Significance
In this study, we reexamined an old mouse mutant named Hammer toe (Hm), which arose spontaneously almost a half century ago and exhibits a limb phenotype with webbing. We revealed that a 150-kb noncoding genomic fragment that was originally located in chromosome 14 has been inserted into a genomic region proximal to Sonic hedgehog (Shh), located in chromosome 5. This inserted fragment possesses enhancer activity to induce Shh expression in the interdigital regions in Hm, which in turn down-regulates bone morphogenetic protein signaling and eventually results in syndactyly and web formation. Since the donor fragment residing in chromosome 14 has enhancer activity to induce interdigital gene expression, the Hm mutation appears to be an archetypal case of enhancer adoption.
Keywords: enhancer adoption, genomic insertion, Hammer toe, Shh, interdigital web
Abstract
Acquisition of new cis-regulatory elements (CREs) can cause alteration of developmental gene regulation and may introduce morphological novelty in evolution. Although structural variation in the genome generated by chromosomal rearrangement is one possible source of new CREs, only a few examples are known, except for cases of retrotransposition. In this study, we show the acquisition of novel regulatory sequences as a result of large genomic insertion in the spontaneous mouse mutation Hammer toe (Hm). Hm mice exhibit syndactyly with webbing, due to suppression of interdigital cell death in limb development. We reveal that, in the Hm genome, a 150-kb noncoding DNA fragment from chromosome 14 is inserted into the region upstream of the Sonic hedgehog (Shh) promoter in chromosome 5. Phenotyping of mouse embryos with a series of CRISPR/Cas9-aided partial deletion of the 150-kb insert clearly indicated that two different regions are necessary for the syndactyly phenotype of Hm. We found that each of the two regions contains at least one enhancer for interdigital regulation. These results show that a set of enhancers brought by the large genomic insertion elicits the interdigital Shh expression and the Hm phenotype. Transcriptome analysis indicates that ectopic expression of Shh up-regulates Chordin (Chrd) that antagonizes bone morphogenetic protein signaling in the interdigital region. Indeed, Chrd-overexpressing transgenic mice recapitulated syndactyly with webbing. Thus, the Hm mutation provides an insight into enhancer acquisition as a source of creation of novel gene regulation.
Gain and loss of cis-regulatory elements (CREs) alter gene regulatory networks and as a consequence can result in morphological variation and sometimes genetic disease (1, 2). Changes in CRE function have restricted effects on specific tissues or organs, whereas mutations in protein-coding sequences can affect multiple tissues and organs where the mutated gene is expressed and often result in drastic phenotypes including embryonic lethality. The restricted effect mentioned above is probably relevant to the fact that functional changes in CREs have contributed to evolution more often than coding sequence mutations (3, 4).
To date, studies of genome variations that cause morphological alteration and disease have focused on coding sequences, which comprise around 1.5% of the whole genome. By contrast, studies of functional change in CREs, which is responsible for phenotype alteration, have been hampered by the fact that sequences of CREs are less well annotated than the coding sequences and because the molecular basis underlying change in CRE function is still poorly understood. In particular, little attention has been paid to structural variations, including insertion, deletion, inversion, and translocation, as a driving force for change in gene regulation. The only exception is the study of transposable elements (TEs): There are many lines of evidence that insertion of a TE harboring enhancer activity into an unrelated genomic region gives rise to a novel expression pattern for the gene neighboring the insertion (5, 6). Some genomic structural variations may reconfigure the connection between CREs and their target coding sequences on the chromosome (7). Recent advances in sequencing technology have revealed a number of structural variants from 100 bp to 1 Mbp in length in human genomes. However, there are very few reports that such variants are associated with a change in gene regulation that leads to morphological change and disease (8, 9).
In this study, we present an example of the acquisition of a novel expression pattern for a developmental gene as a result of structural rearrangement in the genome. Hammer toe (Hm) is an old spontaneous mouse mutant that exhibits soft tissue syndactyly associated with interdigital webbing (10), which is caused by a reduction of interdigital cell death (11). Previous genetic linkage analyses mapped the Hm locus to the region upstream of Sonic hedgehog (Shh). In the developing limb bud, Shh is expressed in a cell population named the zone of polarizing activity (ZPA) in the posterior mesenchyme, and Shh signaling is crucial for anterior–posterior (AP) patterning and determination of digit identity (12). Shh expression in the ZPA is regulated by a limb bud-specific enhancer named MFCS1 (also known as ZRS), which is located 840 kb upstream of the Shh promoter (13–15). Many mutations in the MFCS1 sequence are known to cause preaxial polydactyly, but not syndactyly, in both human and mouse (16, 17).
We extended the linkage analysis and exploration of structural variation in the Hm genome and found that a chromosome 14 (Chr14)-derived 150-kb noncoding DNA fragment is inserted into the interval between the Shh promoter and MFCS1 in the Hm genome and that it elicits ectopic Shh expression in the interdigital region of the developing limb. The ectopic Shh expression in turn down-regulates bone morphogenetic protein (BMP) signaling, leading to a reduction of interdigital cell death and finally resulting in the syndactyly and interdigital webbing phenotype. Thus, we provide unequivocal evidence that interchromosomal translocation is able to generate enhancer activity for a developmental gene, which ultimately leads to morphological alteration.
