Significance
Sequestration of Plasmodium falciparum-infected erythrocytes (IEs) in the brain microvasculature underlies the pathology of cerebral malaria. Parasites that express P. falciparum erythrocyte membrane protein 1 of domain cassette (DC) 8 and DC13 types bind to brain endothelial cells. Recent studies, largely based on recombinant proteins, have identified endothelial protein C receptor (EPCR) as the key receptor for endothelial cell binding. Using DC8- and DC13-expressing IEs, we show that binding of DC13 IEs to brain endothelial cells is not EPCR-dependent and that cytoadhesion of EPCR-binding DC8 IEs to brain endothelial cells is blocked by human serum. This study highlights differences between recombinant protein and native protein in EPCR-binding properties and suggests that other receptors are also required for sequestration in cerebral malaria.
Keywords: cell adhesion, malaria, EPCR, PfEMP1, endothelium
Abstract
Recent advances have identified a new paradigm for cerebral malaria pathogenesis in which endothelial protein C receptor (EPCR) is a major host receptor for sequestration of Plasmodium falciparum-infected erythrocytes (IEs) in the brain and other vital organs. The parasite adhesins that bind EPCR are members of the IE variant surface antigen family Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) containing specific adhesion domains called domain cassette (DC) 8 and DC13. The binding interaction site between PfEMP1 and EPCR has been mapped by biophysical and crystallography studies using recombinant proteins. However, studies examining the interaction of native PfEMP1 on the IE surface with EPCR are few. We aimed to study binding to EPCR by IEs expressing DC8 and DC13 PfEMP1 variants whose recombinant proteins have been used in key prior functional and structural studies. IE binding to EPCR immobilized on plastic and on human brain endothelial cells was examined in static and flow adhesion assays. Unexpectedly, we found that IEs expressing the DC13 PfEMP1 variant HB3var03 or IT4var07 did not bind to EPCR on plastic and the binding of these variants to brain endothelial cells was not dependent on EPCR. IEs expressing the DC8 variant IT4var19 did bind to EPCR, but this interaction was inhibited if normal human serum or plasma was present, raising the possibility that IE–EPCR interaction may be prevented by plasma components under physiological conditions. These data highlight a discrepancy in EPCR-binding activity between PfEMP1 recombinant proteins and IEs, and indicate the critical need for further research to understand the pathophysiological significance of the PfEMP1–EPCR interaction.
The major pathophysiological process implicated in cerebral malaria is the sequestration of mature Plasmodium falciparum-infected erythrocytes (IEs) in the microvasculature of the brain (1–4). Binding to endothelial cell receptors is mediated by members of the variant IE surface protein family, P. falciparum erythrocyte membrane protein 1 (PfEMP1) (5), encoded by var genes (6). PfEMP1 variants are functionally classified into domain cassette (DC) types based on the conserved arrangement of tandem cysteine-rich adhesion domains called Duffy binding-like (DBL) and cysteine-rich interdomain region (CIDR) (7).
Over the past decade, significant progress has been made in linking particular var gene groups to IE adhesion phenotype and clinical disease. It has become apparent that IEs from patients with cerebral and severe malaria predominantly express group A or B/A var genes (8–10). Other studies show that DC8 (group B/A) and DC13 (group A) variants are specifically associated with cerebral and severe malaria (11–13). DC8 and DC13 variants have also been shown to mediate IE binding to human brain endothelial cells (HBECs) (14, 15), potentially explaining the association of parasite DC8/DC13 expression with the clinical development of cerebral malaria, due to sequestration in the brain. Later work showed that DC8- and DC13-expressing IEs bind to endothelial cells from diverse tissues and organs (16).
Binding of IEs to a particular receptor may be the cause of organ-specific pathology, as in the case of pregnancy malaria, where sequestration in the placenta is due to IEs binding to chondroitin sulfate A (CSA) (17). For cerebral and severe malaria, recent work suggests that endothelial protein C receptor (EPCR) may be the key receptor, as DC8 and DC13 recombinant PfEMP1 molecules bind with nanomolar affinity to EPCR (18), and a high-resolution crystal structure of the interaction between EPCR and PfEMP1 has been obtained identifying the DC13-EPCR–binding site (19).
Existing studies on the interaction of P. falciparum with EPCR are largely based on PfEMP1 recombinant proteins, and detailed studies of IEs expressing the DC8 and DC13 PfEMP1 variants are few (20–22). We therefore investigated the role of EPCR in IE adhesion using three P. falciparum parasite lines selected for HBEC binding that predominantly express either a DC8 or DC13 PfEMP1 (14). We show that there is a mismatch between published recombinant protein data and the binding properties of IEs, as IEs from two DC13-expressing parasite lines do not bind to EPCR. IEs from the DC8-expressing parasite line do bind EPCR, but the binding is inhibited by human plasma or serum, raising questions as to the physiological relevance of EPCR as a sequestration receptor in severe malaria.
Results
DC13-Expressing IEs Do Not Bind to EPCR Recombinant Protein in Static and Flow Assays.
Three parasite lines, derived by panning on immortalized HBECs (HBEC-5i), that express the PfEMP1 variants HB3var03 (DC13), IT4var07 (DC13), and IT4var19 (DC8) (14) were used to study IE binding to EPCR. They were chosen because recombinant CIDRα1 domain proteins of these variants bind to EPCR with high affinity, with a Kd reported by Lau et al. (19) as 0.37 nM for HB3var03 (DC13), 1.3 nM for IT4var07 (DC13), and 16 nM for IT4var19 (DC8). Also, HB3var03 and IT4var07 recombinant domain proteins were used to generate the crystal structure of PfEMP1 bound to EPCR (19). The parasite lines were enriched for the variants of interest by panning on HBEC-5i and cell sorting with variant-specific antibodies, such that the “HB3var03” parasite line contained 50–80% of IEs expressing the HB3var03 PfEMP1 on their surface, the “IT4var07” parasite line contained 30–50% of IEs expressing IT4var07 and 1–5% of IEs expressing IT4var19, and the “IT4var19” parasite line contained 50–80% of IEs expressing IT4var19 and <1% of IEs expressing IT4var07 (Fig. S1). The parasite lines are not monovariant, despite frequent selection, because spontaneous var gene switching occurs in vitro, away from the starting variant (23).
