Significance
The coevolution of mycobacteria and amoebae seems to have contributed to shaping the virulence of nontuberculous mycobacteria in macrophages. We identified a pool of genes essential for the intracellular survival of Mycobacterium abscessus inside amoebae and macrophages and discovered a hot spot of transposon insertions within the orthologous ESX-4 T7SS locus. We generated a mutant with the deletion of a structural key ESX component, EccB4. We demonstrate rupture of the phagosomal membrane only in the presence of an intact eccB4 gene. These results suggest an unanticipated role of ESX-4 T7SS in governing the intracellular behavior of a mycobacterium. Because M. abscessus lacks ESX-1, it is tempting to speculate that ESX-4 operates as a surrogate for ESX-1 in M. tuberculosis.
Keywords: TVIISS-ESX4, M. abscessus, survival
Abstract
Mycobacterium abscessus, a rapidly growing mycobacterium (RGM) and an opportunistic human pathogen, is responsible for a wide spectrum of clinical manifestations ranging from pulmonary to skin and soft tissue infections. This intracellular organism can resist the bactericidal defense mechanisms of amoebae and macrophages, an ability that has not been observed in other RGM. M. abscessus can up-regulate several virulence factors during transient infection of amoebae, thereby becoming more virulent in subsequent respiratory infections in mice. Here, we sought to identify the M. abscessus genes required for replication within amoebae. To this end, we constructed and screened a transposon (Tn) insertion library of an M. abscessus subspecies massiliense clinical isolate for attenuated clones. This approach identified five genes within the ESX-4 locus, which in M. abscessus encodes an ESX-4 type VII secretion system that exceptionally also includes the ESX conserved EccE component. To confirm the screening results and to get further insight into the contribution of ESX-4 to M. abscessus growth and survival in amoebae and macrophages, we generated a deletion mutant of eccB4 that encodes a core structural element of ESX-4. This mutant was less efficient at blocking phagosomal acidification than its parental strain. Importantly, and in contrast to the wild-type strain, it also failed to damage phagosomes and showed reduced signs of phagosome-to-cytosol contact, as demonstrated by a combination of cellular and immunological assays. This study attributes an unexpected and genuine biological role to the underexplored mycobacterial ESX-4 system and its substrates.
Mycobacteria and eukaryotic cells have a long-standing history of pronounced host–pathogen coevolution. Although the specialization in parasitism seems most advanced with Mycobacterium tuberculosis, the etiologic agent of tuberculosis, specific host-oriented adaptation processes also exist for several nontuberculous bacteria (NTM) (1, 2). Environmental amoebae represent a natural unicellular eukaryotic host and reservoir for many pathogenic bacteria, such as Legionella, Chlamydia, and Pseudomonas (3–6) and probably also for selected mycobacterial species that can survive within amoebae (7). For example, Mycobacterium marinum, a close relative of M. tuberculosis, contains different sets of virulence determinants that are tailored for specific hosts (8). Moreover, the pathogenic potential of Mycobacterium avium and Mycobacterium abscessus increases through passage in amoebae (9, 10).
M. abscessus is an emerging opportunistic pathogen responsible for muco-cutaneous infections of nosocomial origin (11–13) as well as for severe lung infections, with cystic fibrosis (CF) patients showing a particular susceptibility to infections with this mycobacterium (14–19). The pathophysiological events of M. abscessus infection are tightly associated with the transition from a smooth (S) to a rough (R) morphotype (18, 20–23), which results from a disruption in the biosynthesis and/or transport of glycopeptidolipids to the bacterial surface (24, 25). The R variant is mainly isolated from humans during exacerbation of disease (26) and is more virulent than the S variant in several animal models (23, 26, 27). In addition, the R variant is also hyper-proinflammatory, a property that has been linked to the expression of Toll-like receptor 2 agonists, such as selected lipoproteins, at the bacterial surface (28). The ability of M. abscessus to infect selected host cell types might also be linked to the presence of selected virulence genes of nonmycobacterial origin in its genome that were originally acquired by ancient horizontal gene-transfer events from unrelated microorganisms, such Pseudomonas aeruginosa, Burkholderia spp., or Stenotrophomonas spp. (29). Interestingly, these bacteria can also be found in amoebae (30, 31) and share with M. abscessus the same pathophysiological niche, such as the CF patient airways. Of interest, phospholipase C and the magnesium transporter MgtC, which are often associated with the intracellular survival capacity of diverse pathogens, have been found to be induced in M. abscessus grown inside or in contact with amoebae or macrophages (MΦ) (10, 32). These observations suggest that M. abscessus strains are equipped with a genetic arsenal acquired during their evolution to survive within environmental amoebae. The same genetic program might also help M. abscessus cause infection in human cells. It is thus interesting to identify the genes expressed by M. abscessus inside the amoeba, where it faces the harsh and microbicidal conditions employed by host cells to control bacterial infection. In this study, we sought to identify such amoeba-induced survival (AIS) genes by using an M. abscessus genome-scale Himar mariner transposon (Tn) library (33) generated in the M. abscessus subspecies massiliense clinical isolate 43S. By selecting Tn mutants exhibiting decreased fitness and intraamoebal survival capacity, we identified a selection of AIS gene candidates that were subsequently monitored and validated for their importance in intracellular survival and growth in infected human MΦ. This allowed the definition of a unique “core” of genes required by M. abscessus to survive in both its environmental (amoeba) and human (MΦ) hosts. Importantly, this study unveiled the unanticipated contribution of the ESX-4 type VII secretion system (T7SS) of M. abscessus to the induction of phagosomal rupture and phagosome-to-cytosol contact (34), linked to the increased intracellular survival of the bacterium in the host cell.
Results
Selection of M. abscessus Tn Mutants with Impaired Intracellular Replication.
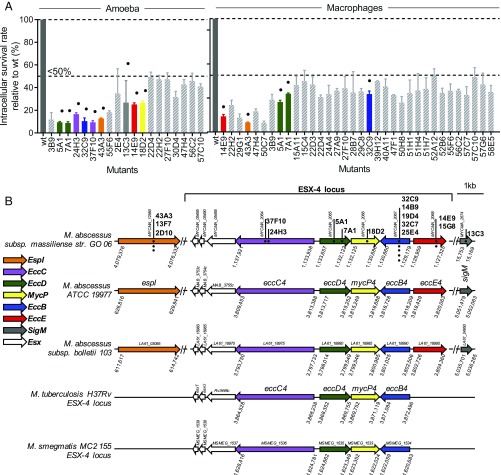
A library of 6,000 Tn mutants generated in an S clinical isolate of M. abscessus subspecies massiliense (Mabs43S) was screened using an intracellular survival assay in Acanthamoeba castellanii, as illustrated in SI Appendix, Fig. S1A. The survival of the control parental Mabs43S strain at 48 h postinfection (hpi) in amoebae was arbitrarily placed at 100%. One hundred thirty-six Tn mutants (2.3% of the total tested) displayed various degrees of intracellular growth inhibition compared with Mabs43S. These 136 mutants were individually screened in A. castellanii for 48 h and in MΦ for 72 h, at a multiplicity of infection (MOI) of 10 bacteria per cell, and intracellular cfus were determined (10). This confirmed the replication-defective phenotype of 47 mutants, which exhibited a survival in amoebae and/or in MΦ of 50% or less that of the controls, as shown by the reduction in cfu counts (Fig. 1A). Importantly, none of these insertions was linked to a significant in vitro growth defect in axenic cultures, indicating that the disrupted genes are specifically required for M. abscessus intracellular growth. Because the first round of screening was performed in amoebae, it was not possible, to identify M. abscessus genes important for intracellular growth exclusively in MΦ as previously reported for M. marinum (8).
