Abstract
Clostridium difficile TcdB (2366 amino acid residues) is an intracellular bacterial toxin that binds to cells and enters the cytosol where it glucosylates small GTPases. In the current study, we examined a putative cell entry region of TcdB (amino acid residues 1753–1851) for short sequences that function as cell-penetrating peptides (CPPs). To screen for TcdB-derived CPPs, a panel of synthetic peptides was tested for the ability to enhance transferrin (Tf) association with cells. Four candidate CPPs were discovered, and further study on one peptide (PepB2) pinpointed an asparagine residue necessary for CPP activity. PepB2 mediated the cell entry of a wide variety of molecules including dextran, streptavidin, microspheres, and lentivirus particles. Of note, this uptake was dramatically reduced in the presence of the Na+/H+ exchange blocker and micropinocytosis inhibitor amiloride, suggesting that PepB2 invokes macropinocytosis. Moreover, we found that PepB2 had more efficient cell-penetrating activity than several other well-known CPPs (TAT, penetratin, Pep-1, and TP10). Finally, Tf assay–based screening of peptides derived from two other large clostridial toxins, TcdA and TcsL, uncovered two new TcdA-derived CPPs. In conclusion, we have identified six CPPs from large clostridial toxins and have demonstrated the ability of PepB2 to promote cell association and entry of several molecules through a putative fluid-phase macropinocytotic mechanism.
Keywords: bacterial toxin, biotechnology, cell-penetrating peptide (CPP), peptides, toxin
Introduction
Intracellular bacterial toxins gain access to the cytosol of eukaryotic cells where they modify, activate, or process substrates (1, 2). Although all intracellular toxins cross the membrane barrier and enter cells, these toxins utilize a wide array of mechanisms to do so. Binary toxins, such as anthrax toxin, use a combination of proteins to deliver an enzymatic component into the cell (3), whereas single-chain A-B toxins, such as diphtheria toxin, translocate using activities encoded within a single protein (4). The study of these toxins has not only led to a better fundamental understanding of their actions but also has provided a suite of tools to study cell biology and to deliver heterologous cargo into cells (5). For example, the cell entry components of anthrax toxin (6), diphtheria toxin (7), botulinum toxin (8, 9), and pseudomonas exotoxin A toxin (10) have been used to deliver heterologous proteins, nucleic acids, and MHC-I peptides into cells (11). Despite being stripped of their toxic components, these proteins can be large, and cargo must be added by constructing in-frame genetic fusions. Moreover, these systems are not particularly useful for the delivery of non-proteinaceous cargo.
As an alternative to toxin-based delivery systems, cell-penetrating peptides (CPPs)2 are used to deliver a variety of cargo into target cells (12). These peptides have originated from diverse sources such as the human immunodeficiency virus transactivator of transcription (TAT) (13), the Drosophila antennapedia homeoprotein (penetratin) (14), and scorpion venom (15, 16). Many CPPs are derived from native protein transduction domains, whereas others are simply a repeating sequence of positively charged residues, such as polyarginine (17). These small peptides mediate the entry of a variety of molecules, including drugs, into cells and do not always require covalent attachment to their cargo. CPP mechanisms of action are not completely defined, but studies focused on TAT suggest that these peptides traverse membranes through various mechanisms including altering membrane curvature, generating pores, and triggering endocytosis (18). More recently a bacterial source of CPPs was described from Yersinia enterocolitica type III effector protein, YopM (19). YopM CPPs are derived from an amino-terminal protein transduction domain (PTD) and function with similar efficiency to TAT. Surprisingly, CPPs have not been described to date in intracellular bacterial toxins, despite extensive studies into their molecular mechanisms of membrane translocation.
In this current work, peptides derived from the large clostridial toxin family of intracellular bacterial toxins were discovered to have cell-penetrating activity, which we broadly defined as peptides that increase the amount of internalized cargo delivered to either the endosome or the cytosol. This discovery stems from work on TcdB2, which is a large clostridial toxin and major virulence factor produced by the NAP1/BI/027 strain of Clostridium difficile. TcdB is a single polypeptide consisting of 2366 amino acid residues that organize into four functional domains associated with glucosyltransferase, autoproteolytic, membrane translocation, and cell-binding activities (20–23). In addition to TcdB2 (toxinotype III), other forms of TcdB exist with varied amino acid sequences; for instance, TcdB1 (common to multiple ribotypes and toxinotype 0) shares 92% amino acid identity with TcdB2, with small sections of these toxins having little sequence identity. This natural amino acid sequence variability between TcdB1 and TcdB2 was used previously to elucidate functional aspects of TcdB and identify regions that influence epitope exposure (24).
Our prior work centered on TcdB2, with a particular focus on amino acid residues 1753–1851 because this region forms critical intramolecular contacts. In an effort to disrupt these intramolecular contacts and inactivate the toxin, a library of 17–20 amino-acid-residue peptides was designed based on amino acid residues 1753–1851 of TcdB2, and this library was then screened for TcdB inhibitory activity yielding four inhibitory peptides (25). Because recent data suggested that amino acid sequences within the 1753–1851 region have protein transduction properties (26–28), we speculated that peptides derived from our inhibitor panel might also function as CPPs. In this current study, we tested our TcdB2 peptide library for CPP activity, explored the cell-penetrating mechanism used by TcdB2-derived peptides, and finally identified putative CPPs in other large clostridial toxins. Collectively, these data reveal the first peptides derived from intracellular bacterial toxins with cell-penetrating activities and suggest that these TcdB-derived peptides represent an entirely new class of CPPs.
