Significance
Large complexes composed of the viral genome encapsidated by viral proteins (capsid or nucleocapsid) need to be transported from the sites of viral genome synthesis to the sites of budding within infected cells. For the highly pathogenic Ebola virus, as for many other viruses, it is not completely understood how this transport is accomplished and which viral components are necessary for transport-competent nucleocapsids. Using live-cell imaging in combination with a surrogate system to analyze nucleocapsid transport under biosafety level 2 conditions, we show that the Ebola virus proteins nucleoprotein, VP35, and VP24 are necessary and sufficient to form transport-competent nucleocapsids. The technical approach developed here is applicable to study the transport of other viruses and to characterize antivirals.
Keywords: Ebola virus, nucleocapsid, transport, live-cell imaging, replicon system
Abstract
The intracytoplasmic movement of nucleocapsids is a crucial step in the life cycle of enveloped viruses. Determination of the viral components necessary for viral nucleocapsid transport competency is complicated by the dynamic and complex nature of nucleocapsid assembly and the lack of appropriate model systems. Here, we established a live-cell imaging system based on the ectopic expression of fluorescent Ebola virus (EBOV) fusion proteins, allowing the visualization and analysis of the movement of EBOV nucleocapsid-like structures with different protein compositions. Only three of the five EBOV nucleocapsid proteins—nucleoprotein, VP35, and VP24—were necessary and sufficient to form transport-competent nucleocapsid-like structures. The transport of these structures was found to be dependent on actin polymerization and to have dynamics that were undistinguishable from those of nucleocapsids in EBOV-infected cells. The intracytoplasmic movement of nucleocapsid-like structures was completely independent of the viral matrix protein VP40 and the viral surface glycoprotein GP. However, VP40 greatly enhanced the efficiency of nucleocapsid recruitment into filopodia, the sites of EBOV budding.
Viruses must safely transport their genome within an infected cell from the site of synthesis to the site of release. To protect the viral genome from recognition by cellular defense mechanisms, it is encapsidated by viral proteins (1). For some DNA viruses, it has been reported that capsid movement in the cytosol can be mediated by one or several capsid-associated proteins and is based on either the actin or the microtubule cytoskeleton (2, 3). Several tegument proteins surrounding the capsid of herpes simplex virus-1 enable capsid transport along microtubules (4), whereas the nucleocapsid proteins p78/83 or VP80 of baculovirus can directly engage the Arp2/3 complex and thus induce actin nucleation (5, 6). The genome of nonsegmented negative-strand RNA viruses (mononegaviruses) is encapsidated by nucleocapsid proteins to form a helical structure, and it is currently unknown which viral factors serve as mediators for the interaction of this structure with the cytoskeleton. Mononegaviruses include important human pathogens, like measles virus, and highly pathogenic zoonotic pathogens, like rabies virus, Marburg virus (MARV), and Ebola virus (EBOV).
EBOV and MARV belong to the filoviruses (family Filoviridae) and cause a severe fever with high case fatality rates. The largest ever EBOV epidemic in West Africa ended in 2016 after almost 3 years and over 11,000 deaths (7), and another EBOV epidemic has been reported in the Democratic Republic of Congo (8). Therefore, it is imperative to improve preparedness against EBOV epidemics. Among other activities, it is necessary to develop antivirals, a process requiring a detailed knowledge of EBOV biology. For example, understanding how EBOV utilize the host cytoskeleton might be important to identify new therapeutic approaches.
The main nucleocapsid protein of EBOV is the nucleoprotein NP, which directly encapsidates the viral genome (9, 10). EBOV nucleocapsids additionally contain the viral protein VP24 and the polymerase cofactor VP35, which are essential structural elements that directly interact with NP to build a long helical nucleocapsid ∼1,000 nm in length and 50 nm in diameter (9, 11–13). Also associated with the nucleocapsid are the viral polymerase L and the transcription factor VP30 (14, 15). Nucleocapsid formation occurs in inclusion bodies in the perinuclear region of EBOV- and MARV-infected cells, which act as virus factories (16–18). In the course of infection, thin rod-like nucleocapsids leave the inclusion bodies and move within the cytosol with varying velocities (100–400 nm/s), driven by actin polymerization (19). Essential for the recruitment of nucleocapsids to the cell periphery and for budding of progeny virions is the filoviral matrix protein VP40 (9, 20–22), which is arranged beneath the plasma membrane in an ordered lattice. VP40-enriched clusters also contain the surface glycoprotein GP, which is inserted into the plasma membrane (23–26). Although interactions between the filoviral VP40 and cytoskeletal proteins have been reported (27–29), there is no direct evidence that VP40 is involved in the intracellular transport of nucleocapsids (30). Thus, the roles of VP40 and the individual EBOV nucleocapsid proteins in the intracellular transport of nucleocapsids remain elusive.