Fig. 1.
Quantitative screening of the Mabs43S Tn library for reduced intracellular survival. (A, Left) A. castellanii cells. (Right) MΦ. A. castellanii cells or J774.2 MΦ were infected with individual selected mutants in triplicate and were cocultured for 2 days in minimum medium or for 3 days of coculture in MΦ medium supplemented with amikacin, respectively. Then cells were lysed, and the bacteria were enumerated by counting cfus. Tn mutants with less than 50% of intracellular survival in amoeba or MΦ (as compared with a 100% survival rate for the parental MabsCIP strain) are shown as means ± SD. Duplicates (i.e., the same Tn insertions) are not shown. Dots represent mutants with Tn insertions in the ESX-4 locus and espI and sigM genes. (B) Dots show the Tn positions within the different locus ESX-4 genes and espI and sigM genes. Identical mutants with the same TA insertion are represented by dots placed vertically. Numbers represent the position of the genes in the genome (NCBI annotations): M. abscessus subspecies massiliense strain GO 06, M. abscessus subspecies abscessus ATCC 19977, M. abscessus subspecies bolletii 103, M. tuberculosis H37Rv, and M. smegmatis MC2 155. The same color code is used in A and B to highlight mutants presenting a Tn insertion in the different the ESX-4 gene loci and the position of this gene within the locus.
Intraamoebal Survival of ESX-4 Tn Mutants Is Strongly Impeded.
Subcloning was undertaken to identify the genomic sequences adjacent to the various Tn insertions. Comparison of these sequences with the M. abscessus ATCC 19977 genome allowed the identification of 47 Tn insertion-disrupted genes (listed in SI Appendix, Table S1). Importantly, 12 (including five duplicates) of the 47 Tn mutants with less than 50% intraamoebal survival presented a Tn insertion in genes belonging to the ESX-4 locus (Fig. 1B and Table 1). This locus encodes one of the two M. abscessus T7SS (29, 35); related systems can also be found in other Actinobacteria and in Gram-positive bacteria of the phylum Firmicutes (36). Among these 12 Tn mutants, nine with Tn insertions in the esx-4 genes [eccE4 (two mutants), eccD4 (two mutants), and eccB4 (five mutants)] showed a 75% reduction in intraamoebal replication. The remaining three mutants with a 50% intraamoebal survival defect presented Tn insertions in the esx-4 genes mycP4 (one mutant) and eccE4 (two mutants) (Fig. 1B and Table 1). Moreover four mutants were identified outside the ESX-4 locus: one mutant in sigM and three mutants in the ESX-secreted protein espI. Of note in M. tuberculosis, espI negatively regulates ESX-1–mediated secretion under conditions where cellular ATP levels are depleted (37). In comparison, part of the sigM regulon included genes encoding two pairs of ESX family proteins and genes adjacent to the ESX-4 secretion locus (38). Supporting the important contribution of the ESX-4 locus to intracellular survival, Tn insertions in espI and sigM also conferred a severe defect in intraamoebal survival (13% and 27%, respectively) (Table 1). The intracellular defects were not due to in vitro growth impairment in axenic cultures or to uptake and/or invasion defects (SI Appendix, Fig. S1 B and C).
Table 1.
Genes lying within or outside the ESX-4 locus and presenting at least one Tn insertion
| Mutant | M. abscessus ATCC 19977 | M. tuberculosis H37Rv | Protein name | Amoeba % survival | MΦ % survival |
| 37F10 | MAB_3756c | Rv3447c | EccC | 9 | >50 |
| 24H3 | MAB_3756c | Rv3447c | EccC | 17 | >50 |
| 5A1 | MAB_3757 | Rv3448 | EccD | 9 | 26 |
| 7A1 | MAB_3757 | Rv3448 | EccD | 9 | 34 |
| 18D2 | MAB_3758 | Rv3449 | MycP | 26 | >50 |
| 32C9* | MAB_3759c | Rv3450c | EccB | 11 | 34 |
| 14E9† | MAB_3760 | EccE | 25 | 14 | |
| 43A3‡ | MAB_0628 | EspI | 13 | 8 | |
| 13C3 | MAB_4938 | Rv3911 | SigM | 27 | >50 |
Mutants with the same TA insertion site found several times: duplications are 14B9, 19D4, 32C7, and 25E4.
Mutants with the same TA insertion site found several times: duplication is 15G8.
Mutants with the same TA insertion site found several times: duplications are 13F7 and 2D10.
To exclude the possible polar effect of Tn in eccD4 and eccC4 on transcription of mycP4 and MAB_3755c, respectively, qRT-PCR was performed (SI Appendix, Table S2). Neither mycP4 nor MAB_3755c mRNA levels were affected by the Tn in eccD4 and eccC4, respectively.
Severely Impaired Intracellular Survival of an eccB4-KO Mutant in the Reference M. abscessus Strain.
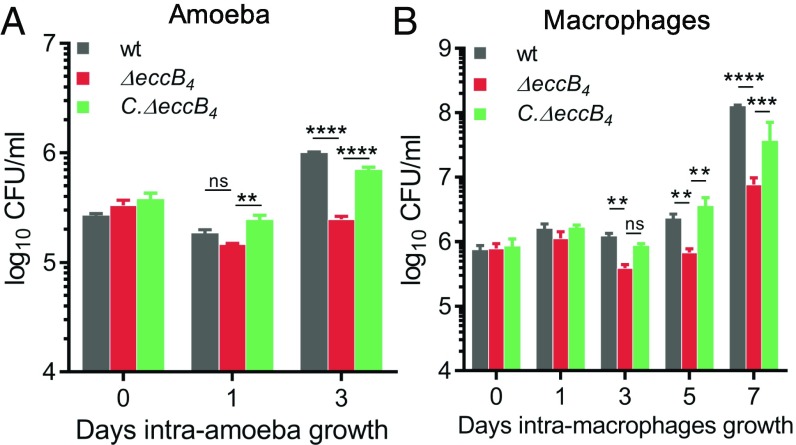
Because Mabs43S was able to grow in presence of both zeocin and hygromycin, and the Himar Tn confers an additional resistance to kanamycin, complementation studies of Tn mutants reared in this background proved challenging. This prompted us to generate an eccB4 (MAB_3759c)-KO mutant in the M. abscessus 104536T type strain (MabsCIP, zeocin sensitive), using a double-crossover strategy as previously reported (10). As illustrated in SI Appendix, Fig. S2A, ΔeccB4 was obtained by allelic exchange of nearly the entire eccB4 coding sequence with a zeocin-resistance cassette (SI Appendix, Fig. S2B) (10, 32). The complemented strain (C.ΔeccB4) was obtained by cloning the eccB4 gene under control of the hsp60 promoter in the integrative plasmid pMVH361, introducing this construct into ΔeccB4, and selecting recombinant clones with zeocin and hygromycin. The intracellular survival of ΔeccB4, C.ΔeccB4, and the parental MabsCIP strains, which had similar growth rates in axenic cultures (SI Appendix, Fig. S2C), was assessed in the amoeba and murine MΦ infection models. Compared with MabsCIP, ΔeccB4 showed a pronounced and reproducible survival defect in amoebae (Fig. 2A) and in MΦ (Fig. 2B), with a 0.5-log reduction in the number of cfus at 3 days postinfection (dpi) in both cell types and a 1.5-log reduction at 7 dpi in MΦ. Complementation with eccB4 restored the wild-type growth and survival, thus excluding the possibility of a polar effect of the mutation on the downstream genes in the mutant strain (Fig. 2). The distinct survival rates were not due to differences in the adherence capacity (SI Appendix, Fig. S3 A and B) or in the internalization (SI Appendix, Fig. S3 C and D) of ΔeccB4 with amoebae or MΦ.
