Abstract
The brilliant cresyl blue (BCB) test is used in both basic biological research and assisted reproduction to identify oocytes likely to be developmentally competent. However, the underlying molecular mechanism targeted by the BCB test is still unclear. To explore this question, we first confirmed that BCB-positive porcine oocytes had higher rates of meiotic maturation, better rates of cleavage and development into blastocysts, and lower death rates. Subsequent single-cell transcriptome sequencing on porcine germinal vesicle (GV)-stage oocytes identified 155 genes that were significantly differentially expressed between BCB-negative and BCB-positive oocytes. These included genes such as cdc5l, ldha, spata22, rgs2, paip1, wee1b, and hsp27, which are enriched in functionally important signaling pathways including cell cycle regulation, oocyte meiosis, spliceosome formation, and nucleotide excision repair. In BCB-positive GV oocytes that additionally had a lower frequency of DNA double-strand breaks, the CDC5L protein was significantly more abundant. cdc5l/CDC5L inhibition by short interference (si)RNA or antibody microinjection significantly impaired porcine oocyte meiotic maturation and subsequent parthenote development. Taken together, our single-oocyte sequencing data point to a potential new role for CDC5L in porcine oocyte meiosis and early embryo development, and supports further analysis of this protein in the context of the BCB test.
Keywords: cell cycle, embryo, gene expression, meiosis, oocyte, brilliant cresyl blue, CDC5L, pig, single cell RNA-seq
Introduction
Mammalian oocytes of better developmental competence are in high demand in both basic and applied research areas of assisted reproductive technology. However, oocyte maturation and development are orchestrated by a complex array of molecular and physiological events, from the differentiation of primordial germ cells into oogonia, to the early growth, meiotic resumption, and maturation of oocytes (1, 2). Currently, in vitro manipulation and maturation of cumulus–oocyte complexes constitute the main channel to produce assisted reproductive technology-ready oocytes in mammals. To augment the developmental competence of oocytes cultured in vitro, a variety of techniques and methods have been pursued, such as the brilliant cresyl blue (BCB)3 test, zona imaging, optimization of culture system, and screening of biomarkers (3, 4).
The BCB staining test is capable of indicating the activity of glucose-6-phosphate dehydrogenase (G6PDH), an effective biomarker originally discovered for oocyte maturation (5, 6), and for selecting oocytes with high developmental competence (7–11). Oocytes discerned by BCB staining had different properties, such as abundance of maternal transcripts (9, 12–16), mitochondrial numbers and function (17–19), lipids content (19), and total cell number and cell survival rates of blastocysts (4, 20). In pigs, the BCB test also seems to be capable of predicting the outcomes of oocyte maturation (21–23), although some caution arises recently (24, 25). Oocytes with better developmental competence could also be used to improve the success rate of production of transgenic pigs with the somatic cell nuclear transfer technique (26, 27). However, the molecular mechanism underlying the effectiveness of BCB staining to distinguish competent and non-competent oocytes remains obscure.
Recently, rapid advancement of single cell biotechnology makes it possible to study oocyte and early embryo development in an unprecedented way (28–33). Application of single cell gene expression profiling demonstrates cellular heterogeneity in early embryo development (34). In addition, simultaneous single cell genome and transcriptome sequencing assist in inferring chromosome crossover events in oocytes, screening embryos of better quality (35), and building the gene regulatory framework for early embryo development (28, 29).
Here, we employed the single-oocyte RNA-seq technique to provide molecular evidence underlying the effectiveness of BCB staining in selecting oocytes of better developmental competence. Genes vital to oocyte maturation and embryo development were found, and enriched in important molecular pathways, such as the cell cycle and spliceosome and nucleotide excision repair. Furthermore, the cell division cycle 5-like (cdc5l) gene was shown to be essential to porcine oocyte maturation and early embryo development.
Results
Porcine oocytes categorized by the BCB test
To explore the effectiveness of the BCB test in selecting oocytes of better maturation and developmental competence, we performed a large-scale experiment by staining a total of 4,790 immature porcine cumulus–oocyte complexes (COCs). We first tested to see if differences existed between two different controls, by comparing COCs incubated with the maturation medium (the normal group) and Dulbecco's phosphate-buffered saline (DPBS) (the BCB-control group) under the same culture conditions, respectively. Between these two control groups, neither the rates of meiotic maturation and death (Fig. S1, A and B) nor the rates of parthenote cleavage and development to blastocyst (Fig. S1, A and C) had significant differences. Therefore, we used only the BCB-control for all remaining experiments.
Ooplasms of 3,653 COCs (74.4%) were stained blue (BCB-positive), indicating low G6PDH activity, whereas the remaining 1,137 COCs (25.6%) were colorless (BCB-negative, of high G6PDH activity) (Fig. 1, A and B). We examined the chromatin configuration of oocytes at germinal vesicle (GV) stage, which were classified by two different methods, as non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) (36), or as GV0–GV4, respectively (37) (Fig. 1C). Except that the percentage of GV4-type oocytes was significantly lower in the BCB-positive group, as compared with the BCB-control and BCB-negative groups (6.7 versus 19.0 and 19.1%; p < 0.01) (Fig. 1D), no significant differences were found for other types of chromatin configurations.
Figure 1.
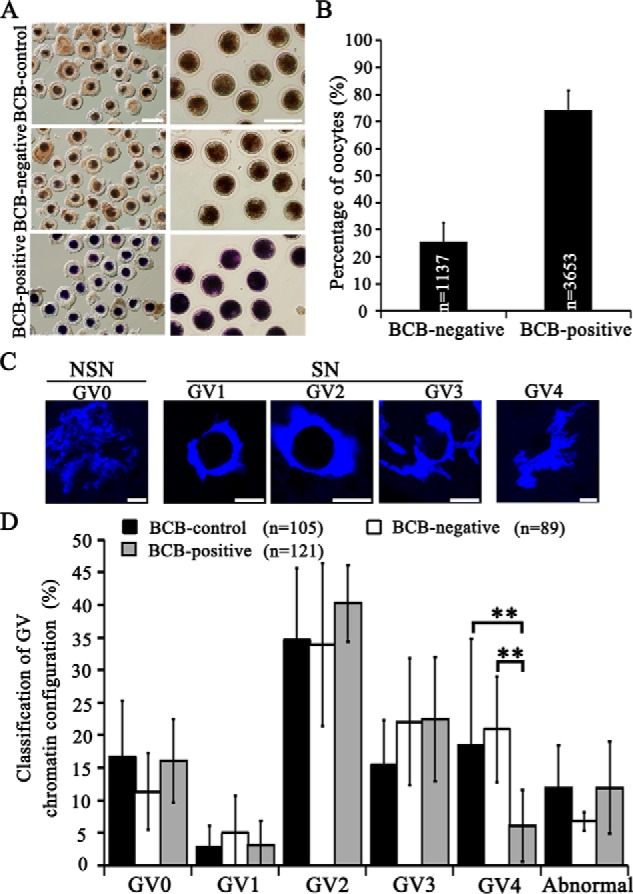
BCB staining to categorize porcine immature GV oocytes and classification of chromatin configuration. A, representative images of porcine COCs after BCB staining (left panel) and oocytes denuded off cumulus cells (right panel). Scale bar, 200 μm. B, the number and percentage of BCB-negative and BCB-positive COCs. C, representative images of chromatin configuration of GV oocyte nuclei. Scale bar, 10 μm. D, percentages of GV oocytes classified into different types of chromatin configuration from BCB-control, BCB-negative, and BCB-positive groups. **, p < 0.01.
During in vitro maturation (IVM), limited cumulus expansion was observed for the BCB-negative group, as compared with the BCB-control and BCB-positive groups (Fig. 2A). After IVM for 44 h, the death rate of the cumulus-denuded oocytes was significantly higher for the BCB-negative group than both BCB-positive and BCB-control groups (59.3 versus 7.8 and 18.3%; p < 0.001) (Fig. 2, A and B). However, the maturation rate was significantly lower for the BCB-negative group, when compared with the BCB-control (52.3 versus 76.8%; p < 0.05) and BCB-positive (52.3 versus 76.5%; p < 0.05) groups (Fig. 2B). Subsequent parthenogenetic activation of mature metaphase II (MII) oocytes showed that the BCB-positive group had significantly higher rates of embryo cleavage (87.1 versus 68.7%; p < 0.01) and blastocyst (59.7 versus 39.2%; p < 0.01) than the BCB-negative group (Fig. 2, C and D). However, as for the average cell number per blastocyst, no significant difference existed (Fig. 2, E and F). Taken together, we confirmed that in pigs, BCB-positive oocytes have lower death rate, and higher rates of maturation and embryo development, indicating better oocyte quality and developmental competence.
Figure 2.
In vitro maturation of COCs classified by BCB staining. A, morphology of COCs (IVM for 24 h) and denuded oocytes (IVM for 44 h), as well as representative images of live (a), MII (b), and dead oocytes (c). Scale bar, 200 μm for the first and second rows. 50 μm for a–c. Black arrows, MII oocytes with the first polar body; gray arrows, dead oocytes with broken ooplasmic membrane. B, percentages of oocytes matured to MII stage, or died during IVM. C, representative images of parthenotes cleaved and developed to blastocysts. Scale bar, 200 μm. D, percentages of parthenotes cleaved and developed to blastocysts for BCB-control, BCB-negative, and BCB-positive groups. E, representative images of Hoechst 33342-stained blastocysts. Scale bar, 200 μm. F, average cell number per blastocyst. **, p < 0.01. ***, p < 0.001.
Single oocyte RNA-seq
To understand the molecular mechanism underlying BCB's ability to discern oocytes with better quality and developmental competence, single cell transcriptome sequencing was performed on three BCB-negative and three BCB-positive cumulus-denuded GV oocytes, respectively. Standard Bioinformatics pipelines were employed to analyze generated short reads (Fig. S2). Mapping rates of short reads, and statistics on their distribution in the pig genome are summarized in Table S1. Of more than 57 million clean reads produced, 63.81% (on average) can be mapped onto the pig reference genome. We obtained a relatively large number of sequence variations, such as single nucleotide polymorphisms, insertions and deletions, possibly due to breed differences between pig samples we used and the reference genome (a Duroc sow).
