Significance
Bardet–Biedl syndrome (BBS) is a rare disease caused by dysfunctional cilia. In bbs mutants, the composition of the ciliary membrane is altered due to defects in the BBSome, a conserved complex of BBS proteins. To determine the molecular function of the BBSome, we used single particle in vivo imaging. Transport of the ciliary membrane protein phospholipase D (PLD) is BBSome-dependent, and PLD comigrates with BBSomes on intraflagellar transport (IFT) trains. PLD accumulates inside cilia after removal of its ciliary export sequence (CES) or in the absence of BBSomes. In conclusion, the BBSome participates directly in ciliary protein transport by serving as an adapter allowing proteins that alone are unable to bind to IFT to be exported from cilia on IFT trains.
Keywords: BBSome, cilia, flagella, ciliopathy, phospholipase D
Abstract
Bardet–Biedl syndrome (BBS) is a ciliopathy resulting from defects in the BBSome, a conserved protein complex. BBSome mutations affect ciliary membrane composition, impairing cilia-based signaling. The mechanism by which the BBSome regulates ciliary membrane content remains unknown. Chlamydomonas bbs mutants lack phototaxis and accumulate phospholipase D (PLD) in the ciliary membrane. Single particle imaging revealed that PLD comigrates with BBS4 by intraflagellar transport (IFT) while IFT of PLD is abolished in bbs mutants. BBSome deficiency did not alter the rate of PLD entry into cilia. Membrane association and the N-terminal 58 residues of PLD are sufficient and necessary for BBSome-dependent transport and ciliary export. The replacement of PLD’s ciliary export sequence (CES) caused PLD to accumulate in cilia of cells with intact BBSomes and IFT. The buildup of PLD inside cilia impaired phototaxis, revealing that PLD is a negative regulator of phototactic behavior. We conclude that the BBSome is a cargo adapter ensuring ciliary export of PLD on IFT trains to regulate phototaxis.
Bardet–Biedl syndrome (BBS) is an inherited cilia-related disorder characterized by a multiorgan phenotype including blindness and obesity (1). The condition results from defects in the assembly, composition, or localization of the BBSome, a conserved eight-subunit protein particle (2). Cilia of bbs mutants over a broad range of species display loss and/or anomalous accumulation of proteins, particularly ciliary membrane proteins (3–11). Examples are the G protein-coupled receptors (GPCRs) somatostatin receptor 3 (Sstr3) and the melanin-concentrating hormone receptor 1 (Mchr1), which are lost from neuronal cilia of Bbs−/− mice and the absence of certain ion channels from cilia of Paramecium bbs RNAi strains (6, 8). In contrast, the dopamine receptor 1 (D1) fails to undergo stimulated exit from neuronal cilia in Bbs−/− mutant mice, and nonouter segment proteins progressively accumulate in the cilia-derived outer segments of rod cells in Bbs−/− mice (7, 10). Thus, BBS or at least certain features of BBS result from improper ciliary signaling due to biochemical defects of the ciliary membrane. The precise molecular activity by which the BBSome influences the protein content of the ciliary membrane and signaling fidelity remains to be determined.
The BBSome cycles through cilia on intraflagellar transport (IFT) trains, multimegadalton protein carriers that move by molecular motors bidirectionally along the axonemal microtubules (4, 12, 13). In contrast to IFT, the BBSome is expendable for ciliary assembly in most systems (3, 4, 13, 14). It has been suggested that the BBSome assists protein transport in and out of cilia by connecting proteins possessing appropriate sorting motifs to IFT (4, 15, 16). However, direct evidence for BBSome-dependent IFT of proteins, as well as the sequence motifs allowing for protein binding to IFT trains in a BBSome-dependent manner, has not been established. BBS proteins are also concentrated at the ciliary base, and changes in protein entry or protein retention in cilia provide an alternative explanation for the observed changes in ciliary protein composition (7, 17, 18). Finally, BBS proteins have been implicated in vesicular traffic to and from the plasma membrane (2, 19–21). Understanding BBSome function could profit from direct monitoring of BBSome-dependent intracellular transport.
In Chlamydomonas reinhardtii, mutations in the BBSome impair phototaxis and cause biochemical defects of the ciliary membrane (4, 22). Phospholipase D (PLD) and an AMP-regulated kinase (AMPK) (XP_001697837, referred to as STPK in ref. 4) accumulate in bbs cilia while the amount of carbonic anhydrase 6 (CAH6) is reduced (5). All three proteins are predicted to be dual fatty acid modified at the N terminus. The ciliary membrane of bbs4 mutants is enriched in phosphatidic acid and diacylglycerol (DAG), indicating increased PLD activity (5). It remains unknown whether maldistribution of PLD contributes to the loss of Chlamydomonas phototaxis in bbs mutants. PLD accumulation in bbs cilia requires hours to reach maximal levels while the protein is removed within minutes from mutant-derived cilia after reintroducing functional BBSomes (5). PLD also becomes trapped in cilia of cells with intact BBSomes when retrograde IFT is defective or IFT is switched off entirely. Thus, PLD can enter cilia in an IFT-independent manner, and the BBSome’s role in avoiding the ciliary buildup of PLD depends on active IFT.
Here, we explored how PLD interacts with the IFT/BBS pathway using in vivo imaging. PLD-mNeonGreen (mNG) moved by IFT in control cilia and comigrated with BBS4 on IFT trains whereas IFT of PLD-mNG was abolished in bbs mutants. Thus, the BBSome functions as a cargo adapter allowing PLD to move by IFT. We determined that the N-terminal 58 residues of PLD are sufficient and necessary for BBSome-dependent transport by IFT. PLD lacking this ciliary export sequence (CES) accumulates in cilia of otherwise WT cells, interfering with their phototactic behavior. We conclude that BBSome-dependent IFT regulates the ciliary level of PLD, a negative regulator of C. reinhardtii phototaxis.
Results
IFT of PLD Is BBSome-Dependent.
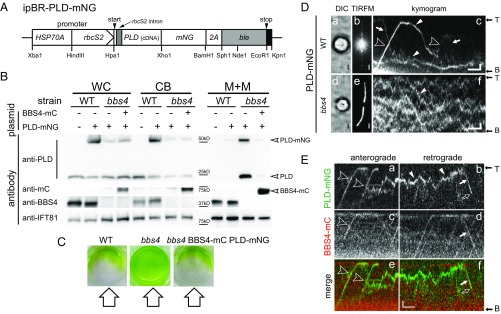
C. reinhardtii bbs mutants accumulate phospholipase D (PLD) in the ciliary membrane (5). To explore how PLD accumulates in bbs cilia, we fused mNeonGreen (mNG) to the C terminus of PLD, preserving the potential acylation sites in its N-terminal region (Fig. 1A). Transformants were analyzed by Western blotting for the expression and distribution of PLD-mNG (Fig. 1B). Since PLD and PLD-mNG are essentially absent from the axonemal fraction, detergent extracts or isolated ciliary membrane was used in most Western blots, allowing us to detect PLD and its derivates in WT-derived ciliary samples, which contain only traces of PLD (Fig. S1A). In control cells, PLD-mNG and endogenous PLD were primarily (>99% of the total protein) present in the cell body (Fig. 1B) (5). In contrast, a substantial amount (∼30%) of the total cellular PLD-mNG was contained in the detergent-soluble fraction of bbs4 cilia, as previously reported for the endogenous PLD (Figs. 1B and 2D). Expression of BBS4-mCherry (BBS4-mC) restored the WT distributions for the endogenous and tagged PLD and rescued the nonphototactic phenotype of the bbs4 PLD-mNG strain (Fig. 1 B and C). PLD-mNG recapitulates the subcellular distribution of endogenous PLD and thus can serve as an in vivo reporter to determine how BBSome deficiency causes a buildup of PLD inside cilia.
