse colon afferent fibers and their sensitization by inflammatory mediators require transient receptor potential vanilloid 1 and acid-sensing ion channel 3. J. Neurosci. 25, 10981–10989 10.1523/JNEUROSCI.0703-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu W.-L., Lin Y.-W., Min M.-Y., and Chen C.-C. (2010) Mice lacking Asic3 show reduced anxiety-like behavior on the elevated plus maze and reduced aggression. Genes Brain Behav. 9, 603–614 [DOI] [PubMed] [Google Scholar]
- 32. Waldmann R., Bassilana F., de Weille J., Champigny G., Heurteaux C., and Lazdunski M. (1997) Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J. Biol. Chem. 272, 20975–20978 10.1074/jbc.272.34.20975 [DOI] [PubMed] [Google Scholar]
- 33. Schuhmacher L.-N., and Smith E. S. (2016) Expression of acid-sensing ion channels and selection of reference genes in mouse and naked mole rat. Mol. Brain 9, 97 10.1186/s13041-016-0279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meng Q. Y., Wang W., Chen X. N., Xu T. L., and Zhou J. N. (2009) Distribution of acid-sensing ion channel 3 in the rat hypothalamus. Neuroscience 159, 1126–1134 10.1016/j.neuroscience.2009.01.069 [DOI] [PubMed] [Google Scholar]
- 35. Park T. J., Lu Y., Jüttner R., Smith E. S., Hu J., Brand A., Wetzel C., Milenkovic N., Erdmann B., Heppenstall P. A., Laurito C. E., Wilson S. P., and Lewin G. R. (2008) Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber). PLos Biol. 6, e13 10.1371/journal.pbio.0060013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith E. S. J., Omerbašić D., Lechner S. G., Anirudhan G., Lapatsina L., and Lewin G. R. (2011) The molecular basis of acid insensitivity in the African naked mole-rat. Science 334, 1557–1560 10.1126/science.1213760 [DOI] [PubMed] [Google Scholar]
- 37. Liu Z., Wang W., Zhang T.-Z., Li G.-H., He K., Huang J.-F., Jiang X.-L., Murphy R. W., and Shi P. (2013) Repeated functional convergent effects of NaV1.7 on acid insensitivity in hibernating mammals. Proc. R. Soc. B Biol. Sci. 281, 20132950–20132950 10.1098/rspb.2013.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. LaVinka P. C., and Park T. J. (2012) Blunted behavioral and C Fos responses to acidic fumes in the African naked mole-rat. PLoS ONE 7, e45060 10.1371/journal.pone.0045060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. LaVinka P. C., Brand A., Landau V. J., Wirtshafter D., and Park T. J. (2009) Extreme tolerance to ammonia fumes in African naked mole-rats: animals that naturally lack neuropeptides from trigeminal chemosensory nerve fibers. J. Comp. Physiol. A 195, 419–427 10.1007/s00359-009-0420-0 [DOI] [PubMed] [Google Scholar]
- 40. Park T. J., Reznick J., Peterson B. L., Blass G., Omerbašić D., Bennett N. C., Kuich P. H. J. L., Zasada C., Browe B. M., Hamann W., Applegate D. T., Radke M. H., Kosten T., Lutermann H., Gavaghan V., et al. (2017) Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 356, 307–311 10.1126/science.aab3896 [DOI] [PubMed] [Google Scholar]
- 41. Omerbašić D., Smith E. S., Moroni M., Homfeld J., Eigenbrod O., Bennett N. C., Reznick J., Faulkes C. G., Selbach M., and Lewin G. R. (2016) Hypofunctional TrkA accounts for the absence of pain sensitization in the African naked mole-rat. Cell Rep. 17, 748–758 10.1016/j.celrep.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kweon H.-J., Kim D.-I., Bae Y., Park J.-Y., and Suh B.-C. (2016) Acid-sensing ion channel 2a (ASIC2a) promotes surface trafficking of ASIC2b via heteromeric assembly. Sci. Rep. 6, 30684 10.1038/srep30684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kweon H.-J., Cho J.-H., Jang I.-S., and Suh B.-C. (2016) ASIC2a-dependent increase of ASIC3 surface expression enhances the sustained component of the currents. BMB Rep. 49, 542–547 10.5483/BMBRep.2016.49.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith E. S., and Lewin G. R. (2009) Nociceptors: a phylogenetic view. J. Comp. Physiol. A 195, 1089–1106 10.1007/s00359-009-0482-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schuhmacher L.-N., Husson Z., and Smith E. S. (2015) The naked mole-rat as an animal model in biomedical research: current perspectives. Open Access Anim. Physiol. 2015, 137–148 10.2147/OAAP.S50376 [DOI] [Google Scholar]
- 46. Garbarino V. R., Orr M. E., Rodriguez K. A., and Buffenstein R. (2015) Mechanisms of oxidative stress resistance in the brain: lessons learned from hypoxia tolerant extremophilic vertebrates. Arch. Biochem. Biophys. 576, 8–16 10.1016/j.abb.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rice P., Longden I., and Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 48. Ziemann A. E., Allen J. E., Dahdaleh N. S., Drebot I. I., Coryell M. W., Wunsch A. M., Lynch C. M., Faraci F. M., Howard M. A., Welsh M. J., and Wemmie J. A. (2009) The amygdala is a chemosensor that detects carbon dioxide and acidosis to elicit fear behavior. Cell 139, 1012–1021 10.1016/j.cell.2009.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benson C. J., Xie J., Wemmie J. A., Price M. P., Henss J. M., Welsh M. J., and Snyder P. M. (2002) Heteromultimers of DEG/ENaC subunits form H+-gated channels in mouse sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 99, 2338–2343 10.1073/pnas.032678399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Diochot S., Baron A., Salinas M., Douguet D., Scarzello S., Dabert-Gay A.-S., Debayle D., Friend V., Alloui A., Lazdunski M., and Lingueglia E. (2012) Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490, 552–555 10.1038/nature11494 [DOI] [PubMed] [Google Scholar]
- 51. Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., and Lazdunski M. (2000) Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J. Biol. Chem. 275, 25116–25121 10.1074/jbc.M003643200 [DOI] [PubMed] [Google Scholar]
- 52. Cushman K. A., Marsh-Haffner J., Adelman J. P., and McCleskey E. W. (2007) A conformation change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J. Gen. Physiol. 129, 345–350 10.1085/jgp.200709757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li C., Wen A., Shen B., Lu J., Huang Y., and Chang Y. (2011) FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotech. 11, 92 10.1186/1472-6750-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen X., and Gründer S. (2007) Permeating protons contribute to tachyphylaxis of the acid-sensing ion channel (ASIC) 1a. J. Physiol. 579, 657–670 10.1113/jphysiol.2006.120733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henikoff S., and Henikoff J. G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 89, 10915–10919 10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]