Results
Hm was reported to be tightly linked to another old mutation named Hemimelic extra toe (Hx) that causes hemimelia and preaxial polydactyly. Only one recombinant was detected between Hx and Hm in a previous study (18). Since a single nucleotide substitution was identified in MFCS1 as the causative mutation of Hx (13, 14), we first sequenced MFCS1 from the Hm genome. The result did not show any kind of mutation in MFCS1.
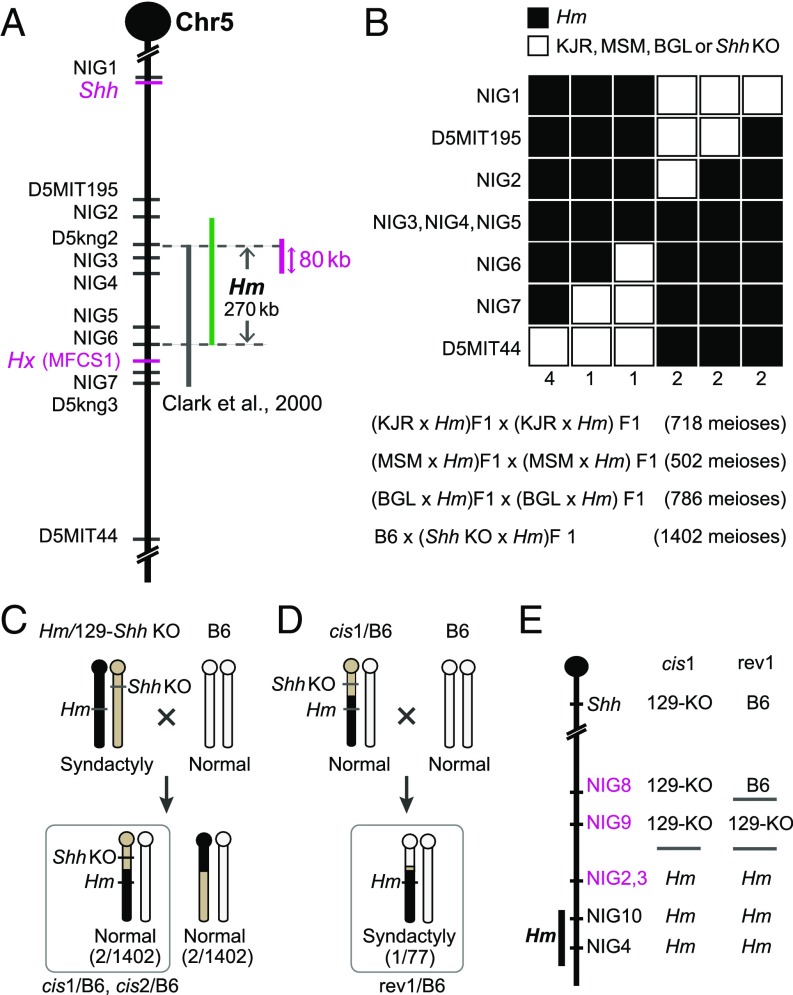
A subsequent linkage study narrowed the critical region of Hm to a 420-kb interval between two DNA markers, D5kng2 and D5kng3 (19) (Fig. 1A). We extended the linkage analysis based on intercrosses of the Hm heterozygotes (Hm/+) that were generated from crosses of Hm/+ and each of three inbred strains—KJR/Ms, BLG/Ms, and MSM/Ms—established from highly polymorphic wild mice (20) and also a backcross involving a Shh-coding knockout (KO) mouse (Fig. 1B). In total, we obtained 12 recombinants from these crosses and thereby assigned the DNA marker NIG6 to the most distal border and NIG2 to the most proximal border of the Hm region (Fig. 1B). This result confined the Hm region to the segment depicted by the green bar in Fig. 1A. By combining the results of the former linkage analysis (19) with these, we further confined Hm to a 270-kb interval between D5kng2 and NIG6 (Fig. 1A).
Fig. 1.
Linkage analysis of the Hm mutation. (A) Genetic map around the Shh locus in mouse Chr5. Vertical black and green bars indicate the Hm region defined by Clark et al. (19) and the present study, respectively. The vertical magenta bar depicts the deleted 80 kb. DNA markers are shown on the left side of the map. (B) Haplotype panel of backcrosses. Microsatellite and DNA marker loci used are listed to the left side of the panel. The number of progeny with each haplotype is listed at the bottom of each haplotype column. Solid squares represent the Hm allele, and open squares represent the alleles of KJR, MSM, BGL, and Shh KO. The mating pairs and the numbers of meioses observed in each backcross are shown below the haplotype panel. (C) Mating scheme and the resultant recombinants between Hm and the Shh coding KO chromosomes. Phenotype and numbers of each recombinant are shown at the bottom. (D) Mating scheme and the revertant recombinants between cis1 and wild-type B6 chromosomes. Phenotype and number of revertants are shown at the bottom. (E) Genotype of two recombinants. Hm, B6, and Shh coding KO on the genetic background of strain 129/SV around the Shh locus are represented by Hm, B6, and 129-KO, respectively.