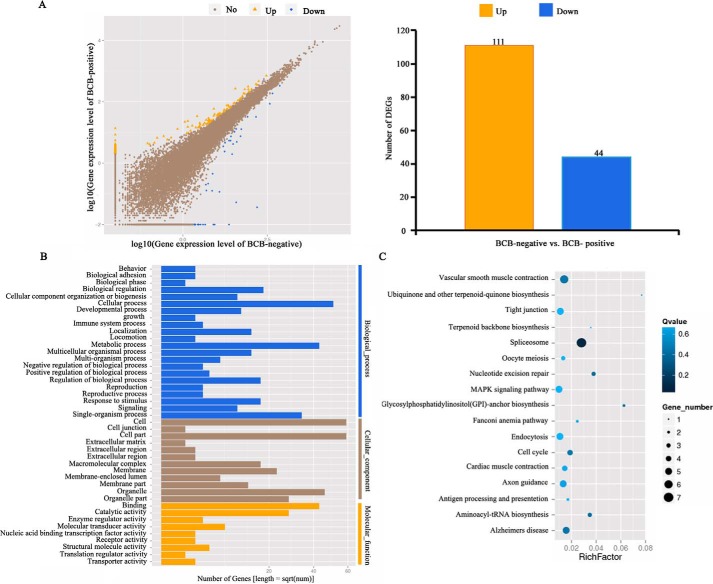
For each sequenced GV oocyte, global gene expression analyses helped to discover the top 10 most abundantly expressed genes, within which 7 genes were common to all 6 sampled oocytes, including dnmt1, ftl, dynll1, and zp3 (Table S2). Between BCB-positive and BCB-negative oocytes, a total of 155 genes (44 down-regulated and 111 up-regulated in BCB-positive oocytes) were differentially expressed (with more than 2-fold changes, p < 0.05) (Fig. 3A; Table S3). We found genes previously reported to be related to cell cycle, or potentially important for oocyte maturation and embryo development, such as cdc5l (38, 39), spata22 (40, 41), rgs2 (42), paip1 (43), melk (44, 45), wee1b (46), and hsp27 (47). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on differentially expressed genes (DEGs) identified a number of important pathways (Fig. 3, B and C; Table 1), such as cell cycle, oocyte meiosis, spliceosome, nucleotide excision repair, metabolic pathway, Janus kinase-signal transducers and activators of transcription pathway, mitogen-activated protein kinase pathway, and tight junction (Fig. S3). Alternative splicing (AS) analysis revealed that intron retention was the most frequent AS event for the largest number of genes (on average 2.99 events per gene), and 5′-UTR splicing was the most rare event (1.62 events per gene) (Fig. S4). Moreover, no statistically significant differences existed for the four AS events (exon skipping, intron retention, 5′-UTR splicing, and 3′-UTR splicing), between BCB-negative and BCB-positive oocytes (p > 0.05).
Figure 3.
Single-oocyte transcriptome sequencing. A, gene expression analysis to identify up- and down-regulated genes in BCB-positive oocytes as compared with BCB-negative oocytes. B, gene enrichment analysis on differentially expressed genes. C, KEGG pathways enriched from differentially expressed genes.
Table 1.
Functional pathways of differentially expressed genes between BCB-negative and BCB-positive immature GV oocytes
| Pathway | No. of DEGs | p value | Q value | |
|---|---|---|---|---|
| 1 | Spliceosome | 7 (6.86%) | 0.00041603 | 0.04950751 |
| 2 | Nucleotide excision repair | 3 (2.94%) | 0.009092659 | 0.38665301 |
| 3 | Aminoacyl-tRNA biosynthesis | 3 (2.94%) | 0.01145166 | 0.38665301 |
| 4 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 2 (1.96%) | 0.01299674 | 0.38665301 |
| 5 | Cell cycle | 4 (3.92%) | 0.02834388 | 0.42338825 |
| 6 | Tight junction | 5 (4.9%) | 0.1001368 | 0.69844209 |
| 7 | Oocyte meiosis | 3 (2.94%) | 0.1222549 | 0.69844209 |
| 8 | MAPKa signaling pathway | 5 (4.9%) | 0.1373397 | 0.69844209 |
| 9 | JAK-STATb signaling pathway | 3 (2.94%) | 0.1782376 | 0.69844209 |
| 10 | Wnt signaling pathway | 3 (2.94%) | 0.1836445 | 0.69844209 |
| 11 | Metabolic pathways | 13 (12.75%) | 0.1845772 | 0.69844209 |
| 12 | Regulation of actin cytoskeleton | 5 (4.9%) | 0.2526815 | 0.69844209 |
a MAPK, mitogen-activated protein kinase.
b JAK-STAT, Janus kinase-signal transducers and activators of transcription.
Single cell transcriptome sequencing can help reveal cellular heterogeneity (34). Several lines of evidence provided by single oocyte RNA-seq also discovered oocyte heterogeneity in the current study. First, correlation analyses on RNA-Seq data could not separate oocytes clearly into two groups (BCB-negative and BCB-positive) (Fig. S5A). Even principal component analysis could only place BCB-negative samples relatively closer, but within the BCB-positive group, one oocyte (named as 15) was still positioned far away from the other two oocytes (named as 6 and 7) (Fig. S5B). Second, for each oocyte sample, different numbers of novel transcripts with coding and noncoding potentials were found (Fig. S5C). Next, a large number of genes were differentially expressed even for two samples within the same group, for example, sample 7 versus sample 15 in the same BCB-positive group (Fig. S5D). Last, after pair-comparison of oocyte samples taken from BCB-negative and BCB-positive groups, 10,000s of common genes were found to be expressed, whereas more than 1000 genes were expressed specifically in only one group (Fig. S6, A–C). Sample-specific gene expression analysis showed that for each oocyte, only a few genes were expressed specifically (Fig. S6D). All these evidence point to the fact that sampled GV oocytes were of heterogeneous origins, even though they belong to the same BCB-negative or BCB-positive group.
CDC5L highly abundant in BCB-positive oocytes with less DNA double strand breaks
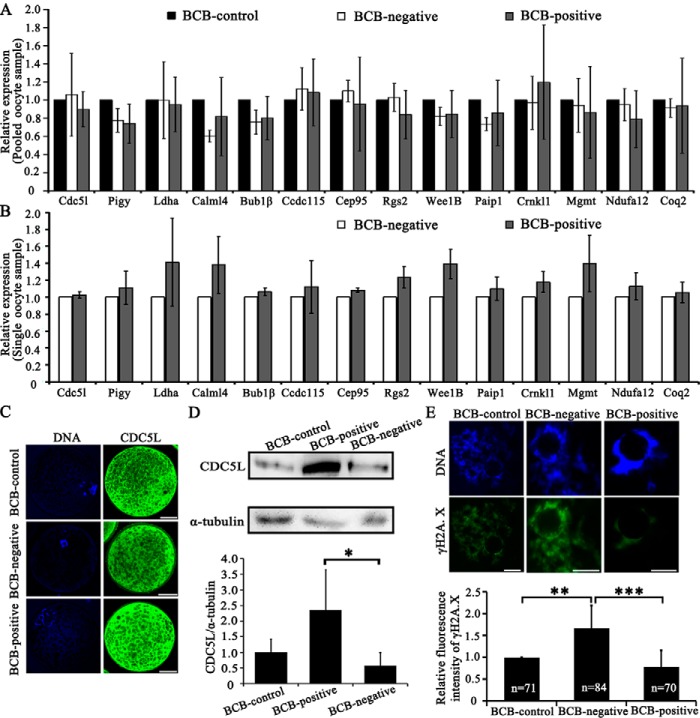
To validate DEGs discovered between BCB-positive and BCB-negative oocytes, cdc5l and 13 other DEGs were selected for RT-qPCR, profiled on pooled or single oocyte samples for the BCB-control, BCB-negative, and BCB-positive groups, respectively (Tables S4–S6). However, no significant expression differences were found for any of these genes, regardless of pooled (Fig. 4A) or single-oocytes (Fig. 4B; Fig. S7). Examination on the subcellular localization by immunostaining showed that the CDC5L protein in porcine GV oocytes was present in both cytoplasm and nuclei areas, except for the nucleolus region (Fig. 4C). Western blots validated significantly higher abundance of CDC5L in BCB-positive oocytes, than BCB-negative oocytes (Fig. 4D).
Figure 4.
Validation of single-oocyte RNA-seq results. A, RT-qPCR to validate 14 genes (cdc5l, pigy, ldha, calml4, bub1β, ccdc115, cep95, rgs2, wee1b, paip1, crnkl1, mgmt, ndufa12, and coq2) using pooled samples, which were selected from differential expressed genes identified by single-oocyte RNA-seq (details in Table S4). B, RT-qPCR using single-oocytes. The graphs presented the average relative expression levels of 9 single-oocytes (three replicates). C, immunostaining of the CDC5L protein, showing its localization in both the cytoplasm and nuclear area (except for nucleolus) of pig GV oocytes. Green, CDC5L; blue, DNA. Scale bar, 50 μm. D, Western blot of the CDC5L protein. Relative expression of CDC5L to α-tubulin in pig GV oocytes was quantified using ImageJ software. E, immunostaining of γH2A.X to indicate the occurrence of DSBs in GV oocytes. Relative fluorescence intensity of γH2A.X was quantified by ImageJ software. Green, γH2A.X; blue, DNA. Scale bar, 10 μm. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Previously, CDC5L was discovered to participate in the formation of the pre-messenger RNA splicing complex, and regulate cell-cycle progression and DNA damage response (39, 48). To see if CDC5L is involved in the nucleotide excision repair pathway as revealed by GO and KEGG enrichment analyses on our single-oocyte RNA-seq data, the γH2A.X level was examined by immunostaining, to see the incidence of DNA double-strand breaks (DSBs). Indeed, the relative fluorescence level of the γH2A.X protein was significantly higher in the BCB-negative group, compared with the BCB-positive group (Fig. 4E). Therefore, BCB-negative oocyte has less CDC5L protein, which possibly contributes to more frequently occurred DSB events.