Results
Peptides derived from TcdB2 facilitate cell association of TfAF488
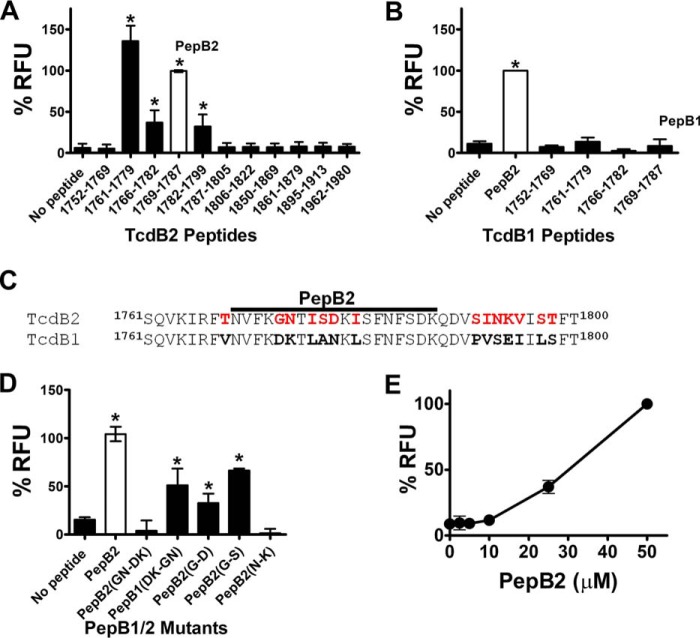
As shown in Fig. 1A, a comprehensive screen for cell-penetrating activity was performed on our peptide library covering amino acid residues 1753–1851 of TcdB2. To assess cell-penetrating activity, an assay was designed in which interactions between fluorescent transferrin (TfAF488) and CHO-K1 cells could be measured. For this assay, we used a low concentration of TfAF488 (250 nm) that did not exhibit detectable levels of cell association, reasoning that candidate peptides would increase TfAF488 to detectable levels on cells. In this screen, cells were exposed to TfAF488 in combination with candidate peptides (50 μm) and incubated for 2 h. After washing cells with PBS to remove non-cell–associated TfAF488, total cellular fluorescence was detected in a microplate reader. To eliminate potential false positives, peptides were discarded from the screen if the peptides were not water-soluble, if the peptides formed autofluorescent precipitants, or if the peptides triggered TfAF488 precipitation. Overall, we discarded four peptides and retained eight for the screen. As shown in Fig. 1A, four of these peptides increased TfAF488–cell interactions with CHO-K1 cells. These peptides spanned a 39-amino-acid-residue (1761–1799) region of TcdB2 and included TcdB2(1761–1779), TcdB2(1766–1782), TcdB2(1769–1787) (PepB2), and TcdB2(1782–1799). We also examined three peptides outside the 1753–1851 region of TcdB2 and did not detect increased interactions between TfAF488 and cells.
Figure 1.
TcdB2 peptides trigger interactions between Tf and cells. A, screen for putative TcdB2-derived CPPs. In this assay, 250 nm TfAF488 was combined with each peptide (50 μm) and applied to CHO-K1 cells to for 2 h. After thorough washing, the amount of cell-associated fluorescence was quantified in a microplate reader. B, screen for putative TcdB1-derived CPPs. The candidate peptides were tested for CPP activity using the same method described for TcdB2 peptides. C, amino acid sequence alignment comparing TcdB1 and TcdB2 from the region where cell uptake–inducing peptides are derived. The PepB2 sequence is denoted by a bold black line, and red residues specify amino acid residues found only in TcdB2. D, variant PepB2 peptides were compared in the TfAF488 assay described above. Variant peptides used in this assay have amino acid residues that differ between TcdB2 and TcdB1 replaced to reflect those found in the reciprocal peptide. In addition, PepB2(G-S) contains glycine replaced with a serine that is found in the same position as in C. sordellii TcsL. E, TfAF488 assay comparing different concentrations of PepB2. All graphs in this figure represent the percentage of PepB2-induced relative fluorescent units (RFU) for each peptide tested. Data are presented as mean (n = 3) ± S.D. Asterisks indicate significant increase above control cells; *, p < 0.05.
PepB2 was selected from the four candidates for use in the remainder of the experimental analyses. To determine the amino acid sequence specificity of PepB2, a peptide derived from the corresponding region in TcdB1 (termed PepB1) was tested in the TfAF488 cell-association assay. As shown in Fig. 1B, PepB1 did not improve TfAF488 association with cells above background. Moreover, two other TcdB1 peptides corresponding to the putative CPPs in TcdB2 did not enhance TfAF488 association with cells (Fig. 1B). Using amino acid sequence differences in the CPP region of TcdB2 and TcdB1 as a guide (Fig. 1C), we examined a series of mutant peptides to identify amino acid residues essential to PepB2 activity. When 5GN6 from PepB2 was replaced with 5DK6 from PepB1 (PepB2-5GN6-5DK6), the resulting peptide no longer exhibited CPP activity (Fig. 1D). Conversely, CPP activity was gained in PepB1 by the reciprocal substitution producing PepB1-5DK6-5GN6 (Fig. 1D). To determine whether Gly-5 is necessary for the activity of PepB2, we substituted Gly-5 with the Asp-5 from PepB1 (PepB2-G5-D5) and found that this single substitution did not prevent cell-associating activities (Fig. 1D). Additionally, replacing Gly-5 with a serine residue (PepB2-G5-S5) did not abolish cell-associating activities. Next, we substituted Asn-6 in PepB2 with the Lys-6 from PepB1 (PepB2-N6-K6) and detected no cell-associating activities, suggesting that Asn-6 is necessary for PepB2 to function as a CPP (Fig. 1D). Finally, we tested PepB2 over a range of concentrations and observed a 60% reduction in activity when the levels of PepB2 were reduced from 50 to 25 μm, and all PepB2 activity was lost when levels were reduced to 10 μm (Fig. 1E).
PepB2 enhances cell association of a variety of molecules
To determine whether PepB2 also enhanced cellular interaction of other molecules, we examined four proteins or protein subdomains known to engage cells through distinctly different mechanisms. Bacillus anthracis protective antigen (PA) binds two cell-surface receptors (ANTXR1 and ANTXR2). TcdB(1852–2366) consists of the combined repetitive oligopeptide (CROP) region of TcdB and is thought to bind carbohydrates. Cholera toxin B-domain (CT-B) is a pentameric oligomer that binds GM1 gangliosides in membrane lipid rafts, and streptolysin-O (SLO) interacts with membrane cholesterol. To avoid cell lysis by SLO, we utilized a pre-pore locked mutant (G130C/S264C, PPLSLO) that binds cells without generating transmembrane channels. Each fluorescently labeled protein was mixed with PepB2 and then applied to CHO-K1 cells for 30 min, at which point the interaction was measured by flow cytometry. As shown in Fig. 2A, PepB2 increased the median fluorescence intensity (MFI) of PAAF647-treated cells by ∼4-fold compared with treatment with PAAF647 in the absence of peptide. CROPAF488 (TcdB(1852–2366)) and CT-BAF488 exhibited only minimal binding in the absence of PepB2, but their levels of cellular interaction increased from 24 to 900 MFI and 12 to 600 MFI, respectively, with PepB2 (Fig. 2A). Conversely, PPLSLOAF488 binding was not enhanced by PepB2 and showed a slight reduction from 1500 to 1200 MFI in the presence of the peptide (Fig. 2A). This flow cytometry assay also demonstrated that PepB2 increased the cellular interactions of TfAF488 from 72 to 1000 MFI (Fig. 2A), similar to the trend observed in the TfAF488 cell-association assay using the plate-reader format (Fig. 1A).