Because Hm is tightly linked to the Shh locus, we examined whether Hm acts in cis on Shh. From the same backcross progeny used in the linkage analysis, we obtained four recombinants between the Shh-coding KO allele and the DNA marker NIG2 (Fig. 1C). Two of these, referred to as cis1/B6 and cis2/B6, have a recombinant chromosome carrying the Hm region and the Shh-coding KO allele. They exhibited normal limb phenotype, indicating a cis-acting nature for Hm. This was confirmed by the finding that genetic restoration of the Shh coding sequence in the cis1 recombinant chromosome yielded the syndactyly phenotype (Fig. 1 D and E). Thus, this cis–trans test showed unequivocally that the regulation by Hm acts in cis on Shh.
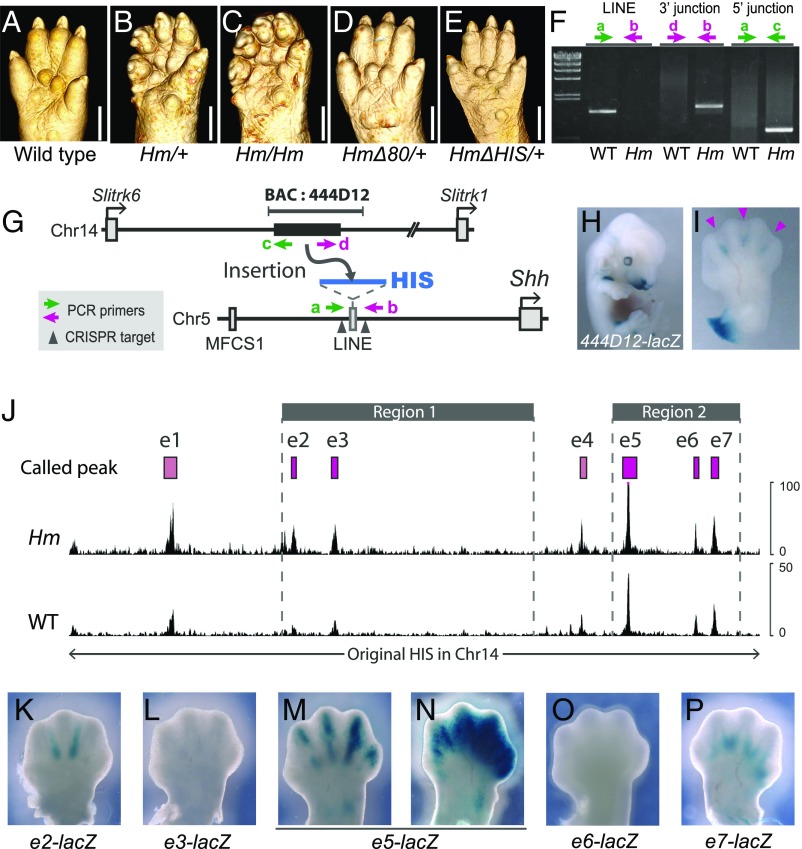
Because the syndactyly of Hm is a dominant phenotype and is likely caused by the gain of a new regulatory activity, we anticipated that removing the mutated region from the Hm genome could rescue its phenotype. Because two Shh enhancers, MFCS4 and MACS1, which are necessary for postnatal viability, are located in the distal side of the 270-kb Hm region between NIG4 and NIG6 (21, 22), we first removed 80 kb that does not contain the two enhancers (Fig. 1A, magenta bar). Mice with the deletion showed normal digits, indistinguishable from those of wild-type mice (Fig. 2 A–D). This result indicated that the causative mutation is contained within the 80 kb on the reference mouse genome (GRCm38/mm10), which does not contain any coding sequence (Fig. 1A).
Fig. 2.