CDC5L is essential for oocyte meiotic maturation and early embryo development
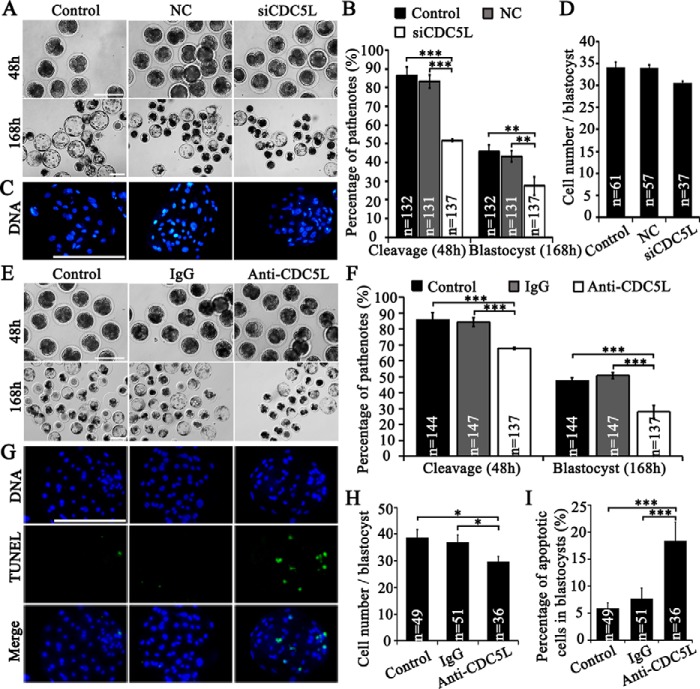
To investigate the role of CDC5L in porcine oocyte maturation, we performed depletion of CDC5L through microinjection of specific cdc5l siRNA and antibody into porcine-denuded GV oocytes followed by co-culture with cumulus cells for 44 h during IVM. We found that no significant differences for the maturation and development rates existed between porcine COCs and denuded GV oocytes co-cultured with cumulus cells (Fig. S8, A–C). Interference efficiency of cdc5l siRNA after microinjection into GV oocytes was validated using RT-qPCR (0.37 versus 1.00 of the control group; p < 0.001; Fig. 5A) and immunostaining (0.65 versus 1.00 of the control group; versus 1.06 of the NC group; p < 0.001; Fig. 5B), on live oocytes collected at 44 h of IVM. Successful injection of anti-CDC5L antibody was verified by immunostaining immediately after injection using only secondary antibody, as compared with control and IgG-injected porcine GV oocytes (Fig. S9, A–C). Western blot analysis verified the antibody specificity, by showing that it can recognize only the CDC5L protein in GV oocytes (Fig. S9D).
Figure 5.
CDC5L's function in meiotic maturation. A, siCDC5L interfering efficiency by RT-qPCR. B, CDC5L immunostaining after siCDC5L microinjection. C, GVBD rates of oocytes after 24 h IVM were decreased after siCDC5L microinjection. D, cell cycle of oocytes after 44 h IVM was perturbed by siCDC5L microinjection. E, anti-CDC5L antibody injection into GV oocytes significantly decreased the GVBD rates of oocytes after 24 h IVM. F, anti-CDC5L antibody injected into GV oocyte-blocked meiotic resumption, induced abnormal chromatin, and decreased oocyte maturation to MII stage. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Scale bar, 50 μm.
After cdc5l siRNA or anti-CDC5L antibody injection into GV oocytes followed by IVM for 24 h, the GVBD rates were shown to decrease significantly in both CDC5L inhibition methods. The GVBD rate of the siCDC5L group was significantly lower than that in the NC injection group (54.2 versus 87.6%; p < 0.001) and the control group (54.2 versus 85.8%; p < 0.001) (Fig. 5C). In parallel, the GVBD rate of the anti-CDC5L antibody group was also significantly lower than that in the IgG injection group (61.8 versus 84.5%; p < 0.001) and control group (61.8 versus 84.4%; p < 0.001) (Fig. 5E). After IVM for 44 h, the percentage of MII oocytes decreased significantly by cdc5l siRNA injection, 46.2 versus 75.2% of the NC group (p < 0.001) and 77.5% of the control group (p < 0.001) (Fig. 5D). The decreased maturation rate was caused by a significant increase of oocytes arrested at the GV stage (13.3 versus 2.9% of the control group; p < 0.01) and with chromosomal abnormality (23.7 versus 5.6% of the NC group; versus 5.1% of the control group; p < 0.001) (Fig. 5D). In parallel, immunoinhibition of CDC5L also induced the lower maturation rate as compared with that in the IgG injection group (42.1 versus 75.0%; p < 0.001) and in the control group (42.1 versus 76.1%; p < 0.001), respectively (Fig. 5F). It also caused a dramatic increase of oocytes arrested at the GV stage (12.2 versus 3.8% of the IgG group; p < 0.05), and also chromosomal abnormality, causing developmental block (17.9 versus 6.5% of the IgG group; versus 4.3% of the control group; p < 0.01) (Fig. 5F). Collectively, these data demonstrate that CDC5L is important for porcine oocyte meiotic maturation.
To examine the importance of maternal CDC5L in early embryo development, we inhibited CDC5L by siRNA and antibody microinjection followed by parthenogenetic activation and embryo culture. First, microinjection of siRNA into GV oocytes induced a significant decrease of cdc5l/CDC5L at both mRNA and protein levels in mature oocytes (Fig. 5, A and B). Further parthenogenetic activation of MII oocytes, in vitro matured from GV oocytes injected with siRNA, showed that the first mitotic cell cycle was delayed, by increasing the percentage of embryos arrested at the 1-cell stage (76.7 versus 61.0% of the NC group; p < 0.01; versus 58.5% of the control group; p < 0.01), and thus decreasing the percentage of 2-cell embryos (20 h post-activation) (23.3 versus 39.0% of the NC group; p < 0.01; versus 39.6% of the control group; p < 0.01) (Table 2). Until later stages of the first (26 h post-activation) and second mitotic cell cycles (39 and 48 h post-activation), the 1-cell parthenote percentage was still higher, whereas 3- and 4-cell parthenote percentages were significantly lower than the control groups (Table 2). As a result, a significant decrease of overall cleavage rates (≥2-cell) at 48 h post-activation was observed (51.8 versus 83.2% of the NC group; versus 86.4% of the control group; p < 0.001; Fig. 6, A and B). In addition, the total blastocyst rates significantly decreased as compared with the NC group (day 6, 26.2 versus 41.2%; p < 0.01; day 7, 27.5% versus 43.2%; p < 0.01) and the control group (day 6, 26.2 versus 42.7%; p < 0.01; day 7, 27.5 versus 45.8%; p < 0.01) (Table 3; Fig. 6, A and B). However, the cell number per blastocyst (day 7) was not affected by CDC5L inhibition (Fig. 6, C and D). Second, immunodepletion of cdc5l by microinjection of anti-CDC5L antibody into MII oocytes significantly increased the percentage of parthenotes arrested at the 1-cell stage, and thus decreased the percentage of 2-cell embryos (26 h post-activation), as compared with the IgG injection group (1-cell, 39.8 versus 26.2%; 2-cell, 38.0 versus 52.8%; p < 0.05) and the control group (2-cell, 38.0 versus 53.4%, p < 0.05) (Table 4). After continuously tracking the second cycle of embryo division, we saw that a number of 4-cell embryos (39 h post-activation) were significantly lower in the anti-CDC5L antibody group in comparison to the IgG (12.9 versus 26.9%; p < 0.01) and control (12.9 versus 26.8%; p < 0.01) groups (Table 4). As a result, a significant decrease of overall cleavage rates (≥2-cell) at 48 h post-activation was observed (67.9 versus 84.2% of the IgG group; versus 85.7% of the control group; p < 0.001; Fig. 6, E and F). When parthenotes in vitro were cultured for 5–7 days, the percentage of expanded blastocysts at days 5, 6, and 7 was significantly lower in the anti-CDC5L antibody group, as compared with the control (day 5, 10.9 versus 18.2%; day 6, 19.1 versus 33.7%; day 7, 20.0 versus 34.2%; p < 0.01) and the IgG (day 5, 10.9 versus 25.5%; day 6, 19.1 versus 37.5%; day 7, 20.0 versus 39.6%; p < 0.01) groups (Table 5). Therefore, a significant decrease of total blastocyst rate at day 7 was observed for the anti-CDC5L antibody group (28.0 versus 50.8% of the IgG group; versus 47.4% of the control group; p < 0.001; Table 5; Fig. 6, E and F). CDC5L antibody microinjection also significantly decreased the total cell number per blastocyst at day 7 (30 versus 37 of the IgG group; versus 39 of the control group; p < 0.05; Fig. 6, G and H), and increased significantly the percentage of apoptotic cells in blastocysts (18.4 versus 7.7% of the IgG group; versus 5.9% of the control group; p < 0.001; Fig. 6, G and I). Taken together, our results indicate that CDC5L insufficiency in mature oocytes induced by both siRNA and antibody immunobinding could delay the progression of the first and second mitotic cell cycles of parthenotes, and thus affected the formation of blastocysts, suggesting that maternal CDC5L is vital for early embryo development in the pig.
Table 2.