Because of its high pathogenicity, EBOV is handled under the highest biosafety conditions [biosafety level (BSL)-4] that complicate and delay research on this virus (31). Here, we present a replicon system that allows analysis of the transport of nucleocapsid-like structures (NCLSs) under BSL-2 conditions. The established live-cell imaging system is based on the ectopic expression of fluorescent viral fusion proteins, which allows the visualization and characterization of NCLS intracytoplasmic transport. Moreover, we investigated the role of VP40 in the transport of nucleocapsids. Notably, this live-cell imaging approach can be utilized to analyze interactions between the nucleocapsid and cellular cytoskeleton, as well as to evaluate antiviral compounds that target the transport of nucleocapsids.
Results
Establishment of a Live-Cell Imaging System for the Analysis of EBOV NCLSs.
Previously established EBOV-specific replicon systems are based on the ectopic expression of viral proteins and an RNA minigenome composed of the nontranscribed 3′ and 5′ regions of the EBOV genome flanking a reporter gene (10, 32, 33). This minigenome is encapsidated by nucleocapsid proteins to form an NCLS, which serves as template for viral replication and transcription (32, 34). In the presence of VP40 and GP, NCLSs are released by budding at the plasma membrane to form transcription- and replication-competent virus-like particles (trVLPs) (32, 33, 35).
To determine whether the transport of NCLSs can be monitored and, if so, which viral components are necessary, we constructed a set of plasmids encoding EBOV nucleocapsid proteins labeled with either GFP or TagRFP. To confirm that the fusion of GFP or TagRFP to the nucleocapsid protein of interest did not impair the protein’s function, the functions of ectopically expressed VP30-GFP, VP35-GFP, or VP24-TagRFP were first tested in the EBOV minigenome system (Fig. S1). While VP30-GFP was able to support minigenome activity (Fig. S1A), VP35-GFP and VP24-TagRFP were not fully functional as the respective wild-type proteins. However, supplementary expression of VP35 partially rescued minigenome activity in the presence of VP35-GFP (Fig. S1B), and supplementary expression of VP24 rescued the regulatory function of VP24-TagRFP on viral transcription (Fig. S1C) (36). Therefore, to tag NCLSs with either VP35-GFP or VP24-TagRFP, we used in all subsequent experiments a combination of the fluorescent fusion protein and its wild-type counterpart.
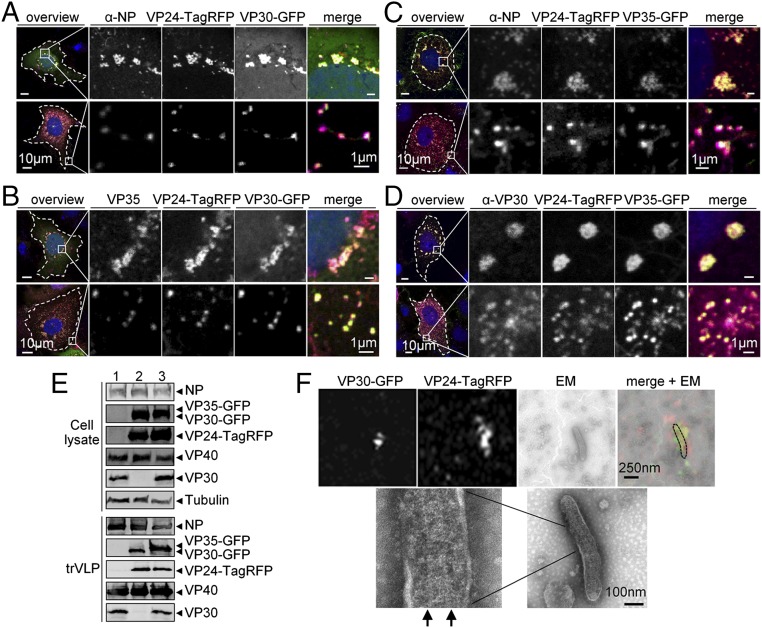
Next, we examined the intracellular distribution of VP30-GFP, VP35-GFP, or VP24-TagRFP in the presence of all components necessary for functional NCLSs in the trVLP assay (i.e., all viral proteins and an EBOV-specific minigenome). Confocal microscopic analyses showed that VP30-GFP and VP24-TagRFP colocalized with NP and VP35 inside NP-induced inclusion bodies, the sites where NCLSs are formed, and in small dot-like structures predominantly visible at the periphery of the transfected cells (Fig. 1 A and B). The presence of NP and VP35 in these peripheral small VP30-GFP+ and VP24-TagRFP+ dot-like structures indicated that they represent NCLSs. Also the combination of VP35-GFP and VP24-TagRFP was detected in the inclusion bodies and small dot-like structures colocalized with NP and VP30 (Fig. 1 C and D). Furthermore, coimmunoprecipitation showed that VP35-GFP and VP24-TagRFP can directly interact with NP (Fig. S2A), and VP35-GFP can interact with VP30 (Fig. S2B). The interaction between VP24 and NP has been described in detail recently (37). The interaction between VP30 and NP as well as between VP30 and VP35 has been previously characterized (15, 38). Altogether, these results indicated that the fluorescent fusion nucleocapsid proteins were recruited by NP into inclusion bodies and incorporated into individual NCLSs.
Fig. 1.