Fig. 1.
IFT of PLD is BBSome-dependent. (A) Schematic presentation of the PLD expression vector ipBR-PLD-mNG. The selectable marker ble was separated from PLD and mNG by a viral 2A sequence encoding a self-cleaving peptide. (B) Western blot analysis comparing whole cells (WC), cell bodies (CB), and the ciliary membrane+matrix (M+M) fractions of control (WT), bbs4, and bbs4 rescued by BBS4-mC. Strains expressing PLD-mNG are marked (+). The membranes were stained with antibodies to PLD, BBS4, mC, and, as a control, IFT81, as indicated. For the M+M fraction, an equivalent of ∼100 cilia per cell body was loaded (50×). (C) Phototaxis assay of control (WT, CC-620), bbs4, and bbs4 BBS4-mC PLD-mNG. The direction of the light is indicated. (D) Brightfield (a and d), TIRF images (b and e), and corresponding kymograms (c and f) of a control (WT) and bbs4 cell expressing PLD-mNG. In kymograms, anterograde transports (from the ciliary base to the tip) result in diagonal trajectories from the bottom left to the top right (open arrowheads); trajectories of retrograde transports run from the top left to the bottom right (arrows). Diffusing PLD-mNG (white arrowheads) and the flagellar base (B) and tip (T) are marked. (Scale bars: 2 s and 1 µm.) (E) Kymograms from two-color imaging of PLD-mNG (a and b; green) and BBS4-mCherry (c and d; red); merged kymograms are shown in e and f. Cotransport transports are marked (open arrowheads for anterograde IFT and white arrows for retrograde IFT). White arrowheads in b: diffusion of PLD-mNG; open arrows in b and f: photobleaching of PLD-mNG. Note that PLD-mNG signals mostly bleached in one step indicative for a single mNG. Fig. 1 E, b, d, and f corresponds to Movie S2. (Scale bars: 2 s and 1 μm.)
Fig. 2.
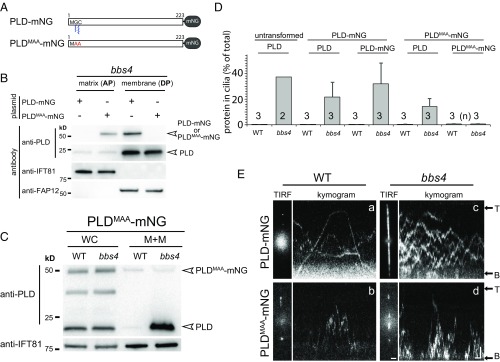
Membrane association of PLD is required for IFT/BBS transport. (A) Schematic presentation of PLD-mNG and PLDMAA-mNG. (B) Western blot analysis of the matrix (AP) and membrane (DP) fractions obtained from isolated bbs4 cilia. The antibodies used for Western blotting are indicated; the matrix protein IFT81 and membrane-associated protein FAP12 were used as loading controls. (C) Western blot analysis comparing the distribution of PLDMAA-mNG and endogenous PLD in whole cell (WC) and the membrane+matrix (M+M) fraction of isolated cilia; loading of M+M fraction was 40×. (D) Cellular distribution of PLD-mNG and PLDMAA-mNG in control (WT) and bbs4 cells. Shown is the share (in percent) of the total protein present in the ciliary membrane+matrix fraction. With exception of the amount of endogenous PLD in bbs4 cilia (n = 2), the data are based on three biological repeats. The SDs are indicated. (E) Still images and kymograms depicting control (WT) and bbs4 cells expressing PLD-mNG (Top) or PLDMAA-mNG (Bottom). The flagellar base (B) and tip (T) are marked. (Scale bars: 1 s and 2 µm.)
In agreement with the Western blotting data, total internal reflection fluorescence (TIRF) imaging of live cells showed PLD-mNG to be abundant in bbs4 cilia while only traces were detected in control cilia (Fig. 1 D, a, b, d, and e). In the latter, PLD-mNG moved bidirectionally by IFT in addition to periods of diffusion; ∼60% of all PLD-mNG transports (i.e., movement by IFT) were retrograde (Fig. 1 D, c, Table 1, and Movie S1). The velocities of PLD transports (2.1 μm/s, SD 0.22 μm/s, n = 88 and 3.2 μm/s, SD 0.54 μm/s, n = 46 for anterograde and retrograde, respectively) were typical for IFT in C. reinhardtii. Despite the high abundance in bbs4 cilia, PLD-mNG moved mostly by diffusion in bbs4 cilia, and IFT of PLD was rarely observed (Fig. 1 D, f) (0.07 transports per minute compared with 1.16 transports per minute in control cells). IFT of PLD-mNG was not observed in cilia of the bbs1, bbs7, and bbs8 mutants (Fig. S1B). A residual BBS complex present in Bbs4 mutant cells could explain the sporadic transports of PLD-mNG (11).
Table 1.
Transport of PLD-mNG
| Strain | Cilia analyzed | Transports | Frequency (transports/min) | Anterograde, % | Retrograde, % | “Entries”, % | “Exits”, % |
| PLD-mNG | 126 | 146 | 1.16 | 36 (n = 53) | 64 (n = 93) | 17 | 30 |
| bbs4 BBS4-mC PLD-mNG | 196 | 204 | 1.04 | 41 (n = 83) | 59 (n = 121) | 14 | 27 |
Transport characteristics of PLD-mNG in a control strain and the bbs4 BBS4-mC rescue strain. Entries refers to trajectories originating near the ciliary base. Exits refers to those advancing to the ciliary base.
Two-color imaging showed cotransport of PLD-mNG and BBS4-mC by IFT (Fig. 1E and Movie S2). Anterograde transport of BBS4-mC occurred with a frequency of 9.3 transports per minute in our PLD-mNG rescue strain, suggesting that only approximately one-sixth of IFT trains carry a BBSome (∼60 anterograde IFT trains per minute in C. reinhardtii) (23). Despite the scarcity of IFT trains with BBSomes, comigration of PLD-mNG and BBS4-mC was observed for 180 of 204 analyzed anterograde and retrograde transports; in the remaining transports, we did not detect a BBS4-mC signal comigrating with PLD-mNG. The signal intensity and photostability of mC is significantly below that of mNG, and a failure to detect small amounts of BBS4-mC or bleaching of BBS4-mC provides a likely explanation for the apparent lack of a BBS4-mC signal in a subset of PLD-mNG transports (24).