Figure 2.
PepB2 increases the cell association of unrelated and diverse molecules. CHO-K1 cells were exposed for 30 min at 37 °C to various fluorescent molecules in the presence and absence of 50 μm of PepB2 and then subjected to flow cytometry analysis. In the bar graphs, flow cytometry results are presented as MFI from three experiments ± S.D. Asterisks indicate significant change. *, p < 0.001. A, cell-binding molecules: 50 nm PAAF647, 100 nm CROPAF488, 100 nm PPLSLOAF488, 90 nm CT-B AF488, or 125 nm TfAF488. B, non-cell–binding molecules: 40 nm SAAF488, 100 μg/ml 10-kDa dextranAF488 (Dex), or 0.2-μm microspheres (MS) at a 1:750,000 dilution.
Next, PepB2 was examined for its capacity to promote cellular association of factors that do not interact with cells. Streptavidin (SA) is a biotin-binding protein not known to directly bind the cell surface. Likewise, dextran (10 kDa) is a repeating glucose structure that does not bind to cells. We also tested 0.2-μm microspheres composed of polystyrene and encapsulating a yellow-green fluorescent dye. As shown in Fig. 2B, each of the molecules interacted poorly with cells in the absence of PepB2 but demonstrated a notable increase in cell association in the presence of the peptide. PepB2 increased SAAF488 MFI levels from 6 to 1500 and similarly increased dextranAF488 MFI from 65 to 1500 (Fig. 2B). PepB2 also enhanced the interactions between microspheres and cells, shifting the MFI from 25 to 900 (Fig. 2B).
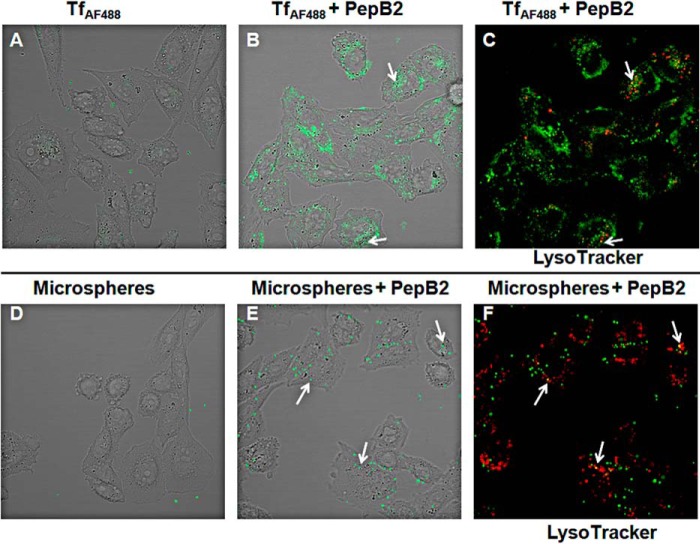
To further evaluate the CPP-like activity of PepB2, confocal microscopy was used to capture images from cross-sections of cells treated with TfAF488 or 0.2-μm microspheres in the presence and absence of PepB2. As shown in Fig. 3, A and B, PepB2 dramatically increased TfAF488 association with cells, whereas TfAF488 interaction with cells could not be detected in the absence of PepB2. Confocal microscopy examination of microspheres also revealed that PepB2 triggered a dramatic increase in the association of microspheres with cells (Fig. 3, D and E). To determine whether PepB2 elicited the uptake of TfAF488 or microspheres, colocalization of these molecules with a lysosomal marker (LysoTracker dye) was examined by confocal microscopy. As shown in Fig. 3, C and F, colocalization of both TfAF488 and microspheres with LysoTracker was observed in the presence of PepB2, suggesting that these molecules entered the cellular endosomal system and eventually reached the lysosome.
Figure 3.
PepB2-induced interactions between cells and cargo as analyzed by confocal microscopy. A-C, confocal microscopy analysis of live CHO-K1 cells exposed for 2 h to 250 nm TfAF488 with and without 50 μm PepB2. D-F, confocal microscopy analysis of live CHO-K1 cells exposed for 2 h to 0.2-μm microspheres at a 1:750,000 dilution with and without 50 μm PepB2. C, same field as in B but showing LysoTracker. F, same field as in E but showing LysoTracker. Arrows highlight TfAF488 or microspheres colocalization with LysoTracker.
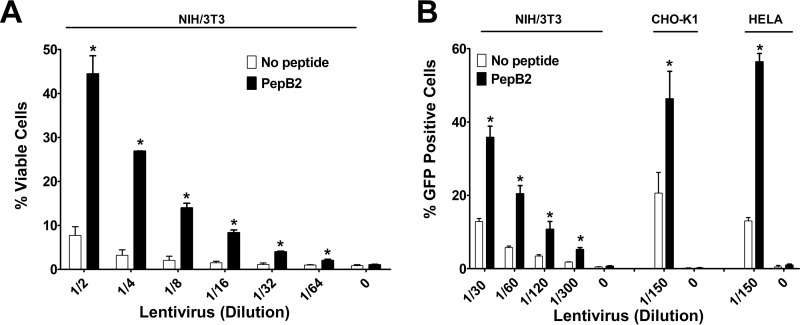
PepB2 enhances cell entry of lentivirus particles
To further verify that PepB2 induces the internalization of cargo into the cells, we tested the ability of PepB2 to enhance the cellular uptake of lentivirus particles. Lentivirus particles are used routinely to deliver genetic material into mammalian cells and possess the machinery necessary for endosomal escape, thus making this system an excellent tool for evaluating cellular internalization. Two approaches were used to test the ability of PepB2 to enhance the uptake of lentivirus particles. First, NIH/3T3 cell resistance to puromycin was used to examine the effects of PepB2 on the cell entry of lentivirus particles carrying the pac gene. Second, GFP expression in NIH/3T3, CHO-K1, or HELA cells was used to evaluate PepB2-mediated cell entry of lentivirus particles carrying the gfp gene. In both experiments, NIH/3T3 cells were treated with increasing dilutions of the lentivirus particles in the presence or absence of 2.5 μm PepB2. As shown in Fig. 4A, when PepB2 was included with the lentivirus particles used at 1/2 dilution, ∼45% of the cells were viable in the presence of puromycin. However, less than 10% were found to be viable in the absence of PepB2. Although the percentage of overall viable cells decreased with higher dilutions of the lentivirus particles, a consistent and substantial enhancing effect from PepB2 was observed. Likewise, PepB2 significantly increased the number of GFP-positive cells across several dilutions of the gfp-encoding lentivirus particles in NIH/3T3 cells (Fig. 4B). PepB2 also boosted the number of GFP-positive cells in both HELA and CHO-K1 cells exposed to gfp-encoding lentivirus particles (Fig. 4B).
