Significance
Myeloid cell leukemia 1 (Mcl-1) is a key cancer survival protein that functions by binding to and blocking the activity of prodeath members of the Bcl-2 family. The prosurvival functionality of Mcl-1 can be inhibited by peptides that compete with the native prodeath factors for interaction with Mcl-1. However, unmodified peptide inhibitors of Mcl-1 are ineffective in cellular assays because they cannot access the cytoplasm. In this work, chemical modification and sequence optimization of Mcl-1 binding peptides generated compounds that have favorable biophysical properties, engage Mcl-1 in a distinctive binding mode, and can enter and selectively kill cancer cells dependent on Mcl-1 for survival. This detailed proof-of-principle study demonstrates how systematic optimization can transform a lead peptide into a drug prototype suitable for diagnostic and therapeutic development.
Keywords: stapled peptide, Mcl-1, apoptosis, BH3 mimetic, inhibitor
Abstract
Bcl-2 family proteins regulate apoptosis, and aberrant interactions of overexpressed antiapoptotic family members such as Mcl-1 promote cell transformation, cancer survival, and resistance to chemotherapy. Discovering potent and selective Mcl-1 inhibitors that can relieve apoptotic blockades is thus a high priority for cancer research. An attractive strategy for disabling Mcl-1 involves using designer peptides to competitively engage its binding groove, mimicking the structural mechanism of action of native sensitizer BH3-only proteins. We transformed Mcl-1–binding peptides into α-helical, cell-penetrating constructs that are selectively cytotoxic to Mcl-1–dependent cancer cells. Critical to the design of effective inhibitors was our introduction of an all-hydrocarbon cross-link or “staple” that stabilizes α-helical structure, increases target binding affinity, and independently confers binding specificity for Mcl-1 over related Bcl-2 family paralogs. Two crystal structures of complexes at 1.4 Å and 1.9 Å resolution demonstrate how the hydrophobic staple induces an unanticipated structural rearrangement in Mcl-1 upon binding. Systematic sampling of staple location and iterative optimization of peptide sequence in accordance with established design principles provided peptides that target intracellular Mcl-1. This work provides proof of concept for the development of potent, selective, and cell-permeable stapled peptides for therapeutic targeting of Mcl-1 in cancer, applying a design and validation workflow applicable to a host of challenging biomedical targets.
Protein–protein interactions that contribute to cell transformation and cancer survival are attractive therapeutic targets. For example, interactions among B-cell lymphoma 2 (Bcl-2) family proteins are critical for controlling cell-death processes and can, when deregulated, block apoptosis despite prodeath signaling (1, 2). Overexpression of Bcl-2 family antiapoptotic proteins, including Bcl-xL, Bcl-2, Bcl-w, Mcl-1, and Bfl-1/A1, is associated with pathologic cell survival, and many cancer cells develop a dependency on one or more family members (3, 4). Mcl-1 in particular is implicated in counteracting apoptosis in cancers, including acute myeloid leukemia, chronic myelogenous leukemia, T-cell lymphomas, multiple myeloma, certain breast, pancreatic, ovarian and cervical cancers, and non–small-cell lung carcinoma (5–11). Indeed, Mcl-1 has been validated as a therapeutic target by studies showing that its inhibitors are cytotoxic to diverse cancer cell lines, reduce tumor volume in multiple myeloma xenografts, and extend lifespan in murine models of lymphoma (12).
X-ray crystallographic and NMR studies have revealed that antiapoptotic proteins Mcl-1, Bcl-2, Bcl-xL, Bcl-w, and Bfl-1/A1 are structurally homologous and contain an extended hydrophobic groove that binds with high affinity to an α-helix corresponding to the Bcl-2 homology 3 (BH3) motif of proapoptotic proteins. This helix-in-groove interaction is often critical to cancer cell survival. Based on molecular mimicry of the BH3 α-helix, a series of small-molecule and engineered peptide inhibitors have been developed that bind competitively to the BH3 groove. For example, venetoclax, a small molecule that selectively inhibits Bcl-2, has been approved for the treatment of chronic lymphocytic leukemia (13). However, overexpression of Bcl-2 paralogs that lie outside the binding spectrum of venetoclax, including Mcl-1, can lead to drug resistance (14, 15). Broader targeting of Bcl-2 family members can have undesired side effects, such as the thrombocytopenia observed upon Bcl-xL inhibition, potentially complicating the development of multitargeting Bcl-2 protein inhibitors (16).
Despite Mcl-1 being one of the most frequently amplified genes in cancer and a major chemoresistance factor (17), development of clinical grade Mcl-1 inhibitors has lagged behind that of Bcl-2 inhibitors. Recently, a potent and selective small molecule antagonist of Mcl-1 was reported to be selectively cytotoxic to Mcl-1–dependent cancer cells in vitro and in vivo (12). This and other Mcl-1 inhibitor molecules that have recently entered phase I testing may provide a new treatment for Mcl-1–driven cancers, pending successful clinical translation. Given the potential translational hurdles, it is prudent to pursue alternative strategies for advancing novel agents for both research and therapeutic purposes. Synthetic peptides have tremendous potential as natural inhibitors of protein–protein interactions, and BH3-based constructs have been demonstrated to selectively engage Mcl-1 and block its antiapoptotic activity in functional assays (18–20). However, unmodified peptides can be poor therapeutic candidates, due to their susceptibility to proteolysis in vivo and limited capacity to enter cells. Thus, applying chemical approaches for optimizing peptide stability and cell permeability of Mcl-1–binding peptides could dramatically expand the utility of these inhibitors for cell-based studies and clinical development.
Prior work on α-helical peptides has established that cross-linking of hydrophobic olefinic side chains spaced 4 or 7 residues apart can “staple” the peptide into a stabilized α-helical conformation (21, 22). When appropriately incorporated into peptide templates, stapling has been shown in numerous examples to promote metabolic stability and cell penetrance via a pinocytotic mechanism (22–25). In certain cases, the installed all-hydrocarbon staple itself fortuitously engaged the surface of the target protein and contributed to an enhanced binding interaction (26–29). SAH-p53-8 is one such stapled peptide that was modeled after the p53 transactivation domain helix that engages both Mdm2 and Mdmx. SAH-p53-8 reactivated p53-dependent apoptosis in nutlin-resistant cancer cells at low micromolar doses in enriched serum-free medium, achieved selective intracellular targeting of p53 complexes in live cells at higher doses in the presence of serum, and suppressed tumor growth in a mouse cancer model (23, 30, 31). Based on this proof of principle, Aileron Therapeutics sought to improve the potency and pharmacology of stapled p53 peptide dual inhibitors of Mdm2/Mdmx. Iterative optimization that incorporated sequences identified by phage display into the SAH-p53-8 backbone led to compounds that were cytotoxic to SJSA-1 osteosarcoma and MCF-7 breast cancer cells at submicromolar doses in the presence of full serum and effectively inhibited tumor growth in xenograft models (32, 33). The clinical grade stapled peptide inhibitor of Mdm2/Mdmx, ALRN-6924, recently completed phase I testing and was both well-tolerated and showed signs of antitumor activity (34). This translational proof of concept provides exciting validation of the stapling technology and its clinical potential. Phase II testing of ALRN-6924 is currently underway.
In 2010, Stewart et al. showed that stapling the BH3 sequence of Mcl-1 itself yielded highly selective, cell-penetrating peptide inhibitors of Mcl-1 function. The staple in the lead compound, termed Mcl-1 stabilized alpha-helix of Bcl-2 domain D (Mcl-1 SAHBD), improved the binding activity by ∼20-fold compared with the corresponding unmodified Mcl-1 BH3 peptide and fortuitously made direct, affinity-enhancing contacts with the Mcl-1 protein surface (28). In other work, Foight et al. screened a combinatorial library based on the Bim BH3 sequence and identified peptides that bind with high affinity to Mcl-1 but interact at least 100-fold more weakly with antiapoptotic paralogs Bcl-2, Bcl-xL, Bcl-w, and Bfl-1/A1 (20). Peptide MS1, which was identified using this screen and is not chemically modified, binds Mcl-1 with a dissociation constant of ∼2 nM as a fluoresceinated analog. MS1 has been used in functional assays to probe cellular dependence on Mcl-1 (7, 35, 36). Subsequent studies showed that stapling MS1 in the position corresponding to the modification in Mcl-1 SAHBD modestly improved binding affinity and that additional mutations discovered in a combinatorial library screen of stapled variants resulted in further enhancement in affinity (37).
Based on the affinities of therapeutically effective inhibitors of Bcl-2 and Bcl-xL (38, 39), a clinically viable Mcl-1 inhibitor is expected to require a subnanomolar dissociation constant for its target. Thus, our goal in this work was to improve the binding affinity, cell permeability, and cancer cell cytotoxicity of stapled peptide candidates, while maintaining Mcl-1 selectivity. We accomplished this goal by synthesizing panels of hydrocarbon-stapled MS1 peptide variants and performing iterative sequence optimization, guided by design principles established through sequence-function analyses of other stapled peptides (40). Remarkably, we identified a distinctive staple position that improved binding affinity and peptide uptake and that independently conferred Mcl-1 specificity to the otherwise broadly targeting Bim BH3 sequence. Optimized stapled peptides were tested in a battery of assays to characterize structure, function, cell permeability, and on-mechanism biological activity in cells, leading to the discovery of inhibitors that are selectively cytotoxic to Mcl-1–dependent cancer cells, in the presence of serum and as single agents.