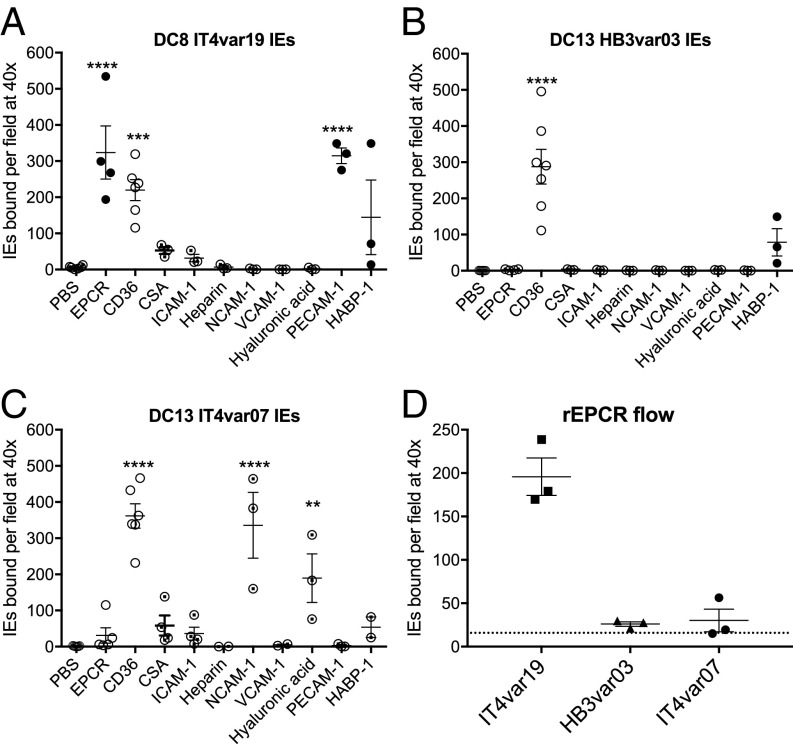
We tested the ability of the three parasite lines to bind to recombinant (r) EPCR in static adhesion assays. DC8 IT4var19 IEs bound well to rEPCR as expected (Fig. 1A). Surprisingly, DC13 HB3var03 and IT4var07 IEs did not bind to rEPCR (Fig. 1 B and C). All parasite lines showed binding to rCD36 significantly above background levels (Fig. 1 A–C), most likely due to subpopulations of parasites in these cultures expressing non-DC8/DC13 PfEMP1 variants of group B and C type, known to bind CD36 (14). To determine if the IEs that bound to rEPCR and rCD36 expressed the DC8 or DC13 variants, we carried out immunofluorescence staining on the bound cells using variant-specific antibodies. IEs bound to rEPCR from the IT4var19 culture were confirmed as expressing the IT4var19/DC8 variant (Fig. 1A and EPCR in Fig. S2), whereas the rCD36-binding IEs from the same culture did not express IT4var19 (Fig. 2A and CD36 in Fig. S2). In the IT4var07 culture, the small numbers of IEs that bound to rEPCR all expressed IT4var19 and not IT4var07, while the rCD36-binding IEs were negative with both antibodies. Similarly, in the HB3var03 culture, the rCD36-binding IEs were negative with antibodies to HB3var03 (Fig. S3A). These data show that DC8 IT4var19 IEs bound to rEPCR in static binding assays but that DC13 IT4var07 and HB3var03 IEs did not bind to rEPCR, despite the nanomolar affinity for rEPCR shown using IT4var07 and HB3var03 recombinant CIDRα1 domain proteins (18, 19).
Fig. 1.
DC8 IT4var19, but not DC13 HB3var03 and IT4var07, IEs bind to rEPCR. IT4var19 IE (A), HB3var03 IE (B), and IT4var07 IE (C) binding to 50 μg/mL receptors absorbed onto plastic dishes tested under static conditions is shown. The filled circles are IEs that stained positive with DC8 or DC13 PfEMP1 homologous antibodies, and the empty circles did not stain with the DC8 or DC13 PfEMP1 antibodies. Half-filled circles are IEs that were not successfully stained with antibodies due to loss of bound IEs during staining. The difference in mean binding values compared with the PBS control from n ≥ 3 independent experiments was analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001. (D) Binding of IT4var19, HB3var03, and IT4var07 IEs to 50 μg/mL rEPCR coated onto a microslide under flow conditions at a shear stress of 1 dyne/cm2. The dotted line shows the background levels of binding seen with an uncoated microslide. Each data point is from an independent experiment, and the mean and SEM are shown.
Fig. 2.
DC8 IT4var19, but not DC13 HB3var03 and IT4var07, IE binding to HBECs is reduced by EPCR antibodies and soluble recombinant protein (rEPCR). IT4var19 IE (A), HB3var03 IE (B), and IT4var07 IE (C) binding to the immortalized HBEC-5i line under static conditions with 20 μg/mL antibody or soluble recombinant protein is shown. (D) Binding of the three parasite lines to primary HBMECs under static conditions with 20 μg/mL soluble rEPCR. Each data point is from an independent experiment, and the mean and SEM are shown. The difference in mean values compared with control (with no added antibody or protein) was analyzed by one-way ANOVA with Tukey’s multiple comparisons test (A–C) and paired t test (D). **P < 0.01. The vertical dotted line indicates experiments performed separately. mAb, monoclonal antibody; pAb, polyclonal antibody.