Figure 4.
PepB2 enhances entry of lentivirus particles into cells. Infections with replication-incompetent lentivirus particles were performed in the presence and absence of 2.5 μm PepB2. A, lentivirus particles contain the pac gene and display resistance to puromycin-induced killing. In this assay, infected NIH/3T3 cells are detected by exposing cells to 2 μg/ml puromycin for 3 days and then measuring cellular viability. The bar graph represents the percent cellular viability. B, lentivirus particles encoding gfp were used to infect NIH/3T3, CHO-K1, or HELA cells in the presence and absence of PepB2. The percentage of GFP-expressing cells was subsequently quantified by flow cytometry. The bar graph corresponds to the percentage of cells that are positive for GFP. Data are presented as mean (n = 3) ± S.D. Asterisks indicate significant increase above controls for each dilution. *, p < 0.01.
PepB2 activity is blocked by inhibitors of macropinocytosis
The ability of PepB2 to increase the cellular association of non-cell–binding cargo (SA, dextran, and microspheres) suggested that PepB2 elicited a cellular response such as macropinocytosis. To test whether PepB2 could trigger macropinocytosis, PepB2-induced cellular interactions were tested in the presence of amiloride or 5-(N-ethyl-N-isopropyl)amiloride (EIPA), which are Na+/H+ exchange blockers that inhibit macropinocytosis. As shown in Fig. 5A, amiloride and EIPA both reduced PepB2-mediated association of TfAF488 with cells, lowering the association by 75% with 5 mm amiloride and by 80% with 100 μm EIPA. Similarly, amiloride and EIPA both reduced PepB2-mediated cellular interactions with microspheres, showing a 50% reduction with 5 mm amiloride and 70% reduction with 100 μm EIPA (Fig. 5B). Other methods (using cytochalasin D and low temperature) for interfering with cellular uptake were also tested to determine their effects on PepB2-induced cellular interactions. First, we tested cytochalasin D, which interferes with certain uptake mechanisms by blocking actin polymerization. As shown in Fig. 5, A and B, 10 μm cytochalasin D reduced PepB2-mediated cell association of TfAF488 and microspheres by 60 and 25%, respectively. Lowering the temperature to near 0 °C halts endocytosis and macropinocytosis. Thus, experiments were preformed next on ice, and the results demonstrated that PepB2-mediated cell association of TfAF488 and microspheres were both decreased by 70% (Fig. 5, A and B).
Figure 5.
Macropinocytosis inhibitors prevent PepB2-induced interactions between cells and cargo. A and B, assay determining whether EIPA, amiloride, cytochalasin D, or temperature reduction prevents PepB2 from increasing cellular interactions with TfAF488 or microspheres. In this assay, CHO-K1 cells were pretreated for 1–2 h with inhibitors. Then, TfAF488 (250 nm) or 0.2-μm microspheres (1:750,000 dilution) were combined with 50 μm PepB2 and applied to CHO-K1 cells for 2 h. After thorough washing, cell-associated fluorescence was quantified in a microplate reader. The bar graphs in this figure represent the percentage of PepB2-induced relative fluorescent units (RFU) for each peptide tested. Data are presented as mean (n = 3) ± S.D. Asterisks indicate significant decrease in fluorescence when comparing inhibitor-treated cells to non-inhibitor–treated control cells. C, measurement of cargo internalization. CHO-K1 cells were exposed for 30 min to SAAF488 in the presence and absence of 50 μm PepB2 on ice or at 37 °C and then analyzed by flow cytometry. Immediately after the initial flow cytometry analysis, extracellular fluorescence was quenched with 0.2% trypan blue and reanalyzed by flow cytometry. The fluorescent signal that remains represents internalized SAAF488. In the bar graphs, flow cytometry results are presented as MFI from 3 experiments ± S.D. Asterisks indicate significant reduction after quenching; *, p < 0.001.
Using macropinocytosis inhibitors, cytochalasin D, or low temperature exposure resulted in a substantial reduction in the ability of PepB2 to induce interactions between cells and cargo, although it did not completely eliminate PepB2-induced cell interactions. Therefore, an additional experiment was performed to determine whether cargo is on the cell surface or internalized when macropinocytosis is blocked by low temperature exposure. In this experiment, we utilized the non-cell–permeable fluorescence quencher trypan blue to discriminate between extracellular and intracellular cargo. As shown in Fig. 5C, using SAAF488 as cargo, we found that cargo was on the cell surface when the experiments were carried out on ice. In a similar experiment at 37 °C, a large amount of SAAF488 was internalized, whereas a significant amount remained on the cell surface (Fig. 5C). Together these data suggest that PepB2 promotes both cell-surface binding and macropinocytosis.
PepB2 CPP activity is more effective than TAT, penetratin, Pep-1, or TP10
Given the novelty of PepB2, we were curious to know how it compared with other commercially available CPPs (TAT, penetratin, Pep-1, and TP10). As shown in Fig. 6A, these CPPs consist of a substantially different amino acid sequence than PepB2. As a further comparison, these CPPs were applied to the cell association assay performed in Fig. 5. When TfAF488 was used as cargo, cellular interactions were not enhanced by TAT, Pep-1, or TP10, whereas penetratin was found to have a marginal level of activity with this cargo (Fig. 6B). Using microspheres as cargo, we found that penetratin, Pep-1, and TP10 improved microsphere interaction with cells but not to the extent observed with PepB2 (Fig. 6C). These results indicate that PepB2 enhanced the cellular interactions of TfAF488 and microspheres with greater efficacy than any of the commercially available CPPs examined in this assay.
Figure 6.
Comparison of PepB2 to cell-penetrating peptides. A, amino acid sequences of PepB2 and other cell-penetrating peptides. B and C, experiments comparing PepB2 with other cell-penetrating peptides. In this experiment, TfAF488 (250 nm) or 0.2-μm microspheres (1:750,000 dilution) were combined with 50 μm peptides and then added to CHO-K1 cells for 2 h. After washing, the amount of cell-associated fluorescence was quantified in a microplate reader. The bar graphs in this figure represent the percentage of PepB2-induced relative fluorescent units (RFU) for each peptide tested. Data are presented as mean (n = 3) ± S.D. Asterisks indicate significant increase above controls. *, p < 0.01.