Results
Staple Insertion Site Dictates Mcl-1 Binding Affinity and Selectivity.
Two Bim-based peptides, MB2 and MS1, were previously engineered to have high affinity and specificity for Mcl-1. Both peptides are based on the BH3 motif of proapoptotic Bim, which binds with low-nanomolar affinity to Mcl-1, Bcl-xL, Bcl-2, Bcl-w, and Bfl-1/A1 (41). In contrast, MB2 and MS1 have high affinity for Mcl-1, but their interactions are at least 100-fold weaker with the other family members, representing the desired profile for an Mcl-1–specific inhibitor. To explore whether and how the properties of MB2 and MS1 could be improved, we generated i, i + 4 and i, i + 7 staple-scanning libraries of a 21-amino acid peptide based on MB2 (SI Appendix, Table S1).
All i, i + 4- and i, i + 7-stapled MB2 peptides demonstrated increased α-helicity, up to fourfold greater than the α-helicity of the unmodified 21-residue MB2 peptide, which was estimated to have an α-helical content of ∼22% (SI Appendix, Table S1). Peptide binding affinities for recombinant Mcl-1ΔNΔC (amino acids 172 to 327, hereafter referred to as “Mcl-1”) were quantified using a fluorescence-polarization (FP) binding assay. Ten of 11 i, i + 7-stapled peptides and 3 of 12 i, i + 4-stapled peptides demonstrated weaker affinity for Mcl-1 (Kd > 1,000 nM) compared with MB2 (Kd = 4.7 nM) (SI Appendix, Table S1). BH3 motifs have four highly conserved hydrophobic residues (positions 2d, 3a, 3d, and 4a, using the heptad notation defined in Table 1), which engage pockets within the canonical Mcl-1 groove. Mutating any of these residues to alanine, glutamate, or lysine weakens binding to Mcl-1 (42). Glycine and aspartate residues at positions 3e and 3f are highly conserved among BH3 motifs and are likewise intolerant of substitution (20, 42, 43). For most staple modifications that substantially reduced binding (9 of 13), one or more of these key positions was altered, consistent with the sequence-function relationship. Two other i, i + 7 staples, in SAH-MB2-17 and SAH-MB2-20, disrupted a salt bridge between arginine at position 3c and glutamate at position 3g that is seen in many structures of Bim BH3 bound to Bcl-2 proteins. Finally, structural modeling showed that an i, i + 7 staple cannot be accommodated between positions 3b and 4b without disrupting a conserved peptide–protein salt bridge (involving aspartate at 3f) (SI Appendix, Fig. S1), explaining the weak binding of SAH-MB2-19. Thus, the effects of staple insertions that impair target engagement can be rationalized based on sequence conservation and protein structure.
Table 1.
Comparative binding affinities and Mcl-1 specificities of SAH-MS1 peptides
| Peptide | Sequence* | IC50Mcl-1, nM† | IC50P, nM† | Helicity, %‡ | ||
| 2 | 3 | 4 | ||||
| fgabcdefgabcdefgabcdefga | ||||||
| MS1 | RPEIWMTQGLRRLGDEINAYYAR | >2,000 | >5,000 | 11 | ||
| SAH-MS1-1 | IWXTQGXRRLGDEINAYYARR | 929 ± 110 | >5,000 | 78 | ||
| SAH-MS1-2 | IWBXQGLXRLGDEINAYYARR | 90 ± 6 | >5,000 | 73 | ||
| SAH-MS1-3 | IWBTQXLRRXGDEINAYYARR | >2,000 | >5,000 | 75 | ||
| SAH-MS1-4 | IWBTQGXRRLXDEINAYYARR | >10,000 | >5,000 | 44 | ||
| SAH-MS1-5 | IWBTQGLXRLGXEINAYYARR | >10,000 | >5,000 | 58 | ||
| SAH-MS1-6 | IWBTQGLRXLGDXINAYYARR | 799 ± 123 | >5,000 | 63 | ||
| SAH-MS1-7 | IWBTQGLRRLXDEIXAYYARR | >10,000 | >5,000 | 80 | ||
| SAH-MS1-8 | IWBTQGLRRLGDXINAXYARR | 860 ± 88 | >5,000 | 75 | ||
| SAH-MS1-9 | IWBTQGLRRLGDEIXAYYXRR | 497 ± 33 | >5,000 | 33 | ||
| Bim SAHB§ | IWIXQELXRIGDEFNAYYARR | 150 ± 21 | >5,000 | 78 | ||
Numbering using the convention (abcdefg)n for heptad repeats 2 to 4 is shown at the top. B, norleucine; X, (S)-4-pentenylalanine.
Stapled peptides were tested in competition fluorescence polarization assays with a 21-mer fluoresceinated Bim-BH3 (25 nM) for binding to recombinant Mcl-1. IC50 values are reported for binding to Mcl-1 (as “IC50Mcl-1”). IC50P indicates affinities for Bcl-xL, Bcl-2, Bcl-w, and Bfl-1. Reported errors are mean ± SD for three experiments performed in duplicate.
The α-helical content was calculated based on circular dichroism analysis of peptides at 25 μM in 50 mM Tris buffer, pH 7.4, as described in Methods.
Bim BH3 (amino acids 146 to 166) with 2e-3b staple.
Binding experiments identified a series of stapled MB2 peptides with high Mcl-1 affinity and selectivity: SAH-MB2-3, SAH-MB2-4, SAH-MB2-8, and SAH-MB2-12 peptides bound to Mcl-1 with dissociation constants below 10 nM (SI Appendix, Table S1). Three stapled peptides, SAH-MB2-3, SAH-MB2-4, and SAH-MB2-12, showed improved binding affinity relative to the parent, unmodified peptide. SAH-MB2-12 was stapled in the same position previously reported for Mcl-1 SAHBD, which likewise demonstrated enhanced Mcl-1 binding affinity (28). For the tightest binder, SAH-MB2-3, cross-linking positions 2e and 3b led to high α-helical content (75%) and high affinity (Kd = 2.1 nM). Comparative FP binding analysis against five Bcl-2 antiapoptotic paralogs confirmed that SAH-MB2-3 retained high selectivity for Mcl-1 (SI Appendix, Table S1). To determine whether the 2e-3b staple itself contributed to the observed selectivity for Mcl-1, we introduced this modification into a Bim BH3 peptide that binds tightly to all five human Bcl-2 family proteins (19, 20). Surprisingly, when the location of the staple in the prototype Bim SAHBA1 peptide (amino acids 146 to 166) (44) was switched to the 2e-3b position, the construct became highly Mcl-1–specific (Table 1), showing that stapling can introduce both gain- and loss-of-function design elements that are beneficial to tailored inhibitor development.
Guided by the MB2 staple-scanning results, we introduced select i, i + 4 cross-links to generate nine stapled-peptide variants of MS1. MS1 exhibits high affinity and specificity for Mcl-1 (Ki = 24 nM) (20) and has been used in diagnostic BH3-profiling assays performed on permeabilized cells (7, 20, 45). SAH-MS1 peptides, together with the tightest Mcl-1 binders from the MB2 panel, were tested in a competition binding assay using a fluorescein isothiocyanate (FITC)-labeled 21-residue Bim BH3 peptide (Table 1 and SI Appendix, Fig. S2 and Table S1). Similar to results obtained for the MB2 panel, the 2e-3b and 4b-4e staple positions yielded the highest affinity constructs in the MS1 panel. The 2e-3b staple produced the highest affinity overall, with an IC50 value more than 20-fold lower than that of MS1. SAH-MS1-2 was also less susceptible to chymotrypsin proteolysis, compared with MS1 (SI Appendix, Fig. S3). Thus, SAH-MS1-2 was selected as the lead stapled peptide template for further optimization.
Optimizing Stapled MS1 Variants for Cellular Delivery.
In advance of cellular application, lead stapled peptide constructs were tested in lactate dehydrogenase (LDH) release assays to screen out membrane lytic constructs (40). We found that 30 μM dosing of SAH-MS1-2 induced nonspecific membrane lysis of mouse embryonic fibroblasts (MEFs), an undesirable property for target-specific drug development (SI Appendix, Fig. S4). Indeed, the combination of high hydrophobicity and positive charge has recently been implicated as a risk factor for lytic activity (40, 46). Substitution of alanine at 2e and arginine at 3b in the MS1 sequence with the hydrophobic staple notably increased hydrophobicity, as assessed by HPLC retention time, compared with the unmodified MS1 peptide (SI Appendix, Fig. S5); SAH-MS1-2 also has a net formal charge of +1. Furthermore, mutation of the WT glutamate at position 2g of Bim BH3 to a hydrophobic stapling amino acid or leucine was previously associated with membrane lysis (40), and we reasoned that the glycine residue at this same position in SAH-MS1-2 might present a similar liability. Thus, the high hydrophobicity, positive net charge and glycine at position 2g were identified as target biophysical features requiring optimization to prevent the membrane toxicity of stapled MS1 peptides.
Introducing G2gE, G2gS, or Y4dR mutations, eliminating a C-terminal arginine residue, and/or extending the sequence at the N terminus with a native glutamate resulted in nine stapled peptides with differences in α-helical content, net charge, and/or hydrophobicity (Table 2 and SI Appendix, Table S2). For example, truncation of the sequence and the double mutation G2gE/Y4dR in SAH-MS1-18 produced a net neutral charge and reduced hydrophobicity, as assessed by HPLC retention time at pH 7.0 (SI Appendix, Fig. S5). Such alterations of biophysical properties eliminated membrane lysis, as evidenced by negative LDH release results for the optimized stapled peptide constructs (SI Appendix, Fig. S4).