Analysis of the incorporation of VP30-GFP, VP35-GFP, and VP24-TagRFP into NCLS. (A–D) Confocal microscopy analysis of the intracellular distribution of the fluorescent fusion proteins in cells expressing all EBOV proteins and an EBOV-specific minigenome. (A and B) Huh-7 cells were transfected with plasmids encoding NP, L, VP35-HA, VP24, VP40, GP, minigenome, T7 polymerase and two fusion proteins, VP24-TagRFP and VP30-GFP. (C and D) Huh-7 cells were transfected with plasmids encoding NP, L, VP35, VP30, VP24, VP40, GP, minigenome, T7 polymerase and two fusion proteins, VP24-TagRFP and VP35-GFP. At 24 h posttransfection, the cells were fixed with 4% paraformaldehyde, stained with NP-, HA-, or VP30-specific antibodies and matching Alexa 680-tagged secondary antibodies, and subjected to confocal microscopy analysis. Intracellular distribution of fluorescent fusion proteins was analyzed by autofluorescence of GFP or TagRFP. Colocalization of fluorescent fusion proteins and wild-type nucleocapsid proteins is shown in the perinuclear located inclusion bodies (Upper) and in small dot-like structures at the cell periphery (Lower). Left panels show cells at low magnification, Right panels show magnified images of the boxed area. (E) Western blot analysis of trVLPs formed in the presence of the fluorescent fusion proteins. HEK293 cells were transfected with plasmids encoding NP, L, VP35, VP30, VP24, VP40, GP, minigenome, T7 polymerase (1), or plasmids encoding NP, L, VP35, VP30-GFP, VP24, VP24-TagRFP, VP40, GP, minigenome, T7 polymerase (2), or plasmids encoding NP, L, VP35, VP35-GFP, VP30, VP24, VP24-TagRFP, VP40, GP, minigenome, T7 polymerase (3). At 72 h posttransfection, cells were lysed, trVLPs were purified from the supernatants of transfected cells by centrifugation through a 20% sucrose cushion. Cell lysates and trVLPs were analyzed by SDS-PAGE and Western blot analysis using NP-, GFP-, TagRFP, VP40-, VP30-, α-tubulin–specific antibodies. (F) Correlative confocal and electron microscopy analyses of the incorporation of the fluorescent fusion proteins into NCLS. The trVLPs purified from the supernatant of HEK293 cells transfected as described in E (2) were adsorbed on Finder grids, the position of VP30-GFP+ and VP24-TagRFP+ particles was recorded by confocal microscopy, then the samples were negatively stained and the presence of NCLSs inside the GFP+ and TagRFP+ particles was analyzed by transmission electron microscopy. Arrows indicate helical structure of NCLS.
Next, we investigated whether the fluorescently tagged NCLSs could be released into the supernatant of cells in the form of trVLPs. At 72 h posttransfection, trVLPs were purified from the supernatant of HEK293 cells expressing all viral proteins and the EBOV-specific minigenome, and at the same time point, cells were lysed for further analysis. Western blot analysis of cell lysates and trVLPs showed that VP30-GFP, VP35-GFP, and VP24-TagRFP were released into the supernatant and purified together with trVLPs, indicating they were incorporated into the released particles (Fig. 1E). This conclusion was further supported by correlative confocal and electron microscopy of released trVLPs, labeled with VP24-TagRFP and VP30-GFP. It was shown that fluorescent signals could be assigned to helical nucleocapsid-like structures in the enveloped particles (Fig. 1F).
The ectopically expressed individual fluorescent nucleocapsid proteins (VP30-GFP, VP35-GFP, VP24-TagRFP, and NP-TagRFP) did not undergo directional transport, neither alone nor upon coexpression with the respective wild-type proteins (Fig. S3). However, ectopic expression of the fluorescent nucleocapsid proteins (VP35-GFP, or VP30-GFP and VP24-TagRFP) in EBOV-infected cells resulted in the formation of motile dual color-labeled rod-like nucleocapsids with the same dimensions previously reported for EBOV nucleocapsids (Fig. S4 A and B and Movies S1 and S2) (19). All fluorescent fusion nucleocapsid proteins labeled the nucleocapsids homogenously, with no site-specific accumulation. The velocities of nucleocapsids varied from 100 nm/s to more than 400 nm/s, with a median velocity of 236 ± 79 nm/s for nucleocapsids labeled with VP30-GFP, 234 ± 84 nm/s (labeled with VP35-GFP), or 201 ± 83 nm/s (labeled with VP24-TagRFP) (Fig. S4C). Moreover, each fluorescent fusion protein was incorporated into purified viral particles (Fig. S4D).
Taken together, the presented data show that fluorescent fusion proteins of VP30, VP35, and VP24 were reliably associated with NP-induced inclusions and individual NCLSs, were released in association with NCLSs, and could be detected within trVLPs. In addition, the fluorescent fusion proteins were suitable for the labeling of nucleocapsids in EBOV-infected cells and were released together with EBOV nucleocapsids.
Analysis of the Intracellular Transport of EBOV NCLSs.