PLD-mNG diffusing inside cilia was picked-up by IFT-bound BBSomes at variable sites along the cilium (Fig. 1E, Fig. S2A, and Movie S2). Such loading events were more abundant in the proximal portion of the cilium (Fig. S1C). This distribution could be explained by rapid loading of PLD-mNG entering the cilia by diffusion onto IFT/BBS trains. While overall sparse, many anterograde PLD-mNG transports were immediately preceded by retrograde transports, indicating stable association or rapid reloading of PLD onto IFT/BBS trains (Figs. 1D and 2E and Fig. S2A). The data suggest that BBSomes cycling via IFT inside cilia are constitutively able to bind PLD with high affinity. A subset of the anterograde PLD-mNG transports originated at the base of the cilium, and a subset of the retrograde transports advanced to the ciliary base via IFT (Fig. S2B and Table 1). Due to the prominent red autofluorescence of the plastid, tracking of BBSomes near the base of the cilium was not achieved. However, PLD-mNG was observed moving with IFT velocities to and from the very base of the cilium, suggesting that a portion of PLD enters and exits cilia in association with IFT. We conclude that PLD transport by IFT is BBSome-dependent and is carried out at least predominately by IFT trains with BBSomes.
Membrane Association of PLD Is a Prerequisite for Transport by the IFT/BBS System.
Studies in a variety of species revealed that BBSome defects affect the distribution of selected ciliary transmembrane proteins. PLD, however, lacks a transmembrane domain but is predicted to be myristoylated on glycine2 and palmitoylated on cysteine3, likely explaining its membrane association (5). Dual acylation has been implicated in ciliary targeting of various proteins (25). To test the importance of membrane anchoring for BBSome-dependent transport of PLD, we substituted N-terminal MGC motif with MAA; we refer to this construct as PLDMAA-mNG (Fig. 2A). Western blotting of fractionated bbs4 cilia revealed that PLDMAA-mNG was present in the aqueous fraction (AP) of Triton X-114 phase partitioning corresponding to the ciliary matrix (Fig. 2B). In contrast, PLD-mNG and endogenous PLD were largely confined to the detergent-soluble fraction (DP) corresponding to the ciliary membrane. Next, we compared the amount of PLDMAA-mNG inside WT and bbs4 cilia obtained from strains expressing similar amounts of the transgene at the whole cell level (Fig. 2C). In contrast to the endogenous PLD, PLDMAA-mNG was present in similar amounts in bbs4 and control cilia, emphasizing that the BBSome-based regulation of its ciliary level was lost (Fig. 2 C and D). Direct imaging of PLDMAA-mNG inside cilia revealed an increased diffusional mobility in comparison with PLD-mNG (Fig. 2E and Movie S3). Dual fatty acid modification likely causes a drag, reducing the mobility of PLD-mNG. The accelerated mobility and the altered solubility support the notion that PLDMAA-mNG has lost its membrane anchor. IFT of PLDMAA-mNG was observed neither in bbs4 nor in WT cilia. We conclude that membrane anchoring of PLD via acylation on its N-terminal MGC motif is required for transport by the IFT/BBS system.
The N-Terminal Region of PLD Is Sufficient for Transport by the IFT/BBS System.
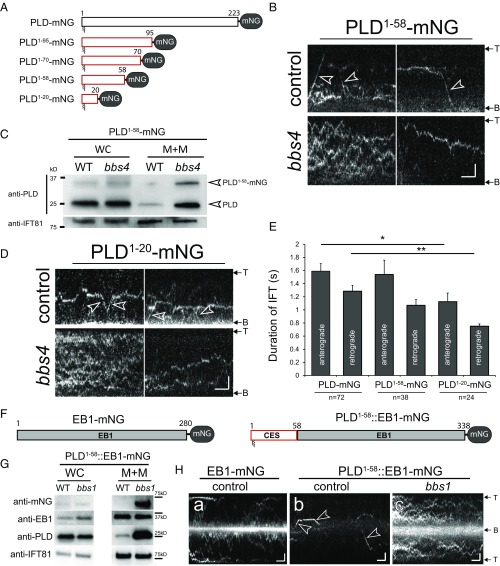
The ciliary membrane of C. reinhardtii is enriched in proteins predicted to undergo myristoylation and palmitoylation on the N-terminal domain (26). However, only a small subset of these proteins (i.e., PLD and AMPK) accumulate in bbs mutant cilia, indicating that membrane association by dual acylation is a prerequisite but not sufficient for BBSome-dependent transport. We performed a series of C-terminal truncations to identify the minimal segment of PLD required for BBSome-dependent transport (Fig. 3A and Table 2). The N-terminal 58 residues of PLD (PLD1-58-mNG) were sufficient for both transport by the IFT/BBS system in WT cilia and accumulation in bbs cilia (Fig. 3 B and C). An even shorter construct, encompassing the N-terminal 20 residues of PLD (PLD1-20-mNG) still accumulated in bbs4 cilia and moved by IFT in control cilia (Fig. 3D). However, the frequency and processivity of PLD1-20-mNG transports were significantly reduced, indicative for a weakened interaction of PLD1-20-mNG with the IFT/BBS system (Fig. 3E and Table 2). Next, we fused PLD1-58 to the N terminus of the microtubule plus-tip end binding protein 1 (EB1), which enters and moves inside cilia in an IFT-independent manner (Fig. 3 F–H) (27). Diffusion of the PLD-EB1 fusion was considerably slower than that of EB1-mNG, suggesting that PLD1-58 anchors EB1-mNG to the ciliary membrane (Fig. 3H, a compared with b and c). In WT cilia, PLD1-58::EB1-mNG frequently moved by IFT and was of low abundance (Fig. 3 G and H, b). In contrast, PLD1-58::EB1-mNG accumulated in bbs1 cilia, and transport by IFT was not observed; the distribution of the endogenous EB1 was unaffected (Fig. 3 G and H, c). We conclude that PLD1-58 is sufficient for BBSome-dependent IFT and mediates BBSome-dependent ciliary export of fused proteins.
Fig. 3.
The N-terminal 58 residues of PLD are sufficient for BBSome-dependent transport. (A) Schematic presentation of full-length and truncated PLD-mNGs. (B) Kymograms of PLD1-58-mNG in control and bbs4 mutant cilia. Arrowheads: transports. (Scale bars: 2 s and 2 µm.) (C) Western blot analysis of whole cell (WC) and ciliary membrane+matrix (M+M) fractions of control (WT) and bbs4 cells showing the accumulation of PLD1-58-mNG in bbs4 cilia. (D) Kymograms of PLD1-20-mNG in control and bbs4 mutant cilia. Arrowheads: transports. (Scale bars: 2 s and 2 μm.) (E) Bar diagram showing the duration of transports of PLD-mNG, PLD1-58-mNG, and PLD1-20-mNG. Error bars indicate SD. The significance based on a two-tailed t test is indicated (*P ≤ 0.05; **P ≤ 0.01). n, number of cilia analyzed. (F) Schematic presentation of EB1-mNG and PLD1-58::EB1-mNG. CES, ciliary export sequence. (G) Western blot analysis of whole cell and ciliary membrane+matrix (M+M) fractions showing the distribution of PLD, endogenous EB1, and PLD1-58::EB1-mNG in WT and bbs1 cells. (H) Kymograms of EB1-mNG in control cilia and of PLD1-58::EB1-mNG in control and bbs1 cilia. In contrast to EB1-mNG, PLD1-58::EB1-mNG moved by IFT in control cilia (arrowheads); IFT was not observed in the bbs1 cilia. (Scale bars: 2 s and 2 μm.)