Distribution of cleaved parthenotes recorded at different time points after microinjection of siCDC5L into GV oocytes followed by maturation and parthenogenetic activation
Control, without microinjection; NC, microinjected negative control; siCDC5L, microinjected cdc5l siRNAs. a-b and A-B indicate significant differences at p < 0.05 and p < 0.01 levels within the same subcolumns, respectively.
| Group | No. embryos | No. 1-cell | No. 2-cell | No. 3-cell | No. 4-cell | No. 5-cell | |
|---|---|---|---|---|---|---|---|
| % ± S.D. | |||||||
| 20 h | Control | 133 | 78 (58.5 ± 2.0%)A | 53 (39.6 ± 2.7%)A | 1 (1.0 ± 1.6%) | 0 | 0 |
| NC | 132 | 80 (61.0 ± 2.5%)A | 52 (39.0 ± 2.5%)A | 0 | 0 | 0 | |
| siCDC5L | 140 | 107 (76.7 ± 3.7%)B | 33 (23.3 ± 3.7%)B | 0 | 0 | 0 | |
| 26 h | Control | 133 | 36 (27.2 ± 1.6%)a | 73 (54.1 ± 7.3%) | 13 (9.4 ± 3.2%)ab | 5 (4.8 ± 8.2%) | 0 |
| NC | 131 | 39 (31.8 ± 13.1%)ab | 65 (48.2 ± 9.7%) | 21 (15.2 ± 5.3%)a | 2 (2.1 ± 3.6%) | 0 | |
| siCDC5L | 137 | 57 (42.6 ± 5.2%)b | 58 (42.3 ± 0.7%) | 9 (6.1 ± 2.9%)b | 0 | 0 | |
| 39 h | Control | 132 | 15 (12.3 ± 7.4%)A | 29 (21.6 ± 3.9%) | 48 (35.9 ± 3.4%) | 37 (28.2 ± 2.4%)A | 0 |
| NC | 131 | 19 (15.4 ± 6.1%)A | 30 (22.4 ± 3.4%) | 50 (37.3 ± 5.3%) | 30 (23.5 ± 4.0%)A | 0 | |
| siCDC5L | 137 | 51 (38.3 6.1%)B | 37 (27.2 ± 4.9%) | 26 (28.7 ± 4.2%) | 8 (5.9 ± 0.5%)B | 0 | |
| 48 h | Control | 132 | 14 (11.6 ± 7.8%)A | 25 (18.5 ± 3.4%) | 34 (25.3 ± 5.9%) | 56 (42.5 ± 3.7%)A | 0 |
| NC | 131 | 19 (15.4 ± 6.1%)A | 36 (26.0 ± 9.9%) | 24 (18.8% ± 3.1%) | 47 (36.4% ± 3.8%)A | 3 (2.0% ± 2.1) | |
| siCDC5L | 137 | 53 (39.5 ± 6.1%)B | 33 (24.2 ± 5.9%) | 26 (19.2 ± 8.3%) | 12 (8.4 ± 3.8v)B | 0 | |
Figure 6.
CDC5L vital to early embryo development. A, parthenotes (48 h) and blastocysts (168 h) after siCDC5L microinjection into oocytes at GV stages. Control, without microinjection; siCDC5L, cdc5l-specific siRNA group. B, total cleavage rates (≥2 cells at 48 h) and blastocyst rates (day 7) of parthenotes after siRNA microinjection. C, representative images of cell number of blastocysts. D, average cell number per blastocyst. E, parthenotes (48 h) and blastocysts (168 h) after antibody microinjection. Control, without injection. F, total cleavage rates (≥2 cells at 48 h) and blastocyst rates (day 7) of parthenotes after antibody microinjection. G, cell number and TUNEL signal of blastocysts at day 7 from control, oocytes injected with IgG and anti-CDC5L antibody. Green, TUNEL signal; blue, DNA. H, average cell number per blastocyst. I, percentages of apoptotic cells in blastocysts at day 7. *, p < 0.05 and ***, p < 0.001. Scale bar, 200 μm.
Table 3.
Blastocysts recorded at different time points after microinjection of siCDC5L into GV oocytes followed by maturation and parthenogenetic activation
Control, without microinjection; NC, microinjected negative control; siCDC5L, microinjected cdc5l siRNAs. a-b and A-B indicate significant differences at p < 0.05 and p < 0.01 levels within the same subcolumns, respectively.
| Group | Total No. embryos | Total No. balstocyst | No. blastocoelic balstocyst | No. expanded blastocyst | No. hatched blastocyst | |
|---|---|---|---|---|---|---|
| % ± S.D. | ||||||
| Day 5 | Control | 132 | 40 (29.7 ± 6.3%) | 18 (13.1 ± 3.8%) | 22 (16.6 ± 4.1%) | 0 |
| NC | 131 | 41 (30.9 ± 2.6%) | 20 (15.0 ± 5.0%) | 21 (15.9 ± 3.6%) | 0 | |
| siCDC5L | 137 | 29 (20.2 ± 8.4%) | 15 (10.4 ± 3.7%) | 14 (9.8 ± 5.1%) | 0 | |
| Day 6 | Control | 132 | 57 (42.7 ± 4.3%)A | 15 (11.4 ± 0.7%) | 34 (25.6 ± 3.6%) | 8 (5.7 ± 2.5%) |
| NC | 131 | 54 (41.2 ± 2.3%)A | 14 (10.9 ± 2.7%) | 34 (25.4 ± 3.1%) | 6 (4.8 ± 1.3%) | |
| siCDC5L | 137 | 35 (26.2 ± 7.0%)B | 11 (9.2 ± 6.0%) | 21 (15.1 ± 3.9%) | 3 (2.0 ± 3.4%) | |
| Day 7 | Control | 132 | 61 (45.8 ± 5.0%)A | 15 (11.4 ± 1.5%) | 35 (26.5 ± 1.1%) | 11 (7.8 ± 4.2%) |
| NC | 131 | 57 (43.2 ± 4.1%)A | 17 (12.9 ± 1.5%) | 32 (24.1 ± 2.0%) | 8 (6.2 ± 2.2%) | |
| siCDC5L | 137 | 35 (27.5 ± 6.8%)B | 13 (10.4 ± 5.0%) | 21 (15.1 ± 3.9%) | 3 (2.0 ± 3.4%) | |
Table 4.
Distribution of cleaved embryos recorded at different time points after microinjection of CDC5L antibody into MII oocytes followed by parthenogenetic activation
Control, without microinjection; NC, microinjected negative control; siCDC5L, microinjected CDC5L siRNAs. a-b and A-B indicate significant differences at p < 0.05 and p < 0.01 levels within the same subcolumns, respectively.
| Group | No. embryos | No. 1-cell | No. 2-cell | No. 3-cell | No. 4-cell | No. 5-cell | |
|---|---|---|---|---|---|---|---|
| % ± S.D. | |||||||
| 20 h | Control | 148 | 88 (59.9 ± 6.5%) | 51 (34.3 ± 0.8%) | 4 (2.7 ± 1.0%) | 0 | 0 |
| IgG | 150 | 85 (57.2 ± 2.8%) | 51 (34.1 ± 5.3%) | 8 (5.0 ± 4.4%) | 0 | 0 | |
| Anti-CDC5L | 140 | 90 (64.3 ± 2.2%) | 32 (22.9 ± 1.1%) | 4 (2.9 ± 1.2%) | 0 | 0 | |
| 26 h | Control | 147 | 37 (26.1 ± 10.5%)a | 78 (53.4 ± 3.8%)a | 13 (8.7 ± 1.1%) | 1 (0.6 ± 1.1%) | 0 |
| IgG | 150 | 39 (26.2 ± 6.6%)a | 79 (52.8 ± 2.6%)a | 15 (10.0 ± 0.9%) | 3 (2.0 ± 2.0%) | 0 | |
| Anti-CDC5L | 139 | 55 (39.8 ± 10.8%)b | 53 (38.0 ± 3.5%)b | 9 (6.5 ± 2.4%) | 1 (7.6 ± 1.3%) | 0 | |
| 39 h | Control | 144 | 15 (10.8 ± 6.4%) | 31 (21.9 ± 8.7%) | 40 (28.5 ± 6.5%) | 40 (26.8 ± 9.2%)A | 7 (4.7 ± 1.8%) |
| IgG | 147 | 14 (9.7 ± 3.2%) | 32 (22.0 ± 6.4%) | 41 (27.9 ± 2.4%) | 40 (26.9 ± 8.0%)A | 6 (4.2 ± 2.4%) | |
| Anti-CDC5L | 138 | 23 (16.7 ± 1.0%) | 32 (23.7 ± 12.2) | 37 (26.6 ± 7.3%) | 18 (12.9 ± 4.9%)B | 4 (2.9 ± 2.5%) | |
| 48 h | Control | 144 | 14 (10.2 ± 7.4%) | 31 (21.7 ± 6.7%) | 37 (25.7 ± 0.2%) | 46 (31.5 ± 4.3%) | 10 (6.9 ± 2.7%) |
| IgG | 147 | 11 (7.7 ± 3.7%) | 30 (20.8 ± 8.9%) | 38 (15.5 ± 6.1%) | 46 (31.2 ± 5.1%) | 7 (4.6 ± 2.7%) | |
| Anti-CDC5L | 137 | 20 (14.5 ± 4.5%) | 23 (17.1 ± 7.8%) | 31 (22.5 ± 5.4%) | 32 (23.4 ± 3.0%) | 7 (5.0 ± 5.1%) | |
Table 5.