To test whether the fluorescent nucleocapsid proteins were suitable for live-cell imaging analysis of EBOV NCLSs, Huh-7 cells were transfected with plasmids encoding VP30-GFP and the other EBOV trVLP assay components (NP, VP35, VP24, L, VP40, GP, the minigenome, and the T7 polymerase). Time-lapse microscopy was started at 20 h posttransfection, when individual VP30-GFP+ NCLSs were recognizable in the cytoplasm (Fig. 2 and Movies S3 and S4). We detected motile fluorescent NCLSs with a median length of 435 ± 105 nm (n = 50), which is approximately half the length of EBOV nucleocapsids in infected cells (9, 19). Maximum-intensity projection of time-lapse images showed that the trajectories of moving particles were of different lengths, and the maximal measured trajectory length was 18.4 µm (Fig. 2A, Left). The velocities of NCLSs varied from 100 nm/s to almost 400 nm/s, with a median velocity of 185 ± 84 nm/s (Fig. 2A, Right). Both the trajectory lengths and velocities of NCLSs were similar to those measured for nucleocapsids in EBOV-infected cells (19).
Fig. 2.
Live-cell imaging analysis of NCLS transport. (A–C) Huh-7 cells were transfected with plasmids encoding VP30-GFP and plasmids encoding the trVLP components (NP, VP35, VP40, GP, VP24, L, EBOV-specific minigenome, T7 polymerase). At 17 h posttransfection, different cytoskeleton-modulating drugs were added to the culture medium: (A) 0.15% DMSO (vehicle), (B) 15 μM nocodazole, or (C) 0.3 μM cytochalasin D, and the cells were incubated for additional 3 h. (D–F) Huh-7 cells were transfected with plasmids encoding VP30-GFP and plasmids encoding NP, VP35, VP24, L, EBOV-specific minigenome, T7 polymerase. At 17 h posttransfection, different cytoskeleton-modulating drugs were added to the culture medium: (D) 0.15% DMSO (vehicle), (E) 15 μM nocodazole, (F) 0.3 μM cytochalasin D, and the cells were incubated for additional 3 h. The pictures show the maximum-intensity projection of time-lapse images of cells recorded for 2 min; images were captured every 2 s. Magnified pictures of the boxed regions are shown in Insets. The graphics show the velocities (n = 20) of the NCLS; the median velocity and the SD are shown in numbers.
Previous studies revealed that the intracellular movement of EBOV nucleocapsids is dependent on the polymerization of actin, but not on tubulin (19). To analyze whether the transport of NCLSs was also dependent on cellular cytoskeleton components, we investigated their movement in the presence of agents that inhibit the polymerization of actin (cytochalasin D) or microtubules (nocodazole). While incubation with nocodazole had no effect (Fig. 2B), cytochalasin D immediately abolished the movement of NCLSs (Fig. 2C). Thus, our live-cell imaging data on the velocity of the NCLSs and the dependence of their movement on actin polymerization are in line with previously published studies of nucleocapsid movement in EBOV-infected cells (19).
Influence of VP40 and GP on the Intracellular Transport of EBOV NCLSs.
To identify the minimal set of viral proteins necessary to support nucleocapsid transport, we analyzed whether the omission of VP40 and GP altered NCLS transport. Huh-7 cells were transfected with plasmids encoding VP30-GFP, NP, VP35, VP24, L, the minigenome, and the T7 polymerase. Live-cell imaging analysis was performed at 20 h posttransfection (Fig. 2D and Movies S5 and S6). The median length of the VP30-GFP+ NCLSs was 465 ± 115 nm (n = 50), which was similar to that observed in cells expressing all of the viral proteins (see above). Maximum-intensity projection of time-lapse images showed that the trajectory length of NCLSs reached 16.3 µm (Fig. 2D, Left) and the median speed of the NCLSs was 228 ± 91 nm/s (Fig. 2D, Right). Treatment of cells with nocodazole did not affect either the length of the trajectories or the speed of the NCLSs (16 µm, 181 ± 76 nm/s) (Fig. 2E). In contrast, when cells were incubated with cytochalasin D, NCLS movement was abolished (Fig. 2F). Together, these experiments show that intracytoplasmic transport of NCLSs is independent of VP40 and GP and has the same characteristics as transport of NCLSs in the presence of all viral proteins.
Identification of Viral Components Necessary for NCLS Transport.
VP30-GFP–labeled moving NCLSs were observed upon ectopic expression of the nucleocapsid components NP, VP35, VP24, L, the minigenome, the T7 polymerase, and VP30-GFP (Fig. 2D). To determine which of the nucleocapsid components were necessary for transport of the NCLSs, we repeated the experiment shown in Fig. 2D, with the omission of each of the nucleocapsid components in turn. Live-cell imaging analysis showed that upon omission of the polymerase L or the EBOV-specific minigenome, NCLS showed normal movement, whereas the absence of either NP, VP35, or VP24 completely blocked NCLS transport (Fig. 3A). In this experimental setting, VP30-GFP was used to monitor NCLS transport, and the role of VP30 itself could not be analyzed. The experiments were therefore repeated as described above, with the NCLSs being labeled with VP35-GFP (in the presence of VP35). Again, the absence of NP and VP24 blocked NCLS transport (Fig. 3B). In absence of VP30, however, NCLSs were still transported (Fig. 3B). The polymerase L and the minigenome had no influence, confirming the results from Fig. 3A. These data indicate that transport of NCLSs requires a complex composed of the NP, VP35, and VP24 proteins.