Table 2.
Transport of PLD-mNG and the truncated derivates
| Strain | Cilia analyzed | Transports | Frequency, transports per minute |
| PLD-mNG | 126 | 146 | 1.16 (SD 0.84) |
| PLD1-95-mNG | 20 | 19 | 0.96 (SD 0.68) |
| PLD1-70-mNG | 14 | 14 | 1.03 (SD 0.73) |
| PLD1-58-mNG | 68 | 75 | 1.1 (SD 0.83) |
| PLD1-20-mNG | 60 | 18 | 0.3 (SD 0.53) |
BBSome Loss Does Not Affect Ciliary Entry of PLD.
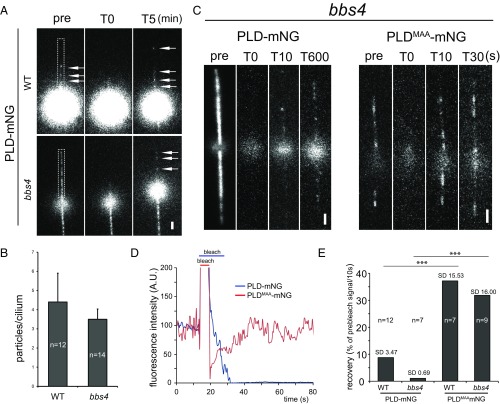
Our data showing that bbs mutants fail to move PLD by IFT do not exclude the possibility that an increased influx of PLD contributes to its accumulation in bbs cilia. BBS proteins are concentrated near the ciliary base, a region thought to regulate protein access to the ciliary compartment (4, 17, 28). The possibility to directly observe PLD-mNG allowed us to determine its rate of entry into control and bbs4 cilia by fluorescence recovery after photobleaching (FRAP) analysis. Before photobleaching, the amount of PLD-mNG present in bbs4 cilia strongly exceeded that of control cilia (Fig. 4A). Postbleach recovery of the PLD-mNG signal progressed at similarly low rates in both strains, with an average of one PLD-mNG particle entering the cilia per minute (Fig. 4 A, B, and E). This is in agreement with earlier data showing that the accumulation of endogenous PLD inside bbs4 cilia is a slow process requiring several hours to reach maximum levels (5). We also analyzed ciliary entry of PLDMAA-mNG, which, due to the loss of its membrane anchor and the lack of transport by IFT, should diffuse freely in and out of cilia, as previously shown for other small soluble proteins (27, 29). Indeed, the signal of PLDMAA-mNG in control and bbs4 cilia recovered quickly to prebleach intensity after photobleaching (∼30 s) (Fig. 4 C–E and Movie S4). The data suggest that the slow entry of PLD into cilia is caused by its membrane anchor rather than its size or other properties. We conclude that the accumulation of PLD in bbs cilia results from a failure to export the protein via IFT rather than from an increased influx of PLD into bbs cilia.
Fig. 4.
PLD entry into cilia is unaffected in bbs mutants. (A) FRAP analysis of PLD-mNG in control (WT) and bbs4 cilia. Shown are single frames before (pre) and after (T0) bleaching of one cilium (indicated by a white box) and after 5 min of recovery. Arrows, PLD-mNG particles. (Scale bar: 2 μm.) (B) The number of PLD-mNG particles in control (WT) and bbs4 cilia after 5-min recovery. (C) FRAP analysis of PLD-mNG and PLDMAA-mNG in bbs4 cilia. Shown are single frames before and after (T0) bleaching of the cilia and at different time points during recovery. Note different time scales. (Scale bars: 2 μm.) (D) Signal recovery after photobleaching for PLD-mNG (blue) and PLDMAA-mNG (red) in bbs4 cilia. The prebleach fluorescence intensity was set to 100. (E) Comparison of FRAP rates for PLD-mNG and PLDMAA-mNG in control (WT) and bbs4 cilia. Data are presented as percentage of the prebleach intensity recovered over 10 s, as calculated based on the postbleach recovery of the signal over a period of 20 to 100 s. The significance based on a two-tailed t test is indicated (***P ≤ 0.001). n, number of cilia.
The N-Terminal Region of PLD Is Required for Transport by the IFT/BBS System.
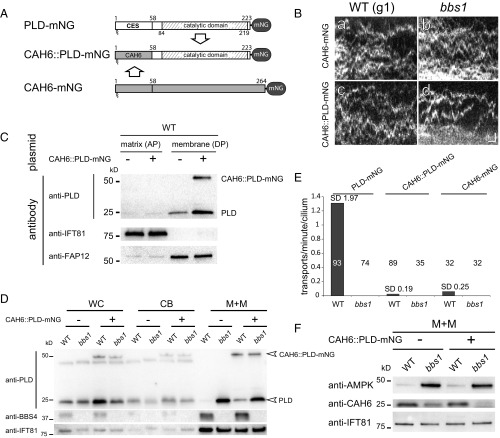
To test whether the N-terminal 58 residues of PLD are also necessary for the BBSome-dependent transport of PLD, we replaced the head domain of PLD with the corresponding N-terminal 58 residues of carbonic anhydrase 6 (CAH6) (Fig. 5A). CAH6 is an abundant membrane-associated ciliary protein predicted to be dual fatty acid-modified on its N-terminal MGC motif. CAH6 is not the ideal choice for this experiment because its levels are reduced in bbs4 flagella (5). However, transformants expressing CAH61-58PLD59-223-mNG (CAH6::PLD-mNG) were obtained while strains expressing fusions between PLD and the head domains of the ciliary dual acylated proteins FAP12 or ODA5-associated adenylate kinase were not detected. Importantly, full-length CAH6-mNG moved predominantly by diffusion in control and bbs cilia, indicating that CAH6 lacks strong binding sites for IFT and the BBSome, allowing us to test PLD for additional IFT/BBS binding sites (Fig. 5 B, a and E).
Fig. 5.
PLD1-58 are necessary for BBSome-dependent ciliary export of PLD. (A) Schematic presentation of the PLD-mNG, CAH6::PLD-mNG, and CAH6-mNG fusion proteins. The ciliary export sequence (CES) and catalytic domain of PLD are indicated. (B) Kymograms (a–d) of control (WT) and bbs1 cells expressing CAH6-mNG (a and b) and or CAH6::PLD-mNG (c and d). (Scale bars: a–d, 1 s and 1 μm) (C) Western blot confirming the presence of CAH6::PLD-mNG in the ciliary membrane fraction (DP); antibodies to IFT81 and FAP12 were used as markers for the aqueous phase (AP) and detergent phase (DP), respectively. (D) Western blot analysis of whole cell (WC), cell body (CB), and the membrane+matrix (M+M) fractions of transformed and untransformed control (WT) and bbs1 cells comparing the distribution of endogenous PLD and CAH6::PLD-mNG. The membrane+matrix fraction of WT and bbs1 cilia contained 15% and 18%, respectively, of the total cellular CAH6::PLD-mNG (n = 1 experiment). (E) Comparison of the IFT frequencies (in transports per minute per cilium) for PLD-mNG, CAH6-mNG, and CAH6::PLD-mNG in control (WT) and bbs1 cells. The number of cilia analyzed is indicated. (F) Western blot analysis of the membrane+matrix (M+M) fractions of transformed and untransformed control (WT) and bbs1 cilia stained with antibodies to CAH6 and AMPK and as a loading control, IFT81.