Blastocysts recorded at different time points after microinjection of CDC5L antibody into MII oocytes followed by parthenogenetic activation
Control, without microinjection; NC, microinjected negative control; siCDC5L, microinjected cdc5l siRNAs. a-b and A-B indicate significant differences at p < 0.05 and p < 0.01 levels within the same subcolumns, respectively.
| Group | Total embryos | Total No. balstocyst | No. blastocoelic balstocyst | No. expanded blastocyst | No. hatched blastocyst | |
|---|---|---|---|---|---|---|
| % ± S.D. | ||||||
| Day 5 | Control | 144 | 41 (29.6 ± 10.6%)AB | 16 (11.4 ± 3.7%) | 25 (18.2 ± 9.2%)AB | 0 |
| IgG | 147 | 50 (34.3 ± 4.8%)A | 13 (8.8 ± 3.0%) | 37 (25.5 ± 6.8%)A | 0 | |
| Anti-CDC5L | 137 | 23 (16.7 ± 4.3%)B | 8 (5.8 ± 1.0%) | 15 (10.9 ± 4.3%)B | 0 | |
| Day 6 | Control | 144 | 65 (45.4 ± 2.3%)A | 15 (10.4 ± 1.5%) | 48 (33.7 ± 4.3%)A | 2 (1.3 ± 1.1%) |
| IgG | 147 | 75 (50.8 ± 2.8%)A | 16 (10.6 ± 5.6%) | 55 (37.5 ± 2.7%)A | 5 (3.4 ± 2.4%) | |
| Anti-CDC5L | 137 | 36 (27.2 ± 5.3%)B | 10 (7.4 ± 3.8%) | 26 (19.1 ± 8.7%)B | 0 | |
| Day 7 | Control | 144 | 68 (47.4 ± 2.8%)A | 14 (9.6 ± 2.2%) | 49 (34.2 ± 1.6%)A | 5 (3.6 ± 1.7%) |
| IgG | 147 | 75 (50.8 ± 2.8%)A | 10 (6.6 ± 4.1%) | 58 (39.6 ± 2.3%)A | 7 (4.7 ± 2.1%) | |
| Anti-CDC5L | 137 | 38 (28.0 ± 5.5%)B | 10 (7.3 ± 2.5%) | 27 (20.0 ± 9.4%)B | 1 (0.7 ± 1.2%) | |
Discussion
BCB test and oocyte quality
In reproductive biology, it is always a challenging task to select oocytes of better quality and developmental competence. In various animal species, the BCB test can color the cytoplasm of immature oocytes of low G6PDH activity in blue, indicating better maturation and developmental competence (4, 7, 20–22). Oocytes with better quality could also improve the production of transgenic pigs by somatic cell nuclear transfer techniques (26, 27). Numerous studies tried to explain why the BCB test is effective in oocyte selection, such as abundance of maternal transcripts (12, 13), mitochondrial number/function (18, 19), and lipid content (19). In line with previous reports (49, 50), we confirmed also that BCB-positive porcine oocytes have a higher maturation rate, better parthenote cleavage, and blastocyst rates.
In addition, chromatins of NSN-type oocytes are transcriptionally active, whereas SN-type oocytes are silent (36, 37). In the present study the percentages of NSN-type and SN-type oocytes showed no obvious difference between BCB-negative and BCB-positive groups, indicating that the transcriptional activity may not link to the maturation ability. Although in the BCB-negative group, a higher percentage of oocytes with the GV4 configuration might indicate that more BCB-negative oocytes are on the way toward atresia, as reported previously (37).
Validation of DEGs and pathways
Single-cell high-throughput sequencing technology has been used to screen human embryos of better quality for transfer purpose (35), profile the global gene expression, and build the transcriptomic landscape and gene regulatory framework of oocytes and early embryos in humans and mice (28, 29). At the same time, it also has advantages in saving rare samples and revealing cellular heterogeneity (34). In the current study, RT-qPCR did not find any significance for transcript levels of 14 selected DEGs between BCB-positive and BCB-negative pooled and single-oocyte samples, suggesting that the RT-qPCR performed using both pooled and single-oocyte samples (different from the RNA-seq samples) might not be effective and sufficient to validate the single-oocyte RNA-seq results. Previous reports on single-oocyte/embryo/blasomere/embryonic stem cell RNA-sequencing did not perform the validation of their results using RT-qPCR (28, 29), or qPCR validation using the same cDNA templates (amplified from the single cell) for RNA-seq (51). Alternatively, validation of findings at the protein level might be another approach, as reported in human blastocysts using immunofluorescence (52). Therefore, combining different approaches to validate the obtained results is important in avoiding the impacts of technical variability and sources of stochastic gene expression coming from single cells.
Using single-oocyte RNA-seq, we identified that the metabolic pathway is potentially involved in oocyte developmental competence. The BCB test can indicate the activity level of G6PDH, which reflects directly metabolic differences. Inherent metabolic differences can influence nuclear and cytoplasmic maturation of oocytes, which has been studied extensively (53–56). Apart from the metabolic pathway, several other signaling pathways were identified, such as cell cycle, oocyte meiosis, spliceosome, and nucleotide excision repair. How the metabolic pathway integrates with other signaling pathways, and exerts its effect on the developmental competence of oocytes, still awaits further investigation.
CDC5L and oocyte maturation
CDC5L has been reported to play a vital role in multiple biological processes. First, as a splicing factor, CDC5L participates in the formation of the spliceosome complex together with multiple other proteins, and functions in pre-mRNA splicing (57). In the present study, BCB-positive oocytes have a higher level of CDC5L protein than BCB-negative oocytes, consistent with the distribution of the transcript abundance detected by single-oocyte RNA-seq. These data indicate that the RNA-splicing role of CDC5L may be associated with the meiotic cell cycle progression of oocytes with different quality. However, as for AS events, no significant difference was found between BCB-negative and BCB-positive groups, although thousands of genes with different types of AS events were detected in each oocyte sample. Second, CDC5L was involved in DNA damage repair (48), and depletion of CDC5L could induce DNA DSB damage in HeLa cells (39). Our findings showed that the DSB incidence occurs more frequently (as revealed by γH2A.X immunostaining) in BCB-negative oocytes than in BCB-positive oocytes, which not only suggested that low CDC5L protein might cause more DNA DSB damages as observed in BCB-negative oocytes, but also explain the nucleotide excision repair pathway found by single-oocyte RNA-seq. Moreover, meiotic resumption of immature oocytes in humans (4) and mice (58) could be hindered by DNA DSB damage, therefore the DNA DSB damage induced partially by the low CDC5L level may help explain the lower maturation rate as observed in BCB-negative oocytes. Third, depletion of CDC5L causes a mitotic catastrophe of HeLa cells through induction of apoptosis (39), and promotes apoptosis in human glioma cell lines (59). Thus the low CDC5L-induced apoptosis might be one reason for the higher death rate in BCB-negative oocytes. Fourth, CDC5L protein has also been shown to regulate the G2/M transition of mitotic cell cycle, and deficiency of CDC5L caused mitotic arrest through induced kinetochore–microtubule attachment defects and chromosome misalignment (39). Our findings showed that depletion of CDC5L by both siRNA and antibody immunobinding in porcine GV oocytes dramatically inhibited the meiotic resumption at 24 h of IVM and the first polar body extrusion at 44 h of IVM through inducing sustained GV arrest and incidence of nuclear abnormality, suggesting that CDC5L is a vital meiotic regulator for GV/M transition. Moreover, depletion of the maternal CDC5L protein in porcine MII oocytes by siRNA microinjection into GV oocytes or anti-CDC5L antibody microinjection into MII oocytes impaired the first and second mitotic cell cycles of parthenote development, and the formation of blastocysts. Therefore, maternal CDC5L plays an essential regulatory role in porcine early embryo development.
Conclusions
Taken together, we confirmed that the BCB test could be used to select porcine immature oocytes with better maturation and developmental competence. Single-oocyte RNA-seq further profiled transcriptome of immature oocytes categorized by the BCB test, and identified 155 genes with significantly different abundances enriched in functionally important signaling pathways. CDC5L is confirmed to be essential to porcine oocyte maturation and early embryo development. Future studies are still needed to investigate the molecular roles of identified genes and pathways in the regulation of oocyte maturation and developmental competence.
Experimental procedures
Chemicals
All reagents used in this study were purchased from Sigma, unless stated otherwise.
Collection of COCs
Porcine ovaries from prepubertal gilts were collected at a local commercial slaughterhouse, as described previously (60). Briefly, ovaries were transported to the laboratory within 3 h in a thermos bottle in physiological saline maintained at 30 °C. Follicular fluids from 3- to 6-mm antral follicles were aspirated using an 18-gauge needle attached to a 10-ml disposable syringe. COCs with at least three layers of intact cumulus cells and uniform ooplasm were selected for further study.
Brilliant cresyl blue test and assessment of chromatin configuration
COCs were washed thoroughly in Hepes-buffered Tyrode medium containing 0.01% polyvinyl alcohol, and then subjected to the BCB test according to a previous protocol reported in pigs with some modifications (24). Briefly, COCs were incubated in Dulbecco's PBS (DPBS) supplemented with 13 μm BCB (BCB medium) for 90 min at 39 °C in a humidified 5% CO2 incubator. Then, COCs were washed twice in DPBS and assessed for BCB staining. Based on the staining intensity, COCs with blue ooplasm were classified as BCB-positive, whereas those with a colorless ooplasm were considered to be BCB-negative (Fig. 1A) (61). To assess the chromatin configuration of pig oocytes at the GV stage stained by BCB, cumulus cells were separated from oocytes by gentle vortexing in 0.1% hyaluronidase in Hepes-buffered Tyrode medium containing 0.01% polyvinyl alcohol. Denuded oocytes were fixed in 4% paraformaldehyde in PBS, stained with 10 μg/ml of Hoechst 33342 in PBS, and examined under a fluorescence microscope (Nikon 80i, Tokyo, Japan). Judging by the degree of chromatin condensation and the disappearance of nucleolus and nuclear membrane, the chromatin configurations of GV oocytes were classified into 5 different categories, GV0–GV4 (62). GV0 in pig oocytes was regarded as corresponding to the NSN configuration in mice and other species, whereas GV1–GV3 were designated as SN configuration (36, 37, 62) (Fig. 1C).
In vitro maturation and parthenogenetic activation
Approximately 50 COCs were transferred into 500 μl of maturation medium covered with mineral oil in a 24-well plate, and cultured in an incubator (39 °C, 5% CO2, and saturated humid air) for 44 h (63). Then, the cumulus cells were detached from oocytes by gentle vortexing in 0.1% hyaluronidase. Under a stereomicroscope, live denuded oocytes with intact ooplasm membranes were picked out and death rates were calculated (number of died oocytes divided by total oocyte number × 100%). Then, MII stage oocytes with the presence of the first polar body were picked out and maturation rates were calculated (number of MII oocytes divided by total live oocyte number × 100%). To assess the oocyte nuclear status, live oocytes collected at 44 h of IVM were fixed, stained with 10 μg/ml of Hoechst 33342 in PBS, and then observed under an inverted fluorescence microscope (Olympus, Tokyo, Japan). Stages of oocyte nuclei were classified as germinal vesicle (GV), pro-metaphase I (pro-MI), metaphase I (MI), anaphase I (AI), telophase I (TI), and MII, respectively (63).