These experiments also revealed that IEs from the DC8 IT4var19 culture showed significant binding to PECAM-1 (Fig. 1A), while IEs from the DC13 IT4var07 culture showed significant binding to NCAM-1 and hyaluronic acid (Fig. 1C). All three parasite lines showed low-level binding to HABP-1 (Fig. 1 A–C), but this was not statistically significant. Staining of the bound cells with variant-specific antibodies showed that IT4var19 IEs bound to PECAM-1 and HABP-1 and that HB3var03 IEs bound to HABP-1 (Figs. S2 and S3). Attempts to stain IEs from the IT4var07 culture bound to NCAM-1, HABP-1, and hyaluronic acid were unsuccessful due to persistent loss of IEs from the plate.
We considered the possibility that rEPCR binding might be absent in static assays and only revealed under flow conditions, as seen with some selectin molecule interactions (24). However, when tested at a shear stress of 1 dyne/cm2, representative of physiological conditions in the microvasculature where sequestration occurs (25), DC8 IT4var19 IEs bound well to an rEPCR-coated microslide but DC13 IT4var07 and HB3var03 IEs showed levels of binding similar to the background levels seen with an uncoated slide (dotted line in Fig. 1D).
In both static and flow assays, we used rEPCR from Sino Biologicals, as used in previous publications (18), and we also tested the rEPCR preparation used for biophysical experiments and crystallization studies (19) (a gift from Matt Higgins, University of Oxford, Oxford, UK). The same results, with binding of DC8 IT4var19 IEs but not DC13 IT4var07 and HB3var03 IEs, were seen with rEPCR from both sources.
DC13-Expressing IEs Do Not Bind to EPCR on HBECs.
We next considered whether IEs could bind to EPCR constitutively expressed on the cell surface, using the immortalized HBEC line HBEC-5i (26). Binding of DC8 IT4var19 IEs to HBEC-5i was inhibited by both monoclonal and polyclonal antibodies to EPCR, and by soluble rEPCR (Fig. 2A). However, the EPCR antibodies and soluble protein had no statistically significant effect on the binding of DC13 HB3var03 and IT4var07 IEs to HBEC-5i (Fig. 2 B and C). Similar results were seen with primary human brain microvascular endothelial cells (HBMECs), with DC8 IT4var19 IE binding being partially reduced by rEPCR, whereas there was no effect on binding of DC13 HB3var03 or IT4var07 IEs (Fig. 2D). Soluble rCD36 had no statistically significant effect on HBEC-5i binding by any of the three parasite lines (Fig. 2 A–C), as expected, because CD36 expression is absent or low on HBECs (26, 27).
To further examine the role of EPCR in IE binding to HBEC-5i, EPCR expression was knocked down using siRNA. The expression of EPCR on the surface of HBEC-5i was reduced by >50% at 48 h after transfection, compared with HBEC-5i transfected with negative control siRNA (a scrambled sequence) (Fig. 3A). Consistent with the antibody inhibition experiments discussed above, DC8 IT4var19 IEs showed significantly reduced binding to the HBEC-5i–EPCR knockdown cells (Fig. 3B), whereas binding of DC13 HB3var03 and IT4var7 IEs was not significantly reduced (Fig. 3 C and D). These data show that EPCR played a major role in the binding of DC8 IT4var19 IEs to brain endothelial cells but that binding of DC13 HB3var03 and IT4var07 IEs to brain endothelium was unaffected by knocking down or inhibiting EPCR, suggesting usage of other endothelial receptors.
Fig. 3.
DC8 IT4var19, but not DC13 HB3var03 and IT4var07, IE binding to the HBEC line HBEC-5i is reduced by EPCR siRNA knockdown. (A) Expression of EPCR on HBEC-5i by immunostaining and flow cytometry. HBEC-5i was transfected with EPCR siRNA (orange) or a negative control siRNA (blue). Forty-eight hours after transfection, HBEC-5i was stained with 1 μg/mL goat polyclonal antibody to EPCR (orange/blue) or goat IgG-negative control (red), followed by Alexa Fluor 488 donkey anti-goat IgG at a 1:1,500 dilution. Data shown are representative of four similar experiments. (B–D) Binding of IEs to HBEC-5i transfected with control siRNA or EPCR siRNA under static conditions. The data shown are the mean and SEM from four independent experiments. The difference in mean values compared with the siRNA control was analyzed by a two-tailed paired t test. *P < 0.05. NS, not significant.
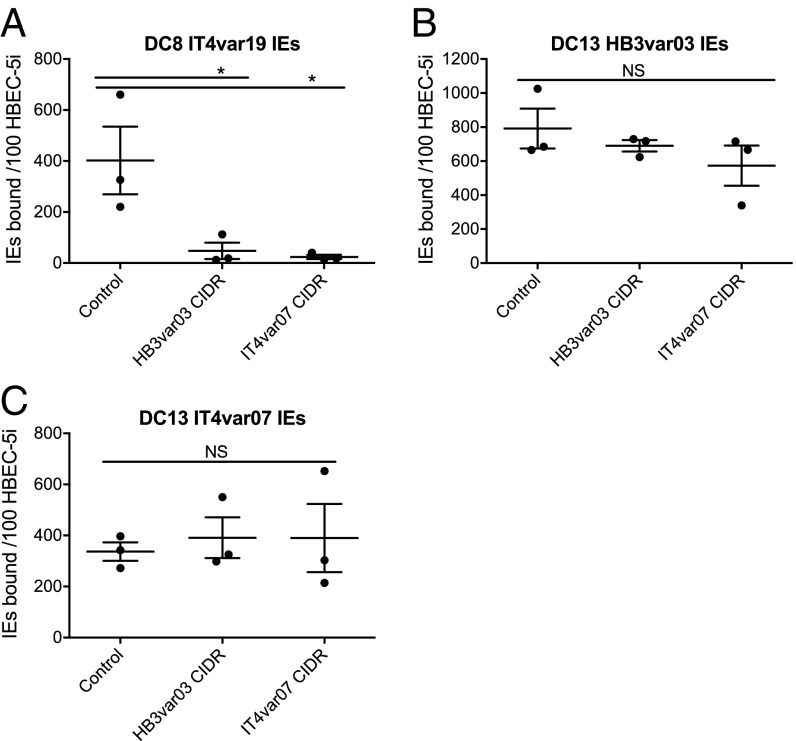
Binding of DC13-Expressing IEs to HBECs Is Not Blocked by EPCR-Binding Recombinant CIDR Proteins.