In Triton X-114 phase partitioning, CAH6::PLD-mNG fractionated with the detergent-soluble ciliary membrane fraction, indicating that membrane association of the fusion protein was preserved (Fig. 5C). In cilia of both bbs1 and control cells, CAH6::PLD-mNG moved by mostly diffusion, qualified by the observation of sporadic transports in control cilia (Fig. 5 B and E). Thus, BBSome-dependent IFT is essentially abolished when the head domain of PLD is replaced with that of CAH6. Western blot analysis showed that the CAH6::PLD-mNG fusion protein was present in similar amounts in the detergent extract of control and bbs cilia, revealing that a BBSome-dependent regulation of its ciliary level is abolished (Fig. 5D). We noted, however, that the amount of endogenous PLD is elevated in control strains expressing CAH6::PLD-mNG (Fig. 5D). Using anti-mNG nanobodies, endogenous PLD coimmunoprecipitated with CAH6::PLD-mNG from ciliary extracts, indicating that CAH6::PLD-mNG and PLD interact (Fig. S1D). PLDs of this type are known to dimerize via their catalytic domains (30). The formation of CAH6::PLD-mNG PLD dimers could explain the increased levels of endogenous PLD, as well as residual IFT of CAH6::PLD-mNG in control cilia (Fig. 5 D and E). C. reinhardtii bbs mutants also accumulated AMPK and reduced CAH6 in cilia (Fig. 5F) (5). In control cells expressing CAH6::PLD-mNG, AMPK levels remained low and those of CAH6 high in cilia, indicating that specifically the distribution of CAH6::PLD-mNG was altered. We conclude that the N-terminal 58 residues of PLD contain a ciliary export sequence (CES) sufficient and necessary for BBSome-dependent IFT.
PLD Is a Negative Regulator of Phototaxis.
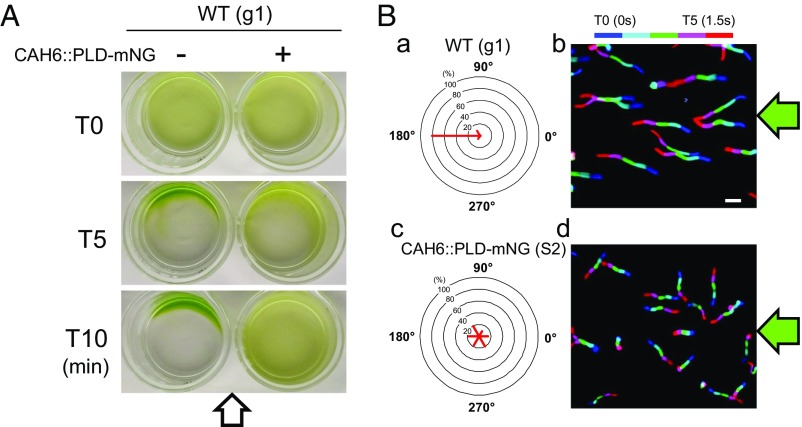
C. reinhardtii bbs mutants are nonphototactic, but it is unclear whether the phenotype is connected to the known biochemical defects of bbs cilia. Due to the absence of the CES, the CAH6::PLD-mNG fusion encompassing PLD’s catalytic domain accumulated in control cilia (Fig. 5D). To test the phototactic capacity of transformants expressing CAH6::PLD-mNG, we used population assays exposing cultures to a light gradient and single cell assays determining the direction of cell locomotion in response to light (Fig. 6). In a screen of 76 transformants using g1, the parental strain of the bbs4 mutant, two independent transformants (S1 and S2) expressing CAH6::PLD-mNG were identified by TIRF microscopy; both positive transformants were nonphototactic while all other transformants showed normal phototaxis (Fig. 6 and Movie S5). Expression of PLD-mNG, in contrast, had no effect on the phototaxis of control cells (Fig. S3A). Four strains (S3–S6) expressing CAH6::PLD-superfolder GFP (sfGFP) were obtained in three transformation experiments encompassing 86 transformants using the WT strain CC-620; one of the strains (S3) had shorter than normal cilia and was not analyzed further. The remaining three strains showed reduced phototaxis, with only a subset of the cells swimming away from the light source in single cell assays (Fig. S3 B and C). We conclude that the expression of CAH6::PLD-mNG is the likely reason for the reduced phototaxis of the otherwise WT strains.
Fig. 6.
PLD is a negative regulator of phototaxis. (A) Population phototaxis assay of control and strain S2 expressing CAH6::PLD-mNG in WT (g1). The direction of the light (arrow) and time of exposure are indicated. (B) Single cell motion analysis of control and strain S2 expressing CAH6::PLD-mNG. The direction of light is indicated (green arrows). The radial histograms (a and c) show the percentage of cells moving in a particular direction relative to the light (six bins of 60° each). (b and d) Composite micrographs showing the tracks of single cells. Each of the five merged frames was assigned a different color (blue, frame 1, and red, frame 5, corresponding to a travel time of 1.5 s). (Scale bar: 50 μm.)
Discussion
In the absence of functional BBSomes, membrane proteins abnormally accumulate in cilia, resulting in ciliary dysfunction and disease (5, 7, 9, 31). However, the specific molecular activity by which the BBSome regulates ciliary membrane composition is under discussion. Our data suggest that the biochemical defects in bbs cilia result from a failure to attach certain cargoes to IFT for motor-driven removal from cilia.
In C. reinhardtii, the most prominent manifestation of BBSome deficiency is the accumulation of PLD inside cilia. We show that IFT of PLD is abolished in bbs mutants and that PLD travels predominately on those IFT trains that also carry BBSomes. Failure to move PLD by IFT correlated with its accumulation inside cilia, indicating that transport by IFT ensures the low levels of PLD typical of WT cilia. However, both anterograde and retrograde IFT of PLD are BBSome-dependent, raising the question how bbs defects result in a buildup of PLD inside cilia. In principle, proteins will accumulate inside cilia when ciliary entry and exit are unbalanced. PLD also enters cilia by diffusion, but the rate of entry into cilia remains low in bbs mutants and we exclude excessive influx as a cause for its accumulation. While only traces of PLD are present in typical WT cilia, an astonishing ∼30 to 50% of the total cellular PLD is present in the cilia of bbs mutants (5). Ciliary accumulation was not observed for the soluble PLDMAA, which diffuses apparently freely in and out of cilia, indicating that membrane association is a prerequisite for PLD accumulation in cilia. However, the surface area of the ciliary membrane is only a fraction (<0.5%) of that of the plasma membrane. Thus, the accumulation of PLD in bbs cilia is not the result of diffusion-driven equilibration between these continuous membrane domains. We conclude that PLD becomes trapped in the ciliary membrane in the absence of transport by the IFT/BBS system. The cilium is separated from the cell body by the transition zone (TZ), which function as a diffusion barrier for soluble and membrane proteins (29, 32, 33). The TZ could work like a fish trap, allowing PLD to enter cilia by diffusion while exit by diffusion is prevented; in the absence of IFT, PLD will accumulate over time. The TZ also functions as a lipid barrier separating the ciliary membrane with its distinct, raft-like composition from the plasma membrane (34, 35). Lipid rafts function in protein sorting by attracting a specific subset of proteins (36). Similarly, PLD could accumulate inside cilia because it prefers the ciliary lipid environment to that of the plasma membrane. IFT trains and associated cargoes move fluidly in and out of cilia typically without pausing at the TZ, indicating that they can surmount the TZ boundary (23, 37). Regardless of how PLD is retained in cilia, we propose that BBSome-mediated association to the motor-driven IFT trains is the mechanism by which PLD is extracted from the ciliary compartment and shuttled through the TZ. When IFT is switched off, PLD accumulates in cilia despite the presence of BBSomes, emphasizing that BBSome function depends on active IFT (5). Defects in BBSome-dependent ciliary export could also explain features of mammalian Bbs mutants, such as the abnormal presence of cell body proteins in the outer segments (7).