MII oocytes were parthenogenetically activated using a protocol as described previously (63). Briefly, MII oocytes were stimulated with two direct current pulses of 1.2 kV/cm for 30 μs using a Electrocell Manipulator (BTX830, USA) in a chamber covered with activation medium (0.28 m mannitol, 0.1 mm CaCl2·2H2O, 0.1 mm MgCl2, 1 mg/ml of BSA, 0.5 mm Hepes). Then oocytes were incubated for 4 h in PZM-3 containing 2.5 mm 6-dimethylaminopurine and 5 μg/ml of cytochalasin B in an incubator (39 °C, 5% CO2 and saturated humid air). Activated embryos were cultured in 500 μl of PZM-3 medium covered with mineral oil for 7 days in an incubator (39 °C, 5% CO2 and saturated humid air). The rates of embryo cleaved and developed to blastocysts were evaluated at different time points post electrostimulation. The embryo rate at different stages was calculated using the number of embryos at different stages divided by the total number of parthenotes. The blastocyst rate was calculated using the number of blastocysts divided by the total number of parthenotes. Blastocysts at day 7 were fixed in 4% paraformaldehyde for 40 min at room temperature, permeabilized in 0.5% Triton X-100 for 2 h at room temperature, incubated in TUNEL reaction medium in dark for 1 h at room temperature (Vazyme Biotech Co., Nanjing, China), and stained using 10 μg/ml of Hoechst 33342. Whole-mount blastocysts were examined under an inverted fluorescence microscope (Olympus, Tokyo, Japan) to determine the number of apoptotic nuclei and total nuclei.
Single-oocyte RNA-seq
BCB-negative and BCB-positive oocytes were denuded off cumulus cells by gentle vortexing in 0.1% hyaluronidase. Following four washes using DPBS containing 1% BSA, the zona pellucida of denuded oocytes was removed using Pronase (0.5% in DPBS). The zona pellucida-free oocytes were washed three times in DPBS containing 1% BSA, and then each single oocyte with a minimal amount of wash buffer (<0.5 μl) was placed into 4.35 μl of cell lysis buffer in a 0.2-ml tube using pulled glass pipette. The same volume of DPBS, 1% BSA wash buffer added into lysis buffer was used as the negative control to ensure the absence of contamination. The whole oocyte lysate of three BCB-negative (numbered as 1, 2, and 3) and three BCB-positive (6, 7, and 15) oocytes were directly submitted for amplification, and the sequencing libraries were prepared as described before (64). Briefly, the first-strand cDNA was synthesized from the whole oocyte lysate using the UP1 primer containing a 24-nt poly(dT) tail at the 3′ end, which allowed all RNAs with poly(A) tails to be reversely transcribed. Then, the first-strand cDNA were amplified by PCR using hot start EX Taq and UP2 primer containing 24-nt poly(dT) at the 3′ end to add a universal tail to the second-strand cDNA. Amplified cDNA were purified from gel and evaluated using Agilent's Bioanalyzer (Agilent Technologies). The prepared cDNAs were sheared into fragments, and used for preparation of deep sequencing libraries. All libraries were sequenced on an Illumina HiSeq 2000 instrument in a single lane.
Raw reads were processed into clean reads by removing low quality and adaptor containing reads. Then, clean reads were mapped onto the pig reference genome (Sscrofa10.2) by Burrows-Wheeler Aligner (BWA) (65). After performing the assessment of sequence saturation and read distribution on reference genes, gene expression levels were calculated as FPKM by Tophat and Cufflinks (66), using the number of reads uniquely mapped to the specific gene and the total number of uniquely mapped reads for each sample. DEGs were identified by RSEM (RNA-Seq by Expectation Maximization) (67), and DAVID was used to enrich the GO biological process and KEGG pathways of DEGs (68).
RT-qPCR
To validate the RNA-seq results using pooled or single-oocyte samples, denuded GV oocytes from different groups were treated with Pronase (0.5% in DPBS) to remove zona pellucida. After rinsing twice with DPBS, 30 pooled oocytes (3 replicates) were lysed directly using the Single Cell Lysis Kit (Ambion®, Life Technologies) according to the manufacturer's recommendations and our previous description (60), and single-oocytes (3 single-oocyte for each replicate, total 3 replicates) were lysed directly in 0.2% Triton X-100 with RNase inhibitor (69). To validate siRNA knockdown efficiency, 20 pooled denuded oocytes were used to extract total RNA using a RNeasy mini kit (Qiagen, Shanghai, China). The lysates or total RNAs were then used for reverse transcription using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) in a 20-μl system. Quantitative PCR was conducted in a 10-μl reaction volume including 1 μl of cDNA templates, primers, and Roche FastStart Universal SYBR Green master mix (Roche Molecular Systems), using a 7500 real-time PCR detection system (Applied Biosystems, Carlsbad, CA). Primers were designed by primer-blast (Table S6). Thermal cycling parameters were set up as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Transcripts were quantified in triplicate assays, and Actin was used as the reference gene. Relative abundance was calculated using the comparative Ct (2−ΔΔCt) method (63).
Western blot
Denuded GV oocytes (a total of 100) were pooled, snap frozen in liquid nitrogen, and stored at −80 °C. Western blot analysis was performed according to our previously reported method with some modifications (60). Briefly, samples were lysed using SDS sample buffer, boiled for 4 min, separated on SDS-PAGE gel, and transferred onto nitrocellulose membrane. The membrane was blocked for 1 h in 5% low-fat milk, incubated overnight at 4 °C in 0.5% low-fat milk with 1:2000 rabbit anti-CDC5L polyclonal antibody (A5560, ABclonal, Nanjing, China), and then 1 h at room temperature with anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:10,000). Finally, membranes were processed using a chemiluminescence detection system. Detected bands of predicted size for CDC5L and tubulin (an internal control for normalization) were quantified using ImageJ software.
Immunocytochemistry staining
Denuded oocytes were fixed for 40 min at room temperature using 4% paraformaldehyde in PBS, permeabilized with 1% Triton X-100 overnight in PBS at 4 °C, blocked with 1% BSA in PBS at room temperature for 1 h, then incubated in rabbit anti-CDC5L polyclonal antibody (1:50) overnight at 4 °C. After washing three times in PBS supplemented with 0.01% Triton X-100 and 0.1% Tween 20 (PBST), oocytes were incubated in FITC-conjugated goat anti-rabbit IgG (H+L) secondary antibody (1:150) for 1 h at room temperature. Following three washes, samples were stained with 10 μg/ml of Hoechst 33342 at room temperature for 10 min and then mounted onto glass slides in ProLong Diamond Antifade Mountant reagent (Life Technologies). For γH2A.X immunostaining, oocytes were permeabilized in 1% Triton X-100 at room temperature for 30 min, blocked using 3% BSA, incubated in mouse anti-γH2A.X antibody (1:1000, Abcam, Cambridge, UK) overnight at 4 °C and FITC-conjugated goat anti-mouse IgG(H+L) for 1 h. Samples incubated without the primary or secondary antibodies were treated as negative controls. Fluorescent images were taken under a laser-scanning confocal microscope (Leica, Germany) to assess the subcellular localization of proteins.
siRNA and antibody microinjection
siRNA targeting of the pig cdc5l gene was designed and synthesized by GenePharma Co. (Shanghai, China). The sense sequence of cdc5l siRNA was (5′-3′) GCUGCCCUCCAGAAGAGAATT, and the antisense sequence was (5′-3′) UUCUCUUCUGGAGGGCAGCTT. The sense sequence of negative control (NC) was (5′-3′) UUCUCCGAACGUGUCACGUTT and the antisense sequence was (5′-3′) ACGUGACACGUUCGGAGAATT. Rabbit anti-CDC5L antibody and rabbit IgG protein were from ABclonal Co. (Nanjing, China). Microinjection of siRNA or antibody into porcine-denuded GV and MII stage oocytes was performed using the procedure as described previously (70). Briefly, oocytes in the manipulation medium supplemented with 7.5 μg/ml of cytochalasin B and 0.3% BSA were microinjected with ∼10 pl of siRNA or antibody using sterile femtotips and the FemtoJet express microinjector (Eppendorf). The injection parameters were 200 hectopascal injection pressure, 100 hectopascal compensation pressure, and 0.3-s injection time. The same amount of siRNA NC or rabbit IgG protein was injected into oocytes as the control. After microinjection, denuded GV oocytes were co-cultured with cumulus cells in the maturation medium and MII oocytes were parthenogenetically activated.
Statistical analysis
Statistical analyses were performed in SPSS19 (SPSS Inc., Chicago, IL), and SAS9.1 (SAS Institute Inc., Cary, NC). Each experiment was repeated at least three times. The differences in the percentages of chromatin configuration, GVBD, nuclear status, maturation, death, embryo cleavage, blastocysts, and apoptotic cells in blastocysts were compared using the χ-square test or Fisher's exact test followed by multiple comparisons with Bonferroni correction (SPSS19). One-way analysis of variance was used to analyze the data of cell number per blastocyst and protein levels detected by Western blots (SPSS19). Differences in gene expression detected by RT-qPCR were analyzed using the linear mixed model procedure (SAS9.1). Statistical differences were determined to be significant at p < 0.05, 0.01, and 0.001 levels. Data are displayed as mean ± S.D.
Author contributions
Z. Q. D. and C. X. Y. designed, coordinated the study, and wrote the paper. X. M. L. performed immunostaining, microinjection, and embryo culture. Y. K. W. collected RNA-seq samples and performed the RT-qPCR validation. Y. H. L. performed Western blot analysis. X. X. Y. performed the single-oocyte RT-qPCR. P. C. W. and X. L. provided technical assistance and contributed to the data analysis. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We are grateful for the help and support provided by other lab members in Dr. Yang's group.