Genomic insertion caused the Hm mutation. (A–E) Micro-CT images of right hind limb from (A) wild-type, (B) Hm/+, (C) Hm/Hm, (D) HmΔ80/+, and (E) HmΔHIS/+ at P0 (day of birth). (Scale bars, 1 mm.) (F) PCR-amplified DNA fragments from wild-type (WT) and Hm genomes. Green and magenta arrows indicate primer pairs for the amplifications shown in G. Leftmost lane is λ-HindIII as DNA marker. (G) Schematic diagram of genomic insertion of a fragment derived from Chr14 into Chr5 in the Hm mutation. The original sequence of HIS in Chr14 is marked by a black bar (Chr14: 110087232–110237755, GRCm38). Green and magenta indicate primers used for PCR with genomic DNAs of WT and Hm mice. Arrowheads indicate target positions of the CRISPR/Cas9 system, used to induce the ΔHIS modification. (H) LacZ expression in a BAC transgenic mouse with RP23-444D12-LacZ at E13.5. (I) Ventral view of LacZ expression in the left forelimb of the BAC transgenic mouse in H. Arrowheads indicate LacZ expression in interdigital regions. (J) ATAC-sequencing in the 150 kb of original HIS region in chr14 from the Hm homozygote (upper track) and wild-type mouse (lower track). Gray bars indicate regions 1 and 2. Magenta bars indicate seven called peaks in HIS. (K–O) Ventral view of LacZ expression in the left forelimb of a transgenic mouse with e2-LacZ (K) (n = 3/12), e3-LacZ (L) (n = 0/6), e5-LacZ (M and N) (n = 5/6), e6-LacZ (O) (n = 0/7), and e7-LacZ (P) (n = 1/8) at E13.5. Each n indicates the number of LacZ expression in autopod per transgenic mouse.
Next, to explore the genomic variation responsible for Hm in this 80 kb, we amplified genomic fragments using tiling PCR primer pairs from Hm-homozygous and wild-type mice (Fig. S1A). We found that no fragment was amplified with the primer pair F specifically from the Hm genome, suggesting a structural variation in a long interspersed nuclear element (LINE) sequence that resides in the 80 kb. Subsequent inverse PCR identified the structural variation in this LINE sequence (Fig. S1B). Sequencing revealed that the fragment amplified by the inverse PCR contains abnormal junctions of Chr5 and Chr14 (Fig. S1C). We confirmed the presence of these junctions by genomic PCR (Fig. 2F). These results indicated that a 150-kb fragment residing in the intergenic region between Slitrk1 and Slitrk6 in Chr14 is inserted into the region upstream of Shh in Chr5 (Fig. 2G). No coding sequences or noncoding RNAs are annotated in this 150-kb fragment. We termed this fragment the Hm inserted sequence (abbreviated as HIS).
To determine whether the HIS insertion directly causes the syndactyly phenotype of Hm, we removed the LINE sequence containing HIS from the Hm genome using the CRISPR/Cas9 system (Fig. 2G and Dataset S2). A mouse line with this deletion showed normal digits that were indistinguishable from those of wild-type mice (Fig. 2E). The homozygote of the HIS-deleted Hm mutant was viable and did not show any abnormal phenotype in the whole limb or in other organs. This result indicated that the original LINE sequence in the Hm region in Chr5 has no significant function in mouse development. Thus, we conclude that insertion of HIS was the direct cause of Hm.
To test whether the original HIS in the normal mouse Chr14 has enhancer activity, we performed a transgenic reporter assay using BAC clone RP23-444D12, which was isolated from the B6 genome library and harbors the HIS in Chr14 (Fig. 2G). We made a transgene construct, introducing a LacZ reporter gene into the BAC clone. Transgenic mouse embryos with this construct showed reporter expression in the interdigital region at E13.5 (Fig. 2 H and I). The LacZ signals were more abundant on the ventral side than on the dorsal side. This difference in signal intensity along the dorsoventral axis correlates well with the fact that interdigital cell death in the Hm limb is more severe on the dorsal side than on the ventral side (11). Therefore, HIS in Chr14 of strain B6 appears to possess enhancer activity to elicit gene expression in the interdigital regions of mouse limbs.
To assess the function of the original HIS in wild type, we removed HIS from Chr14 using the CRISPR/Cas9 system. Mice homozygous for the deletion did not show any developmental abnormality, including limb phenotype (Dataset S2), suggesting no significant role for HIS in Chr14 in morphogenesis.
To find regulatory region(s) of HIS, we generated a series of partial deletion of HIS in the Hm genome by the CRISPR/Cas9 system (Fig. S2). Observation of the phenotype of these deletion mutants indicated that the middle part of HIS (region 1) was necessary to cause the syndactyly phenotype of Hm. Moreover, another part of HIS (region 2), which resides at the Shh side and shares no sequence similarity with region 1, was also required for syndactyly.
Next, to identify enhancers in HIS, we searched open chromatin regions by using ATAC-sequencing (ATAC-seq) with distal autopod cells from E13.5 embryos. We found seven peaks in the 150 kb of HIS (magenta bars of e1 to e7 in Fig. 2J). Among them, two peaks (e1 and e4) were not included in regions 1 and 2. We carried out transgenic reporter assays with the five peaks excluding e1 and e4 by inserting each fragment upstream of the hsp promoter and LacZ sequence (Dataset S3). Out of the five peaks to be tested, we found three autopod enhancers (Fig. 2 K–P). The e2 peak induced interdigital expression of LacZ (Fig. 2K). The e5 peak directed the expression of LacZ mainly in digits sometimes with a broad signal in the entire autopod, including interdigit regions (Fig. 2 M and N). The e7 peak induced interdigital expression of LacZ, although the signal was observed in only one embryo (Fig. 2P). The other two peaks induced no LacZ signals in the limb (Fig. 2 L and O).