Many proteins of the ciliary membrane and axoneme travel by IFT, but how cargoes and IFT interact remains often unknown. Here, we show that the membrane-associated head domain of PLD is sufficient and necessary for BBSome-dependent IFT and ciliary export. PLD’s ciliary export sequence (CES) is not conserved, raising the question of how the BBSome identifies its other putative cargoes, which include membrane-associated proteins, GPCRs, ion channels, single pass cytokine receptors, and melanosomes (6, 8, 20, 38, 39). The BBSome binding site of certain GPCRs encompasses portions of intracellular loop 3 (i3) and the C-terminal region (10, 15, 16). Further, the i3 motif bestows BBSome-dependent translocation of a plasma membrane protein into the ciliary membrane. Sstr3′s i3 contains a cysteine that is critical for BBSome-dependent transport and is predicted to be palmitoylated; a cysteine in a similar position is also present in i3 of D1, which exits cilia in a BBSome-dependent manner (15, 40). Hypothetically, BBSome binding could require a cargo motif layered onto the membrane by lipidation. Similar to IFT proteins, the BBSome subunits are ∼40% identical between Chlamydomonas and human. Domains involved in BBSome intraparticle protein–protein interactions are likely to be conserved best while the BBSome surface is likely to be more variable and could have coevolved with the specific transport needs of different organisms.
C. reinhardtii bbs mutants accumulate PLD and lack proper phototaxis (4, 22). PLD lacking its CES accumulates in WT cilia, diminishing phototaxis, indicating that PLD is a negative regulator of phototaxis. The cilia of bbs mutants fail to perform the steering movements required for phototactic orientation because they do not respond properly to changes in the ciliary calcium concentration that are triggered by photocurrents originating in the cell body (22, 41). Of note, demembranated bbs cell models still fail to respond appropriately to changes in free calcium, suggesting that bbs mutations ultimately affect axonemal function (4, 22). Lipidomics of bbs4 cilia revealed a strong (>5×) increase of DAG, a downstream product of PLD activity (5). DAG is an activator of protein kinase C, a known regulator of ciliary motility in mammals (42). In Chlamydomonas, atypical phosphorylation of dynein subunits, particularly of inner arm dynein I1, has been linked to the loss of phototaxis (43, 44). While details remain to be explored, proteins that regulate dynein activity are possible targets of a signaling chain from membrane-associated PLD to the axoneme.
The physiological role of PLD in cilia and phototaxis, however, is unclear. The limited breadth of the biochemical defects in bbs cilia suggests a specific role of the BBSome in the transport of PLD (and that of a few other proteins) rather than all-purpose scavenging of proteins leaking accidentally into cilia. In phototaxis, cells move toward or away from a light source, seeking positions to optimize photosynthetic revenue; the behavior is modulated: e.g., by the circadian clock and the cell’s energy and redox state (45–47). Late in the light phase, for example, cells are typically less responsive to phototactic cues while cells preincubated in the dark often display particularly strong phototaxis. Changes in IFT-BBSome or BBSome-PLD interactions could alter the ciliary level of PLD to modulate phototactic responsiveness. Indeed, some BBSome-dependent transports have been shown to be conditional depending on the state of the cargo (10). In Caenorhabditis elegans cilia, for example, IFT of RAB28 depends on both GTP binding and the BBSome (39).
The BBSome is conserved in organisms with cilia, and it is tempting to speculate that it also performs a conserved role, despite the apparent heterogeneity of its putative cargoes. In addition to PLD, C. reinhardtii bbs mutants accumulate AMPK, a key regulator of cellular energy production. Similarly, IMCD3 cells null for the putative BBSome-IFT docking factor IFT27 accumulate an AMPK and BBS proteins in cilia (48). In mammalian systems, the BBSome has been implicated in the transport of the leptin receptor, the Sstr3, the Mchr1, and the D1, all of which contribute to the control of appetite and energy homeostasis and are implicated with circadian patterns (8, 10, 20, 49, 50). We hypothesize that the BBSome is part of an ancient pathway transporting proteins in and out of cilia to modulate ciliary responses, such as swimming behavior or signal output in reaction to the nutritional state of the cell.
Materials and Methods
Strains and Culture Conditions.
C. reinhardtii strains used in this study are listed in Table S1. Cells were grown in modified minimal medium (22) and maintained at 21 °C with a light/dark cycle of 14:10 h; larger cultures were aerated with air enriched by 0.5% CO2.
Transgenic Strain Generation.
To express mNG-tagged PLD, the PLD cDNA was synthesized (Genewiz) and inserted into the HpaI and XhoI sites of the ipBR vector, placing PLD downstream of the HSP70A-rbcS2 fusion promoter and upstream of the mNG gene followed by the 2A sequence, the selectable marker ble, and the tubulin terminator (Fig. 1A) (51). To enhance expression efficiency, the first intron of rbcS2 was inserted into the PLD cDNA in position 162 downstream from ATG (52). Primers nos. 1 and 2 (Table S2) were used to generate PLDMAA by PCR, and the resulting fragment was digested with HpaI and XhoI and inserted into ipBR. To generate C-terminally truncated PLDs, we used primer no. 3 in combination with primer 4 (PLD1-95), primer 5 (PLD1-70), or primer 6 (PLD1-58); the PLD1-20 cDNA was synthesized (Genewiz). Fragments were cloned as described above. To express CAH6-mNG, primers nos. 7 and 8 were used to PCR amplify the coding portions of the gene using CC-620 genomic DNA as a template; the PCR fragment was cloned into ipBR as described above. The plasmid encoding the CAH6::PLD fusion protein was made as follows: PCR with primers nos. 8 and 9 was used to obtain a genomic fragment encoding CAH61-58; the PCR fragment was digested with HpaI and BglII, and the PLD59-223 cDNA fragment was amplified using primers nos. 10 and 11 and digested with BglII and XhoI. Both fragments were ligated simultaneously into ipBR digested with the HpaI and XhoI. To express the PLD::EB1-mNG fusion, EB1 was amplified from genomic DNA and ligated downstream of PLD1-58. The final plasmids were linearized with KpnI and transformed into C. reinhardtii by electroporation. Transformants were selected on Tris-acetate-phosphate (TAP) plates containing 5 to 10 μg/mL zeocin (Invitrogen), and clones expressing mNG-tagged proteins were identified by TIRF microscopy. Expression efficiency of constructs in the ipBR vector was moderate to low, and ∼50 or more transformants had to be screened to identify a strain expressing the transgene. Positive strains were reselected on zeocin plates when expression levels of the transgenes decreased. To express mC-tagged BBS4, we replaced GFP with mC DNA in pKL3-BBS4-GFP using BamH1 digestion and ligation (4). The AphVII gene conferring resistance to hygromycin was then inserted into the pKL3′s unique HindIII site, and the plasmid was transformed into bbs4 PLD-mNG. After selection of transformants on hygromycin plates (10 μg/mL), transformants expressing BBS4-mC were identified by a phototaxis assay and confirmed by TIRF microscopy and Western blotting.