Table 2.
Comparative binding affinities and net charge of optimized SAH-MS1-2 peptides
| Peptide | Sequence* | IC50Mcl-1, nM† | IC50p, nM† | +/−‡ | ||
| 2 | 3 | 4 | ||||
| abcdefgabcdefgabcdefga | ||||||
| SAH-MS1-2 | IWBXQGLXRLGDEINAYYARR | 90 ± 6 | >5,000 | +1 | ||
| SAH-MS1-10 | IWBXQGLXRLGDEINAYYAR | 105 ± 16 | >5,000 | 0 | ||
| SAH-MS1-11 | EIWBXQGLXRLGDEINAYYAR | 60 ± 5 | >5,000 | −1 | ||
| SAH-MS1-12 | EIWBXQGLXRLGDEINAYYA | 84 ± 8 | >5,000 | −2 | ||
| SAH-MS1-13 | IWBXQELXRLGDEINAYYARR | 55 ± 5 | >5,000 | 0 | ||
| SAH-MS1-14 | IWBXQSLXRLGDEINAYYARR | 80 ± 5 | >5,000 | +1 | ||
| SAH-MS1-15 | IWBXQSLXRLGDEINAYYAR | 180 ± 13 | >10,000 | 0 | ||
| SAH-MS1-16 | IWBXQELXRLGDEINAYYAR | 64 ± 6 | >5,000 | −1 | ||
| SAH-MS1-17 | IWBXQGLXRLGDEINARYAR | 87 ± 5 | >5,000 | +1 | ||
| SAH-MS1-18 | IWBXQELXRLGDEINARYAR | 25 ± 7 | >5,000 | 0 | ||
Mutated residues are indicated in bold.
Binding experiments and abbreviations are as in Table 1.
The +/− column indicates formal net charge based on standard titration states of residues at pH 7. B, norleucine; X, (S)-4-pentenylalanine;
Importantly, sequence changes that eliminated lytic activity had little to no effect on Mcl-1 binding specificity and even yielded several stapled peptides with improved affinity for Mcl-1 (Table 2). For example, peptides SAH-MS1-11, SAH-MS1-13, SAH-MS1-14, SAH-MS1-16, and SAH-MS1-18 all competed with FITC-Bim BH3 for Mcl-1 binding, with IC50 values lower than that of SAH-MS1-2. The highest affinity stapled peptide, SAH-MS1-18, showed a 3.6-fold improvement in IC50, which can be attributed to glutamate at 2g and arginine at 4d. Glycine at 2g in MS1, in comparison with the glutamate at 2g in Bim BH3, is known to destabilize interactions with the other antiapoptotic members of the Bcl-2 family, providing specificity, but at the cost of weakening Mcl-1 binding activity (20). In SAH-MS1-18, the 2e-3b hydrocarbon staple itself provides Mcl-1 selectivity, allowing glutamate to be reintroduced at this site to improve affinity and eliminate membrane lytic activity.
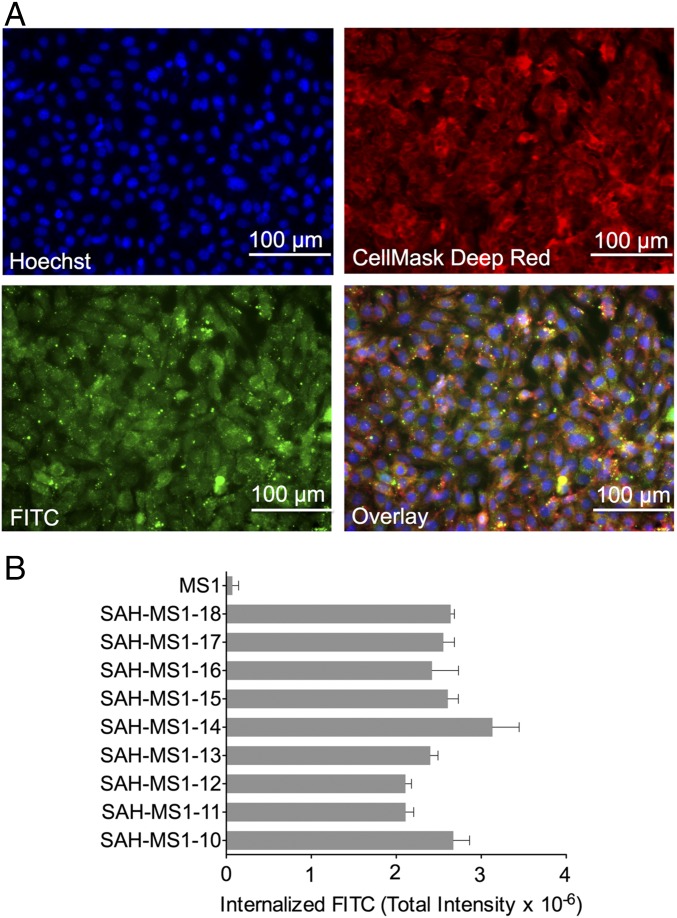
To investigate cellular uptake of stapled MS1 variants, FITC was appended to the N termini of the peptides, and total internalized FITC intensity was quantified in WT MEFs. MS1 was not internalized, but MS1-stapled variants bearing the 2e-3b hydrocarbon staple demonstrated pinosomal and cytosolic fluorescence, confirming cellular entry of FITC-labeled stapled peptides in the presence and absence of 5% FBS (Fig. 1 and SI Appendix, Fig. S6). This same staple position was associated with the second most penetrant of 18 stapled Bim BH3 peptides previously generated for a comparative cellular uptake analysis (40). Based on the binding and cellular uptake results, SAH-MS1-14 and SAH-MS1-18 peptides were prioritized for further study.
Fig. 1.
Cellular uptake of SAH-MS1 peptides. (A) MEFs were treated with FITC-SAH-MS1-18 peptide (green) in the presence of 5% FBS and counterstained with Hoechst 33342 (blue) and Cellmask Deep Red (red). (B) Cellular uptake was measured by high-content epifluorescence microscopy analysis. Total internalized fluorescence intensity values are mean ± SD for experiments performed in duplicate wells with four image acquisitions per well. Three replicates were performed with independent preparations of cells and peptide treatments, with similar results.
Selective Cytotoxicity of Mcl-1–Targeting Stapled Peptides in Mcl-1–Dependent Cancer Cells.
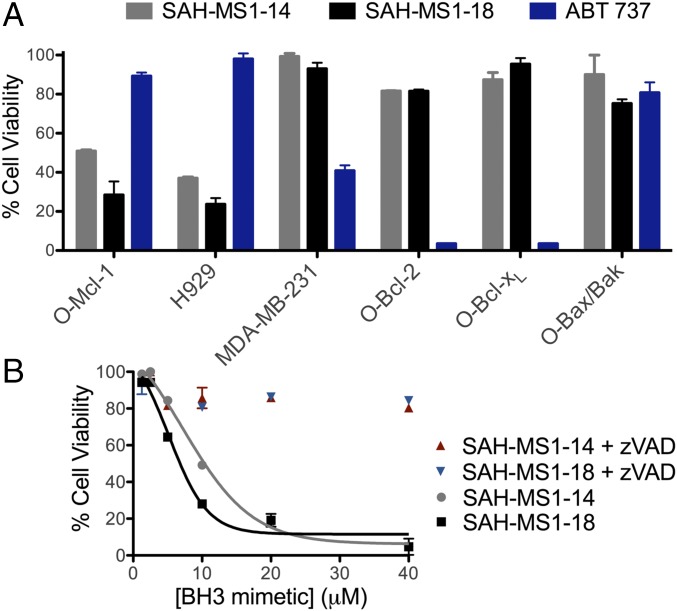
Cancer cells can avoid cell death by overexpressing antiapoptotic proteins to counteract proapoptotic signaling (47). To determine whether SAH-MS1-14 and SAH-MS1-18 could selectively overcome an Mcl-1–driven apoptotic blockade, we performed cytotoxicity assays using p185+Arf−/− B-ALL cell lines engineered to be dependent on Mcl-1, Bcl-xL or Bcl-2 (here called O-Mcl-1, O-Bcl-xL and O-Bcl-2, respectively), in addition to Mcl-1–dependent multiple myeloma (H929) and Bcl-xL–dependent MDA-MB-231 breast cancer cell lines (Fig. 2 and SI Appendix, Table S3) (48–50). In advance of viability testing, we screened lead SAH peptides in each cell line for membrane lytic activity by LDH release assay. Neither SAH-MS1-14 nor SAH-MS1-18 exhibited lytic activity in any of the cell lines tested (SI Appendix, Fig. S7).
Fig. 2.
Selective cytotoxicity of SAH-MS1-14 and SAH-MS1-18 in Mcl-1–dependent cancer cells. (A) Cell viability was measured after 24-h incubation with SAH-MS1-14, SAH-MS1-18, or ABT 737 at 10-µM dosing in 10% serum, in the indicated cancer cell lines. (B) O-Mcl-1 cell viability was measured after 24-h incubation with MS1-SAH-14 or MS1-SAH-18 in the presence or absence of the caspase inhibitor, zVAD. Fitted EC50 values are 10 ± 0.4 and 5 ± 0.5 µM for SAH-MS1-14 and SAH-MS1-18, respectively. For A and B, error bars are mean ± SD for experiments performed in duplicate three times with independent preparations of cells and BH3 mimetic treatments.