Fig. 2.
Intracellular survival of ΔeccB4 and C.ΔeccB4 during coculture. Intracellular survival kinetics of wild-type, ΔeccB4, and C.ΔeccB4 strains determined by cfu counts during infection within amoeba A. castellanii (A) or J774.2 MΦ (B) at a MOI of 10:1. Histograms with error bars represent means ± SD that are representative data of three independent experiments. P values were generated by ANOVA, with the Dunnett’s test using GraphPad prism program: ns, nonsignificant; **P < 0.01, ***P < 0.001, ****P < 0.0001.
We next took advantage of the in vivo attenuated phenotype of ΔeccB4 to analyze and dissect the contribution of ESX-4 to the intracellular outcome of MabsCIP.
M. abscessus EccB4 Is Involved in the Blockage of Phagosome Acidification.
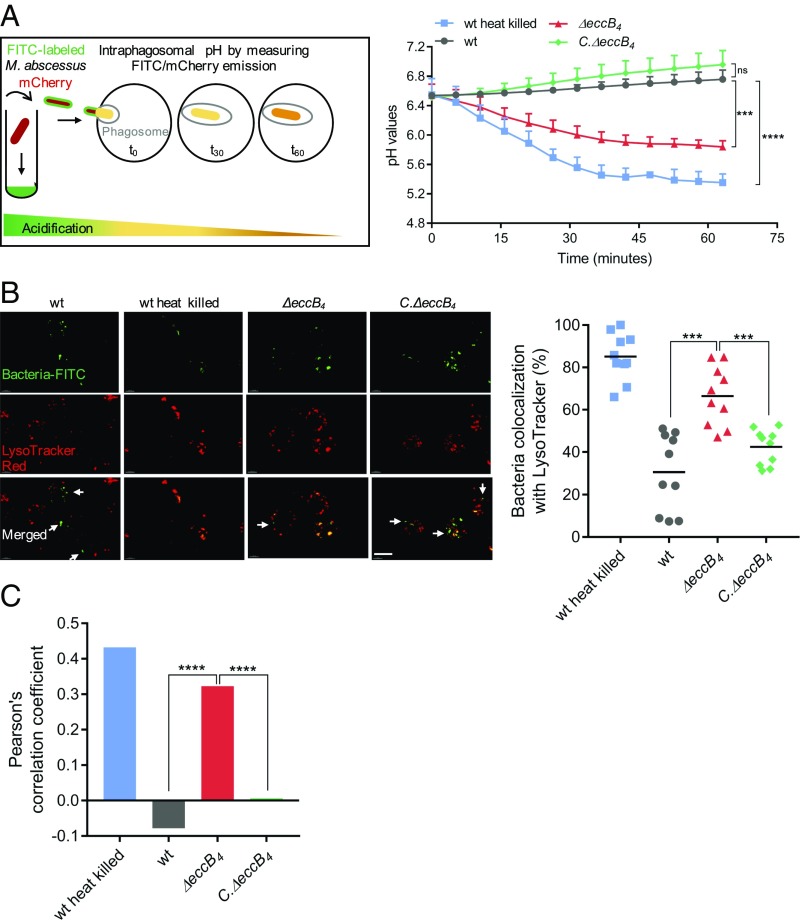
Inhibition of phagosome acidification is a well-characterized property of slow-growing pathogenic mycobacteria (SGM) (39, 40). Using pH-sensitive and pH-resistant dyes, we recently demonstrated that the RGM MabsCIP strain is able to block a decrease in the intraphagosomal pH as a means of promoting its intracellular survival (34). By a similar approach, we found here that ΔeccB4 has a reduced capacity to block phagosome acidification (Fig. 3A). The intraphagosomal pH curve of the ΔeccB4 mutant was comparable to that of heat-killed MabsCIP, whereas the intraphagosomal pH curve of C.ΔeccB4 was similar to that of the wild-type strain (Fig. 3A).
Fig. 3.
Evaluation of phagosomal acidification in MΦ infected with different M. abscessus strains. (A, Left) Differentiated THP-1 cells were infected with FITC-labeled and mCherry-expressing (fluorescent protein) wild-type (wt) M. abscessus (MabsCIP), ΔeccB4, C.ΔeccB4, or heat-killed MabsCIP strains. Fluorescent signals were measured sequentially using excitation at 485 nm (FITC) and 565 nm (mCherry). The pH at each time point was extrapolated from a standardized pH curve. (Right) The intraphagosomal pH of the different MΦ strains was measured every 5 min for more than 1 h. (B) Analysis of the bacterial compartment by confocal microscopy using a fluorescent marker for acidic compartments. (Left) Colocalization of FITC-labeled wild-type MabsCIP, heat-killed wild-type MabsCIP, ΔeccB4, and C.ΔeccB4 with the acidotropic dye LysoTracker Red in infected J774.2 MΦ. Arrows indicate intracellular mycobacteria not colocalizing with LysoTracker Red. (Scale bar, 10 µm.) (Right) The percentage of FITC-labeled strains colocalized with LysoTracker was quantified by counting at least 100 infected cells in 10 different fields. Horizontal bars show the mean percentages of bacilli colocalized with red acidic compartments in MΦ infected with heat-killed wild-type, wild-type, ΔeccB4, and C.ΔeccB4 strains. (C) PCC of images of internalized FITC-labeled mycobacteria and LysoTracker Red marker in J774.2 cells. Data are representative of three independent experiments and represent means ± SD. P values were determined by ANOVA with the Dunnett’s test using GraphPad prism program (A and C) and unpaired t test (B); ns, nonsignificant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Confocal microscopy of LysoTracker Red-labeled cells infected with FITC-stained bacilli showed that a significantly higher number of phagosomes containing ΔeccB4 colocalized with the LysoTracker dye compared with phagosomes containing the wild-type or C.ΔeccB4 strains (Fig. 3B). Results of the colocalization studies are shown as plots (Fig. 3B) and as Pearson’s correlation coefficient (PCC) of the quantified colocalization signals (Fig. 3C).
Together, these findings suggest that the impaired intracellular replication of ΔeccB4 is linked to the progressive acidification of ΔeccB4-containing compartments.
ΔeccB4 Is Unable to Damage the Phagosomal Membrane.