Recombinant PfEMP1 domains CIDRα1.1 of DC8 IT4var19 and CIDRα1.4 of DC13 HB3var03 and IT4var07 have been shown previously to bind to EPCR with high affinity (18, 19), and their binding site, which overlaps with the binding site of protein C to EPCR, has been identified (19). We tested the ability of CIDR recombinant proteins to block binding of IEs to HBEC-5i. The CIDRα1.4 domains of DC13 HB3var03 and IT4var07 were produced as soluble proteins of the expected molecular mass in Escherichia coli (Fig. S4), but CIDRα1.1 of DC8 IT4var19 had degraded fragments and aggregates, so it was not used further (not shown). Consistent with the above experiments, the two EPCR-binding CIDRα1.4 proteins from DC13 HB3var03 and IT4var07 inhibited binding of DC8 IT4var19 IEs to HBEC-5i (Fig. 4A) but had no significant effect on binding of DC13 HB3var03 and IT4var07 IEs (Fig. 4 B and C). These data are consistent with the known EPCR-binding activity of the CIDRα1.4 proteins (18, 19), and suggest that other PfEMP1 domains apart from CIDRα1.4, or other parasite adhesins, mediate binding of HB3var03 and IT4var07 IEs to HBEC-5i.
Fig. 4.
DC8 IT4var19, but not DC13 HB3var03 and IT4var07, IE binding to the HBEC cell line HBEC-5i is reduced by recombinant PfEMP1 CIDR proteins. IT4var19 IE (A), HB3var03 IE (B), and IT4var07 IE (C) binding to HBEC-5i under static conditions with 50 μg/mL soluble recombinant PfEMP1 protein is shown. The data shown are the mean and SEM from three independent experiments. The difference in mean binding values compared with control (with no added protein) was analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. *P < 0.05. NS, not significant.
The PfEMP1 Sequences of the DC8 and Two DC13-Expressing IEs Match the Database.
The N-terminal segment (NTS)-DBLα-CIDRα1 domains of DC8 IT4var19, DC13 HB3var03, and IT4var07 IEs used for the experiments were sequenced from cDNA. The sequences for the HB3var03 and IT4var19 IEs were identical to NTS-DBLα-CIDRα1 sequences on the VarDom 1.0 Server (www.cbs.dtu.dk/services/VarDom/) (7) (Figs. S5 and S6). IT4var07 IEs had two nucleotide changes that resulted in the following amino acid changes: I268M in the DBLα domain and T678I in the CIDRα1 domain (Fig. S7). These changes are outside the binding site and do not involve the residues essential for EPCR binding (19). Many other CIDRα1 domains that bind to EPCR have isoleucine rather than threonine at the equivalent amino acid position (19). It is therefore unlikely that these changes account for the lack of EPCR binding by the IEs.
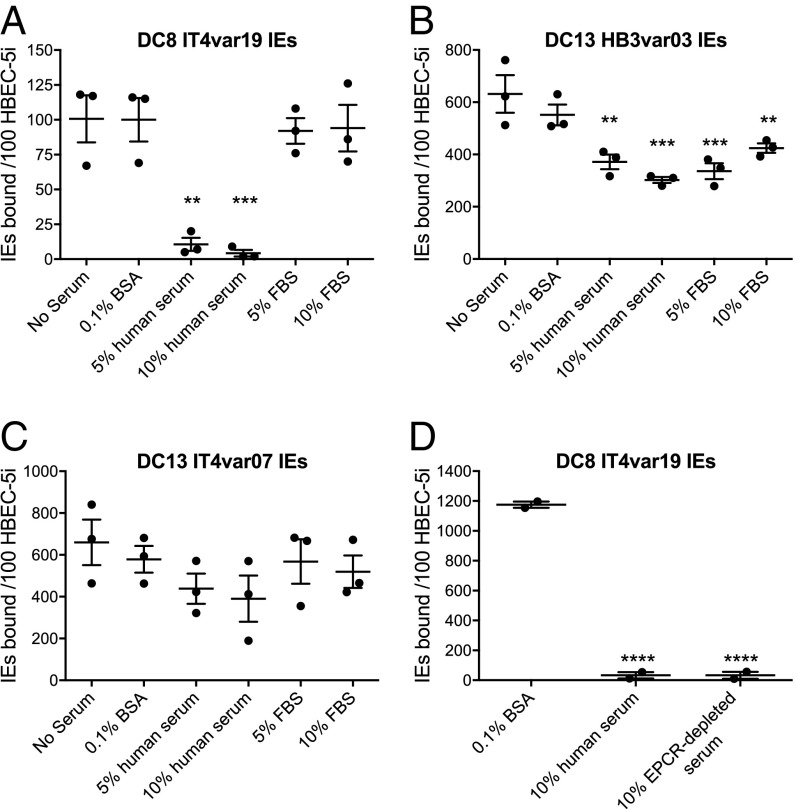
Human Serum and Plasma Inhibit Binding of DC8 IT4var19 IEs to HBEC-5i and EPCR.
Previous studies on EPCR binding by P. falciparum IEs in vitro have used binding medium containing FBS or BSA to block nonspecific binding (18, 19, 21, 22). To further examine the physiological relevance of EPCR binding by DC8 IT4var19 IEs, we examined binding to HBEC-5i in the presence of human serum. We found that binding was greatly reduced when 5% or 10% pooled normal human serum was included in the binding medium, an effect not seen with FBS (Fig. 5A). Some reduction in the binding of DC13 HB3var03 IEs to HBEC-5i occurred in the presence of both human and bovine serum, but this was less marked than with DC8 IT4var19 IEs (Fig. 5B). Serum had no significant effect on binding of DC13 IT4var07 IEs to HBEC-5i (Fig. 5C). Heat-inactivated serum from a different pool of six Scottish donors showed the same inhibitory effect on DC8 IT4var19 IE binding to HBEC-5i (Fig. S8A). Freshly prepared serum and plasma from two different Scottish donors also abolished binding of IT4var19 IEs to HBEC-5i (Fig. S8B), as did serum from five individual donors different from those used in the serum pools above (Fig. S8C).