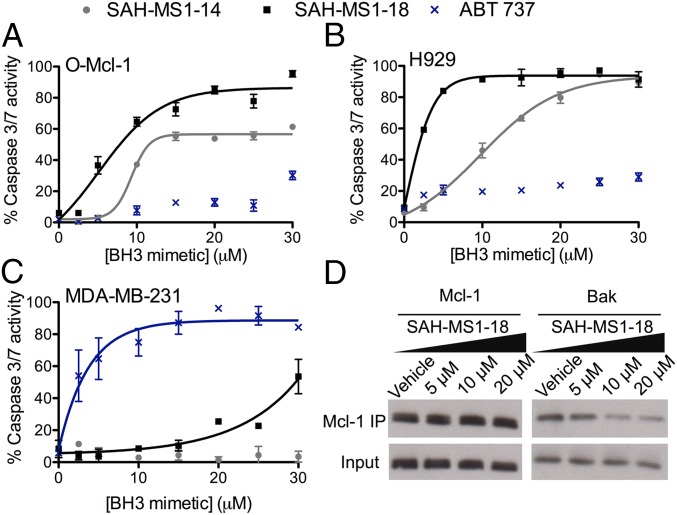
We next compared cancer cell viability following treatment with stapled peptide inhibitors of Mcl-1 or ABT-737, a selective Bcl-2/Bcl-xL inhibitor (39). In a CellTiter-Glo assay performed in 10% serum, ABT-737 selectively reduced the viability of O-Bcl-2, O-Bcl-xL, and MDA-MB-231 cells, with little to no effect on Mcl-1–dependent cells, as expected. In contrast, both acetylated and FITC-labeled variants of SAH-MS1-14 and SAH-MS1-18 were dose-dependently cytotoxic to Mcl-1–dependent H929 and O-Mcl-1 cells but had little-to-no effect on the Bcl-2/Bcl-xL–driven cancer lines (Fig. 2A and SI Appendix, Figs. S8 and S9). Importantly, the stapled peptides did not kill B-ALL cells lacking Bax/Bak (O-Bax/Bak) (Fig. 2 and SI Appendix, Fig. S8F), and cotreatment with the caspase inhibitor zVAD abolished cytotoxicity in Mcl-1–dependent cells, indicating that the peptides trigger caspase-dependent apoptotic activity (Fig. 2B). Consistent with this mechanism of action, SAH-MS1-14 and SAH-MS1-18 selectively induced caspase 3/7 activation in Mcl-1–dependent cell lines, but not in MDA-MB-231 cells (Fig. 3), with potencies that were consistent between cell viability and caspase assays. We further investigated whether the observed Mcl-1–dependent cytotoxicity correlated with stapled peptide disruption of inhibitory Mcl-1/Bak complexes. We treated intact O-Mcl-1 cells with SAH-MS1-18 and monitored Mcl-1 association with Bak by immunoprecipitation (IP) and Western blot (Fig. 3D). SAH-MS1-18 dose-responsively dissociated Mcl-1/Bak complexes in the same dose range that impaired cell viability and induced caspase 3/7 activation in Mcl-1–dependent cells. No change in total protein level of Mcl-1 was observed, indicating that the decrease in Bak was not a result of Mcl-1 down-regulation. Taken together, these data support a mechanism in which SAH-MS1-14 and SAH-MS1-18 selectively inhibit Mcl-1 to induce Bax/Bak-dependent apoptotic cell death.
Fig. 3.
Stapled peptide treatment selectively activates caspase 3/7 in Mcl-1–dependent cancer cells and dissociates Mcl-1/Bak complexes. Dose-responsive activation of caspase 3/7 by SAH-MS1 peptides in the Mcl-1–dependent cancer cells O-Mcl-1 (A) and H929 (B), but little-to-no activation in the Bcl-2–dependent MDA-MB-231 cell line (C). Cells were treated with stapled peptides at the indicated concentrations in media containing 10% FBS. ABT-737, a selective inhibitor of Bcl-2/Bcl-xL, exhibits the opposite caspase 3/7 activation profile, consistent with the differential binding specificities of SAH-MS1 peptides and the small molecule. Error bars are mean ± SD for experiments performed in duplicate three times with independent preparations of cells and BH3 mimetic treatments. (D) Coimmunoprecipitation of the Mcl-1/Bak complex from O-Mcl-1 cells treated with SAH-MS1-18 in the presence of 10% FBS. SAH-MS1-18 dose-responsively disrupted the Mcl-1/Bak complex at concentrations that induced cell death. Vehicle, 0.1% DMSO.
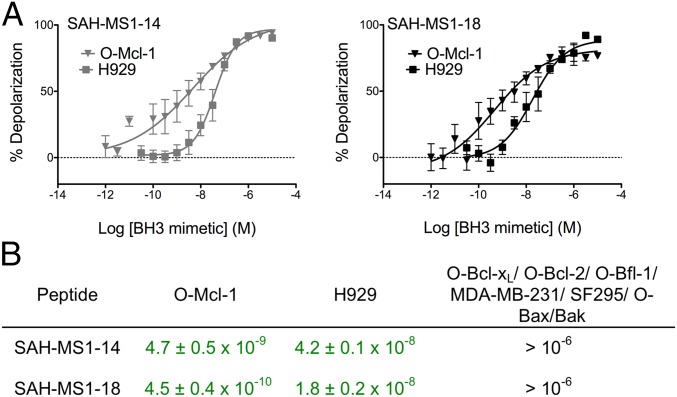
To further verify the specificity of action of SAH-MS1-14 and SAH-MS1-18 in inducing mitochondrial permeabilization, we performed whole cell BH3 profiling on permeabilized O-Mcl-1, O-Bcl-xL, O-Bcl-2, O-Bfl-1, H929, MDA-MB-231, and SF295 cells. SF295 cells require an activator peptide that can directly trigger Bax or Bak to induce pore formation; mitochondria in permeabilized SF295 cells did not depolarize in response to Hrk, Bad, or a short Puma peptide, but did respond to Bim BH3 (SI Appendix, Fig. S10). Permeabilized cells were stained with the JC1 dye to monitor mitochondrial membrane integrity in response to increasing doses of BH3 peptides (51). BH3 profiling performed with native BH3 peptides documented cell line dependencies on the individual antiapoptotic proteins (SI Appendix, Fig. S11). Strikingly, SAH-MS1-14 and SAH-MS1-18 induced mitochondrial membrane depolarization in the permeabilized Mcl-1–dependent O-Mcl-1 and H929 cells at nanomolar and picomolar dosing, reflecting a >100-fold window of specificity compared with the EC50 values recorded for the Bcl-2/Bcl-xL–dependent lines or for SF295 cells (Fig. 4 and SI Appendix, Fig. S12). As expected, based on the intended mechanism of action, neither SAH-MS1-14 nor SAH-MS1-18 demonstrated mitochondrial depolarization activity in Bax/Bak-null cells (O-Bak/Bax) across this broad dosing range. Although other SAH-MS1 constructs also exhibited an impressive degree of Mcl-1 selectivity in the BH3 profiling assay (SI Appendix, Table S4), SAH-MS1-14 and SAH-MS1-18 were the most potent, consistent with their optimal performance in fluorescence polarization binding assays (Table 2).
Fig. 4.
Stapled peptides selectively depolarize mitochondria in Mcl-1–dependent cancer cells as assayed by BH3 profiling. (A) Percent mitochondrial membrane depolarization in permeabilized H929 or O-Mcl-1 cells treated with SAH-MS1 peptides. Data are mean ± SD for at least three independent measurements. (B) EC50 values (peptide concentration in M) for mitochondrial membrane depolarization induced by SAH-MS1-14 and SAH-MS1-18 peptides in the indicated cell lines (Mcl-1–dependent cell line data highlighted in green).
Because SAH-MS1-14 and SAH-MS1-18 are related in sequence to Bim BH3, which is an activator peptide that can directly trigger Bax to induce membrane pore formation (51), the activity of the stapled peptides was tested in a liposomal dye-release assay. This assay has been shown to recapitulate the functional effect of peptides that induce Bax-mediated pore formation in mitochondria (52, 53). Addition of SAH-MS1-14 to Bax in the presence of liposomes did not lead to dye release. In contrast, SAH-MS1-18 showed activity that was intermediate between the positive control Bim SAHBA2 peptide and the vehicle control (SI Appendix, Fig. S13). Taken together, our data support a mechanism in which the stapled peptides primarily function as sensitizers, consistent with their low to undetectable activity in BH3 profiling of cell lines that are not dependent on Mcl-1.
Structural Analysis of SAH-MS1-14 and SAH-MS1-18 Interactions with Mcl-1.