Fig. 3.
Identification of nucleocapsid components necessary for NCLS transport. (A) Huh-7 cells were transfected with plasmids encoding VP30-GFP and NP, VP35, VP24, L, EBOV-specific minigenome as well as T7 polymerase. With exception of VP30-GFP, each of the plasmids (or the combination of minigenome and T7 polymerase) was omitted in turn, as indicated. (B) Huh-7 cells were transfected with plasmids encoding VP35-GFP and NP, VP35, VP30, VP24, L, EBOV-specific minigenome, as well as T7 polymerase. With exception of VP35-GFP and VP35, each of the plasmids (or the combination of minigenome and T7 polymerase) was omitted in turn, as indicated. The cells were analyzed by live-cell imaging at 20 h posttransfection. The panels show the maximum-intensity projection of time-lapse images recorded for 2 min; images were captured every 3 s. Magnified pictures of the boxed regions are shown in Insets. (Scale bar in Insets, 2 μm.)
Transport Characteristics of NCLSs Formed only by NP, VP35, and VP24.
To analyze whether these three components were also sufficient to form transport-competent NCLSs, NP, VP35, and VP24 alone were coexpressed together with VP35-GFP in Huh-7 cells. At 20 h posttransfection, cells were subjected to live-cell imaging analysis (Fig. 4A and Movies S7 and S8). The detected motile NCLSs had essentially the same dimensions as observed in the previous experiments (440 ± 81 nm, n = 50). The NCLS trajectories had a maximal length of 14.8 µm and the median velocity of the NCLSs was 187 ± 67 nm/s. When the cells were incubated with nocodazole, the median speed of the NCLSs was 213 ± 65 nm/s (Fig. 4B, Lower). The incubation of cells with cytochalasin D stopped the movement of NCLSs (Fig. 4C). These data indicate that NCLSs formed by NP, VP35, and VP24 alone displayed long-distance movement that was dependent on actin polymerization. Thus, the movement of NCLSs formed by NP, VP35, and VP24 alone was highly similar to the movement of nucleocapsids in EBOV-infected cells; this leads to the conclusion that these three nucleocapsid proteins are essential and sufficient to mediate NCLS transport.
Fig. 4.
Live-cell imaging analysis of NCLS formed by NP, VP35 and VP24. (A–C) Huh-7 cells were transfected with plasmids encoding NP, VP24, VP35-GFP and VP35. At 17 h posttransfection, different cytoskeleton-modulating drugs were added to the culture medium: (A) 0.15% DMSO (vehicle), (B) 15 μM nocodazole, or (C) 0.3 μM cytochalasin D for 3 h; consequently cells were subjected to live-cell imaging analysis. The pictures show the maximum-intensity projection of time-lapse images of cells recorded for 2 min; images were captured every 3 s. Magnified pictures of the boxed regions are shown in Insets. (Scale bar in Insets, 2 μm.) The graphics show the velocities (n = 20) of the NCLS, the median velocity and the SD are shown in numbers.
Influence of VP40 on the Recruitment of NCLSs to Budding Sites.
Previous analyses revealed that MARV nucleocapsids moving in the cell body did not contain detectable amounts of VP40 (30). However, the budding of nucleocapsids at either the side or the tip of filopodia, the preferred site of filoviral release, was dependent on the presence of VP40 and its interaction with nucleocapsids (19, 39, 40). In line with these results, filopodia in MARV-infected cells exclusively contained nucleocapsids that were associated with VP40, suggesting that VP40-association was a prerequisite for the nucleocapsids to enter filopodia (30). Our results here demonstrate that EBOV VP40 is dispensable for the intracytoplasmic transport of NCLSs. However, it remained unclear whether VP40 is needed for the entry of nucleocapsids into filopodia.
To answer this question, we constructed a plasmid encoding a VP40-TagRFP fusion protein, which was characterized by Western blot, minigenome, and trVLP assays (Fig. S5), showing that VP40-TagRFP was targeted to the plasma membrane upon coexpression with VP40 (Fig. S5A), and colocalized with NP upon coexpression with trVLP components (Fig. S5B). Whereas VP40-TagRFP was unable to inhibit transcription/replication in the minigenome assay and to form trVLPs, these two functions were rescued in the presence of wild-type VP40 (Fig. S5 C and D).