Fig. 5.
DC8 IT4var19, but not DC13 HB3var03 and IT4var07, IE binding to the HBEC line HBEC-5i is abolished by human serum. IT4var19 IE (A), HB3var03 IE (B), and IT4var07 IE (C) binding to HBEC-5i under static conditions with pooled human or FBS is shown. (D) Binding of IT4var19 IEs to HBEC-5i with sEPCR-depleted pooled human serum. Each data point is from an independent experiment, and the mean and SEM are shown. The difference in mean values compared with the “No Serum” control (A–C) or “0.1% BSA” control (D) was analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
The level of soluble EPCR (sEPCR) in serum/plasma from healthy individuals ranges from ∼50–500 ng/mL (28, 29). Therefore, we investigated whether sEPCR in serum might be inhibiting the binding of IT4var19 IEs to HBEC-5i. A serum pool from six Scottish donors was depleted of sEPCR with antibody-coated beads, and the depletion was confirmed by ELISA using an rEPCR standard curve. The starting serum pool contained 57 ng/mL sEPCR, whereas EPCR was below the limit of detection (<0.3 ng/mL) after depletion. The EPCR-depleted serum inhibited DC8 IT4var19 IE binding to HBEC-5i to the same extent as the normal serum pool (Fig. 5D).
EPCR is known to bind lipid (phosphatidylcholine) from serum in its hydrophobic groove (30), so we also tested whether delipidated serum affected binding. Delipidated serum showed the same inhibitory activity as normal human serum (Fig. S8D). Human cord blood serum and plasma also showed the same inhibitory effect on DC8 IT4var19 IE binding to HBEC-5i as adult sera (Fig. S8D).
To test directly whether serum blocks binding of ITvar19 IEs to EPCR, in vitro static binding assays to recombinant receptors were carried out. Addition of 10% pooled human serum to the binding medium caused a significant reduction in binding to rEPCR (Fig. 6). The presence of human serum did not significantly alter binding to CD36, ICAM-1, or CSA, but did significantly increase binding to HABP-1 (Fig. 6).
Fig. 6.
Human serum inhibits adhesion of DC8 IT4var19-expressing IEs to rEPCR. IT4var19 IE binding to 50 μg/mL receptors absorbed onto plastic dishes tested under static conditions in binding medium with 10% pooled human serum or 0.1% BSA (No serum) is shown. The mean and SEM from three independent experiments are shown. The differences in mean values compared with the No serum controls were analyzed by a two-tailed paired t test for each receptor. *P < 0.05; **P < 0.01. NS, not significant.
High-Molecular-Weight Serum Components, Including IgG, Block Binding of DC8 IT4var19 IEs to HBEC-5i.
To further investigate the nature of the adhesion-blocking component in serum, we fractionated serum into >100-kDa and <100-kDa fractions. The adhesion-blocking activity was present in the >100-kDa fraction (Fig. S8E). We found that purified human IgG, but not purified IgM, could partially block binding of IT4var19 IEs to HBEC-5i (Fig. S8F). However, IgG-depleted serum retained full inhibitory activity (Fig. S8F), suggesting that serum components other than IgG also have adhesion-blocking effects.
Discussion
PfEMP1 variants are large complex multidomain proteins that, through their binding to receptors expressed on host endothelial cell surfaces, play a critical role in determining the outcome of P. falciparum infections. However, to date, most of our understanding of the binding specificity and affinity of PfEMP1 molecules for their host receptors comes from studies of individual recombinant PfEMP1 domains. Individual domains may show different properties when expressed in the context of the complete PfEMP1 protein (31). Therefore, to more fully understand PfEMP1–receptor interactions, it is essential to study the binding of full-length native PfEMP1 variants. Here, we focused on two PfEMP1 variant types containing CIDRα1 domains that are essential components of DC8 and DC13, which bind similarly to the endothelial protein EPCR in recombinant protein studies (18, 19). The results presented here show that the binding properties of PfEMP1 expressed on the IE surface can be remarkably different from those indicated by recombinant protein experiments. DC8-expressing IEs did bind to EPCR-expressing endothelial cells, and the binding was blocked by EPCR-specific antibodies or by siRNA knockdown of EPCR. In contrast, DC13-expressing IEs did not bind to EPCR protein in static assays, and their binding to endothelial cells was not dependent on the expression of EPCR, as it was not affected by the addition of EPCR-specific antibodies or by knockdown of EPCR. How might these observations be explained? The native full-length DC13 PfEMP1 may have significantly reduced affinity for EPCR. This possibility is consistent with the observations of Avril et al. (22), who demonstrated binding of DC13-expressing IEs to CHO cells expressing high levels of EPCR, which may increase the avidity of binding, compensating for the lower affinity of DC13 PfEMP1 for EPCR.
A second explanation for the discrepancy in results is that native PfEMP1 molecules on the surface of IEs adopt a different conformation from that of PfEMP1 recombinant domains, such that the EPCR-binding site is masked on IEs. If this is true, it is possible that allosteric changes (e.g., upon engaging with another receptor) might alter PfEMP1 conformation to expose or interfere with the EPCR-binding domain. In such a scenario, IE binding to EPCR on endothelial cells might depend on the particular combination of host receptors available. Other recent studies describe the importance of dual-binding phenotypes, suggesting that DC13 variants interact with both EPCR and ICAM-1 to bind to endothelial cells (22, 32). Other authors show that some DC8 PfEMP1 variants bind to HABP-1 (also known as gC1qR) and suggest that this receptor is important for IE sequestration in severe malaria (33).