We determined the crystal structures of SAH-MS1-14 and SAH-MS1-18 in complex with Mcl-1 at 1.9 Å and 1.4 Å resolution (SI Appendix, Table S5), respectively, to probe how hydrocarbon stapling achieved the observed affinity enhancements. The two structures are very similar and show that the peptides engage Mcl-1 at the canonical BH3 binding groove, as expected, through interaction with portions of helices α3, α4, α5 (BH1), α8 (BH2), and α2 (BH3). Hydrophobic residues norleucine at 2d, leucine at 3a, leucine at 3d, and isoleucine at 4a contact Mcl-1 in a manner similar to that observed for the corresponding residues of Bim BH3 (42, 54). However, both structures showed extensive additional hydrophobic interactions between the staple moiety itself, including the cross-linking olefin seen in (Z) configuration, and the Mcl-1 protein (Fig. 5 A and B). These supplementary contacts cover 130 Å2 of Mcl-1 surface (SI Appendix, Fig. S14). To test the consequences of these hydrophobic interactions on binding activity, we chemically modified the olefin group in SAH-MS1-2 to generate SAH-MS1-2Ox. Dihydroxylation of the double bond converted the hydrophobic olefin into a polar group, and we predicted this would weaken interactions with the binding site composed of two valine residues and a histidine in Mcl-1 (Fig. 5B). Competition binding assays indicated that SAH-MS1-2Ox remained highly selective for Mcl-1, but the IC50 increased ∼fourfold compared with SAH-MS1-2, consistent with the modification disturbing the hydrophobic packing between the 2e-3b staple and Mcl-1 (SI Appendix, Fig. S15).
Fig. 5.
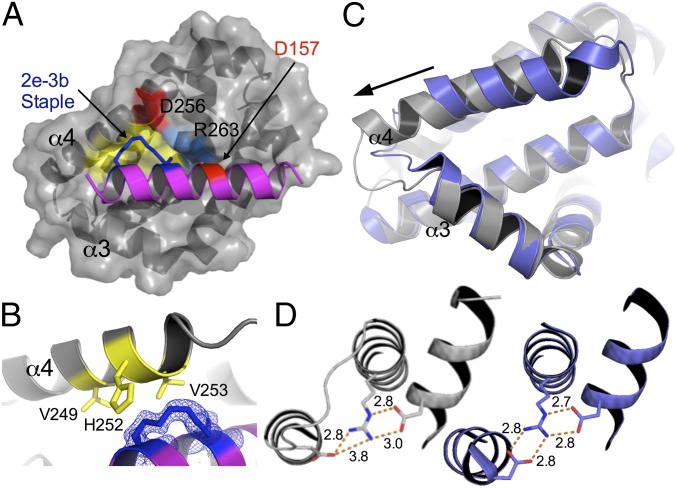
SAH-MS1-18 interaction induces a conformational change in Mcl-1. (A) Crystal structure of SAH-MS1-18 (ribbon, PDB ID code 5W89) in complex with Mcl-1 (surface). The components of the staple are shown as blue sticks. A hydrophobic patch that involves Mcl-1 residues Val-249, Met-250, His-252, and Val-253 is colored yellow. The polar network involving Mcl-1 residues Asp-256 and Arg-263 interacting with Asp-157 in SAH-MS1-18 is colored red for Asp residues and blue for Arg-263. (B) The hydrocarbon staple in SAH-MS1-18 contributes additional hydrophobic contacts to the BH3 helix-in-groove interaction (coloring as in A). (C) In the complex between SAH-MS1-18 and Mcl-1 (gray), α-helix 4 is significantly displaced, as indicated by the arrow, compared with the position of this helix in Mcl-1 bound to Bim BH3 (PDB ID code 2PQK, blue). BH3 peptides are not shown. (D) Loop rearrangement maintains a three-residue salt-bridge network between Asp-256 and Arg-263 in Mcl-1 and Asp-157 in SAH-MS1-18; the three-residue network in Mcl-1:SAH-MS1-18 (gray) is shown at Left, and the corresponding three-residue network in Mcl-1:Bim BH3 (PDB ID code 2PQK, blue) is shown at Right.
Comparison of the structures of the stapled peptides in complex with Mcl-1 with the structure of Bim BH3 bound to Mcl-1 [Protein Data Bank (PDB) ID code 2PQK] (54) revealed an unexpected local change in the Mcl-1 structure. Although modeling suggests that the 2e-3b staple can be accommodated with little to no change in the geometry of the Mcl-1:Bim BH3 complex (SI Appendix, Fig. S16), we found that the binding groove that engages the SAH-MS1 stapled peptides is actually resculpted, as shown in Fig. 5C. Apo Mcl-1 undergoes a groove-opening conformational change upon binding native BH3 peptides, and the structures of Mcl-1 bound to the 2e-3b stapled peptides showed a further opening that accommodates the hydrocarbon staple. Specifically, Mcl-1 α-helix 4 (α4) was displaced in the SAH-MS1 complexes relative to its position in the Mcl-1:Bim BH3 complex such that the distance between Ala-227 at the amino terminus of α3 and Val-253 at the carboxyl terminus of α4 increased from 15.6 Å (Cα-to-Cα atom) in the Bim BH3-bound state to 16.8 Å in the SAH-MS1-18–bound state (Fig. 5C). The loop between helices 3 and 4 was also remodeled, lengthening α4 in the SAH-MS1 complexes relative to the structure of this helix in the Bim BH3 complex. This rearrangement could potentially be disfavored if it disrupted a critical three-residue salt-bridge network formed between Mcl-1 Asp-256 and Arg-263, and Asp-157 at position 3f in the SAH peptide. However, the loop conformational change maintained these interactions, albeit in an altered structural context (Fig. 5D). Interestingly, the structural change induced by the staple was localized to only part of the complex. Most regions of the Mcl-1 protein, as well as the C-terminal end of the stapled BH3 peptide, showed little change in geometry between the SAH-MS1-14/18 and Bim BH3 complexes. Of note, the peptide and displaced helix made minimal crystal contacts, so the observed changes in structure relative to the Mcl-1:Bim BH3 complex are unlikely to be affected by the lattice. Pairwise structural comparisons with 28 other complexes of Mcl-1 bound to BH3 peptides showed that the SAH-MS1 binding mode is unique among conformations previously observed (SI Appendix, Fig. S17).
Discussion
Natural peptides have striking protein-targeting benefits, yet established liabilities when considered as therapeutic candidates. The capacity of peptides to mimic native protein interaction motifs is appealing, and their larger number of potential contacts compared with small molecules is attractive for engineering affinity and specificity. Furthermore, powerful library-screening techniques can assist in the discovery of high-affinity peptide ligands. These benefits have led to the development of prototype peptide inhibitors of several antiapoptotic Bcl-2 family proteins (19, 20, 43, 55). Native BH3 peptides and improved variants discovered using library screening and rational design have been deployed as reagents to probe disease mechanisms and to diagnose the dependency of specific cancers on individual antiapoptotic Bcl-2 proteins (7, 35, 36).
Peptides are often considered poor drug candidates due to the challenges of intracellular delivery and in vivo proteolysis. Chemical modifications that reinforce peptide structure and stability have led to renewed interest in peptides as therapeutics (56), and hydrocarbon-stapled BH3 helices were an early success story (57). In the first cellular application of hydrocarbon-stapled peptides, constructs modeled after the Bid BH3 helix efficiently activated the apoptotic signaling cascade in vitro and in vivo (22, 44). A stapled peptide inhibitor of Mcl-1, derived from the native BH3 region of Mcl-1 itself, sensitized cancer cells to TNF-related apoptosis-inducing ligand (TRAIL) or Fas ligand (FasL)-induced apoptosis (28). Key principles to emerge from this early work and follow-up analyses are that modification of natural or designed sequences with a chemical staple alters the structural and biophysical properties of peptides, providing an opportunity to optimize peptides to arrive at biologically active, cell-penetrating drug prototypes (30, 33, 40). Here, we tackled the challenge of converting an engineered peptide inhibitor of Mcl-1, previously applied in biochemical studies and antiapoptotic dependency analyses in permeabilized cells, into a chemically modified agent that selectively induces mitochondrial apoptosis in Mcl-1–dependent cells by specifically relieving an Mcl-1–mediated apoptotic blockade.
Transforming MS1 into a specific and cell-permeable Mcl-1 inhibitor required a systematic and iterative approach that was guided by prior sequence-function analyses of stapled BH3 peptides (28, 40). An initial staple scan revealed the optimal staple positions for enhancing α-helicity and binding affinity, and, importantly, these substitutions preserved Mcl-1–binding specificity. Then, in preparation for cellular proof-of-concept studies, we screened our constructs for membrane disruptive effects, identifying sequence vulnerabilities that were adjusted to eliminate unwanted lytic activity. Application of lead constructs in BH3 profiling and apoptosis analyses, using cells with characterized antiapoptotic dependencies, led to our demonstration of selective cytotoxicity in Mcl-1–dependent cancer cells.
Our data are consistent with a mechanism which involves SAH-MS1-14 and SAH-MS1-18 acting as apoptotic sensitizers, selectively inhibiting the BH3-binding activity of Mcl-1. SAH-MS1-18, but not SAH-MS1-14, modestly activates Bax in liposomal assays and may contribute to Bax activation in cells. However, SAH-MS1-18 did not induce mitochondrial outer membrane permeabilization in BH3 profiling of five cell lines not dependent on Mcl-1, and no cytotoxicity was observed in non–Mcl-1–dependent cells, indicating that any direct Bax activation is below the threshold for inducing apoptosis in these cells. Thus, the proapoptotic activity of the optimized stapled peptide constructs relies on targeted Mcl-1 inhibition.