Cell-associating peptides derived from other large clostridial toxins
Collectively, our data indicated that TcdB2 encodes a region spanning amino acid residues 1761–1800 that promotes the cellular interactions of heterologous molecules. Although TcdB1 did not appear to encode CPP sequences (Fig. 1B), we wondered whether the corresponding regions in other large clostridial toxins might encode amino acid sequences exhibiting CPP activity. To assess this possibility, another panel of synthetic peptides based on sequences from the C. difficile TcdA and Clostridium sordellii TcsL were tested in the TfAF488 cell association assay. The amino acid sequence identity between TcdB2 and TcdA across this region is only 30% (Fig. 7A), and the same region in TcsL exhibits 68% identity with TcdB2, which is the same as the identity between TcdB1 and TcdB2 in this portion of the toxin (Fig. 7A).
Figure 7.
Identification of putative CPPs from other large clostridial toxins. A, amino acid sequence alignment comparing large clostridial toxins in the region where CPPs are derived in TcdB2. In this region, the percent identify of each of these toxins is compared with TcdB2. The gray shading indicates amino acid residues shared by at least one toxin and TcdB2. The yellow shading reveals amino acid residues shared by all four toxins. B and C, screen determining whether TcsL- or TcdA-derived peptides increase interactions between TfAF488 and cells, using the same approach as described in the legend for Fig. 1. The bar graphs in this figure represent the percentage of PepB2-induced relative fluorescent units (RFU) for each peptide tested. Data are presented as mean (n = 3) ± S.D. Asterisks indicate significant increase above control cells; *, p < 0.05.
As shown in Fig. 7B, similar to the results obtained using peptides from TcdB1, the corresponding peptides from TcsL did not enhance TfAF488 interactions with cells. However, when a series of peptides were tested from TcdA, two peptides spanning amino acid residues 1755–1795 significantly enhanced the interactions between TfAF488 and cells (Fig. 7C). Two of these peptides (TcdA(1755–1774) and TcdA(1774–1795)) displayed ∼50% of the CPP activity that was observed with PepB2. Finally, we expanded the analysis of TcdA into the CROP region and tested 12 other peptides from this region of the toxin. None of the CROP-based peptides caused increased the TfAF488 association of the cell above background, further confirming the importance of amino acid residues 1753–1795 as a source of CPPs.
Discussion
CPPs were discovered previously from several biological sources, including viruses (13), scorpion venoms (15, 16), a bacterial effector protein (19), and cellular proteins that naturally cross membranes (14). Other than designed synthetic peptides, almost all current CPPs come from proteins or cytolytic peptides that naturally interact with and cross membranes. Surprisingly, CPP or peptides that otherwise mediate cell association or trigger cellular uptake have not been described for intracellular bacterial toxins, despite their highly evolved ability to cross membranes during the intoxication of target cells. The experimental data from this study show that peptides derived from a putative PTD of TcdB2(1753–1851) dramatically enhanced the cell association of a variety of unrelated proteins and other molecules. This effect was observed not only in CHO-K1 cells but also in NIH/3T3 (lentivirus, Fig. 4), HELA (lentivirus, Fig. 4), and HAP1 (data not shown) cells. Four putative CPPs were identified from our panel of peptides derived from amino acid residues 1753–1851 of TcdB2 (Fig. 1). One of these peptides, representing the 1769–1787 region of TcdB2 (PepB2), was capable of enhancing the cell association of various molecules regardless of whether the cargo had the ability to bind cells (Fig. 2). Perhaps the most striking effect was observed with the enhanced entry of lentivirus particles, where PepB2 greatly increased virus infection as measured by puromycin resistance and GFP expression (Fig. 4). Collectively, these findings indicate that a putative PTD region of TcdB2 contains sequences that can be used to enhance the cell entry and cell association of a variety of different molecules.
Candidate CPPs were identified in TcdB2 and TcdA but not in TcdB1 or TcsL (Fig. 1 and Fig. 7). Clearly, the latter two toxins must enter the cells, and the lack of identifiable CPPs does not suggest that these toxins lack sequences with protein transduction properties. CPPs may not have been detected in TcdB1 or TcsL simply because these toxins lack a sequence where 17–20 amino-acid-residue peptides can be derived with the characteristics necessary to fold into a structure suitable for CPP activity. Thus, peptides with cell-penetrating activity may possibly be derived from TcdB1 and TcsL if longer peptides were designed or if slight modifications were made to the peptides. Conversely, although TcdB1 and TcsL sequences are sufficient to aid in toxin cell entry, the sequences may lack enough protein transduction activity to produce an effective CPP. Along this line, previous work has shown that TcdB1 is less efficient at cell entry than TcdB2 (29), so the lack of CPPs from TcdB1 may not be completely unexpected. TcsL is also more limited in its efficiency of cell entry and in some cases requires a pH pulse from the cell surface to achieve higher levels of membrane translocation (30).
The capacity of PepB2 to enhance the cell association of a broad range of cargo could be an extremely important trait. As demonstrated in Fig. 6, PepB2 may be a more robust platform than other commercially available CPPs, especially when direct coupling between peptides and cargo is not used. For instance, when Tf was utilized as a cargo, PepB2 was much more efficient than TAT, penetratin, Pep-1, or TP10 (Fig. 6). With microspheres as cargo, penetratin, Pep-1, and TP10 promoted cell association at about 30–50% of the level of PepB2, whereas TAT did not increase interactions (Fig. 6). The microspheres used in this study have a sulfate-coated surface giving them the capacity to passively bind protein through hydrophobic interactions. Therefore, the ability of penetratin, Pep-1, and TP10 to induce microsphere cell association is possibly linked to the ability of these microspheres to bind these CPPs.
Because PepB2 does not need to specifically bind cargo and because PepB2 promotes the cell association of non-cell–binding cargo, we postulated that PepB2 activates fluid-phase macropinocytosis. Using inhibitors of macropinocytosis (amiloride and EIPA), the CPP activity of PepB2 was blocked, indicating that macropinocytosis is necessary for CPP activity (Fig. 5). Other approaches (using cytochalasin D and low temperature) for impeding cellular uptake were able to limit PepB2-mediated cell association of TfAF488 and microspheres (Fig. 5), which also suggests that PepB2 may enable a mechanism such as macropinocytosis. However, these results also do not exclude the possibility that PepB2 acts as a molecular tether that binds cargo to cells. Previous work has shown that PepB2 forms large oligomeric structures (25), and possibly these large oligomers could serve as a binding surface for the diverse set of molecules used in these studies. This idea is supported by our experiment in Fig. 5C, which indicates that PepB2 promotes cell-surface binding. Thus, although macropinocytosis appears to be central to PepB2 activity, physical interaction between PepB2 and cargo is likely also to play a role in the activities of PepB2.