Another possible explanation for the mismatch between published recombinant protein data and the adhesion properties of IEs is that a human serum/plasma component might bind to PfEMP1 or EPCR in our assays and block the binding interaction site. We found that IEs expressing a DC8 variant (IT4var19) did bind to rEPCR and to brain endothelial cells via EPCR in serum-free medium (Figs. 1–4). However, addition of 5–10% normal human serum or plasma to the binding medium inhibited these adhesion interactions (Figs. 5 and 6 and Fig. S8). A difference between our data and other published work on EPCR using DC8-expressing IEs is that previous binding studies have used FBS (18, 19) or BSA (21, 22) in the binding medium, neither of which blocks IE adhesion to EPCR (Fig. 5). Our data suggest that the serum factor that blocks binding of DC8-expressing IEs is unlikely to be sEPCR, as sEPCR-depleted serum showed the same inhibitory effect (Fig. 5D). EPCR has an MHC class 1 structural fold and, under physiological conditions, contains lipid in its hydrophobic groove (30). However, delipidated serum showed the same inhibitory effect as normal human serum (Fig. S8). Fractionation experiments showed that the serum component(s) responsible for inhibiting EPCR binding are >100 kDa and may include nonimmmune IgG (Fig. S8). However, IgG-depleted serum fully inhibited binding, suggesting that other high-molecular-weight components might affect the ability of DC8-expressing IEs to bind to EPCR under physiological exposure to plasma in vivo.
Previous work has shown that distinct DC8 and DC13 PfEMP1 subsets identified on the basis of sequence similarity have evolved (7), which share biological functions such as the interaction with EPCR demonstrated in recombinant protein studies (18, 19), and that parasite expression of these subsets is associated with severe and cerebral malaria (11, 13, 34, 35). Our findings do not undermine these important observations, and they do not contradict work suggesting that the loss of EPCR on brain endothelial cells may contribute to coagulopathy and impaired endothelial barrier integrity in cerebral malaria (36). Our results do, however, raise questions regarding the identification of EPCR as the major host receptor for IE sequestration in the brain during cerebral malaria. Our data, together with published studies showing that DC8- and DC13-expressing IEs bind to other receptors such as HABP-1 (33) and ICAM-1 (22, 32), support a reinterpretation of the cerebral malaria paradigm to one that downplays the role of a single receptor in the brain and, instead, emphasizes that IEs interact with multiple host receptors to bring about sequestration, as suggested by other authors (20, 37, 38). It remains possible that even if sequestration in severe malaria is mediated by other receptors, PfEMP1 might engage with EPCR on endothelial cells to modify EPCR’s endothelial-protective functions (39). However, further work is needed to determine if such functional interactions do occur, especially in the presence of human plasma.
Materials and Methods
Ethics Statement.
Human erythrocytes and sera were obtained following receipt of informed consent from blood donors, with approval from the Scottish National Blood Transfusion Service Committee for the Governance of Blood and Tissue Samples for Nontherapeutic Use (reference SNBTS 12-35). In addition, anonymous blood donations were obtained from the Interstate Blood Bank, Memphis, TN.
P. falciparum Culture and Parasite Lines.
Parasites were cultured as described elsewhere (14). The parasite lines HB3-HBEC and IT-HBEC were generated previously by panning on HBEC-5i (40). Their var gene transcriptional profiles were determined by variant surface antigen (VSA)-supplemented microarray and reverse transcriptase PCR and sequencing (14). Specific antibodies to HB3var03 were used to detect the PfEMP1 variant on the surface of IEs by immunofluorescence assay (IFA) and flow cytometry as described elsewhere (14, 22). The IT-HBEC parasite line had two predominant variants: one DC13 encoded by IT4var07 and one DC8 encoded by IT4var19 (14). Antibodies raised to IT4var07 and IT4var19 were used in fluorescence-activated cell sorting to generate two separate parasite lines, IT4var19 and IT4var07. The IT4var07 line underwent rapid var gene switching, so that even with frequent panning, it was not possible to maintain more than 30–50% of IEs expressing IT4var07. The three parasite lines studied here do not form rosettes or platelet-mediated clumps (14), and do not bind nonimmune IgM or IgG on IEs.
Static Adhesion Assays to Receptor Molecules Immobilized on Plastic.
The receptor-binding assays were done as previously described (14) with 50 μg/mL rEPCR, PECAM-1, ICAM-1, CD36, NCAM-1, HABP-1, or VCAM-1 or with purified heparin sodium salt, CSA, or hyaluronic acid spotted onto plastic dishes. IEs were suspended at 2% hematocrit (Ht) in bicarbonate-free DMEM-F12 Ham (D8900; Sigma)/0.1% BSA (pH 7.2–7.4), hereafter called “binding medium.” The detailed method is provided in SI Materials and Methods.
IFA with PfEMP1 Antibodies on the Bound IEs.
Binding assays were carried out as above, and after washing off unbound cells, 25 μg/mL purified polyclonal IgG from a rabbit immunized with NTS-DBLα1 of HB3var03 (14), IT4var07 (22), IT4var19 (22), or IgG from a nonimmunized rabbit and diluted in PBS/1% BSA was added to the spot and incubated for 30 min. The dishes were washed twice with PBS for 5 min. Then, 50 μL of a 1:500 dilution of Alexa Fluor 488 goat anti-rabbit IgG (A-11034; Invitrogen) in PBS/0.1% BSA/1 μg/mL DAPI was added to the spot and incubated for 30 min in the dark. Dishes were washed twice as above and air-dried; DABCO glycerol mountant (290734; Sigma) or Fluoromount (F4680; Sigma) was then added to the spot, and the samples then sealed with a coverslip for viewing by fluorescence microscopy with a 100× objective. Pictures were taken with a YenCam digital camera.
Flow Adhesion Assay with rEPCR.