This work was supported by National Natural Science Foundation of China Grant 31472098 and a Start-up grant from the Northeast Agricultural University (to C. X. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9 and Tables S1–S6.
- BCB
- brilliant cresyl blue
- G6PDH
- glucose-6-phosphate dehydrogenase
- CDC5L
- cell division cycle 5-like
- COC
- cumulus-oocyte complex
- GV
- germinal vesicle
- GVBD
- germinal vesicle breakdown
- NSN
- non-surrounded nucleolus
- SN
- surrounded nucleolus
- IVM
- in vitro maturation
- MII
- metaphase II
- DEG
- differentially expressed gene
- GO
- Gene Ontology
- AS
- alternative splicing
- qPCR
- quantitative PCR
- DSB
- double-strand break
- NC
- negative control
- KEGG
- Kyoto Encyclopedia of Genes and Genomes.
References
- 1. Moussa M., Shu J., Zhang X. H., and Zeng F. (2015) Maternal control of oocyte quality in cattle “a review.” Anim. Reprod. Sci. 155, 11–27 10.1016/j.anireprosci.2015.01.011 [DOI] [PubMed] [Google Scholar]
- 2. Sha Q. Q., Dai X. X., Dang Y., Tang F., Liu J., Zhang Y. L., and Fan H. Y. (2017) A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 144, 452–463 10.1242/dev.144410 [DOI] [PubMed] [Google Scholar]
- 3. Mohammadi-Sangcheshmeh A., Held E., Rings F., Ghanem N., Salilew-Wondim D., Tesfaye D., Sieme H., Schellander K., and Hoelker M. (2014) Developmental competence of equine oocytes: impacts of zona pellucida birefringence and maternally derived transcript expression. Reprod. Fertil. Dev. 26, 441–452 10.1071/RD12303 [DOI] [PubMed] [Google Scholar]
- 4. Coticchio G., Dal Canto M., Guglielmo M. C., Albertini D. F., Mignini Renzini M., Merola M., Lain M., Sottocornola M., De Ponti E., and Fadini R. (2015) Double-strand DNA breaks and repair response in human immature oocytes and their relevance to meiotic resumption. J. Assist. Reprod. Genet. 32, 1509–1516 10.1007/s10815-015-0547-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Schepper G. G., van Noorden C. J., and Houtkooper J. M. (1987) Age-related changes of glucose-6-phosphate dehydrogenase activity in mouse oocytes. Histochem. J. 19, 467–470 10.1007/BF01675415 [DOI] [PubMed] [Google Scholar]
- 6. Alcoba D. D., Conzatti M., Ferreira G. D., Pimentel A. M., Kussler A. P., Capp E., von Eye Corleta H., and Brum I. S. (2016) Safety of brilliant cresyl blue staining protocols on human granulosa and cumulus cells. Zygote 24, 83–88 10.1017/S0967199415000052 [DOI] [PubMed] [Google Scholar]
- 7. Rodríguez-González E., López-Béjar M., Velilla E., and Paramio M. T. (2002) Selection of prepubertal goat oocytes using the brilliant cresyl blue test. Theriogenology 57, 1397–1409 10.1016/S0093-691X(02)00645-3 [DOI] [PubMed] [Google Scholar]
- 8. Catalá M. G., Izquierdo D., Uzbekova S., Morató R., Roura M., Romaguera R., Papillier P., and Paramio M. T. (2011) Brilliant Cresyl Blue stain selects largest oocytes with highest mitochondrial activity, maturation-promoting factor activity and embryo developmental competence in prepubertal sheep. Reproduction 142, 517–527 10.1530/REP-10-0528 [DOI] [PubMed] [Google Scholar]
- 9. Tabandeh M. R., Golestani N., Kafi M., Hosseini A., Saeb M., and Sarkoohi P. (2012) Gene expression pattern of adiponectin and adiponectin receptors in dominant and atretic follicles and oocytes screened based on brilliant cresyl blue staining. Anim. Reprod. Sci. 131, 30–40 10.1016/j.anireprosci.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 10. Mohapatra S. K., Sandhu A., Neerukattu V. S., Singh K. P., Selokar N. L., Singla S. K., Chauhan M. S., Manik R. S., and Palta P. (2015) Buffalo embryos produced by handmade cloning from oocytes selected using brilliant cresyl blue staining have better developmental competence and quality and are closer to embryos produced by in vitro fertilization in terms of their epigenetic status and gene expression pattern. Cell Reprogram. 17, 141–150 10.1089/cell.2014.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salviano M. B., Collares F. J., Becker B. S., Rodrigues B. A., and Rodrigues J. L. (2016) Bovine non-competent oocytes (BCB-) negatively impact the capacity of competent (BCB+) oocytes to undergo in vitro maturation, fertilisation and embryonic development. Zygote 24, 245–251 10.1017/S0967199415000118 [DOI] [PubMed] [Google Scholar]
- 12. Torner H., Ghanem N., Ambros C., Hölker M., Tomek W., Phatsara C., Alm H., Sirard M. A., Kanitz W., Schellander K., and Tesfaye D. (2008) Molecular and subcellular characterisation of oocytes screened for their developmental competence based on glucose-6-phosphate dehydrogenase activity. Reproduction 135, 197–212 10.1530/REP-07-0348 [DOI] [PubMed] [Google Scholar]
- 13. Janowski D., Salilew-Wondim D., Torner H., Tesfaye D., Ghanem N., Tomek W., El-Sayed A., Schellander K., and Hölker M. (2012) Incidence of apoptosis and transcript abundance in bovine follicular cells is associated with the quality of the enclosed oocyte. Theriogenology 78, 656–669 10.1016/j.theriogenology.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 14. Ashry M., Lee K., Mondal M., Datta T. K., Folger J. K., Rajput S. K., Zhang K., Hemeida N. A., and Smith G. W. (2015) Expression of TGFβ superfamily components and other markers of oocyte quality in oocytes selected by brilliant cresyl blue staining: relevance to early embryonic development. Mol. Reprod. Dev. 82, 251–264 10.1002/mrd.22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Em S., Kataria M., S. B., and Yadav P. (2014) Expression profile of developmentally important genes between hand-made cloned buffalo embryos produced from reprogramming of donor cell with oocytes extract and selection of recipient cytoplast through brilliant cresyl blue staining and in vitro fertilized embryos. J. Assist. Reprod. Genet. 31, 1541–1552 10.1007/s10815-014-0316-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salimi M., Salehi M., Masteri Farahani R., Dehghani M., Abadi M., Novin M. G., Nourozian M., and Hosseini A. (2014) The effect of melatonin on maturation, glutathione level and expression of HMGB1 gene in brilliant Cresyl Blue (BCB) stained immature oocyte. Cell J. 15, 294–301 [PMC free article] [PubMed] [Google Scholar]
- 17. Spikings E. C., Alderson J., and St. John J. C. (2007) Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol. Reprod. 76, 327–335 10.1095/biolreprod.106.054536 [DOI] [PubMed] [Google Scholar]
- 18. Fakruzzaman M., Bang J. I., Lee K. L., Kim S. S., Ha A. N., Ghanem N., Han C. H., Cho K. W., White K. L., and Kong I. K. (2013) Mitochondrial content and gene expression profiles in oocyte-derived embryos of cattle selected on the basis of brilliant cresyl blue staining. Anim. Reprod. Sci. 142, 19–27 10.1016/j.anireprosci.2013.08.012 [DOI] [PubMed] [Google Scholar]
- 19. Castaneda C. A., Kaye P., Pantaleon M., Phillips N., Norman S., Fry R., and D'Occhio M. J. (2013) Lipid content, active mitochondria and brilliant cresyl blue staining in bovine oocytes. Theriogenology 79, 417–422 10.1016/j.theriogenology.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 20. Keefe D., Kumar M., and Kalmbach K. (2015) Oocyte competency is the key to embryo potential. Fertil. Steril. 103, 317–322 10.1016/j.fertnstert.2014.12.115 [DOI] [PubMed] [Google Scholar]
- 21. Wongsrikeao P., Otoi T., Yamasaki H., Agung B., Taniguchi M., Naoi H., Shimizu R., and Nagai T. (2006) Effects of single and double exposure to brilliant cresyl blue on the selection of porcine oocytes for in vitro production of embryos. Theriogenology 66, 366–372 10.1016/j.theriogenology.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 22. Opiela J., and Kątska-Ksiazkiewicz L. (2013) The utility of Brilliant Cresyl Blue (BCB) staining of mammalian oocytes used for in vitro embryo production (IVP). Reprod. Biol. 13, 177–183 10.1016/j.repbio.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 23. Grupen C. G. (2014) The evolution of porcine embryo in vitro production. Theriogenology 81, 24–37 10.1016/j.theriogenology.2013.09.022 [DOI] [PubMed] [Google Scholar]
- 24. Pawlak P., Warzych E., Chabowska A., and Lechniak D. (2014) Differences in cytoplasmic maturation between the BCB+ and control porcine oocytes do not justify application of the BCB test for a standard IVM protocol. J. Reprod. Dev. 60, 28–36 10.1262/jrd.2013-092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos E. C., Sato D., Lucia T. Jr., and Iwata H. (2015) Brilliant cresyl blue staining negatively affects mitochondrial functions in porcine oocyte. Zygote 23, 352–359 10.1017/S0967199413000610 [DOI] [PubMed] [Google Scholar]