A mixture of VP40-TagRFP in combination with wild-type VP40 (referred to as VP40 for simplicity in the next paragraph) was then used to analyze the impact of VP40 on the distribution of NCLSs at the cell periphery. The number of VP30-GFP–labeled NCLSs moving within filopodia per cell was counted in the presence or absence of VP40. On average, 10 NCLSs per cell were observed in the absence of VP40, whereas 49 NCLSs were detected in cells expressing VP40 (Fig. 5A). Thus, VP40 significantly increased the number of intrafilopodial NCLSs, although VP40 was not absolutely necessary for the entry of NCLSs into filopodia. Live-cell imaging analysis of intrafilopodial transport of NCLSs was performed in the presence or absence of VP40 (Fig. 5 B–D). The median speed of intrafilopodial NCLSs in the presence or absence of VP40 was 56 ± 32 and 57 ± 46 nm/s, respectively. These results suggest that VP40 has no significant impact on the movement of NCLSs inside filopodia. Taken together, our data from this study show that VP40 was not essential for the intracytoplasmic transport of NCLSs. In addition, VP40 was dispensable for the entry of NCLSs into filopodia, although it increased the number of intrafilopodial NCLSs.
Fig. 5.
The influence of VP40 on the NCLS transport at the cell periphery. Huh-7 cells were transfected with plasmids encoding VP30-GFP, NP, VP35, VP24 (1), or with plasmids encoding VP30-GFP, NP, VP35, VP24, and a mixture of VP40-TagRFP and VP40 (2). Live-cell imaging analysis was performed at 20 h posttransfection. (A) The number of motile NCLS inside the filopodia was counted. The median number of moving NCLSs within filopodia per cell and the SD are shown. ***P < 0.001. (B) The trajectories and direction of moving NCLSs inside the filopodia are shown by white and black arrowheads, as well as red and orange lines. (C) The trajectories and direction of moving NCLSs inside filopodia are shown by white and black arrowheads as well as orange and white lines. (D) The graphics show the velocities of NCLSs (n = 30) inside filopodia. The numbers indicated the median speed and the SD.
Discussion
In the present study, we identified NP, VP35, and VP24 to be the viral components that are essential and sufficient for the intracytoplasmic transport of NCLSs, indicating they are also necessary for the transport of nucleocapsids in EBOV-infected cells.
Transport of viral capsids is best investigated with DNA viruses (e.g., herpes simplex type 1 and baculovirus), in which several proteins have been reported to mediate capsid transport. The functions of these proteins were identified using virus deletion mutants (4, 6, 41). Although reverse genetics systems for filoviruses are available, analogous experiments are difficult to perform, because the few nucleocapsid proteins of filoviruses serve multiple essential functions in replication, morphogenesis, and the anti-IFN response and deletion of the corresponding genes results in nonreplicating viruses (17, 30, 42, 43). Using a trVLP system and a reductionist approach, we determined the essential EBOV components for transport of NCLSs employing live-cell imaging. The used trVLP system has limitations; for example, it does not fully mimic the capacity of EBOV-infected cells to regulate viral transcription/replication and translation of viral proteins (33, 34). Nevertheless this system was suitable for our purpose to investigate the role of individual viral proteins for the transport of NCLSs because we could freely remove and combine the different viral proteins and analyze the effects on NCLSs movement. Visualization of NCLSs was achieved by incorporation of fluorescent nucleocapsid fusion proteins. It could not be excluded that labeling with VP35-GFP or VP24-TagRFP impaired NCLSs intracellular movement in comparison with labeling with VP30-GFP, which was previously used for visualization of motile nucleocapsids (30). To test this, we separately monitored movement of NCLSs labeled with each fluorescent fusion protein. All differently labeled motile NCLSs had comparable velocities, suggesting their transport was not significantly influenced by tagging (Figs. 2–4). This was supported by the finding that EBOV nucleocapsids, which were labeled with either of the fluorescent fusion proteins, all showed a similar pattern of movement in infected cells (Fig. S4).
NP, VP35, and VP24 are also the components that form the helical EBOV nucleocapsid (11–13). Filoviral VP24 has no analog among the mononegaviruses, and the function of this protein has only been unfolded recently (32, 37, 44–47). It has been shown that EBOV VP24 inhibits transcription and replication of the minigenome (36). The finding that VP24 was essential for transport suggests that only nucleocapsids in a “locked” state, and therefore unable to serve as templates for transcription and replication, are transported (36, 44). Whether VP24 binds to preformed nucleocapsids after they served as templates or is incorporated into nucleocapsids concomitant with replication and encapsidation of the viral RNA remains elusive. The ultrastructural studies on the formation of nucleocapsids based on ectopically expressed NP, VP35, and VP24 are consistent with both of these scenarios (9, 12, 13, 48, 49).
How the connection between the EBOV nucleocapsids and cellular transport machineries is mediated is currently not understood. Because neither NP, VP35, nor VP24 was able to support its own movement inside the cell, it is suggested that the protein–protein interactions driving the formation of the helical nucleocapsid also result in conformational changes in one or more of the three proteins that expose hidden binding sites for cytoskeleton-interacting proteins. While interactions between individual viral and cytoskeleton proteins were detected, the impact of these interactions on the transport dynamics of viral protein complexes in living cells is so far unclear. For example, while it has been reported that dynein light chain protein 8 interacts with VP35 (50), our study suggests that this interaction is not sufficient to transport VP35 expressed alone (Fig. S3B). This is supported by the finding that the P protein of rabies virus, a VP35 analog, also interacts with dynein light chain protein 8 (51), and blocking this interaction did not impair viral transport (52).