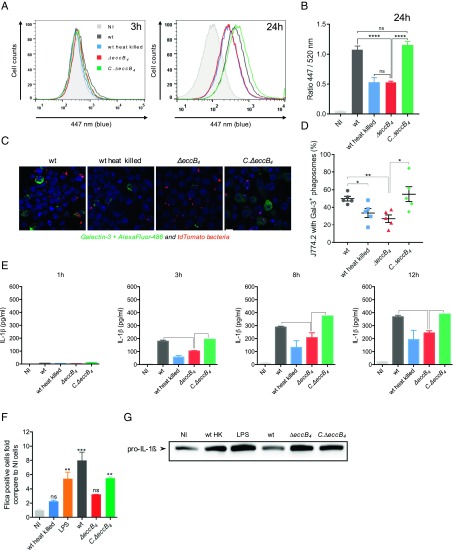
MabsCIP was recently shown to share important pathogenic features with SGM (34). These include the potential to induce damage to and disruption of the phagosome membrane, thereby establishing phagosome-to-cytosol communication (34). Since EccB4 appears to be involved in the inhibition of phagosome acidification, which is a prerequisite for the induction of phagosomal rupture (41), we evaluated the capacity of ΔeccB4 to induce a phagosomal rupture. To this end, we applied the previously developed FRET-based phagosomal rupture assay (34, 42), which depends on the activity of the M. abscessus endogenous β-lactamase (43). In this assay, when the β-lactamase reaches the cytosol, it cleaves the cephalosporin core of the coumarin-cephalosporin-fluorescein (CCF-4) dye present in the cytosol, resulting in inhibition of the FRET from coumarin to fluorescein and the subsequent switch of fluorescence emission from green to blue. In contrast to MabsCIP, and similarly to heat-killed MabsCIP, the ΔeccB4 strain was unable to inhibit FRET signaling, as revealed by the absence of a green-to-blue fluorescent shift at 24 hpi (Fig. 4A). The wild-type phenotype was fully restored in C.ΔeccB4, thus supporting the involvement of EccB4 in establishing phagosome-to-cytosol communication in M. abscessus. These results were corroborated by calculating the ratio of mean fluorescence intensities (MFI) of the blue (447 nm) versus green (520 nm) signals (Fig. 4B).
Fig. 4.
Phagosomal rupture detected by CCF-4 FRET-based flow cytometry, galectin-3 immunostaining confocal microscopy, and IL-1β activation and production. (A) Results are depicted as signal overlays per group with 1,000,000 events per condition acquired in wild-type (dark gray line), heat-killed wild-type (blue line), ΔeccB4 (red line), C.ΔeccB4 (green line), and noninfected (NI, light gray filled curve) strains. (B) Histograms represent the ratio of blue MFI at 447 nm and green MFI at 520 nm and SD from at least three independent experiments. (C) Representative confocal images of MΦ infected with different tdTomato M. abscessus wild-type, heat-killed wild-type, ΔeccB4, and C.ΔeccB4 strains (red) and stained for galectin-3 (green). (Scale bar, 10 µm.) (D) The percentage of infected MΦ containing at least one positively stained phagosome (Gal-3+) was determined. The values represent the mean ± SEM of 100 infected cells in five different fields. (E) Kinetics of IL-1β production during THP-1 infection by wild-type, heat-killed wild-type, ΔeccB4, and C.ΔeccB4 strains. (F) Caspase-1 activation was measured with the FAM-PLICA caspase-1 assay. THP-1 cells were infected or not infected (NI) with wild-type, heat-killed wild-type, ΔeccB4, and C.ΔeccB4 strains at a MOI of 10 for 3 h. As a positive control of apoptosis induction, cells were incubated with LPS 500 nM for 8 h at 37 °C. The adherent cells were gently dissociated with TrypLE Express reagent; then the cells were washed twice and resuspended in RPMI medium with 10% FBS before staining with 30× FAM-VAD-FMK for the CD11b antibody and were analyzed by flow cytometry. Results are expressed as the percentage of cells with the caspase-1 p10 and p12 active forms. (G) Western blot analysis for pro–IL-1β was performed after cell lysis (conditions are as in F). P values were generated by ANOVA, with the Dunnett’s test (B and D) or unpaired t test (C) using GraphPad prism program: ns, nonsignificant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Additionally, fluorescent-labeled galectin-3 was used to visualize the remnants of ruptured phagosomal membranes following infection of J774 MΦ with the different strains. An intense labeling with galectin-3 was observed upon infection with MabsCIP and C.ΔeccB4. In contrast, the percentage of galectin-3–positive cells infected with either ΔeccB4 or heat-killed MabsCIP was significantly lower (Fig. 4 C and D). This indicates that, in the absence of functional EccB4, only reduced signs of phagosomal lysis are observed.
Recently, synergy between the phthiocerol dimycocerosate and the ESX-1 pathways has been proposed as a cause of phagosomal membrane rupture in M. tuberculosis (44), prompting us to analyze and compare the lipid profiles in ΔeccB4 and MabsCIP. No obvious differences in the polar or apolar lipid fractions or in the mycolic acid pattern were noticed (SI Appendix, Fig. S4).
Overall, these results strongly suggest that M. abscessus uses the ESX-4 machinery to interfere with or damage the phagosomal membrane, inducing phagosomal rupture which ultimately may allow the escape of MabsCIP into the cytosol.
Recent studies reported that, during infection of differentiated human THP-1 cells, M. tuberculosis engages distinct cytosolic pattern recognition receptor systems, namely the cyclic GMP-AMP synthase (cGAS)–IFN-I axis and the inflammasome–IL-1β pathway. The relative abundance of ESX-1 effectors determines which pathway is triggered (45–47). We therefore quantified IL-1β and IFN-β in the culture supernatant of THP-1 cells by ELISA. A time-dependent increase in IL-1β production was observed for both the parental and complemented strains. In contrast, reduced levels of IL-1β were observed in the supernatant of MΦ infected with ΔeccB4, further substantiating a functional involvement of EccB4 in phagosome-to-cytosol communication (Fig. 4E). We confirmed that the difference in IL-1β production was due to caspase-1 activation and not to an altered pro–IL-1β transcription in ΔeccB4 compared with MabsCIP (Fig. 4 F and G). In contrast, we were not able to detect IFN-β in the supernatant of THP-1 cells infected with either MabsCIP or ΔeccB4 for up to 40 hpi.
EccB4-Dependent Secretome.
A proteomic-based approach was used to analyze and compare the concentrated culture supernatants from MabsCIP and ΔeccB4 on four replicates to unveil proteins possibly missing or present in diminished quantity in the ΔeccB4 mutant relative to the MabsCIP strain. Three comparisons were made using two M. abscessus proteomes (UP000007137 and UP0000038470) and a M. tuberculosis proteome (UP000001584), with the restriction that a full and comprehensive proteome comparison could not be done for the M. tuberculosis proteome due to low homologies between peptides. Based on the two M. abscessus proteomes, 152 and 166 differently expressed or secreted proteins were identified between MabsCIP and ΔeccB4 (SI Appendix, Tables S3 and S4). The analysis highlighted the following proteins of particular interest: (i) three PE/PPE proteins (MAB_0664, MAB_0047, and MAB_4141) that are not part of ESX-4 locus were decreased in ΔeccB4; (ii) four putative MmpL transporters, two MmpS proteins, and four Mce-like proteins were diminished in ΔeccB4; and (iii) secretion of four lipoproteins (LpqY, LprC, LppL, and LprN) was decreased in the ΔeccB4 mutant (SI Appendix, Tables S3 and S4). For the ESX-4–encoded proteins, a decrease of EccB4 in ΔeccB4 was noticed (P = 0.24; SI Appendix, Fig. S5A). Importantly, the presence of EsxT (MAB_3753c) and EsxU (MAB_3754c) in the supernatant was reduced by 52 and 27%, respectively, in the ΔeccB4 mutant compared with MabsCIP. While there was a trend for less secretion to be observed, the decrease was not statistically significant (P = 0.49 and P = 0.82, respectively) (SI Appendix, Fig. S5A).