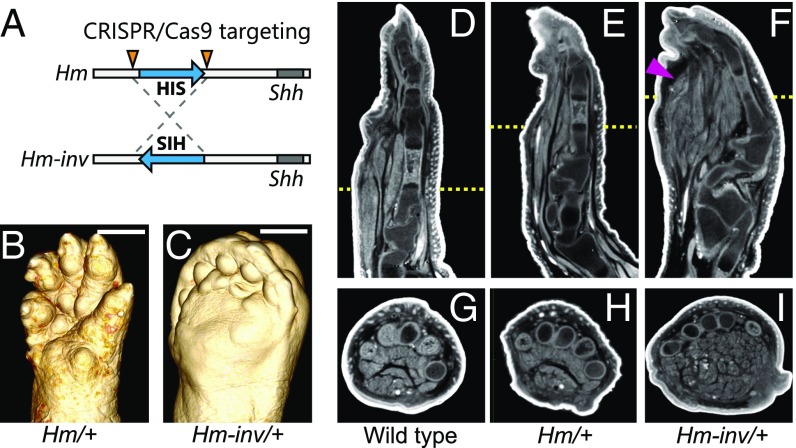
In the CRISPR/Cas9-mediated deletion experiment of HIS, we obtained G0 mice with a more severe syndactyly phenotype than that in the original Hm heterozygote. Sequencing of PCR products amplified from the Hm genome with a primer pair for the 3′ junction indicated that a CRISPR/Cas9-mediated inversion of HIS had occurred in the G0 mouse (Fig. 3A and Dataset S2). We named this HIS inverted allele Hm-inv and found that mice heterozygous for Hm-inv showed neonatal lethality; in contrast, both heterozygotes and homozygotes of the original Hm are fully viable. The Hm-inv heterozygotes exhibited swollen soles and a cleft palate, which were not observed in the original Hm heterozygotes (Fig. 3B). Sagittal and transverse sections of X-ray micro-computed tomography (micro-CT) images showed that the volume of muscle fiber in the Hm-inv heterozygotes was increased in the swollen sole (Fig. 3 D–I). On the other hand, each phalange was normal (Fig. 3 D–I).
Fig. 3.
Regulatory activity of HIS depends on its orientation. (A) Schematic diagram of inversion of HIS by the CRISPR/Cas9 system. Arrowheads indicate target sites and are identical to the arrowheads in G. (B and C) Micro-CT images of right hindlimb of (B) Hm/+ and (C) Hm-inv/+ mice at P0. (Scale bars, 1 mm.) The image in B is identical to that of Fig. 2B. (D–F) Sagittal sections of micro-CT images of right hindlimb of (D) wild type, (E) Hm/+, and (F) Hm-inv/+. The red arrowhead in F indicates increased muscle fiber. (G–I) Transverse sections of micro-CT images at the positions shown by the yellow dotted bars in D–F.
We examined the interaction between Hm and MFCS1, because MFCS1 is a sole enhancer necessary and sufficient for Shh expression in normal limb development (21). We eliminated MFCS1 from the Hm genome using the CRISPR/Cas9 system (Fig. S3A). Among the G0 pups generated, two individuals exhibited truncated limbs, resembling the phenotype of the MFCS1 KO mouse, indicating that MFCS1 was successfully eliminated from both chromosomes in these two pups (Fig. S3B). We then crossed the G0 male to B6 and established a mouse line in which the MFCS1-eliminated chromosome (ΔMFCS1) is located at the cis-position relative to Hm. All mice of this line (Hm, ΔMFCS1/+) clearly exhibited syndactyly, similar to the original Hm heterozygotes (Fig. S3C). Therefore, the presence of MFCS1 in cis relative to Hm is not necessary to generate the syndactyly phenotype of Hm. In other words, the limb enhancer MFCS1 is dispensable for Hm action.