Interestingly, our staple scan of MB2 and MS1 sequences led to the observation that the 2e-3b staple is sufficient to confer selective binding for Mcl-1 in the context of the Bim BH3 sequence. Prior mutational studies uncovered point mutations that can contribute to Mcl-1 binding affinity and specificity (19, 42). However, in this case, the staple itself contributed to specificity, yielding peptide constructs that were also cell-penetrant and protease-resistant. The optimal location of the staple in our lead peptides at the 2e-3b position is consistent with a prior study, which determined that hydrocarbon staples placed at the hydrophobic/hydrophilic interface of an amphipathic α-helix were especially effective at promoting cell uptake (40).
Structural studies provided insight into the unique mechanism by which high-affinity binding was achieved by SAH-MS1-14 and SAH-MS1-18. Both peptides induce a conformational change in the α3-α4 region of Mcl-1, leading to a gain in hydrophobic interaction surface. Supplementary hydrophobic contacts afforded by a peptide staple have been observed previously in the interactions between SAH-p53-8 and Mdm2, ATSP-7041 and Mdmx, Mcl-1 SAHBD and Mcl-1, SP1 and ERβ LBD, and SP2 and ERα (26–28, 33). In these examples, fortuitous hydrophobic interactions between the staple and the protein surface are believed to support complex formation. The structures presented here demonstrate an example of how site-specific introduction of a hydrocarbon staple can induce a conformational change in the target, leading to enhanced binding affinity for Mcl-1.
Foight et al. previously showed that an alanine-to-threonine mutation at position 2e in Bim imparts specificity for Mcl-1 and noted that position 2e of Bim is oriented differently in the binding grooves of Bcl-xL and Bfl-1, compared with that of Mcl-1 (20). This structural distinction was proposed as a possible explanation for increased tolerance of position 2e substitutions by Mcl-1 and a reason for the Mcl-1 specificity of peptide MS1. Our solved structures show that Mcl-1 can undergo a conformational change to bind large α,α-disubstituted nonnatural amino acid moieties at positions 2e and 3b. A similar change might occur to accommodate the Thr residue at 2e in MS1. If the other antiapoptotic Bcl-2 proteins cannot undergo a similar change, this difference in conformational flexibility could account for the observed specificity of SAH-MS1-14 and SAH-MS1-18, and perhaps MS1 as well.
Although features of peptide optimization can resemble traditional medicinal chemistry approaches used for small molecule development, a stepwise iterative approach to converting a lead peptide into a stapled derivative with cellular activity has the potential to yield drug prototypes in an accelerated fashion (40, 58). Combinatorial peptide library synthesis and screening methods, which can be applied to stapled peptides, have the potential to streamline peptide optimization even further (37). Broad application of these strategies to develop and test ever more diverse sets of stapled peptides will continue to elucidate and refine design principles, providing a new arsenal of stapled peptide prototypes for clinical development. Indeed, a focused, iterative strategy of stapled peptide optimization has now led to the first series of clinical trials of a stapled peptide designed to inhibit intracellular protein interactions in cancer (34). Our work suggests that optimization of stapled BH3 peptides for selective antiapoptotic targeting in cancer is feasible and holds promise for clinical translation.
Methods
Stapled Peptide Synthesis and Characterization.
SAH peptides were synthesized on rink amide resin LL (0.24 mmol/g). The solid phase synthesis resin was swollen with 1‐methyl‐2‐pyrrolidinone (NMP) for 15 min. Piperidine (20% vol/vol) in NMP was used for Fmoc deprotection (30 min). The resin beads were exposed to a 0.5-M solution of amino acid in NMP, followed by a 0.5-M solution of 2‐(6‐chloro‐1H‐benzotriazole‐1‐yl)‐1,1,3,3‐tetramethylaminium hexafluorophosphate (HCTU) coupling agent. The resin was then treated with N,N‐diisopropyl ethylamine (DIEA), and the reaction was carried out for 30 to 60 min with shaking under nitrogen. Typically, for standard couplings, a fourfold excess of amino acids and coupling reagent and eight equivalents of DIEA relative to resin loading were used. After completion of the peptide synthesis, the amino terminus was either acetylated by reaction with acetic anhydride and DIEA in NMP (1:0.1:5) or subjected to alternative derivatization (e.g., FITC‐β‐Ala, biotin‐β‐Ala). The olefin metathesis step was carried out by first swelling the resin with 1,2-dichloroethane, followed by exposure to a 10-mM solution of Grubbs’ first-generation catalyst in 1,2‐dichloroethane (DCE) for 2 h. The stapling reaction was carried out twice. The peptides were then cleaved off the resin and deprotected using 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% tri-isopropylsilane (TIS). The crude peptides were purified by reverse-phase HPLC on a C18 column with a linear water:acetonitrile gradient, and the identity of the peptides was confirmed by MALDI-TOF mass spectrometric analysis. The peptide concentrations were quantified by amino acid analysis. The sequences of all SAH peptides used in this study are listed in SI Appendix, Tables S6 and S7.
Dihydroxylation of the staple olefin following ring-closing metathesis was performed by suspending resin-bound stapled peptide in acetone/water/t-BuOH (17:1.5:1 vol/vol), N-methylmorpholine N-oxide (NMO) (1.2 mmol) and K2OsO4·2H2O (1.1 mmol). The reaction was stirred overnight at room temperature under argon. Product formation was monitored by MALDI mass spectrometry. The peptide was cleaved using 95% TFA/2.5% H2O/2.5% TIS and purified by HPLC to yield dihydroxylated peptide as a white solid. The peptide concentration was quantified by UV absorbance at 280 nm.
Protein Production.
Recombinant Bcl-xLΔC (residues 1 to 209), Mcl-1ΔNΔC (residues 172 to 327), Bcl-wΔC (residues 1 to 164), Bcl-2ΔC (residues 1 to 217), and BFL-1/A1ΔC (residues 1 to 151) bearing a c-myc tag at the amino terminus were expressed in Escherichia coli BL21 from a pSV282 vector (pSVM). This vector encodes the Bcl-2 proteins as maltose binding protein (MBP) fusions, which, upon tobacco etch virus (TEV) protease cleavage, yielded an N-terminally c-myc–tagged protein no longer fused to MBP. Bacterial cells were cultured in ampicillin-containing Luria broth, and protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside. The bacterial pellet was resuspended in lysis buffer (50 mM Tris and 100 mM NaCl, pH 8.0). Cell debris was removed by centrifugation, and sodium chloride was added to the supernatant to a final concentration of 0.5 M. The supernatant was applied to an Ni-nitrilotriacetic acid agarose (Qiagen) column, equilibrated in Tris buffer (20 mM Tris and 500 mM NaCl, pH 8.0). After washing the column, His-tagged MBP fusion proteins were eluted with buffer containing 500 mM imidazole. Eluted fractions were pooled and dialyzed against TEV cleavage buffer (50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT, pH 8.0) overnight at 4 °C. The dialyzed MBP fusions at 1 mg/mL were mixed with TEV protease at a ratio of 50:1 (wt/wt) and incubated overnight at room temperature. The TEV-cleaved reaction mix was centrifuged to remove any insoluble precipitate and purified using a second Ni-nitrilotriacetic acid column to separate the c-myc–tagged Bcl-2 protein from His-tagged MBP and His-tagged TEV protease. Proteins were at least 95% pure by Coomassie-stained SDS/PAGE, and the masses were verified by MALDI. Full-length human Bax was expressed and purified as described using the pTYB1 vector in BL21 (DE3) Escherichia coli (53, 59). Pellets were resuspended in 20 mM Tris, 250 mM NaCl, pH 7.2, and lysed by two passes through a microfluidizer (Microfluidics) chilled to 4 °C. The lysate was clarified by centrifugation at 48,000 × g and purified on chitin affinity resin (New England Biolabs). The intein–chitin-binding domain tag was cleaved by incubation in 50 mM DTT at 4 °C. Full-length protein was isolated by size-exclusion chromatography (Superdex 75 10/300; 20 mM Hepes, 150 mM KCl, pH 7.2) using an FPLC system (GE Healthcare Life Sciences).
CD Spectroscopy.
Circular dichroism spectra were acquired on an Aviv 420 spectrophotometer. Samples were prepared in 10 mM phosphate buffer, pH 7.4, at a peptide concentration of 25 μM. Data were recorded at 25 °C from 190 nm to 260 nm in a 1-mm path length quartz cell using 1-nm wavelength increments and a response time of 1 s. The data were converted to per-residue molar ellipticity [θ] (degrees per square centimeter per decimole per residue) and smoothed using Igor Pro software. Percent α-helicity was calculated as described previously (37).
Peptide Proteolysis Assay.
Protease reaction samples contained 20 μM peptide and chymotrypsin (0.1 mg/L, from bovine pancreas, 40.0 units/mg) in 20 mM phosphate buffer, pH 7.4. Aliquots of 5 μL were removed at fixed time points and quenched with 95 μL of acetonitrile containing 0.1% TFA. All samples were immediately subjected to analytical HPLC. The amount of intact peptide remaining as a function of time was quantified by integration of the appropriate peak area.
Direct and Competition Fluorescence Polarization Assays.