To further explore these findings, the esxU-esxT operon was placed downstream of the hsp60 promoter in a replicative plasmid to generate a His-tagged EsxT fusion protein. The construct was introduced in MabsCIP and ΔeccB4, and expression of His-EsxT was analyzed by immunoblotting (SI Appendix, Fig. S5B). As shown in SI Appendix, Fig. S5B, His-EsxT was not detected in the culture supernatant of either MabsCIP or the ΔeccB4 mutant. However, His-EsxT was detected in the cytosol and the cell membrane —but not in the cell wall—extracts of both MabsCIP and the ΔeccB4 mutant (SI Appendix, Fig. S5B), but its expression levels were lower in the ΔeccB4 construct (SI Appendix, Fig. S5B) despite similar amount of proteins loaded (SI Appendix, Fig. S5C). In addition, the esxT transcript levels remained unchanged, as indicated by qRT-PCR analyses performed on both strains (ratio of 1.15 compared with sigA). Taken together, these results confirm the decreased expression/secretion of ESX-4 substrates, such as PE/PPE proteins and EsxU/T, in the ΔeccB4 mutant.
Discussion
Mycobacteria and free-living amoebae have coevolved for a long time, as inferred from the ability of selected mycobacteria to survive not only in trophozoites but also in amoebal cysts (48). In the environment, interactions with amoebae seem to have contributed to shaping the virulence and pathogenic potential of NTM, as illustrated recently for M. avium and M. abscessus (9, 10). Here we have identified a pool of genes essential for the intracellular survival/growth of M. abscessus inside amoebae and MΦ (SI Appendix, Table S1). Our major finding is the discovery of a hot spot of Tn insertions within the ESX-4 T7SS locus, with 12 individual mutants with insertions in the eccC4, eccD4, mycP4, eccB4, and eccE4 genes identified (Table 1).
Interestingly, the M. abscessus ESX-4 locus differs from that of other mycobacterial species, such as M. tuberculosis or Mycobacterium smegmatis, in that it contains the ESX conserved component EccE4 (Fig. 1B) (35). To further explore the function of ESX-4 in the intracellular survival of M. abscessus, we generated a deletion mutant in one of the key structural ESX-4 components, namely EccB4, using an allelic exchange strategy. This approach confirmed the essential role of the eccB4 gene in intraamoebal and intra-MΦ replication/survival of M. abscessus. The replication/survival defect of ΔeccB4 resulted neither from decreased adhesion to the host cell surface nor from defective phagocytosis but from the intracellular fate of the mycobacteria.
In contrast to most other RGM, MabsCIP can replicate inside the MΦ thanks to the inhibition of phagosomal acidification (10, 34). We recently proved that the M. abscessus MAB_4780 locus, which encodes a dehydratase that is involved in the metabolism of mycolic acids, participates in the blockage of phagolysosomal fusion (49). The present study demonstrates that EccB4 is also a major player in this process. Previous electron microscopy studies have suggested the occurrence of MabsCIP in various intracellular compartments and the presence of lesions in the phagosomal membrane that may ultimately lead to membrane rupture of the phagosome containing the S variant of MabsCIP (34). These features may result in the unique ability of MabsCIP as an RGM to replicate inside the cells, thereby avoiding the antibacterial response of the host MΦ. Unexpectedly, this phenotype was also lost in the absence of a functional EccB4. The combination of the FRET assay and the use of a fluorescent galectin-3 (FG3) probe highlighted the rupture of the phagosomal membrane, which occurred only in presence of an intact eccB4 gene. FG3 was originally used to visualize vacuole lysis induced by extraphagosomal pathogens (50, 51) and has been used successfully to confirm M. tuberculosis-induced phagosomal damage (42). In addition, the establishment of a phagosome-to-cytosol contact by wild-type MabsCIP was also supported by the measurements of IL-1β secretion. Indeed, recent reports have suggested that mycobacterial access to the cytosolic compartment has several consequences for the innate immune response, including enhanced activation of the inflammasome and release of functional IL-1β (45, 46). Quantification of IL1-β produced by THP-1 cells infected with different M. abscessus mutants demonstrates the involvement of EccB4 in triggering the inflammasome, further emphasizing a functional link between ESX-4 and phagosome-to-cytosol contact. However, the IFN-β results differ from those obtained in M. tuberculosis, as production of IFN-β was not observed during the M. abscessus infection; a possible explanation is the potential absence of M. abscessus DNA release into the cytosol. In the case of M. tuberculosis, mycobacterial dsDNA, liberated from the bacteria by still unknown mechanisms and translocated to the cytosol, most likely through the ESX-1–mediated phagosomal membrane rupture, is recognized as being responsible for the activation of the IFN-I axis (46).
Whether ESX-4 plays a direct role in phagosomal acidification and membrane permeabilization or is involved in a signaling event that occurs before these processes is currently being addressed. This phenomenon is reminiscent of analogous studies associating the crucial role of ESX-1 and its substrates with the phagosomal rupture induced by M. tuberculosis or M. marinum (46). Here, we demonstrated decreased expression of M. abscessus ESX-4 substrates such as EsxT in the ΔeccB4 mutant, showing that such compounds have a similar role in the M. abscessus-induced phagosomal membrane damage. Taken together, these results suggest an unanticipated role for the ESX-4 T7SS, and particularly EccB4, in governing the intracellular behavior of a mycobacterium. Because M. abscessus lacks ESX-1, it is tempting to speculate that ESX-4 operates as a surrogate of ESX-1. Indeed, in pathogenic SGM the induction of phagosomal rupture is linked to the genomic presence of the ESX-1 locus (42, 52). The ESX loci arose by gene duplication in the phylogenetic order of ESX-4, ESX-1, ESX-3, ESX-2, and ESX-5 (53). M. abscessus possesses only ESX-3 and ESX-4 loci (29, 35) and lacks ESX-1 found in other RGM, including M. smegmatis. The importance of ESX-4 for conjugal recipient activity in M. smegmatis has been reported recently (54). Of importance, induction of esx-4 transcripts in the recipient requires a functional recipient ESX-1 secretion system (54). M. smegmatis lacks EccE4 compared with M. abscessus. Whether M. abscessus requires a membrane-damaging system, such as the one formed by the ESX-1 substrates, or whether a functional ESX-4 locus is sufficient to promote conjugal transfer for M. abscessus remains to be explored.
In contrast to a recent report (53), we failed to obtain Tn insertions in the ESX-3 locus, which brings into question the essentiality of this locus in M. abscessus. The ESX-4 is considered the most ancestral T7SS in mycobacteria (35), and it is also the smallest (9,870 bp) in M. tuberculosis, comprising only seven genes (55, 56). However, in M. abscessus the ESX-4 locus is larger (12,745 bp) and possesses a supplementary gene, referred to as “eccE4,” which is absent in the ESX-4 loci of all the other mycobacteria, including SGM species (Fig. 1B) (35). The presence of the complete set of eccB4-eccC4-eccD4-eccE4 genes encoding the structural components of the ESX machine is reminiscent of other ESX loci, including ESX-5, whose structural organization was recently unveiled (55, 57). Of importance is the interdependence of the different Ecc components for the formation of a functional nano-machine, as previously shown by the lack of stable expression of eccB5 when eccC5 or eccD5 was mutated (55). Of note, the related Mycobacterium chelonae and Mycobacterium immunogenum species possesses an ESX-4 locus similar to that of the M. abscessus complex with an intact eccE4 gene, for which the closest paralogues so far identified are eccE5 and eccE1 of M. tuberculosis (35). The ubiquitous absence of gene eccE4 from the ESX-4 loci of almost all interrogated mycobacterial species suggests that in M. abscessus, where this gene is exceptionally present, its gene product EccE4 might be involved in the secretion of proteins such as the ESX-4–associated EsxU and EsxT substrates across the cell envelope (58). The ESX-4 loci usually also lack also the pe/ppe and espG genes that may play important physiological functions and participate in host–pathogen interactions (58, 59). However, our comparative analysis revealed lower amounts of PE/PPE proteins in the supernatant of the ΔeccB4 mutant. This situation is reminiscent of the specific role of ESX-5 from M. marinum in the secretion of dedicated PPE and PE-PGRS proteins (58). Whether these proteins are directly or indirectly secreted via ESX-4 in M. abscessus and whether they play a role in intracellular survival is currently unknown and will have to be investigated in further studies. In addition, EsxT, a very likely substrate of ESX-4, was identified as being associated with the membrane fraction, and decreased levels of EsxT were found in the membrane of ΔeccB4, supporting the proteomic data. Whether EsxU and EsxT fulfill functions in M. abscessus similar to that of EsxA/B in M. tuberculosis will deserve further studies.