- 26. Su J., Wang Y., Li R., Peng H., Hua S., Li Q., Quan F., Guo Z., and Zhang Y. (2012) Oocytes selected using BCB staining enhance nuclear reprogramming and the in vivo development of SCNT embryos in cattle. PLoS ONE 7, e36181 10.1371/journal.pone.0036181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan Y., Spate L. D., Redel B. K., Tian Y., Zhou J., Prather R. S., and Roberts R. M. (2017) Quadrupling efficiency in production of genetically modified pigs through improved oocyte maturation. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.1703998114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xue Z., Huang K., Cai C., Cai L., Jiang C. Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y. E., Liu J. Y., Horvath S., and Fan G. (2013) Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., and Tang F. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- 30. Macaulay I. C., and Voet T. (2014) Single cell genomics: advances and future perspectives. PLoS Genet. 10, e1004126 10.1371/journal.pgen.1004126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smallwood S. A., Lee H. J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S. R., Stegle O., Reik W., and Kelsey G. (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 10.1038/nmeth.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reyes J. M., Chitwood J. L., and Ross P. J. (2015) RNA-Seq profiling of single bovine oocyte transcript abundance and its modulation by cytoplasmic polyadenylation. Mol. Reprod. Dev. 82, 103–114 10.1002/mrd.22445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar P., Tan Y., and Cahan P. (2017) Understanding development and stem cells using single cell-based analyses of gene expression. Development 144, 17–32 10.1242/dev.133058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaw L., Sneddon S. F., Zeef L., Kimber S. J., and Brison D. R. (2013) Global gene expression profiling of individual human oocytes and embryos demonstrates heterogeneity in early development. PLoS ONE. 8, e64192 10.1371/journal.pone.0064192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hou Y., Fan W., Yan L., Li R., Lian Y., Huang J., Li J., Xu L., Tang F., Xie X. S., and Qiao J. (2013) Genome analyses of single human oocytes. Cell 155, 1492–1506 10.1016/j.cell.2013.11.040 [DOI] [PubMed] [Google Scholar]
- 36. Bonnet-Garnier A., Feuerstein P., Chebrout M., Fleurot R., Jan H. U., Debey P., and Beaujean N. (2012) Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int. J. Dev. Biol. 56, 877–887 10.1387/ijdb.120149ab [DOI] [PubMed] [Google Scholar]
- 37. Sun M. J., Zhu S., Li Y. W., Lin J., Gong S., Jiao G. Z., Chen F., and Tan J. H. (2016) An essential role for the intra-oocyte MAPK activity in the NSN-to-SN transition of germinal vesicle chromatin configuration in porcine oocytes. Sci. Rep. 6, 23555 10.1038/srep23555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Attner M. A., Miller M. P., Ee L. S., Elkin S. K., and Amon A. (2013) Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. U.S.A. 110, 14278–14283 10.1073/pnas.1311845110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mu R., Wang Y. B., Wu M., Yang Y., Song W., Li T., Zhang W. N., Tan B., Li A. L., Wang N., Xia Q., Gong W. L., Wang C. G., Zhou T., Guo N., Sang Z. H., and Li H. Y. (2014) Depletion of pre-mRNA splicing factor Cdc5L inhibits mitotic progression and triggers mitotic catastrophe. Cell Death Dis. 5, e1151 10.1038/cddis.2014.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. La Salle S., Palmer K., O'Brien M., Schimenti J. C., Eppig J., and Handel M. A. (2012) Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 86, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishishita S., Matsuda Y., and Kitada K. (2014) Genetic evidence suggests that Spata22 is required for the maintenance of Rad51 foci in mammalian meiosis. Sci. Rep. 4, 6148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang M. X., Shi Y., Sun Z. G., Zhang Z., and Zhu Y. (2016) Inhibition of the binding between RGS2 and β-tubulin interferes with spindle formation and chromosome segregation during mouse oocyte maturation in vitro. PLoS ONE 11, e0159535 10.1371/journal.pone.0159535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gray N. K., Coller J. M., Dickson K. S., and Wickens M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19, 4723–4733 10.1093/emboj/19.17.4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Badouel C., Körner R., Frank-Vaillant M., Couturier A., Nigg E. A., and Tassan J. P. (2006) M-phase MELK activity is regulated by MPF and MAPK. Cell Cycle 5, 883–889 10.4161/cc.5.8.2683 [DOI] [PubMed] [Google Scholar]
- 45. Heyer B. S., Warsowe J., Solter D., Knowles B. B., and Ackerman S. L. (1997) New member of the Snf1/AMPK kinase family, Melk, is expressed in the mouse egg and preimplantation embryo. Mol. Reprod. Dev. 47, 148–156 10.1002/(SICI)1098-2795(199706)47:2%3C148::AID-MRD4%3E3.0.CO%3B2-M [DOI] [PubMed] [Google Scholar]
- 46. Han S. J., Chen R., Paronetto M. P., and Conti M. (2005) Wee1B is an oocyte-specific kinase involved in the control of meiotic arrest in the mouse. Curr. Biol. 15, 1670–1676 10.1016/j.cub.2005.07.056 [DOI] [PubMed] [Google Scholar]
- 47. Liu J. J., Ma X., Cai L. B., Cui Y. G., and Liu J. Y. (2010) Downregulation of both gene expression and activity of Hsp27 improved maturation of mouse oocyte in vitro. Reprod. Biol. Endocrinol. 8, 47 10.1186/1477-7827-8-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang N., Kaur R., Akhter S., and Legerski R. J. (2009) Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 10, 1029–1035 10.1038/embor.2009.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Egerszegi I., Alm H., Rátky J., Heleil B., Brüssow K. P., and Torner H. (2010) Meiotic progression, mitochondrial features and fertilisation characteristics of porcine oocytes with different G6PDH activities. Reprod. Fertil. Dev. 22, 830–838 10.1071/RD09140 [DOI] [PubMed] [Google Scholar]
- 50. Ishizaki C., Watanabe H., Bhuiyan M. M., and Fukui Y. (2009) Developmental competence of porcine oocytes selected by brilliant cresyl blue and matured individually in a chemically defined culture medium. Theriogenology 72, 72–80 10.1016/j.theriogenology.2009.02.015 [DOI] [PubMed] [Google Scholar]
- 51. Bai L., Li M., Sun J., Yang X., Lu Y., Lu S., and Lu K. (2016) RNA-Seq profiling of intact and enucleated oocyte SCNT embryos reveals the role of pig oocyte nucleus in somatic reprogramming. PLoS ONE 11, e0153093 10.1371/journal.pone.0153093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blakeley P., Fogarty N. M., del Valle I., Wamaitha S. E., Hu T. X., Elder K., Snell P., Christie L., Robson P., and Niakan K. K. (2015) Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165 10.1242/dev.123547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alvarez G. M., Ferretti E. L., Gutnisky C., Dalvit G. C., and Cetica P. D. (2013) Modulation of glycolysis and the pentose phosphate pathway influences porcine oocyte in vitro maturation. Reprod. Domest. Anim. 48, 545–553 10.1111/rda.12123 [DOI] [PubMed] [Google Scholar]
- 54. Gutnisky C., Dalvit G. C., Thompson J. G., and Cetica P. D. (2014) Pentose phosphate pathway activity: effect on in vitro maturation and oxidative status of bovine oocytes. Reprod. Fertil. Dev. 26, 931–942 10.1071/RD12397 [DOI] [PubMed] [Google Scholar]
- 55. Gu L., Liu H., Gu X., Boots C., Moley K. H., and Wang Q. (2015) Metabolic control of oocyte development: linking maternal nutrition and reproductive outcomes. Cell Mol. Life Sci. 72, 251–271 10.1007/s00018-014-1739-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gao Y. Y., Chen L., Wang T., Nie Z. W., Zhang X., and Miao Y. L. (2016) Oocyte aging-induced Neuronatin (NNAT) hypermethylation affects oocyte quality by impairing glucose transport in porcine. Sci. Rep. 6, 36008 10.1038/srep36008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ajuh P., Kuster B., Panov K., Zomerdijk J. C., Mann M., and Lamond A. I. (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19, 6569–6581 10.1093/emboj/19.23.6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma J. Y., Ou Yang Y. C., Wang Z. W., Wang Z. B., Jiang Z. Z., Luo S. M., Hou Y., Liu Z. H., Schatten H., and Sun Q. Y. (2013) The effects of DNA double-strand breaks on mouse oocyte meiotic maturation. Cell Cycle 12, 1233–1241 10.4161/cc.24311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen W., Zhang L., Wang Y., Sun J., Wang D., Fan S., Ban N., Zhu J., Ji B., and Wang Y. (2016) Expression of CDC5L is associated with tumor progression in gliomas. Tumour Biol. 37, 4093–4103 10.1007/s13277-015-4088-5 [DOI] [PubMed] [Google Scholar]
- 60. Yang C. X., Wright E. C., and Ross J. W. (2012) Expression of RNA-binding proteins DND1 and FXR1 in the porcine ovary, and during oocyte maturation and early embryo development. Mol. Reprod. Dev. 79, 541–552 10.1002/mrd.22059 [DOI] [PubMed] [Google Scholar]
- 61. El Shourbagy S. H., Spikings E. C., Freitas M., and St. John J. C. (2006) Mitochondria directly influence fertilisation outcome in the pig. Reproduction 131, 233–245 10.1530/rep.1.00551 [DOI] [PubMed] [Google Scholar]
- 62. Sun X. S., Liu Y., Yue K. Z., Ma S. F., and Tan J. H. (2004) Changes in germinal vesicle (GV) chromatin configurations during growth and maturation of porcine oocytes. Mol. Reprod. Dev. 69, 228–234 10.1002/mrd.20123 [DOI] [PubMed] [Google Scholar]
- 63. Li X., Wang Y. K., Song Z. Q., Du Z. Q., and Yang C. X. (2016) Dimethyl sulfoxide perturbs cell cycle progression and spindle organization in porcine meiotic oocytes. PLoS ONE 11, e0158074 10.1371/journal.pone.0158074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang F., Barbacioru C., Nordman E., Li B., Xu N., Bashkirov V. I., Lao K., and Surani M. A. (2010) RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 5, 516–535 10.1038/nprot.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 69. Trombetta J. J., Gennert D., Lu D., Satija R., Shalek A. K., and Regev A. (2014) Preparation of single-cell RNA-Seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–17 10.1002/0471142727.mb0422s107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huan Y., Xie B., Liu S., Kong Q., and Liu Z. (2015) A novel role for DNA methyltransferase 1 in regulating oocyte cytoplasmic maturation in pigs. PLoS ONE 10, e0127512 10.1371/journal.pone.0127512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.