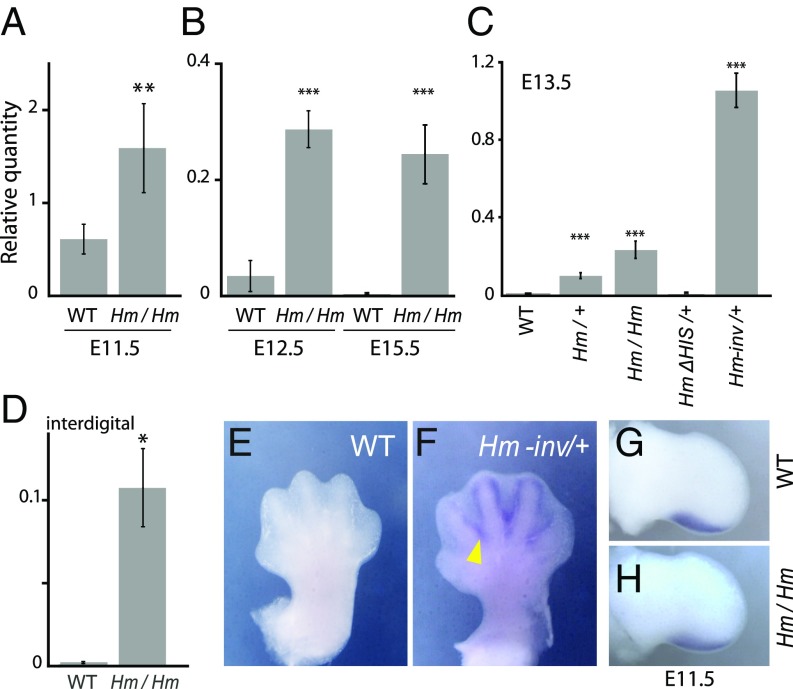
To determine the effect of HIS on Shh regulation, we analyzed the expression level of Shh mRNA, using qRT-PCR, in the Hm mutant limb at different stages from E11.5 to E15.5. We first examined Shh expression at the early embryonic stage (E11.5), when Shh expression remains in the ZPA. Expression was observed in both the Hm homozygote and the wild-type embryo, but its level was 2.6-fold higher in the Hm homozygote (Fig. 4A). After Shh expression ceases in the ZPA, interdigital cell death proceeds from E12.5 to E15.5. During these later stages, significantly higher Shh expression was observed in the Hm limb than in the wild-type limb (Fig. 4B).
Fig. 4.
Ectopic Shh expression in Hm. (A–D) Expression level of Shh in the autopod was measured by qRT-PCR. Expression level was normalized as 1/1,000 of the value for Actb. (A) E11.5 limb bud, n = 8; (B) E12.5 and E15.5 autopod, n = 8; (C) E13.5 autopod, n = 8 for each genotype; and (D) E13.5 interdigital region, n = 3. (E and F) Ventral views of in situ hybridization of Shh in E13.5 forelimb of wild-type (E) and Hm-inv/+ (F) mice. The yellow arrowhead in F indicates interdigital expression of Shh. (G and H) In situ hybridization of Shh in E11.5 hind limb of wild-type (G) and Hm/Hm (H) mice. *P < 0.05, **P < 0.001, ***P < 0.0001 in Welch’s t test.
We examined the dose effect of the Shh expression level on phenotypic severity at E13.5. We found that the expression level correlated well with dose of the Hm allele, and the Hm-inv allele showed an extremely high level of Shh expression (Fig. 4C). When HIS was removed from Chr5, the heterozygote (HmΔHIS/+) did not show ectopic expression of Shh. Thus, the level of ectopic Shh expression correlated with the severity of syndactyly.
Next, we analyzed the localization of ectopic Shh expression in the autopod. RT-PCR of mRNA prepared from the interdigital region at E13.5 showed ectopic expression of Shh in the Hm homozygote but not in the wild-type embryo (Fig. 4D). This ectopic Shh expression was not detected by in situ hybridization in the Hm homozygote, probably due to its lower expression level. On the other hand, in situ hybridization of the Hm-inv/+ embryo clearly showed ectopic Shh expression in the interdigital region (Fig. 4 E and F, yellow arrowhead), reflecting the high level of Shh mRNA (Fig. 4C). Shh expression was more abundant on the ventral side than the dorsal side, consistent with the LacZ expression pattern of the BAC transgenic mouse (Fig. 2I), as well as with the biased interdigital cell death along the dorsoventral axis.
There was no obvious difference in the Shh expression pattern in the limb bud between the Hm homozygote and wild-type embryo at the early stage of limb development (E11.5) (Fig. 4 G and H). This result is consistent with the finding that Hm did not exhibit abnormality in digit number. Taken together, these results indicate that HIS acts as a new regulatory region that induces Shh expression in the interdigital region at the later stages of limb development when Shh expression has normally ceased in the ZPA.