Although we have tentatively classified PepB2 as a CPP, PepB2 has very little in common with these peptides other than to trigger or enhance uptake into cells. The amino acid sequence of commonly studied CPPs exhibits strong cationic, aromatic, or hydrophobic characteristics or a combination of these characteristics. For example, at neutral pH the net charges of TAT and penetratin are +8 and +7, whereas PepB2 has a net charge of +1. PepB2 is better classified among the amphipathic CPPs, such as the neuropeptide-derived CPP transportan (27 amino acid residues), which contains 15 hydrophobic and 4 charged amino acid residues (31). In comparison, PepB2 (19 amino acid residues) contains 6 hydrophobic and 4 charged amino acid residues, providing some amphipathic characteristics but not to the same extent as transportan. Further examination of the primary amino acid sequence of PepB2 has uncovered a potential N-glycosylation motif (Asn-X-(Thr/Ser)), which is not observed in TAT, penetratin, Pep-1, or TP10. Our previous work indicates that PepB2 folds into a β-hairpin (25), a conformation that is not associated with TAT, penetratin, Pep-1, or TP10. Thus, although PepB2 exhibits some attributes of CPPs, its physical characteristics place it in a class of its own. Indeed, the experimental evidence from this study suggests that PepB2 could trigger uptake in a way entirely different from that described for CPPs. Depending on the outcome of future experiments, a more accurate term for describing PepB2 may be as a micropinocytosis-enabling peptide.
The cell-penetrating activities of PepB2 may reflect a previously undefined event that occurs when TcdB intoxicates cells. Models described to date suggest that TcdB utilizes receptor-mediated endocytosis to enter target cells (32). After engaging cell-surface receptors, TcdB is taken up into endocytic vesicles and translocates into the cytosol following endosomal acidification. Yet how engaging the cell-surface receptors or the cell surface in general causes TcdB to enter cells is not understood. Whether this is a passive process or something that is actively triggered by TcdB is also not known. Indeed, very little is known about any intracellular bacterial toxin in this regard. Previous work by Chen et al. (28) shows that deleting amino acid residues 1756–1780 causes TcdB to accumulate within endocytic vesicles and not translocate into the cytosol; this leads to TcdB degradation in LAMP5-positive endosomes, suggesting that this region of TcdB is involved in promoting the escape of the toxin into the cytosol. Whether the exact PepB2 region of TcdB2 is involved in a similar process is not clear, but possibly this portion of the toxin both initiates cell entry and mediates escape from endocytic vesicles.
We recently reported that PepB2 is a potent inhibitor of TcdB and triggers conformational instability in the toxin by binding to repeat amino acid sequences in the CROP domain (25). Because of the toxin instability created by PepB2, TcdB is unable to properly engage cells and undergo the structural rearrangements necessary for toxin delivery into the cytosol. To date, we have no evidence that the inhibitor and CPP activities of PepB2 are related. In fact, we noted a reduction in the cell-binding kinetics of TcdB in the presence of PepB2 in our previous study, which is unlike the cell-penetrating activities of PepB2 described in this current work. Our working model describing TcdB inhibition by PepB2 suggests that the PepB2 sequence in TcdB(1769–1787) binds the CROP domain and these interactions are disrupted by the addition of the PepB2 peptide. This model correlates with our previous report showing that that the PepB2 sequence in TcdB(1769–1787) influences the exposure of neutralizing epitopes in the proximal CROP region (24). Considering the collective data, we hypothesized that the PepB2 region of TcdB plays an important role in cell entry while also engaging proximal regions of the toxin that are critical for maintaining an organized structure.
Experimental procedures
Peptides and other reagents
All peptides, including known CPPs (TAT, penetratin, Pep-1, and TP10), were synthesized and purified to greater than 90% purity by GenScript (Piscataway, NJ). The assessment of peptide quality was made by HPLC and MS. TfAF488 (catalog no. T13342), CT-BAF488 (catalog no. C22841), SAAF488 (catalog no. S11223), 10-kDa dextranAF488 (catalog no. D22910), and 0.2-μm yellow-green fluorescent sulfate microspheres (catalog no. F8848) were purchased from Thermo Fisher Scientific. PAAF647, expressed and purified using a previously described method (33), was labeled with Alexa Fluor 647 using maleimide chemistry. The CROP domain of TcdB was expressed and purified as described previously (24) and then labeled with Alexa Fluor 488 on primary amines. Pre-pore locked SLOAF488 is a site-specific mutant of streptolysin-O containing two mutations (G130C and S264C) that restricts the protein to a pre-pore conformation. The protein and its activity were described previously (34) and is a gift from R. K. Tweten. Amiloride (catalog no. A7410), EIPA (catalog no. A3085), and cytochalasin D (catalog no. C2618) were purchased from Sigma-Aldrich.
Cell uptake assay
In these experiments, CHO-K1 cells (ATCC) were cultured at 37 °C in the presence of 6% CO2 with F12-K medium containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Prior to these assays, CHO-K1 cells were seeded in 96-well plates at a density of 1.0 × 104 cells/well and allowed to grow for 48 h. Next, fluorescent molecules were combined with 50 μm peptide and applied to CHO-K1 cells for 2 h. The cells were then washed three times with PBS, and cell-associated fluorescence was quantified in a microplate reader (TECAN Infinite F200 PRO). Readings were taken from five separate sections of each well. Fluorescence was detected with an excitation wavelength of 485 nm and emission wavelength of 535 nm.
Flow cytometry
CHO-K1 cells were cultured at 37 °C in the presence of 6% CO2 with F12-K medium containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Prior to flow cytometry analysis, cells were detached using a non-enzymatic cell dissociation buffer (Thermo Fisher Scientific, catalog no. 13150-016). Next, 4.0 × 105 cells were suspended in 300 μl of medium (F12-K with 10% FBS) and exposed to fluorescent molecules with and without PepB2 (50 μm) for 30 min at 37 °C. The cells were then placed on ice, and cell-associated fluorescence was quantified using a FACSCalibur flow cytometer (University of Oklahoma Health Sciences Center). All data were analyzed using FLOWJO software (Tree Star, Inc., San Carlos, CA). To distinguish between extracellular and intracellular cargo, extracellular fluorescence was quenched with non-cell–permeable trypan blue (0.2% solution in PBS). The quenched cells were then analyzed by flow cytometry and compared with signal prior to quenching.