Both competition and direct-binding fluorescence anisotropy assays were performed in 20 mM NaPO4, 50 mM NaCl, 1 mM EDTA, 0.001% Triton X-100 (v/v), and 5% DMSO (vol/vol), pH 7.8. For competition FP binding assays, a 21-mer N-terminally fluoresceinated Bim BH3 peptide (FITC-βA-IWIAQELRRIGDEFNAYYARR, βA = beta-alanine) was used as the competitor peptide, and Bcl-2 proteins and fluoresceinated Bim BH3 peptide were added to final concentrations of 50 nM and 25 nM, respectively, in a final volume of 100 μL. The fluorescence polarization assay was performed in 96-well plates that were incubated in the dark at ∼25 °C. Fluorescence polarization was measured after 2 h on a microplate reader (SpectraMax M5 Microplate Reader; Molecular Devices), using excitation and emission wavelengths of 485 nm and 535 nm. All experiments were performed in duplicate three times. Competition fluorescence anisotropy data were fit to a complete competitive binding model (60) using Prism software (Graphpad). For direct binding assays, a 12-point dilution of receptor protein was added to the wells of a Corning 96-well, black, polystyrene, nonbinding surface plate, followed by the fluoresceinated peptide (10 nM in 120 μL total volume). Plates were incubated for 2 h and then read on a SpectraMax M5 plate reader (Molecular Devices) at 25 °C. Binding equilibrium was confirmed by comparing curves read at 2 and 24 h. Fluorescence anisotropy data for direct binding experiments were fit to the direct binding model of reference (60) using Prism. Curves were fit based on three replicate experiments that each included duplicate measurements, conducted over 3 d, and the error bars represent the SD among these replicates.
Cell Culture.
B-ALL (Bax−/−/Bak−/− and Mcl-1, Bcl-xL, Bcl-2 and Bfl-1-reconstituted p185+Arf−/−Mcl-1del)3,4, MDA-MB-231 and H929 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (ATCC) supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, 0.1 mM minimal essential medium (MEM) nonessential amino acids, and 50 mM β-mercaptoethanol. Mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified eagle medium (DMEM) high glucose (Invitrogen) supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, 2 mM l-glutamine, 50 mM Hepes, 0.1 mM MEM nonessential amino acids, and 50 mM β-mercaptoethanol. SF295 cells were grown in RPMI media with 10% FBS, 2 mM l-glutamine, and 10 mL/L 100× penicillin/streptomycin.
Cellular BH3 Profiling Assay.
Assay plates were produced by serial dilution of each peptide from 200 μM to 0.2 nM using 10-fold dilutions in derived from trehalose experimental buffer (DTEB): 150 mM trehalose, 50 mM KCl, 20 μM EDTA, 20 μM EGTA, 0.1% BSA, 5 mM succinate, 10 mM Hepes-KOH, pH 7.5, containing 0.005% wt/vol digitonin, 10 mM 2-mercaptoethanol, 2 μM JC-1, and 20 mg/mL oligomycin. Triplicate wells for each peptide were made for each cell line by adding 15 μL of the peptide dilutions to each well of a black, untreated 384-well plate. Control wells containing no peptide or 20 μM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), a chemical uncoupler of oxidative phosphorylation, were included for zero and complete depolarization, respectively. Multiple plates were produced from the same stock and frozen at −80 °C for later use. Frozen plates were brought to room temperature before use, cells were suspended in DTEB at a density of 1.34 × 106 cells per milliliter, and 15 μL of cell suspension was added to each well of the dilution series to yield wells ranging from 0.1 nM to 100 μM peptide with 20,000 cells per well. Fluorescence of JC-1 aggregates was measured at 590 nm, with 545 nm excitation on a Tecan Safire2 at 5-min intervals for 3 h. The area under each signal-vs.-time curve was calculated and normalized to the untreated and FCCP values to produce the percent depolarization. Curves were plotted as log [peptide] vs. percent depolarization, with sigmoidal dose−response curves fitted using Graphpad Prism 6.
ImageXpress Microscopy Analysis.
For high-content fluorescence microscopy analysis, mouse embryonic fibroblasts (MEFs) were plated in black, clear-bottom plates overnight at a density of 2 × 104 cells per well in DMEM supplemented with 10% (vol/vol) FBS, 1% penicillin/streptomycin, and 1% glutamine. The following day, cells were treated with 1 μM FITC-labeled peptides or the equivalent amount of vehicle (0.1% DMSO) for 4 h in phenol red-free DMEM with 5% FBS, and then stained with Hoechst 33342 and CellMask Deep Red (CMDR) (Invitrogen) for 10 min. The media was aspirated, and cells were fixed with 4% (wt/vol) paraformaldehyde for 10 min, washed three times with PBS, and imaged by ImageXpress Microscopy (IXM) (high-throughput epifluorescence microscope; Molecular Devices). Data were collected for four sites per well at 20× magnification, with each treatment performed in duplicate, and then analyzed and quantified using MetaXpress software. Image and data analysis were performed as reported (40).
Lactate Dehydrogenase Release Assay.
MEFs were plated in 96-well format (2 × 104 cells per well), and, after overnight incubation, full media was replaced with DMEM containing 10% FBS. B-ALL cells were plated in 96-well format (2 × 104 cells per well) in RPMI containing 10% FBS. Serial dilutions of SAH from a 10-mM DMSO stock, or vehicle, were added to the cells in a final volume of 100 μL and incubated at 37 °C for 2 h. The plate was spun down at 478 × g for 5 min at 4 °C, and 80 μL of cell culture media was transferred to a clear plate (Corning), incubated with 80 μL of LDH reagent (Roche) for 15 min while shaking, and absorbance was measured at 490 nm on a microplate reader (SpectraMax M5 Microplate Reader; Molecular Devices). Duplicate wells were read, and values were averaged for each biological replicate.
Cell Viability Assay.
H929 and B-ALL cells (2 × 104 per well) were seeded in 96-well opaque plates in a volume of 50 μL in RPMI containing 10% FBS. A 2× concentrated plate of the indicated serial dilutions of peptide (10 mM DMSO stock) or DMSO (0.4%) in RPMI with 10% FBS in a volume of 50 μL was then transferred to the cells; the plate was incubated at 37 °C for 24 h, and cell viability was assayed by addition of CellTiter-Glo reagent, according to the manufacturer’s protocol (Promega). MDA-MB-231 cells (1.5 × 104) cells were plated, and, after overnight incubation, the media was removed and replaced with 100 μL of media containing a serial dilution of the indicated doses of SAH peptides. Cell viability was measured at 24 h, as above, in the presence or absence of the pan-caspase inhibitor z-VAD (50 μM), which was administered to cells 30 min before treatment with stapled peptides. Duplicate wells were read, and values were averaged for each biological replicate.
Caspase-3/7 Activation Assay.
Cancer cells were treated with SAH peptides as described above for cell viability assays, and caspase-3/7 activation was measured at 6 h by addition of the Caspase-Glo 3/7 chemiluminescence reagent in accordance with the manufacturer’s protocol (Promega). Luminescence was detected by a SpectraMax M5 microplate reader (Molecular Devices). Duplicate wells were read, and values were averaged for each biological replicate.
Coimmunoprecipitation.
To monitor the Mcl-1/Bak complex, p185+Arf−/− B-ALL cancer cells dependent on FLAG-tagged Mcl-1 (O-Mcl-1) (10 × 106 cells) were incubated with vehicle (DMSO) or SAH-MS1-18 at the indicated concentrations (final DMSO concentration = 0.1%) for 2 h, followed by PBS wash at 4 °C, and incubation on ice in 300 μL of lysis buffer [50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% (vol/vol) Nonidet P-40, complete protease inhibitor tablet]. Mcl-1 complexes were then immunoprecipitated at 4 °C overnight using anti-Flag M2 agarose beads. The beads were pelleted and washed with lysis buffer five times at 4 °C, and proteins were eluted by boiling the beads at 90 °C for 10 min in lithium dodecyl sulfate (LDS)/DTT loading buffer. Immunoprecipitates were subjected to electrophoresis and Western blot analysis using anti-Bak (1:1,000) (NT; CalBiochem) and anti–Mcl-1 (1:500) (S-19; Santa Cruz Biotechnology Inc.) antibodies.
HPLC Analysis.
HPLC data were recorded on a Shimadzu LC-10AS liquid chromatograph equipped with a Phenomenex-Luna C18 2.0 × 30-mm column using an SPD-10AV UV-Vis detector reading absorbance at 220 nM. The flow rate was 1 mL/min, and a 20-min continuous gradient was used to transition from 100% solvent A/0% solvent B to 0% solvent A/100% solvent B. Solvent A was 10% acetonitrile/90% H2O/10 mM ammonium acetate; solvent B was 10% H2O/90% acetonitrile/10 mM ammonium acetate.
Liposomal Release Assay.
Large unilamellar vesicles (LUVs) with a lipid composition representing that of the outer mitochondrial membrane were formed by liposome extrusion as previously described (61). Lipid films containing a 48:28:10:10:4 molar ratio of phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, dioleoyl phosphatidylserine, and tetraoleolyl cardiolipin (Avanti Polar Lipids) were formed by evaporation of solvent, initially under nitrogen gas and then by overnight vacuum, followed by storage at −80 °C under nitrogen. Lipid films were hydrated in 1 mL of assay buffer (10 mM Hepes, 200 mM KCl, 5 mM MgCl2, pH 7.0) and mixed with the fluorophore and quencher pair, 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) (12.5 mM) and p-xylene-bis-pyridinium bromide (DPX) (45 mM). Liposomes were formed by five freeze–thaw cycles followed by extrusion through a 100-nm polycarbonate membrane and purified using a Sepharose CL-2B size-exclusion column. For measurement of Bax activation, Bax (500 nM) was mixed with or without 250 nM stapled peptides and Bim SAHBA2 (EIWIAQELRXIGDXFNAYYA) as the positive control (53) in the presence of liposomes. The assay was carried out in black opaque 384-well plates (30 μL per well). ANTS and DPX release was monitored over time at room temperature in a spectrofluorometer (Tecan Infinite M1000) using an excitation wavelength of 355 nm, an emission wavelength of 540 nm, and a bandwidth of 20 nm. Maximal release was determined by the addition of Triton X-100 to a final concentration of 0.2% (vol/vol). Percent release was calculated as [(F − F0)/(F100 − F0)] × 100, where F is the observed fluorescence at a given time, and F0 and F100 represent baseline and maximal fluorescence, respectively.