A μ-Slide I 0.8 Luer (80191; ibidi GmBH) was coated with 50 μg/mL rEPCR for 2 h in a humid box at 4 °C. The EPCR was then removed, replaced with PBS/1% BSA, and incubated overnight at 4 °C. An ibidi pump system (10902; ibidi GmBH) was set up, and IEs at 1% Ht and 50–90% parasitemia in binding medium were flowed unidirectionally through the EPCR-coated microslide at a shear stress of 1 dyne/cm2. After 5 min, the number of IEs bound under continuous flow was counted per 10 fields viewed with a 40× objective.
HBEC Lines and Culture.
HBEC-5i, an immortalized HBEC line (26, 40), was cultured as previously described (14). The brain endothelial characteristics of HBEC-5i were confirmed by staining positive in the IFA for cytoplasmic von Willebrand factor (vWF) and membrane Glut-1, and negative for the fibroblast marker smooth muscle actin (SMA) and for CD36. As previously reported, HBEC-5i is negative for PECAM-1/CD31, a protein usually found on endothelial cells (26). Primary HBECs (HBMECs) from ScienCell (SC-1000), which stain positively for cytoplasmic vWF and membrane PECAM-1, were cultured as described (14) and were used up to passage 7.
HBEC-Binding Assays and Inhibition with Antibodies and Recombinant Proteins.
For binding assays, HBEC-5i or HBMECs were seeded onto gelatin-coated 48-well plates to reach 80–90% confluency on the day of the assay. Gelatin-purified pigmented trophozoites at 50–90% parasitemia were suspended in binding medium (as described above) at 2% Ht. To test the inhibitory effect of antibodies, HBEC-5i was preincubated with 20 μg/mL goat polyclonal antibody to human EPCR (AF2245; R&D Systems), rat monoclonal antibody to human EPCR (clone RCR-252, HM2145; Hycult), or isotype control rat IgG1 (HI3001; Hycult) or polyclonal goat IgG (AB-108-C; R&D Systems) for 20 min before addition of IEs. To test the inhibitory effect of recombinant proteins, IEs were resuspended at 2% Ht with 20 μg/mL soluble rEPCR or rCD36 in binding medium, or the HBEC-5i was preincubated with 50 μg/mL recombinant CIDRα1.4 proteins of DC13 HB3var03 or IT4var07 in binding medium/0.1% BSA for 20 min. After addition of IEs to the HBEC-5i, the plate was incubated for 75 min at 37 °C with resuspension of cells by tilting the plate after 30 and 60 min. The cells were washed by removing the supernatant and adding warm binding medium using a plastic Pasteur pipette and gently rocking the plate. The washing was repeated until there were very few or no unbound IEs remaining. The cells were fixed with 2% glutaraldehyde in PBS for 1 h and stained with 5% Giemsa for 10 min.
Binding of IEs to EPCR siRNA-Transfected HBEC-5i.
HBEC-5i at 60–80% confluency was transfected with an equal volume of 10 μM EPCR siRNA (sc-39932; Santa Cruz Biotechnology) and Lipofectamine RNAiMAX Transfection Reagent (13778075; Life Technologies), each diluted with Opti-MEM reduced serum medium (31985062; Life Technologies) at a ratio of 3:50. HBEC-5i transfected with control A siRNA (sc-37007; Santa Cruz Biotechnology) was used as a negative control. Details of the transfection are provided in SI Materials and Methods. For the binding assay, gelatin-purified pigmented trophozoites at 2% Ht in 190 μL of binding medium/0.1% BSA were added to each of two wells in a 48-well plate containing EPCR siRNA-transfected HBEC-5i or control siRNA-transfected HBEC-5i and incubated for 60 min at 37 °C. The cells were gently resuspended after 15 and 30 min and washed, fixed, and stained as above.
HBEC-5i–Binding Inhibition with Serum and Plasma.
Binding assays were carried out as above with gelatin-purified pigmented trophozoites (50–90% parasitemia with the exception of IT4var19 IEs in Fig. 5A, where 5% parasitemia was used) in binding medium at 2% Ht with added 5% or 10% (vol/vol) pooled human serum from multiple donors (Scottish National Blood Transfusion Service), EPCR-depleted human serum (described in SI Materials and Methods), FBS (10500064; Thermo Fisher Scientific), or 0.1% BSA (A0336; Sigma–Aldrich). Additional serum components and fractionation are described in SI Materials and Methods.
Statistical Analyses and Graphing.
Statistical analyses and graphs were prepared using GraphPad Prism (v7.0b; GraphPad Software, Inc.). Technical replicates based on counts of duplicate spots or wells were averaged to generate a binding value for each receptor or condition in each experiment. Statistical analysis was carried out on the binding values from n ≥ 2 independent experiments using two-tailed paired t tests or one-way ANOVA with Dunnett’s or Tukey’s multiple comparisons test. Asterisks indicate *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, and NS indicates no statistical significance (P > 0.05).
Supplementary Material
Acknowledgments
This work was funded by a studentship award from the Darwin Trust of Edinburgh, the Wellcome Trust (Grants 084226 and 095831), and the NIH Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712879115/-/DCSupplemental.