In conclusion, we describe here peculiar properties of the ESX-4 locus in M. abscessus and demonstrated its crucial role in the intracellular survival and pathogenic potential of mycobacteria. Our findings open perspectives for better defining the biological role of the ESX-4 system in the light of the evolution of mycobacterial pathogens.
Materials and Methods
Strains, Cells, and Culture Conditions.
M. abscessus subspecies massiliense 43S (Mabs43S) is a smooth clinical isolate from Korea used to generate the Tn mutant library. The CIP 104536T type strain of M. abscessus subspecies abscessus (MabsCIP) was also used to generate the eccB4-deletion mutant. Mycobacterial strains and mutants were routinely grown aerobically at 37 °C in Middlebrook 7H9 medium (Sigma-Aldrich) supplemented with 0.2% glycerol, 1% glucose, and 250 mg/L kanamycin (Thermo Fisher Scientific) when necessary, with 25 mg/L zeocin (Thermo Fisher Scientific) for the ΔeccB4 strain, and with 25 mg/L zeocin plus 250 mg/L hygromycin (Thermo Fisher Scientific) for C.ΔeccB4. Reporter strains and markers were used as described previously (23, 34). A. castellanii (ATCC 30010) was grown at room temperature without CO2 in peptone yeast extract glucose (PYG) broth for the amplification of the strain. J774.2 and THP-1 MΦ cell lines were grown and used as described (32, 34).
Cell Infection Assays.
The A. castellanii culture was washed four times in 10 mL of AC buffer [4 mM MgSO4, 0.4 M CaCl2, 0.1% sodium citrate dehydrate, 0.05 mM Fe(NH4)2(SO4)2-6H2O, 2.5 mM NaH2PO3, 2.5 mM K2HPO3, pH 6.5] and distributed into 96-well plates at a concentration of 5 × 104 amoebae per well. Following 1 h of incubation at 32 °C, the amoebae monolayer was inoculated with 5 µL of a 4-day culture of each Tn mutant. After 1.5 h of incubation at 32 °C, extracellular mycobacteria were removed by one washing in AC buffer followed by 2 h of incubation in presence of 100 µg/mL amikacin, followed by two successive washes in AC buffer. Plates were incubated at 32 °C, with the addition of 107 heat-inactivated Escherichia coli (70 °C for 60 min) every 24 h (10). After 2 days of coculture, the A. castellanii monolayer was disrupted with 0.1% SDS for 30 min at 32 °C. Thirty microliters of a 1/25 dilution were dropped onto a blood agar plate to perform the screen. Mutants with a 50% or greater reduction in cfus were selected (SI Appendix, Fig. S1). For the second screen, cfu counts were performed to confirm the intracellular behavior, as reported earlier (10). Infection of J774.2 and/or THP-1 MΦ and cfu counts were performed as described (10, 34). Adhesion and internalization assays were carried out in 24-well plates, as described (SI Appendix, Fig. S3) (60).
Subcloning and Sequencing of Tn-Adjacent Genomic Regions.
Genomic DNA was extracted from each mutant, ClaI restricted, and subcloned in pUC19 (Invitrogen, Thermo Fisher Scientific) (61). Genomic DNA was sequenced using a 5′ primer in the Tn (TTGACGAGTTCTTCTGA). Genomic sequence comparisons were made using the BLAST server [genome database from the National Center for Biotechnology Information (NCBI) website]. Sequence data were used to design primers to confirm the exact position of the Tn insertion by PCR amplification on Mabs43S genomic DNA and sequencing. The web interface MaGe (Magnifying Genomes) was used to automatically assign the gene product function of the genome of M. abscessus CIP 104536T and of the mutated gene responsible of the resistance to degradation in MΦ cells (SI Appendix, Table S1).
ΔEccB4 Mutant Construction and Complementation.
Disruption of the eccB4 gene was performed using the recombineering system as described previously (10, 61). Complementation was performed after amplifying and cloning eccB4 into the integrative plasmid pMVH361 as described (SI Appendix, Fig. S2) (10).
Analysis of Phagosomal Acidification by pH Measurements.
M. abscessus strains expressing the fluorescent protein mCherry were labeled with FITC (Sigma-Aldrich) and dispensed into 24-well plates containing J774.2 MΦ at a MOI of 10:1. Plates were centrifuged at low speed, allowing mycobacteria to reach the monolayer, and were incubated 15 min at 37 °C before fluorescence measurements (34). Fluorescent signals were then measured by sequential excitation at 485 nm (FITC) and 565 nm (mCherry) using a Tecan Infinite M200 multimode plate reader. A standard pH curve was established to correlate the fluorescent signal with the pH by measuring the emission levels from fluorescent M. abscessus phagocytosed by MΦ after placing infected cell monolayers in standard series of buffers from pH 4.6–7.6 (25).
Analysis of Phagosomal Acidification by LysoTracker Colocalization.
Fully confluent monolayers of MΦ in 24-well plates were infected with FITC-labeled M. abscessus, and analysis of fluorescent signals was performed. The acidotropic dye LysoTracker Red DND-99 (Life Technologies) was used as a marker of phagosome acidification and maturation. J774.2 MΦ were distributed into 12-well plates containing glass coverslips and were infected with FITC-labeled M. abscessus (100 mg/mL of FITC for 2 h at room temperature). At the end of the infection, LysoTracker was added to each well. The percentage of FITC-labeled mycobacteria that colocalized with LysoTracker-labeled phagosomes was determined by analyzing more than 100 infected cells from at least 10 random fields with a Leica SP8-X confocal microscope. Pictures analyses and PCC evaluation were done with IMARIS software.
Galectin-3 Immunostaining to Detect Phagosomal Membrane Damage.
MΦ infected with fluorescent tdTomato-expressing M. abscessus strains were fixed on glass coverslips. Phagosomal membrane damage was revealed by immunostaining using a purified mouse anti-human Galectin-3–specific monoclonal antibody (555746; BD Pharmingen) used at 1/200 dilution. The antibody was visualized using secondary antibody conjugated to Alexa Fluor 488 (Invitrogen). An infected cell was considered positive when at least one of its mycobacteria-containing phagosomes was stained positive with the phagosomal damage marker. The majority of specific galectin-3–positive signals were associated to membranous structures surrounding bacteria‐containing phagosomes. The percentages of infected cells having at least one M. abscessus-containing phagosome that was positive for galectin-3 were determined from at least 100 infected cells found in five different fields. All images were analyzed with IMARIS software.
Flow Cytometry Analysis to Study Phagosomal Rupture and Caspase-1 Activation.
CCF-4 FRET measurements to study phagosomal rupture were performed as recently described (42).