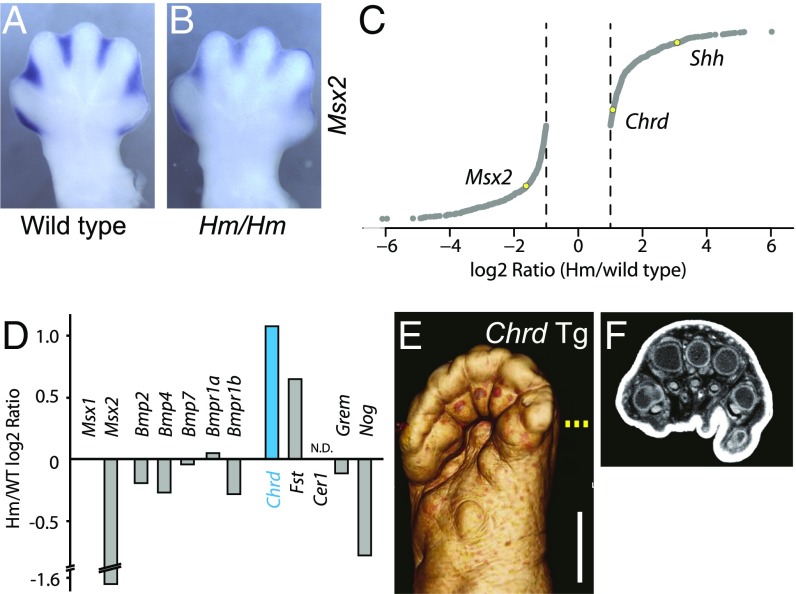
BMP signaling positively regulates interdigital cell death (23). To investigate whether the ectopic Shh expression in Hm affects BMP signaling, we examined the expression level of Msx2, a BMP downstream target (24, 25). In situ hybridization showed clearly that Msx2 was down-regulated in the Hm mutant (Fig. 5 A and B). This result suggested that the ectopic Shh expression suppresses interdigital cell death through BMP down-regulation. To uncover the molecular changes of BMP signaling in the developing Hm limb, we analyzed the interdigital transcriptome by microarray and found a total of 892 differentially expressed genes, including Shh and Msx2 (|log2(Hm/WT)| > 1; Fig. 5C). We focused on those involved in BMP signaling. In contrast to Msx2, Msx1 expression level was unchanged (Fig. 5D). There was no marked change in expression level of the BMP and BMP receptor genes. On the other hand, among BMP antagonists, only Chordin (Chrd) was included in the 892 differentially expressed genes. To test the possibility that the up-regulation of Chrd suppresses BMP signaling in the Hm limb, we generated a transgenic mouse that overexpresses Chrd in the limb. We observed soft tissue syndactyly, similar to the Hm phenotype, in the transgenic mouse (Fig. 5 E and F, n = 3/6). This successful phenocopying of syndactyly by the ectopic expression of Chrd suggests that Chrd changes may also mediate the down-regulation of BMP signaling in the Hm limb.
Fig. 5.
Down-regulation of BMP signaling in Hm. (A and B) Ventral view of in situ hybridization of a BMP-downstream gene, Msx2, in E13.5 hind limb. (C) Differentially expressed genes in the Hm interdigital region compared with the wild-type interdigital region by microarray. Yellow dots indicate Shh, Chrd, and Msx2. (D) Fold change of expression level of BMP signaling genes by microarray. (E) Micro-CT image of a ventral view of the right hind limb of a Chrd-transgenic mouse at P0. (Scale bar, 1 mm.) (F) Transverse section of the micro-CT image in E. Sectioning position is shown by the yellow dotted line in E.
Discussion
Enhancer Adoption by Genomic Insertion in Hm.
Genetic mechanisms underlying enhancer acquisition are currently categorized into three types: de novo acquisition by accumulation of nucleotide substitutions, insertion of a TE, and acquisition by chromosomal rearrangement (3). The third type, in which a preexisiting enhancer for one gene is captured by another gene, is termed enhancer adoption (26). While de novo acquisition requires a long time for a new enhancer to emerge, the latter two mechanisms, insertion of a TE and chromosomal rearrangement, can take place rapidly and saltationally. Many cases of enhancer acquisition by TE insertion have been reported (27, 28). For example, retrotransposition of a short interspersed element created a new enhancer to regulate Fgf8 expression in mouse forebrain and may have contributed to the evolution of the mammalian neuronal networks (29). By contrast, there are very few reports that enhancer adoption due to chromosomal rearrangement has occurred in evolution (30, 31).
This study revealed that a 150-kb genomic fragment, which was derived from Chr14, was inserted into the region upstream of Shh in Chr5 of the Hm genome. As a consequence, Shh was ectopically expressed in the interdigital region and led to syndactyly and web formation. This syndactyly phenotype is very similar to the previously reported case of enhancer adoption by Shh (26). Removing HIS from Chr5 in the Hm genome restored normal Shh expression and limb morphology, which demonstrated that the insertion of HIS is the direct cause of syndactyly with webbing. Thus, this study provides direct evidence that enhancer adoption can cause morphological alteration. Our BAC-transgenic reporter assay revealed that HIS in Chr14 of wild-type mouse also has enhancer activity in the interdigital region (Fig. 2 H and I). The microarray assay detected expression of Slitrk6, which neighbors HIS, in the interdigital region of wild-type mouse (Dataset S5), implying that HIS at the original position in Chr14 has tissue-specific enhancer activity. On the other hand, Slitrk6 KO mice show a reduction of innervation in the cochlea but no external abnormality (32). Given that removing HIS from mouse Chr14 did not cause any visible phenotype, any regulatory activity of the original HIS in Chr14 has little impact on limb development.