Confocal microscopy
CHO-K1 cells were seeded in 8-well chambered coverglass slides (Nunc) at a density of 6.3 × 103 cells/well in F12-K medium containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin and allowed to attach overnight. Cells were then exposed to TfAF488 (250 nm) or 0.2-μm microspheres (1:750,000 dilution) in the presence and absence of 50 μm PepB2 for 2 h. For experiments with LysoTracker, 75 nm LysoTracker Red DND-99 (Thermo Fisher Scientific, catalog no. L7528) was included in the medium. The medium was removed and replaced with phenol red free F12-K medium containing 10% FBS, and images were captured on a Zeiss LSM-710 confocal microscope.
Lentivirus infection
Lentivirus particles containing a puromycin resistant gene (pac gene) were prepared by the following method using these three plasmids: pCMV-VSV-G (Addgene plasmid 8454, gift from Bob Weinberg) (35), psPAX2 (Addgene plasmid 12260, gift from Didier Trono), and lentiCRISPRv1 (Addgene plasmid 52961, gift from Feng Zhang) for the expression of the pac gene (36). These plasmids were co-transfected into 293FT cells (Thermo Fisher Scientific, catalog no. R70007) using the calcium phosphate transfection method. 293FT cells were cultured in DMEM containing 10% FBS and supplemented with 0.1 mm MEM non-essential amino acids, 1 mm sodium pyruvate, and 2 mm l-glutamine. Forty-eight hours after transfection, lentivirus particles released into the medium were harvested and used for infection experiments. Lentivirus particles containing the GFP-encoding gene were purchased from Santa Cruz Biotechnology (catalog no. sc-108084).
Lentivirus infections were carried out using NIH/3T3, HELA, or CHO-K1cells. The NIH/3T3 and HELA cells were cultured in DMEM containing 10% FBS. CHO-K1 cells were cultured in F12-K medium containing 10% FBS. NIH/3T3 cells were seeded in 96-well plates at a density of 2.5 × 103 cells/well for the experiment testing puromycin resistance. NIH/3T3, HELA, or CHO-K1 cells were seeded in 24-well plates at a density of 2.5 × 104 cells/well for experiments measuring GFP. The lentivirus particles with and without PepB2 were added to the cells, and the resulting plate of cells was centrifuged for 2 h at 1200 × g at 25 °C. The cells containing the lentivirus mixture were placed in the tissue culture incubator overnight, and the following day the mixture was removed and replaced with fresh medium. For puromycin resistance, cells were exposed to 2 μg/ml puromycin for 3 days, and then cellular viability was measured using the cell-counting kit 8 (CCK-8) assay (Sigma), a WST-8 dye-based colorimetric reaction. For GFP expression experiments, cells were cultured for 24 h after infection. Cells were then removed from the tissue culture plate with trypsin, and the percentage of GFP-positive cells was determined using a FACSCalibur flow cytometer (University of Oklahoma Health Sciences Center). The resulting data were analyzing with FLOWJO software (Tree Star, Inc.).
Author contributions
J. L. L. conceived, designed, and executed the experiments; analyzed the data; and wrote the paper. G. D. H. executed the experiments and analyzed the data. J. D. B. analyzed the data and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank the Oklahoma Medical Research Foundation Imaging Core Facility for providing microscopy expertise.
This work was supported by National Institutes of Health Grants R21AI121925 and R01AI119048 (to J. D. B.) and by Grant HR13-070 from the Oklahoma Center for the Advancement of Science and Technology (OCAST) (to J. L. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CPP
- cell-penetrating peptide
- Tf
- transferrin
- TAT
- transactivator of transcription
- PTD
- protein transduction domain
- PA
- protective antigen
- CROP
- combined repetitive oligopeptide
- CT-B
- cholera toxin B-domain
- SLO
- streptolysin-O
- PPLSLO
- pre-pore locked streptolysin-O
- SA
- streptavidin
- MFI
- median fluorescence intensity
- EIPA
- 5-(N-ethyl-N-isopropyl)amiloride.
References
- 1. Watson P., and Spooner R. A. (2006) Toxin entry and trafficking in mammalian cells. Adv. Drug Deliv. Rev. 58, 1581–1596 10.1016/j.addr.2006.09.016 [DOI] [PubMed] [Google Scholar]
- 2. Falnes P. O., and Sandvig K. (2000) Penetration of protein toxins into cells. Curr. Opin. Cell Biol. 12, 407–413 10.1016/S0955-0674(00)00109-5 [DOI] [PubMed] [Google Scholar]
- 3. Collier R. J., and Young J. A. (2003) Anthrax toxin. Annu. Rev. Cell Dev. Biol. 19, 45–70 10.1146/annurev.cellbio.19.111301.140655 [DOI] [PubMed] [Google Scholar]
- 4. Simon N. C., Aktories K., and Barbieri J. T. (2014) Novel bacterial ADP-ribosylating toxins: Structure and function. Nat. Rev. Microbiol. 12, 599–611 10.1038/nrmicro3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beilhartz G. L., Sugiman-Marangos S. N., and Melnyk R. A. (2017) Repurposing bacterial toxins for intracellular delivery of therapeutic proteins. Biochem. Pharmacol. 142, 13–20 [DOI] [PubMed] [Google Scholar]
- 6. Rabideau A. E., and Pentelute B. L. (2016) Delivery of non-native cargo into mammalian cells using anthrax lethal toxin. ACS Chem. Biol. 11, 1490–1501 10.1021/acschembio.6b00169 [DOI] [PubMed] [Google Scholar]
- 7. Auger A., Park M., Nitschke F., Minassian L. M., Beilhartz G. L., Minassian B. A., and Melnyk R. A. (2015) Efficient delivery of structurally diverse protein cargo into mammalian cells by a bacterial toxin. Mol. Pharm. 12, 2962–2971 10.1021/acs.molpharmaceut.5b00233 [DOI] [PubMed] [Google Scholar]
- 8. Bade S., Rummel A., Reisinger C., Karnath T., Ahnert-Hilger G., Bigalke H., and Binz T. (2004) Botulinum neurotoxin type D enables cytosolic delivery of enzymatically active cargo proteins to neurones via unfolded translocation intermediates. J. Neurochem. 91, 1461–1472 10.1111/j.1471-4159.2004.02844.x [DOI] [PubMed] [Google Scholar]