The FAM FLICA Caspase-1 Assay Kit (ImmunoChemistry Technologies) was used to evaluate the presence of catalytically active forms of caspase 1 p10 and p12. THP-1 cells were infected at a MOI of 10 bacteria per cell for 3 h. As a positive control of apoptosis induction, cells were incubated with LPS (500 nM) for 8 h at 37 °C. The adherent cells were gently dissociated with TrypLE Express reagent (Gibco) as recommended. The cells were washed twice and resuspended in RPMI medium with 10% FBS before staining with 30× FAM-VAD-FMK for 30 min at 37 °C. for specific gating, the cells were also stained with the CD11b antibody (42) before being fixed with 5% paraformaldehyde. Cells were analyzed by flow cytometry. Results are expressed as the percentage of cells with the caspase 1 p10 and p12 active forms. The same extracts were used to evaluate mature and precursor IL-1β (1:1,000; Abcam ab2105,) by Western blot analysis.
ELISA Analysis.
Supernatants from THP-1 MΦ/M. abscessus cocultures were collected and assayed for human IL-1β secretion using the IL1-β kit DY201-05 (Bio-Techne) and for IFN-β secretion using the human IFN-β kit 09341410-1 (tebu-bio), according to the manufacturer’s recommendations.
EsxU and EsxT Secretion Analysis.
MAB_3754c and MAB_3753c were cloned in the PvuI and HindIII sites of pMVH361. Primers were designed to incorporate a His-tag at the C terminus of the MAB_3753c protein. A stop codon was also introduced between the two genes. The plasmid was transformed in the wild-type and ΔeccB4 strains.
Bacterial cultures were recovered at an OD600 of 0.8, resuspended in PBS containing PMSF and 1 mM EDTA (1 mL/g), sonicated, and subjected to fractionation. After a first centrifugation at 1,000 × g to eliminate nonlysed bacteria or debris, supernatants were centrifugated at 15,000 × g for 30 min to obtain the cell wall fraction. The supernatant was then submitted to ultracentrifugation at 100,000 × g for 3 h to separate the plasma membranes from the cytosolic compartment. Equal protein amounts were subjected to SDS/PAGE and immunoblotting and were revealed using mice anti-His antibodies conjugated with peroxidase (Sigma-Aldrich) (SI Appendix, Fig. S5).
Secretome Analysis.
M. abscessus CIP and ΔeccB4 strains were grown in 7H9 medium to OD600 = 0.8 (four biological duplicates). Then cells were diluted with Sauton medium plus glucose (1%), the appropriate antibiotics (see Strains, Cells, and Culture Conditions, above), and 0.05% Tween 80 to an OD600 of 0.1. The diluted cells were incubated until OD600 = 0.6–0.7 was reached. Then bacteria were centrifuged, washed three times with cold PBS 1×, diluted again Sauton’s medium plus glucose (1%) and antibiotics, and incubated to an OD600 of 0.8–1. Culture supernatants then were concentrated 100× with Amicon Ultra centrifugal filters (3-kDa cutoff), and the protein concentration was measured with the Pierce protein BCA assay. Briefly, secretomes were diluted with ammonium bicarbonate and incubated for 4 h with 0.2 µg trypsin/LysC (Promega) at 37 °C. Samples then were loaded onto a homemade C18 Sep-Pak–packed stage tip for desalting. Samples were eluted using 40% ACN/0.1% formic acid and were vacuum concentrated to dryness. Desalted samples were reconstituted in 2% ACN/0.3% TFA that contained iRT peptides (Biognosys) for quality control. Samples were then analyzed by nanoLC-MS/MS for protein identification. Spectra were recorded on a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific) and analyzed with Sequest through Proteome Discoverer 2.0 using the UniProt M. abscessus UP000038470 and UP000007137 databases. All data were further processed using myProMS software (SI Appendix, Fig. S5 and Tables S3 and S4) (62).
Lipid Extractions and Analysis by TLC.
For lipid analyses M. abscessus was grown in 7H9 broth, collected by centrifugation, and subjected to lyophilization. Apolar and polar lipid fractions or methyl esters of fatty acids (FAME) and mycolic acids (MAME) were obtained from 50 mg of freeze-dried mycobacterial cells as previously described (63). Lipid fractions were applied to aluminum TLC silica gel 60 F254 plates (Merck) and were separated using solvent systems as described in the text. After solvent separation, TLCs with apolar lipids and FAME and MAME were treated with a mist of 5% phosphomolybdic acid (dissolved in ethanol) and charred by heating to reveal lipid spots or bands. TLCs with polar lipids were treated with a mist of 0.1% orcinol (dissolved in 20% H2SO4) before charring (SI Appendix, Fig. S4).
RNA Extraction and RT-PCR Analysis.
Bacterial RNA was extracted from various M. abscessus strains as described previously (10). Briefly, total RNA was extracted using TRIzol reagent and a bead beater. PCR was performed with a CFX96 thermal cycler (Bio-Rad). The primer sequences used for qRT-PCR are listed in SI Appendix, Table S5. Controls without reverse transcriptase were done on each RNA sample to rule out DNA contamination. The sigA gene was used as an internal control (SI Appendix, Table S2) (10).
Supplementary Material
Acknowledgments
We thank G. Arras for bioinformatic analysis of proteomic data and F. Dingli for help during sample preparation for mass spectrometry, F. Le Chevalier for his advice on proteomics, and B. Maury for confocal microscopy analysis. This work was supported by the French Cystic Fibrosis Patients Association Vaincre la Mucoviscidose Grant RF20150501377, French Research National Agency Program DIMIVYR Grant ANR-13-BSV3-0007-01(to J.-L.H. and L.K.), and Fondation pour la Recherche Médicale Grant DEQ20150331719 (to L.K.). The Région Ile-de-France (Domaine d’Intérêt Majeur Maladies Infectieuses et Emergentes) funded postdoctoral fellowships (to V.L.M.) and for mass spectrometry analysis (to D.L.). L.L. is a doctoral fellow of the Ministère de l’Enseignement Supérieur et de la Recherche. This work was presented at the American Society for Microbiology Conference on Tuberculosis: Past, Present and Future (American Society for Microbiology, New York, April 2017).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.A.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713195115/-/DCSupplemental.