Long-distance cytoplasmic movement of the nucleocapsid complex was independent of the viral matrix protein VP40 and the glycoprotein GP. The matrix proteins of mononegaviruses have been considered as primary factors that induce the redistribution of viral nucleocapsids from perinuclear inclusion bodies to the plasma membrane (53–55). The role of the filovirus matrix proteins for nucleocapsid transport has been debated for several years (16, 30, 48). The present study shows almost identical velocities for intracytoplasmic EBOV NCLSs in the absence or presence of VP40, indicating that a complex composed of three proteins—NP, VP35, and VP24—is sufficient to hook onto the actin cytoskeleton machinery that provides the driving force for nucleocapsid movement (Fig. 4). On the other hand, VP40 could significantly increase the number of NCLSs inside the filopodia, which is consistent with its role as a facilitator of budding and release. The exact molecular mechanism by which VP40 facilitates this effect on peripherally located nucleocapsids is currently under investigation.
The transport of filovirus helical nucleocapsids seems to follow a clear directionality, which is probably determined by the structural polarity of the nucleocapsids that show characteristic “pointed” and “barbed” ends (49). For other viruses, the arrangement of proteins within the nucleocapsid has been shown to potentially influence their motility. For example, positioning of p78/83 or VP80 at one end of the baculovirus nucleocapsid has been shown to be important for the directional transport of nucleocapsids (5, 6). The filovirus proteins NP, VP35, and VP24, however, are homogenously distributed over the whole nucleocapsid, indicating that the structural polarity of the EBOV nucleocapsid is sufficient for directional transport (49). Future studies have to identify mechanisms of interaction between EBOV nucleocapsids and cytoskeletal proteins.
Experimental Procedures
Experimental procedures for cell culture and virus infection, as well as conventional microscopy, have been performed as described previously (30). More detailed information on the experimental procedures, including live-cell imaging microscopy, confocal laser scanning microscopy, virological methods, and all reagents, is provided in SI Experimental Procedures.
All experiments involving gene-modified organisms have been approved by the Regierungspräsidium Gießen, Gießen, Germany. Experiments with infectious Ebola virus have been carried out in the BSL-4 facility, Philipps-University Marburg, according to national regulations with the approval of the Regierungspräsidium Gießen, Gießen, Germany.
Supplementary Material
Acknowledgments
We thank Olga Dolnik, Sandro Halwe, Alexander Koehler, Marc Ringel, and Gordian Schudt for fruitful discussions and assistance; Viktor E. Volchkov for providing Ebola virus anti-VP24 antibodies; and Takeshi Noda for critical reading and discussing of the manuscript. Biosafety level 4 work would not have been possible without the supervision of Markus Eickmann, Olga Dolnik, and technical support from Michael Schmidt and Gotthard Ludwig. The work was supported by overseas research fellowships of the Uehara memorial foundation and Japan Society for the Promotion of Science (to Y.T.) and by the Deutsche Forschungsgemeinschaft through the SFB 1021 (to L.K. and S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712263115/-/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8:465–472. doi: 10.1016/s0966-842x(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 3.Smith GA, Enquist LW. Break ins and break outs: Viral interactions with the cytoskeleton of mammalian cells. Annu Rev Cell Dev Biol. 2002;18:135–161. doi: 10.1146/annurev.cellbio.18.012502.105920. [DOI] [PubMed] [Google Scholar]
- 4.Radtke K, et al. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goley ED, et al. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science. 2006;314:464–467. doi: 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- 6.Marek M, Merten OW, Galibert L, Vlak JM, van Oers MM. Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J Virol. 2011;85:5350–5362. doi: 10.1128/JVI.00035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO 2016 Ebola Situation Report, 10 June. Available at who.int/csr/disease/ebola/en/. Accessed July 1, 2017.
- 8.WHO 2017 Ebola outbreak situation report. Available at www.who.int/emergencies/ebola-DRC-2017/en/. Accessed July 15, 2017.