Isolation of Cilia.
To isolate flagella, cells were concentrated and washed with 10 mM Hepes (pH 7.4) and resuspended in 10 mL of HMS (10 mM Hepes, pH 7.4, 5 mM MgSO4, and 4% sucrose wt/vol) (53). Then, 2 mL of dibucaine (25 mM in H2O; Sigma) were added, and cells were deciliated by vigorous pipetting on ice. Next, 20 mL of 0.7 mM EGTA (pH 8.0) in HMS were added, and the cell bodies were removed by centrifugation (1,150 × g, 3 min, 4 °C; Sorvall Legend XTR; Thermo Scientific). The supernatant was underlaid with a sucrose cushion (10 mL of 25% sucrose in HMS) and centrifuged (1,700 × g, 4 °C, 10 min) to remove the remaining cell bodies. The flagella were harvested from the supernatant by centrifugation (27,000 × g, 4 °C, 20 min) (Avanti JXN-26; Beckman Coulter), resuspended in HMEK+PI (30 mM Hepes, 5 mM MgSO4, 0.5 mM EGTA, 25 mM KCl, 1% protease inhibitor mixture P9599; Sigma-Aldrich), and extracted with Nonidet P-40 Alternative (1% final concentration, CAT no. 492016; CalBiochem) for 20 min on ice. The axonemal and membrane+matrix fractions were separated by centrifugation (27,000 × g, 4 °C, 15 min). Alternatively, flagella were extracted with 1% final concentration Triton X-114 (CAT no. 9036-19-5; Sigma-Aldrich), the axonemes were removed by high-speed centrifugation (see above), and the supernatant was incubated briefly at 30 °C to induce phase separation. The aqueous phase (AP) and detergent phase (DP) were separated by centrifugation (6,000 × g for 5 min at room temperature). The aqueous phase was treated with 1% Triton X-114, the detergent phase was diluted with HMEK+PI, and the phase separation was repeated to yield the final AP and DP. Protein in the DP was precipitated using methanol/chloroform to remove detergent.
Immunoprecipitation.
Cilia isolated from strains expressing CAH6::PLD-mNG and as a control untransformed WT and bbs1 strains were resuspended in HMEK+PI supplemented with 50 mM NaCl and lysed by adding Nonidet P-40 to 1%. The axonemes were removed by centrifugation (27,000 × g, 4 °C, 15 min), and the supernatants were incubated for 1 h at 5 °C with agitation with anti-mNG agarose beads (Allele Biotechnology) pretreated with 5% BSA. The beads were then washed with 150 mM NaCl, 50 mM NaCl and finally 0 mM NaCl in HMEK, and proteins were eluted using 1 M glycine, pH 2.5. The eluted proteins and other fractions were then analyzed by Western blotting.
Antibodies and Western Blotting.
Isolated whole cell, cell body, and flagella samples were incubated at 85 °C for 10 min in Laemmli SDS sample buffer. Protein samples were separated by SDS/PAGE using Precast Gels (Bio-Rad TGX) and transferred to PVDF membrane. Membranes were blocked in 3.5% nonfat milk and stained following standard procedures incubating the primary antibodies overnight at 4 °C and the secondary antibodies for ∼90 min at room temperature. The following antibodies were used in this study: rabbit polyclonal anti-PLD (1:1,000; ref. 5), anti-BBS4 (1:800; ref. 4), anti-CAH6 (1:1,000; ref. 54), anti-FAP12 (1:1,000; courtesy of Doug Cole, University of Idaho, Moscow, ID), anti-mNG (1:1,000; ChromoTek), and anti-AMPK (1:1,000). The polyclonal antibody to AMPK was raised against the peptide “PSEKYEVQGQSSKPC” in rabbit (Genscript) and affinity-purified using the peptide linked to carrier protein immobilized on PVDF membrane. The mouse monoclonal antibody anti-IFT81 was diluted 1:200 (55). Rabbit polyclonal anti-mCherry (CAT no. 5993-100; CBiovision) was diluted 1:1,000. Secondary antibodies (anti–mouse or anti–rabbit IgG conjugated to horseradish peroxidase; Invitrogen) were diluted 1:2,000, and FemtoGlow WesternPLUS (Michigan Diagnostics) was used for developing. A ChemiDoc MP imaging system and Image Lab software (Bio-Rad Laboratories) were used for documentation and quantification of signals.
In Vivo Microscopy.
For through-the-objective TIRF imaging, we used a Nikon Eclipse Ti-U inverted microscope equipped with a 60×/1.49 N.A. objective and 75-mW 561-nm and 40-mW 488-nm diode lasers (Spectraphysics; refs. 56 and 57). The excitation light was filtered with a Nikon GFP/mCherry TIRF filter cube, and emission was separated by using an image-splitting device (Dual-View2; Photometrics). Samples for in vivo imaging were prepared as follows: 8 to 10 μL of cells were placed inside of a ring of petroleum jelly or vacuum grease onto a 24 × 60-mm no. 1.5 coverslip and allowed to settle for ∼1 to 3 min. Then a 22 × 22-mm no. 1.5 coverslip with an 8- to 10-μL drop of buffer (10 mM Hepes, 6.25 mM EGTA, pH 7.4) was inverted onto the larger cover glass to form a sealed observation chamber. Photobleaching of the flagella was accomplished either by bleaching of the entire flagellum by increasing the output of the 488-nm laser to ∼10% for ∼5 s or by bleaching a specific area of the flagellum using a focused 488-nm laser beam (23). Images were recorded mostly at 10 fps using the iXon X3 DU897 electron multiplying charge-coupled device (EMCCD) camera (Andor) and the Elements software package (Nikon). ImageJ (National Institutes of Health) with the Multiple Kymogram plug-in (European Molecular Biology Laboratory) was used to analyze the recording and generate kymograms. Kymograms, individual frames, and movies were cropped and adjusted for brightness and contrast in ImageJ and Photoshop CC (Adobe); Illustrator CC 2017 (Adobe) was used to assemble the figures.
FRAP and Fluorescence Intensity Analysis.
For FRAP analysis, videos were opened in ImageJ, and the region of interest (ROI) was selected using the rectangle tool. The fluorescence intensity inside the selected region was determined using the plot z axis tool, and the data were exported into Excel. The fluorescence intensity in the ROI was corrected for the background fluorescence. The highest intensity value before the bleaching event was set to 100%, and the recovery of fluorescence in percentage of the prebleached value was calculated.
Phototaxis Assays.