References
- 1.Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boritsch EC, et al. A glimpse into the past and predictions for the future: The molecular evolution of the tuberculosis agent. Mol Microbiol. 2014;93:835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
- 3.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corsaro D, Michel R, Walochnik J, Müller K-D, Greub G. Saccamoeba lacustris, sp. nov. (Amoebozoa: Lobosea: Hartmannellidae), a new lobose amoeba, parasitized by the novel chlamydia ‘Candidatus Metachlamydia lacustris’ (Chlamydiae: Parachlamydiaceae) Eur J Protistol. 2010;46:86–95. doi: 10.1016/j.ejop.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Fritsche TR, Gautom RK, Seyedirashti S, Bergeron DL, Lindquist TD. Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol. 1993;31:1122–1126. doi: 10.1128/jcm.31.5.1122-1126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croxatto A, Murset V, Chassot B, Greub G. Early expression of the type III secretion system of Parachlamydia acanthamoebae during a replicative cycle within its natural host cell Acanthamoeba castellanii. Pathog Dis. 2013;69:159–175. doi: 10.1111/2049-632X.12065. [DOI] [PubMed] [Google Scholar]
- 7.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weerdenburg EM, et al. Genome-wide transposon mutagenesis indicates that Mycobacterium marinum customizes its virulence mechanisms for survival and replication in different hosts. Infect Immun. 2015;83:1778–1788. doi: 10.1128/IAI.03050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakala N’Goma JC, et al. Mycobacterium abscessus phospholipase C expression is induced during coculture within amoebae and enhances M. abscessus virulence in mice. Infect Immun. 2015;83:780–791. doi: 10.1128/IAI.02032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace RJ, Jr, Brown BA, Griffith DE. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu Rev Microbiol. 1998;52:453–490. doi: 10.1146/annurev.micro.52.1.453. [DOI] [PubMed] [Google Scholar]
- 12.Duarte RS, et al. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol. 2009;47:2149–2155. doi: 10.1128/JCM.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leão SC, et al. Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil. Future Microbiol. 2010;5:971–980. doi: 10.2217/fmb.10.49. [DOI] [PubMed] [Google Scholar]
- 14.Olivier KN, et al. Nontuberculous Mycobacteria in Cystic Fibrosis Study Group Nontuberculous mycobacteria. I: Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 15.Roux A-L, et al. Jean-Louis Herrmann for the OMA Group Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol. 2009;47:4124–4128. doi: 10.1128/JCM.01257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith DE, Girard WM, Wallace RJ., Jr Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis. 1993;147:1271–1278. doi: 10.1164/ajrccm/147.5.1271. [DOI] [PubMed] [Google Scholar]
- 17.Qvist T, et al. Comparing the harmful effects of nontuberculous mycobacteria and gram negative bacteria on lung function in patients with cystic fibrosis. J Cyst Fibros. 2016;15:380–385. doi: 10.1016/j.jcf.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jönsson BE, et al. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol. 2007;45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant JM, et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous Mycobacterium. Science. 2016;354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullen AR, Cannon CL, Mark EJ, Colin AA. Mycobacterium abscessus infection in cystic fibrosis. Colonization or infection? Am J Respir Crit Care Med. 2000;161:641–645. doi: 10.1164/ajrccm.161.2.9903062. [DOI] [PubMed] [Google Scholar]
- 21.Kreutzfeldt KM, et al. Molecular longitudinal tracking of Mycobacterium abscessus spp. during chronic infection of the human lung. PLoS One. 2013;8:e63237. doi: 10.1371/journal.pone.0063237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catherinot E, et al. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J Clin Microbiol. 2009;47:271–274. doi: 10.1128/JCM.01478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernut A, et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci USA. 2014;111:E943–E952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawlik A, et al. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol. 2013;90:612–629. doi: 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 25.Bernut A, et al. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol Microbiol. 2016;99:866–883. doi: 10.1111/mmi.13283. [DOI] [PubMed] [Google Scholar]
- 26.Catherinot E, et al. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun. 2007;75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernut A, Herrmann J-L, Ordway D, Kremer L. The diverse cellular and animal models to decipher the physiopathological traits of Mycobacterium abscessus infection. Front Cell Infect Microbiol. 2017 doi: 10.3389/fcimb.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux A-L, et al. Overexpression of proinflammatory TLR-2-signalling lipoproteins in hypervirulent mycobacterial variants. Cell Microbiol. 2011;13:692–704. doi: 10.1111/j.1462-5822.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- 29.Ripoll F, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falkinham JO, 3rd, Hilborn ED, Arduino MJ, Pruden A, Edwards MA. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ Health Perspect. 2015;123:749–758. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cateau E, et al. Stenotrophomonas maltophilia and Vermamoeba vermiformis relationships: Bacterial multiplication and protection in amoebal-derived structures. Res Microbiol. 2014;165:847–851. doi: 10.1016/j.resmic.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Le Moigne V, et al. MgtC as a host-induced factor and vaccine candidate against Mycobacterium abscessus infection. Infect Immun. 2016;84:2895–2903. doi: 10.1128/IAI.00359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubin EJ, et al. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux A-L, et al. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 2016;6:160185. doi: 10.1098/rsob.160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumas E, et al. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol Evol. 2016;8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The enigmatic Esx proteins: Looking beyond mycobacteria. Trends Microbiol. 2017;25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M, et al. EspI regulates the ESX-1 secretion system in response to ATP levels in Mycobacterium tuberculosis. Mol Microbiol. 2014;93:1057–1065. doi: 10.1111/mmi.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman S, et al. Mycobacterium tuberculosis SigM positively regulates Esx secreted protein and nonribosomal peptide synthetase genes and down regulates virulence-associated surface lipid synthesis. J Bacteriol. 2006;188:8460–8468. doi: 10.1128/JB.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturgill-Koszycki S, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 41.Simeone R, Bottai D, Frigui W, Majlessi L, Brosch R. ESX/type VII secretion systems of mycobacteria: Insights into evolution, pathogenicity and protection. Tuberculosis (Edinb) 2015;95:S150–S154. doi: 10.1016/j.tube.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Simeone R, et al. Cytosolic access of Mycobacterium tuberculosis: Critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 2015;11:e1004650. doi: 10.1371/journal.ppat.1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soroka D, et al. Characterization of broad-spectrum Mycobacterium abscessus class A β-lactamase. J Antimicrob Chemother. 2014;69:691–696. doi: 10.1093/jac/dkt410. [DOI] [PubMed] [Google Scholar]
- 44.Augenstreich J, et al. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017;19:e12726. doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- 45.Majlessi L, Brosch R. Mycobacterium tuberculosis meets the cytosol: The role of cGAS in anti-mycobacterial immunity. Cell Host Microbe. 2015;17:733–735. doi: 10.1016/j.chom.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Wassermann R, et al. Mycobacterium tuberculosis differentially Activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe. 2015;17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Gröschel MI, et al. Recombinant BCG expressing ESX-1 of Mycobacterium marinum combines low virulence with cytosolic immune signaling and improved TB protection. Cell Rep. 2017;18:2752–2765. doi: 10.1016/j.celrep.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 48.Adékambi T, Ben Salah S, Khlif M, Raoult D, Drancourt M. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl Environ Microbiol. 2006;72:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halloum I, et al. Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent. Proc Natl Acad Sci USA. 2016;113:E4228–E4237. doi: 10.1073/pnas.1605477113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paz I, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 51.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim YS, et al. Mycobacterium abscessus ESX-3 plays an important role in host inflammatory and pathological responses during infection. Microbes Infect. 2017;19:5–17. doi: 10.1016/j.micinf.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Gray TA, et al. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science. 2016;354:347–350. doi: 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houben ENG, et al. Composition of the type VII secretion system membrane complex. Mol Microbiol. 2012;86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 56.Abdallah AM, et al. Type VII secretion–Mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 57.Beckham KSH, et al. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol. 2017;2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- 58.Abdallah AM, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 59.Singh VK, et al. A unique PE_PGRS protein inhibiting host cell cytosolic defenses and sustaining full virulence of Mycobacterium marinum in multiple hosts. Cell Microbiol. 2016;18:1489–1507. doi: 10.1111/cmi.12606. [DOI] [PubMed] [Google Scholar]
- 60.Tanne A, et al. A murine DC-SIGN homologue contributes to early host defense against Mycobacterium tuberculosis. J Exp Med. 2009;206:2205–2220. doi: 10.1084/jem.20090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medjahed H, Singh AK. Genetic manipulation of Mycobacterium abscessus. Curr Protoc Microbiol. 2010;Chapter 10:Unit 10D.2. doi: 10.1002/9780471729259.mc10d02s18. [DOI] [PubMed] [Google Scholar]
- 62.Poullet P, Carpentier S, Barillot E. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics. 2007;7:2553–2556. doi: 10.1002/pmic.200600784. [DOI] [PubMed] [Google Scholar]
- 63.Besra GS. Mycobacteria Protocols. Humana; Totowa, NJ: 1998. Preparation of cell-wall fractions from mycobacteria; pp. 91–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.