- 9. Wilson B. A., and Ho M. (2014) Cargo-delivery platforms for targeted delivery of inhibitor cargos against botulism. Curr. Top. Med. Chem. 14, 2081–2093 10.2174/1568026614666141022094517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng X., Zhang G., Zhang L., Feng Y., Li Z., Wu G., Yue Y., Li G., Cao Y., and Zhu P. (2015) Developing a novel gene-delivery vector system using the recombinant fusion protein of Pseudomonas exotoxin A and hyperthermophilic archaeal histone HPhA. PLoS ONE 10, e0142558 10.1371/journal.pone.0142558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ballard J. D., Collier R. J., and Starnbach M. N. (1998) Anthrax toxin as a molecular tool for stimulation of cytotoxic T lymphocytes: Disulfide-linked epitopes, multiple injections, and role of CD4+ cells. Infect. Immun. 66, 4696–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guidotti G., Brambilla L., and Rossi D. (2017) Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 38, 406–424 10.1016/j.tips.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 13. Giacca M. (2004) The HIV-1 Tat protein: A multifaceted target for novel therapeutic opportunities. Curr. Drug Targets Immune Endocr. Metabol. Disord. 4, 277–285 10.2174/1568008043339767 [DOI] [PubMed] [Google Scholar]
- 14. Dupont E., Prochiantz A., and Joliot A. (2015) Penetratin story: An overview. Methods Mol. Biol. 1324, 29–37 10.1007/978-1-4939-2806-4_2 [DOI] [PubMed] [Google Scholar]
- 15. Poillot C., Dridi K., Bichraoui H., Pêcher J., Alphonse S., Douzi B., Ronjat M., Darbon H., and De Waard M. (2010) d-Maurocalcine, a pharmacologically inert efficient cell-penetrating peptide analogue. J. Biol. Chem. 285, 34168–34180 10.1074/jbc.M110.104919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poillot C., Bichraoui H., Tisseyre C., Bahemberae E., Andreotti N., Sabatier J. M., Ronjat M., and De Waard M. (2012) Small efficient cell-penetrating peptides derived from scorpion toxin maurocalcine. J. Biol. Chem. 287, 17331–17342 10.1074/jbc.M112.360628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt N., Mishra A., Lai G. H., and Wong G. C. (2010) Arginine-rich cell-penetrating peptides. FEBS Lett. 584, 1806–1813 10.1016/j.febslet.2009.11.046 [DOI] [PubMed] [Google Scholar]
- 18. Mishra A., Lai G. H., Schmidt N. W., Sun V. Z., Rodriguez A. R., Tong R., Tang L., Cheng J., Deming T. J., Kamei D. T., and Wong G. C. (2011) Translocation of HIV TAT peptide and analogues induced by multiplexed membrane and cytoskeletal interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 16883–16888 10.1073/pnas.1108795108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gomarasca M., F. C. Martins T., Greune L, Hardwidge P. R., Schmidt M. A., and Rüter C. (2017) Bacterium-derived cell-penetrating peptides deliver gentamicin to kill intracellular pathogens. Antimicrob. Agents Chemother. 61, e02545–e02516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carter G. P., Rood J. I., and Lyras D. (2012) The role of toxin A and toxin B in the virulence of Clostridium difficile. Trends Microbiol. 20, 21–29 10.1016/j.tim.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 21. Davies A. H., Roberts A. K., Shone C. C., and Acharya K. R. (2011) Super toxins from a super bug: Structure and function of Clostridium difficile toxins. Biochem. J. 436, 517–526 10.1042/BJ20110106 [DOI] [PubMed] [Google Scholar]
- 22. Jank T., Belyi Y., and Aktories K. (2015) Bacterial glycosyltransferase toxins. Cell. Microbiol. 17, 1752–1765 10.1111/cmi.12533 [DOI] [PubMed] [Google Scholar]
- 23. Pruitt R. N., and Lacy D. B. (2012) Toward a structural understanding of Clostridium difficile toxins A and B. Front. Cell. Infect. Microbiol. 2, 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larabee J. L., Krumholz A., Hunt J. J., Lanis J. M., and Ballard J. D. (2015) Exposure of neutralizing epitopes in the carboxyl-terminal domain of TcdB is altered by a proximal hypervariable region. J. Biol. Chem. 290, 6975–6985 10.1074/jbc.M114.612184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larabee J. L., Bland S. J., Hunt J. J., and Ballard J. D. (2017) Intrinsic toxin-derived peptides destabilize and inactivate Clostridium difficile TcdB. MBio 8, e00503–e00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hunt J. J., Larabee J. L., and Ballard J. D. (2017) Amino acid differences in the 1753-to-1851 region of TcdB influence variations in TcdB1 and TcdB2 cell entry. mSphere 2, e00268–e00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Shi L., Li S., Yang Z., Standley C., Yang Z., ZhuGe R., Savidge T., Wang X., and Feng H. (2013) A segment of 97 amino acids within the translocation domain of Clostridium difficile toxin B is essential for toxicity. PLoS ONE 8, e58634 10.1371/journal.pone.0058634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen S., Wang H., Gu H., Sun C., Li S., Feng H., and Wang J. (2016) Identification of an essential region for translocation of Clostridium difficile toxin B. Toxins (Basel) 8, E241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lanis J. M., Barua S., and Ballard J. D. (2010) Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 6, e1001061 10.1371/journal.ppat.1001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qa'Dan M., Spyres L. M., and Ballard J. D. (2001) pH-enhanced cytopathic effects of Clostridium sordellii lethal toxin. Infect. Immun. 69, 5487–5493 10.1128/IAI.69.9.5487-5493.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pooga M., Hällbrink M., Zorko M., and Langel U. (1998) Cell penetration by transportan. FASEB J. 12, 67–77 [DOI] [PubMed] [Google Scholar]
- 32. Orrell K. E., Zhang Z., Sugiman-Marangos S. N., and Melnyk R. A. (2017) Clostridium difficile toxins A and B: Receptors, pores, and translocation into cells. Crit. Rev. Biochem. Mol. Biol. 52, 461–473 10.1080/10409238.2017.1325831 [DOI] [PubMed] [Google Scholar]
- 33. Larabee J. L., Maldonado-Arocho F. J., Pacheco S., France B., DeGiusti K., Shakir S. M., Bradley K. A., and Ballard J. D. (2011) Glycogen synthase kinase 3 activation is important for anthrax edema toxin-induced dendritic cell maturation and anthrax toxin receptor 2 expression in macrophages. Infect. Immun. 79, 3302–3308 10.1128/IAI.05070-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bolz D. D., Li Z., McIndoo E. R., Tweten R. K., Bryant A. E., and Stevens D. L. (2015) Cardiac myocyte dysfunction induced by streptolysin O is membrane pore and calcium dependent. Shock 43, 178–184 10.1097/SHK.0000000000000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., and Novina C. D. (2003) Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 10.1261/rna.2192803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]