- 9.Bharat TA, et al. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe S, Noda T, Kawaoka Y. Functional mapping of the nucleoprotein of Ebola virus. J Virol. 2006;80:3743–3751. doi: 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan W, et al. Structure and assembly of the Ebola virus nucleocapsid. Nature. 2017;551:394–397. doi: 10.1038/nature24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartlieb B, Modrof J, Mühlberger E, Klenk HD, Becker S. Oligomerization of Ebola virus VP30 is essential for viral transcription and can be inhibited by a synthetic peptide. J Biol Chem. 2003;278:41830–41836. doi: 10.1074/jbc.M307036200. [DOI] [PubMed] [Google Scholar]
- 15.Biedenkopf N, Hartlieb B, Hoenen T, Becker S. Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: Impact on viral transcription and replication. J Biol Chem. 2013;288:11165–11174. doi: 10.1074/jbc.M113.461285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoenen T, et al. Inclusion bodies are a site of ebolavirus replication. J Virol. 2012;86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolnik O, Stevermann L, Kolesnikova L, Becker S. Marburg virus inclusions: A virus-induced microcompartment and interface to multivesicular bodies and the late endosomal compartment. Eur J Cell Biol. 2015;94:323–331. doi: 10.1016/j.ejcb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Schudt G, et al. Transport of Ebolavirus nucleocapsids is dependent on actin polymerization: Live-cell imaging analysis of Ebolavirus-infected cells. J Infect Dis. 2015;212(Suppl 2):S160–S166. doi: 10.1093/infdis/jiv083. [DOI] [PubMed] [Google Scholar]
- 20.Noda T, Watanabe S, Sagara H, Kawaoka Y. Mapping of the VP40-binding regions of the nucleoprotein of Ebola virus. J Virol. 2007;81:3554–3562. doi: 10.1128/JVI.02183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda T, et al. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolnik O, Kolesnikova L, Stevermann L, Becker S. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol. 2010;84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker S, Klenk HD, Mühlberger E. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology. 1996;225:145–155. doi: 10.1006/viro.1996.0582. [DOI] [PubMed] [Google Scholar]
- 24.Mittler E, Kolesnikova L, Strecker T, Garten W, Becker S. Role of the transmembrane domain of marburg virus surface protein GP in assembly of the viral envelope. J Virol. 2007;81:3942–3948. doi: 10.1128/JVI.02263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beniac DR, Booth TF. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci Rep. 2017;7:46374. doi: 10.1038/srep46374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Booth TF, Rabb MJ, Beniac DR. How do filovirus filaments bend without breaking? Trends Microbiol. 2013;21:583–593. doi: 10.1016/j.tim.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Ruthel G, et al. Association of ebola virus matrix protein VP40 with microtubules. J Virol. 2005;79:4709–4719. doi: 10.1128/JVI.79.8.4709-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, et al. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol. 2013;87:7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Z, Harty RN. Packaging of actin into Ebola virus VLPs. Virol J. 2005;2:92. doi: 10.1186/1743-422X-2-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schudt G, Kolesnikova L, Dolnik O, Sodeik B, Becker S. Live-cell imaging of Marburg virus-infected cells uncovers actin-dependent transport of nucleocapsids over long distances. Proc Natl Acad Sci USA. 2013;110:14402–14407. doi: 10.1073/pnas.1307681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falzarano D, Geisbert TW, Feldmann H. Progress in filovirus vaccine development: Evaluating the potential for clinical use. Expert Rev Vaccines. 2011;10:63–77. doi: 10.1586/erv.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoenen T, et al. Infection of naive target cells with virus-like particles: Implications for the function of ebola virus VP24. J Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biedenkopf N, Hoenen T. Modeling the Ebolavirus life cycle with transcription and replication-competent viruslike particle assays. Methods Mol Biol. 2017;1628:119–131. doi: 10.1007/978-1-4939-7116-9_9. [DOI] [PubMed] [Google Scholar]
- 34.Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: Reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011;91:195–208. doi: 10.1016/j.antiviral.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoenen T, et al. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol. 2010;84:7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology. 2010;403:56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Banadyga L, et al. Ebola virus VP24 interacts with NP to facilitate nucleocapsid assembly and genome packaging. Sci Rep. 2017;7:7698. doi: 10.1038/s41598-017-08167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biedenkopf N, Schlereth J, Grünweller A, Becker S, Hartmann RK. RNA binding of Ebola virus VP30 is essential for activating viral transcription. J Virol. 2016;90:7481–7496. doi: 10.1128/JVI.00271-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9:939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 40.Welsch S, et al. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 2010;6:e1000875. doi: 10.1371/journal.ppat.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douglas MW, et al. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J Biol Chem. 2004;279:28522–28530. doi: 10.1074/jbc.M311671200. [DOI] [PubMed] [Google Scholar]
- 42.Halfmann P, et al. Generation of biologically contained Ebola viruses. Proc Natl Acad Sci USA. 2008;105:1129–1133. doi: 10.1073/pnas.0708057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enterlein S, et al. Rescue of recombinant Marburg virus from cDNA is dependent on nucleocapsid protein VP30. J Virol. 2006;80:1038–1043. doi: 10.1128/JVI.80.2.1038-1043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196(Suppl 2):S284–S290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- 45.Reid SP, et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang AP, et al. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012;8:e1002550. doi: 10.1371/journal.ppat.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page A, et al. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep. 2014;6:1026–1036. doi: 10.1016/j.celrep.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 48.Noda T, et al. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubota T, et al. Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J Virol. 2009;83:6952–6956. doi: 10.1128/JVI.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raux H, Flamand A, Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan GS, Preuss MA, Williams JC, Schnell MJ. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc Natl Acad Sci USA. 2007;104:7229–7234. doi: 10.1073/pnas.0701397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenard J. Negative-strand virus M and retrovirus MA proteins: All in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 54.Runkler N, Pohl C, Schneider-Schaulies S, Klenk HD, Maisner A. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol. 2007;9:1203–1214. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 55.Dancho B, McKenzie MO, Connor JH, Lyles DS. Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J Biol Chem. 2009;284:4500–4509. doi: 10.1074/jbc.M808136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.