Population phototaxis assays were performed by placing a cell suspension (∼107 cells per milliliter) harvested during the first half of the light phase into a glass beaker or Petri dish, followed by illumination with bright light from one side for ∼5 min. Images were taken with a standard digital camera (PowerShot S95; Canon). For the single cell phototaxis assay, 20-µL of cell suspensions were placed in a chambered plastic slide (14-377-259; Fisherbrand) and observed using an inverted microscope (Eclipse 55i; Nikon) and nonphototactic red light illumination. After 5 s of illumination with phototactic active green light, a video was recorded at ∼3 fps using an MU500 camera (Amscope). The swimming tracks were analyzed in ImageJ by merging five sequential images each displayed in a different color, allowing us to determine the angle and the direction of a cell’s movements. ImageJ and Excel 2010 were used to analyze the data and generate polar histograms with 60° bins. As controls, we used g1 (nit1, agg1, mt+), a strain selected for strong negative phototaxis, and CC-620, a subclone of the 137c WT strain (22). Cells were washed into fresh M-medium before phototaxis assays.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award R01GM110413. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713226115/-/DCSupplemental.
References
- 1.Sheffield VC. The blind leading the obese: The molecular pathophysiology of a human obesity syndrome. Trans Am Clin Climatol Assoc. 2010;121:172–181; discussion 181–182. [PMC free article] [PubMed] [Google Scholar]
- 2.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 3.Mykytyn K, et al. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lechtreck KF, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lechtreck KF, et al. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valentine MS, et al. Paramecium BBS genes are key to presence of channels in Cilia. Cilia. 2012;1:16. doi: 10.1186/2046-2530-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta P, et al. Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc Natl Acad Sci USA. 2015;112:E4400–E4409. doi: 10.1073/pnas.1510111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eguether T, et al. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;31:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domire JS, et al. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2011;68:2951–2960. doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo S, et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and smoothened. PLoS Genet. 2011;7:e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blacque OE, et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams CL, et al. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun. 2014;5:5813. doi: 10.1038/ncomms6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klink BU, et al. A recombinant BBSome core complex and how it interacts with ciliary cargo. eLife. 2017;6:e27434. doi: 10.7554/eLife.27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansley SJ, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starks RD, et al. Regulation of insulin receptor trafficking by Bardet Biedl syndrome proteins. PLoS Genet. 2015;11:e1005311. doi: 10.1371/journal.pgen.1005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo DF, et al. The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet. 2016;12:e1005890. doi: 10.1371/journal.pgen.1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langousis G, et al. Loss of the BBSome perturbs endocytic trafficking and disrupts virulence of Trypanosoma brucei. Proc Natl Acad Sci USA. 2016;113:632–637. doi: 10.1073/pnas.1518079113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pazour GJ, Sineshchekov OA, Witman GB. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J Cell Biol. 1995;131:427–440. doi: 10.1083/jcb.131.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wingfield JL, et al. IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife. 2017;6:e26609. doi: 10.7554/eLife.26609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorn K. Genetically encoded fluorescent tags. Mol Biol Cell. 2017;28:848–857. doi: 10.1091/mbc.E16-07-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godsel LM, Engman DM. Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 1999;18:2057–2065. doi: 10.1093/emboj/18.8.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris JA, Liu Y, Yang P, Kner P, Lechtreck KF. Single-particle imaging reveals intraflagellar transport-independent transport and accumulation of EB1 in Chlamydomonas flagella. Mol Biol Cell. 2016;27:295–307. doi: 10.1091/mbc.E15-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Q, et al. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol. 2012;14:950–957. doi: 10.1038/ncb2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kee HL, et al. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magotti P, et al. Structure of human N-acylphosphatidylethanolamine-hydrolyzing phospholipase D: Regulation of fatty acid ethanolamide biosynthesis by bile acids. Structure. 2015;23:598–604. doi: 10.1016/j.str.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nager AR, et al. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 2017;168:252–263.e14. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musgrave A, et al. Evidence for a functional membrane barrier in the transition zone between the flagellum and cell body of Chlamydomonas eugametos gametes. Planta. 1986;167:544–553. doi: 10.1007/BF00391231. [DOI] [PubMed] [Google Scholar]
- 33.Jensen VL, Leroux MR. Gates for soluble and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol. 2017;47:83–91. doi: 10.1016/j.ceb.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Tyler KM, et al. Flagellar membrane localization via association with lipid rafts. J Cell Sci. 2009;122:859–866. doi: 10.1242/jcs.037721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gealt MA, Adler JH, Nes WR. The sterols and fatty-acids from purified flagella of Chlamydomonas-reinhardi. Lipids. 1981;16:133–136. [Google Scholar]
- 36.Bagnat M, Simons K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol Chem. 2002;383:1475–1480. doi: 10.1515/BC.2002.169. [DOI] [PubMed] [Google Scholar]
- 37.Wren KN, et al. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum Mol Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 39.Jensen VL, et al. Whole-organism developmental expression profiling identifies RAB-28 as a novel ciliary GTPase associated with the BBSome and intraflagellar transport. PLoS Genet. 2016;12:e1006469. doi: 10.1371/journal.pgen.1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witman GB. Chlamydomonas phototaxis. Trends Cell Biol. 1993;3:403–408. doi: 10.1016/0962-8924(93)90091-e. [DOI] [PubMed] [Google Scholar]
- 42.Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol. 2007;69:401–422. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- 43.King SJ, Dutcher SK. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VanderWaal KE, et al. bop5 mutations reveal new roles for the IC138 phosphoprotein in the regulation of flagellar motility and asymmetric waveforms. Mol Biol Cell. 2011;22:2862–2874. doi: 10.1091/mbc.E11-03-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi T, Watanabe M. Photosynthesis modulates the sign of phototaxis of wild-type Chlamydomonas reinhardtii. Effects of red background illumination and 3-(3′,4′-dichlorophenyl)-1,1-dimethylurea. FEBS Lett. 1993;336:516–520. doi: 10.1016/0014-5793(93)80867-t. [DOI] [PubMed] [Google Scholar]
- 46.Wakabayashi K, Misawa Y, Mochiji S, Kamiya R. Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2011;108:11280–11284. doi: 10.1073/pnas.1100592108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruce VG. Mutants of the biological clock in Chlamydomonas reinhardi. Genetics. 1972;70:537–548. doi: 10.1093/genetics/70.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mick DU, et al. Proteomics of primary cilia by proximity labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green JA, et al. Recruitment of β-arrestin into neuronal cilia modulates somatostatin receptor subtype 3 ciliary localization. Mol Cell Biol. 2015;36:223–235. doi: 10.1128/MCB.00765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kettner NM, et al. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22:448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasala BA, et al. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- 52.Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. [Google Scholar]
- 53.King SM. Large-scale isolation of Chlamydomonas flagella. Methods Cell Biol. 1995;47:9–12. doi: 10.1016/s0091-679x(08)60783-9. [DOI] [PubMed] [Google Scholar]
- 54.Moroney JV, et al. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth Res. 2011;109:133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- 55.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lechtreck KF. In vivo imaging of IFT in Chlamydomonas flagella. Methods Enzymol. 2013;524:265–284. doi: 10.1016/B978-0-12-397945-2.00015-9. [DOI] [PubMed] [Google Scholar]
- 57.Lechtreck KF. Methods for studying movement of molecules within cilia. Methods Mol Biol. 2016;1454:83–96. doi: 10.1007/978-1-4939-3789-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.