References
- 1.Marchiafava E, Bignami A. On Summer-Autumnal Fever. The New Sydenham Society; London: 1894. [Google Scholar]
- 2.Taylor TE, Molyneux ME. The pathogenesis of pediatric cerebral malaria: Eye exams, autopsies, and neuroimaging. Ann N Y Acad Sci. 2015;1342:44–52. doi: 10.1111/nyas.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am J Pathol. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 4.Milner DA, Jr, et al. Quantitative assessment of multiorgan sequestration of parasites in fatal pediatric cerebral malaria. J Infect Dis. 2015;212:1317–1321. doi: 10.1093/infdis/jiv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baruch DI, et al. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: Conserved function with variant sequence. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- 6.Su X-Z, et al. A large and diverse family gene family (var) encodes 200-350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythocytes. Cell. 1995;82:89–99. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 7.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes–Divide and conquer. PLOS Comput Biol. 2010;6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchgatter K, Portillo HdA. Association of severe noncerebral Plasmodium falciparum malaria in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine residues. Mol Med. 2002;8:16–23. [PMC free article] [PubMed] [Google Scholar]
- 9.Kyriacou HM, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warimwe GM, et al. Prognostic indicators of life-threatening malaria are associated with distinct parasite variant antigen profiles. Sci Transl Med. 2012;4:129ra45. doi: 10.1126/scitranslmed.3003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavstsen T, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA. 2012;109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertin GI, et al. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One. 2013;8:e68368. doi: 10.1371/journal.pone.0068368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernabeu M, et al. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proc Natl Acad Sci USA. 2016;113:E3270–E3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claessens A, et al. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci USA. 2012;109:E1772–E1781. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avril M, et al. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci USA. 2012;109:E1782–E1790. doi: 10.1073/pnas.1120534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avril M, Brazier AJ, Melcher M, Sampath S, Smith JD. DC8 and DC13 var genes associated with severe malaria bind avidly to diverse endothelial cells. PLoS Pathog. 2013;9:e1003430. doi: 10.1371/journal.ppat.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 18.Turner L, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau CK, et al. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe. 2015;17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillrie MR, et al. Diverse functional outcomes of Plasmodium falciparum ligation of EPCR: Potential implications for malarial pathogenesis. Cell Microbiol. 2015;17:1883–1899. doi: 10.1111/cmi.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampath S, et al. Plasmodium falciparum adhesion domains linked to severe malaria differ in blockade of endothelial protein C receptor. Cell Microbiol. 2015;17:1868–1882. doi: 10.1111/cmi.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avril M, Bernabeu M, Benjamin M, Brazier AJ, Smith JD. Interaction between endothelial protein C receptor and intercellular adhesion molecule 1 to mediate binding of Plasmodium falciparum-infected erythrocytes to endothelial cells. MBio. 2016;7:e00615. doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters JM, Fowler EV, Krause DR, Cheng Q, Gatton ML. Differential changes in Plasmodium falciparum var transcription during adaptation to culture. J Infect Dis. 2007;195:748–755. doi: 10.1086/511436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finger EB, et al. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature. 1996;379:266–269. doi: 10.1038/379266a0. [DOI] [PubMed] [Google Scholar]
- 25.Cooke BM, Morris-Jones S, Greenwood BM, Nash GB. Adhesion of parasitized red blood cells to cultured endothelial cells: A flow-based study of isolates from Gambian children with falciparum malaria. Parasitology. 1993;107:359–368. doi: 10.1017/s0031182000067706. [DOI] [PubMed] [Google Scholar]
- 26.Wassmer SC, Combes V, Candal FJ, Juhan-Vague I, Grau GE. Platelets potentiate brain endothelial alterations induced by Plasmodium falciparum. Infect Immun. 2006;74:645–653. doi: 10.1128/IAI.74.1.645-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner GDH, et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 28.Stearns-Kurosawa DJ, Burgin C, Parker D, Comp P, Kurosawa S. Bimodal distribution of soluble endothelial protein C receptor levels in healthy populations. J Thromb Haemost. 2003;1:855–856. doi: 10.1046/j.1538-7836.2003.t01-4-00115.x. [DOI] [PubMed] [Google Scholar]
- 29.Uitte de Willige S, et al. Haplotypes of the EPCR gene, plasma sEPCR levels and the risk of deep venous thrombosis. J Thromb Haemost. 2004;2:1305–1310. doi: 10.1046/j.1538-7836.2004.00855.x. [DOI] [PubMed] [Google Scholar]
- 30.Oganesyan V, et al. The crystal structure of the endothelial protein C receptor and a bound phospholipid. J Biol Chem. 2002;277:24851–24854. doi: 10.1074/jbc.C200163200. [DOI] [PubMed] [Google Scholar]
- 31.Dahlbäck M, Nielsen MA, Salanti A. Can any lessons be learned from the ambiguous glycan binding of PfEMP1 domains? Trends Parasitol. 2010;26:230–235. doi: 10.1016/j.pt.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Lennartz F, et al. Structure-guided identification of a family of dual receptor-binding PfEMP1 that is associated with cerebral malaria. Cell Host Microbe. 2017;21:403–414. doi: 10.1016/j.chom.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magallón-Tejada A, et al. Cytoadhesion to gC1qR through Plasmodium falciparum erythrocyte membrane protein 1 in severe malaria. PLoS Pathog. 2016;12:e1006011. doi: 10.1371/journal.ppat.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jespersen JS, et al. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRα1 domains. EMBO Mol Med. 2016;8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mkumbaye SI, et al. The severity of Plasmodium falciparum infection is associated with transcript levels of var genes encoding endothelial protein C receptor-binding P. falciparum erythrocyte membrane protein 1. Infect Immun. 2017;85:e00841. doi: 10.1128/IAI.00841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moxon CA, et al. Loss of endothelial protein C receptors links coagulation and inflammation to parasite sequestration in cerebral malaria in African children. Blood. 2013;122:842–851. doi: 10.1182/blood-2013-03-490219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho M. EPCR: Holy grail of malaria cytoadhesion? Blood. 2014;123:157–159. doi: 10.1182/blood-2013-12-541318. [DOI] [PubMed] [Google Scholar]
- 38.Bernabeu M, Smith JD. EPCR and malaria severity: The center of a perfect storm. Trends Parasitol. 2017;33:295–308. doi: 10.1016/j.pt.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohan Rao LV, Esmon CT, Pendurthi UR. Endothelial cell protein C receptor: A multiliganded and multifunctional receptor. Blood. 2014;124:1553–1562. doi: 10.1182/blood-2014-05-578328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claessens A, Rowe JA. Selection of Plasmodium falciparum parasites for cytoadhesion to human brain endothelial cells. J Vis Exp. 2012:e3122. doi: 10.3791/3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.