X-Ray Crystallography.
Crystals of Mcl-1 in complex with SAH-MS1-14 and SAH-MS1-18 peptides were grown in hanging drops over a reservoir containing 25% PEG 3350, 50 mM Tris, pH 9, and 0.2 M ammonium acetate. Mcl-1 [408 μM in 20 mM Tris, 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 5 mM zinc sulfate] was mixed with peptide at a 1:1 molar ratio. The hanging drops contained 2 to 3.5 μL of complex solution mixed with 0.5 μL of reservoir solution. Crystals were cryoprotected by transferring into 25% PEG 400 in 50% well solution. Diffraction data were collected at the Advanced Photon Source at the Argonne National Laboratory, NE-CAT beamline 24-IDE. The Mcl-1:SAH-MS1-14 and Mcl-1:SAH-MS1-18 datasets were integrated and scaled to 1.9 Å and 1.4 Å, respectively, using HKL2000 and phased by molecular replacement using PHENIX rigid body refinement with chain A of structure 3MK8 as a search model (62, 63). The peptide was built into the difference density from the rigid-body refinement, and the structure was refined using iterative rounds of model rebuilding using PHENIX and COOT (63, 64). Structures are deposited in the PDB with identifiers 5W8F (SAH-MS1-14 complex) and 5W89 (SAH-MS1-18 complex).
Supplementary Material
Acknowledgments
We acknowledge the Drennan laboratory and especially T. Grell for collecting X-ray diffraction data. We thank the laboratory of J. Opferman (St. Jude Children's Research Hospital) for providing the cell lines of defined antiapoptotic dependence used in the BH3 profiling and cell viability assays. We thank V. Frappier for structure comparisons; and R. Guerra, S. Escudero, and A. L. Edwards for valuable discussions and input regarding the coimmunoprecipitation experiment. We thank members of the A.E.K. laboratory for providing helpful discussions; and A. Jasanoff and A. Barandov [Massachusetts Institute of Technology (MIT)] for use of chemistry facilities. We thank R. Cook for use of a MALDI mass spectrometer. We acknowledge S. Rudnicki [Institute of Chemistry and Cell Biology-Longwood (ICCB-L)] for assistance with IXM screening and analysis. This work was supported by the Koch Institute/MIT–Dana-Farber/Harvard Cancer Center Bridge Project and used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Additional support was provided by NIH Grant R35 CA197583 and a Leukemia and Lymphoma Scholar Award (to L.D.W.), NIH Grant R01 GM110048 (to A.E.K.), and NIH Grant R50 CA211399 (to G.H.B.).
Footnotes
Conflict of interest statement: L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
This article is a PNAS Direct Submission. D.R.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5W89 and 5W8F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712952115/-/DCSupplemental.
References
- 1.Bhola PD, Letai A. Mitochondria – judges and executioners of cell death sentences. Mol Cell. 2016;61:695–704. doi: 10.1016/j.molcel.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 3.Opferman JT. Attacking cancer’s Achilles heel: Antagonism of anti-apoptotic BCL-2 family members. FEBS J. 2016;283:2661–2675. doi: 10.1111/febs.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Certo M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Gong J-N, et al. Hierarchy for targeting prosurvival BCL2 family proteins in multiple myeloma: Pivotal role of MCL1. Blood. 2016;128:1834–1844. doi: 10.1182/blood-2016-03-704908. [DOI] [PubMed] [Google Scholar]
- 6.Kelly GL, et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28:58–70. doi: 10.1101/gad.232009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodwin CM, Rossanese OW, Olejniczak ET, Fesik SW. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death Differ. 2015;22:2098–2106. doi: 10.1038/cdd.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser SP, et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26:120–125. doi: 10.1101/gad.182980.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Wei G, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell. 2012;21:547–562. doi: 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belmar J, Fesik SW. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol Ther. 2015;145:76–84. doi: 10.1016/j.pharmthera.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotschy A, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 13.Roberts AW, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374:311–322. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 15.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason KD, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 17.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EF, et al. Novel Bcl-2 homology-3 domain-like sequences identified from screening randomized peptide libraries for inhibitors of the pro-survival Bcl-2 proteins. J Biol Chem. 2009;284:31315–31326. doi: 10.1074/jbc.M109.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta S, et al. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foight GW, Ryan JA, Gullá SV, Letai A, Keating AE. Designed BH3 peptides with high affinity and specificity for targeting Mcl-1 in cells. ACS Chem Biol. 2014;9:1962–1968. doi: 10.1021/cb500340w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 22.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaBelle JL, et al. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:2018–2031. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takada K, et al. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips C, et al. Design and structure of stapled peptides binding to estrogen receptors. J Am Chem Soc. 2011;133:9696–9699. doi: 10.1021/ja202946k. [DOI] [PubMed] [Google Scholar]
- 27.Baek S, et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc. 2012;134:103–106. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 28.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang YS, et al. Stapled α–helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wachter F, et al. Mechanistic validation of a clinical lead stapled peptide that reactivates p53 by dual HDM2 and HDMX targeting. Oncogene. 2017;36:2184–2190. doi: 10.1038/onc.2016.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 33.Chang YS, et al. Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meric-Bernstam F, et al. Phase I trial of a novel stapled peptide ALRN-6924 disrupting MDMX- and MDM2-mediated inhibition of WT p53 in patients with solid tumors lymphomas. J Clin Oncol. 2017;35(15_suppl):2505. [Google Scholar]
- 35.Ryan J, Montero J, Rocco J, Letai A. iBH3: Simple, fixable BH3 profiling to determine apoptotic priming in primary tissue by flow cytometry. Biol Chem. 2016;397:671–678. doi: 10.1515/hsz-2016-0107. [DOI] [PubMed] [Google Scholar]
- 36.Touzeau C, et al. BH3 profiling identifies heterogeneous dependency on Bcl-2 family members in multiple myeloma and predicts sensitivity to BH3 mimetics. Leukemia. 2016;30:761–764. doi: 10.1038/leu.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezaei Araghi R, Ryan JA, Letai A, Keating AE. Rapid optimization of Mcl-1 inhibitors using stapled peptide libraries including non-natural side chains. ACS Chem Biol. 2016;11:1238–1244. doi: 10.1021/acschembio.5b01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 39.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 40.Bird GH, et al. Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices. Nat Chem Biol. 2016;12:845–852. doi: 10.1038/nchembio.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 42.Boersma MD, Sadowsky JD, Tomita YA, Gellman SH. Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: Application to the Bim BH3 domain. Protein Sci. 2008;17:1232–1240. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutta S, et al. Potent and specific peptide inhibitors of human pro-survival protein Bcl-xL. J Mol Biol. 2015;427:1241–1253. doi: 10.1016/j.jmb.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walensky LD, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Deng J, et al. Bruton’s tyrosine kinase inhibition increases BCL-2 dependence and enhances sensitivity to venetoclax in chronic lymphocytic leukemia. Leukemia. 2017;31:2075–2084. doi: 10.1038/leu.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q, et al. Development of a lytic peptide derived from BH3-only proteins. Cell Death Discov. 2016;2:16008. doi: 10.1038/cddiscovery.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 48.Leverson JD, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax) Cell Death Dis. 2015;6:e1590. doi: 10.1038/cddis.2014.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koss B, et al. Defining specificity and on-target activity of BH3-mimetics using engineered B-ALL cell lines. Oncotarget. 2016;7:11500–11511. doi: 10.18632/oncotarget.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci USA. 2010;107:12895–12900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng J, et al. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 53.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fire E, Gullá SV, Grant RA, Keating AE. Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–519. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jenson JM, Ryan JA, Grant RA, Letai A, Keating AE. Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1. eLife. 2017;6:e25541. doi: 10.7554/eLife.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rezaei Araghi R, Keating AE. Designing helical peptide inhibitors of protein-protein interactions. Curr Opin Struct Biol. 2016;39:27–38. doi: 10.1016/j.sbi.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walensky LD, Bird GH. Hydrocarbon-stapled peptides: Principles, practice, and progress. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bird GH, Gavathiotis E, LaBelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol. 2014;9:831–837. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: Coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 60.Roehrl MHA, Wang JY, Wagner G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 61.Pitter K, Bernal F, LaBelle J, Walensky LD. Dissection of the BCL‐2 family signaling network with stabilized α‐helices of BCL‐2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellotto S, Chen S, Rentero Rebollo I, Wegner HA, Heinis C. Phage selection of photoswitchable peptide ligands. J Am Chem Soc. 2014;136:5880–5883. doi: 10.1021/ja501861m. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.