The result of a series of partial deletion experiments clearly showed that at least two regions in HIS are necessary for the syndactyly phenotype of Hm (Fig. S2). One of them, region 1, contains the interdigital enhancer e2 (Fig. 2K). On the other hand, region 2 contains two enhancers, e5 and e7, which induce reporter expression broadly in autopod and specifically in interdigits, respectively (Fig. 2 M, N, and P). These results imply that neither one of the three enhancers represents the total regulatory activity of HIS. Instead, it is most likely that a cooperative activity of these enhancers is necessary for the interdigital regulation of Shh in Hm. It is consistent with recent reports that many genes are regulated by multiple enhancers (33–35). Thus, in general, insertion of a large genomic fragment such as HIS can cause enhancer adoption, bringing a set of enhancers, which eventually leads to morphological change.
Recent whole-genome sequencing analyses have revealed a vast number of structural variants, a median of 18.4 Mbp per diploid genome, in human individuals. It is interesting to note that they are enriched around expression-quantitative trait loci (36). In addition, potential enhancers exist throughout the vertebrate genome (37). Taking all these findings into account, it is possible that numerous sequences in human genomes have potential enhancer activity and that enhancer adoption by chromosomal rearrangement occurs frequently in human populations. Recent reports that certain genetic diseases are caused by enhancer adoption (26, 38) support this assertion.
The limb enhancer MFCS1 is known to topologically interact with the Shh promoter beyond the long distance (39), and we found a small or negligible effect of HIS on the enhancer activity of MFCS1. This implies that frequent enhancer adoption can occur at the genome-wide scale, adding novel expression domains without disturbing original enhancer activity.
We found that the HIS inversion in Chr5 dramatically changed Shh expression level and resulted in a much severer morphological defect than that of the original Hm heterozygote. In addition to syndactyly, the inversion caused other abnormalities including increased muscle volume in the autopod and cleft palate, the latter of which is perhaps the cause of perinatal lethality. Few studies to date have documented orientation-dependent enhancer activity (40, 41). Recently, it was reported that orientation-dependent CTCF binding sites determine topological DNA looping for promoter–enhancer interaction and thereby exert proper gene regulation (42). We could not identify any CTCF binding sites in the original HIS in Chr14 by in silico searching of public databases. It is possible that another unknown orientation-sensitive sequence, rather than CTCF binding sites, exists in HIS.
Ectopic Shh Expression and Downstream Targets.
Ectopically expressed Shh in the Hm mutant up-regulates Chrd expression and down-regulates Msx2, a BMP signaling marker (Fig. 5). BMP signaling positively regulates interdigital cell death (43, 44). Since CHRD antagonizes BMP signaling (45), it is reasonable that up-regulation of Chrd via ectopic Shh expression results in inhibition of interdigital cell death. Indeed, our transgenic expression of Chrd clearly recapitulated syndactyly, consistent with earlier findings that ectopic expression of Gremlin (Grem) and Noggin (Nog), two other BMP antagonists, also inhibited interdigital cell death (46, 47). This molecular model of syndactyly in Hm is consistent with previous reports that ectopic expression of Shh suppresses interdigital cell death (26, 48).
Proper spatiotemporal gene regulation is essential for executing the developmental program. When the ectopic expression level of Shh was elevated by the inversion of HIS, digit patterning did not change. It is known that ectopic Shh expression in the anterior region of the early stage limb bud causes polydactyly (14, 17). In another mutant, Short digits (Dsh), ectopic Shh expression in the joint region causes brachydactyly with shortened digit length (49). Thus, both the timing and the pattern of ectopic expression of Shh are crucial for proper limb development. A change of Shh expression induces different downstream targets of Shh. In the early limb bud (approximately E9.0 to E12), in contrast to CHRD, another BMP antagonist, GREM, is induced under the control of SHH signaling (50). This sensitivity of GREM to SHH signaling disappears approximately when Shh expression ceases in the ZPA, which results in termination of the SHH–GREM–FGF feedback loop (51). This switching of sensitivity from early to later stages in limb development likely alters the downstream pathway of SHH signaling. In the Hm limb, we could not detect any difference in Fgf4 expression, suggesting that early developmental events relying on the SHH–GREM–FGF feedback loop terminated properly in the Hm limb.
Reduction of interdigital cell death, which leads to interdigital web formation, arose recurrently in independent lineages of different taxa. In this convergent evolution, gain of Grem expression was purportedly a molecular mechanism by which interdigital webbing emerged in the limbs of duck and bat (46, 52). We propose that gain of expression of Chrd, another BMP antagonist, may also have contributed to interdigital web formation in nature.
Materials and Methods
Microarray data have been deposited in the GEO of NCBI under accession no. GSE98424 and are available in Dataset S5. ATAC-seq data have been deposited in the DRA of DDBJ under accession no. DRA006161. An extended description of the materials and methods is provided in SI Materials and Methods.
Animal experiments in this study were approved by the Animal Care and Use Committee of National Institute of Genetics (NIG).
Supplementary Material
Acknowledgments
We thank Y. Mizushina, K. Fukunaga, A. Kondo, Y. Suzuki, and H. Nakazawa for generating transgenic mice and mutants; Dr. A. McMahon for the Shh riboprobe and Dr. R. Hill for the Msx2 riboprobe; and the staff of Comparative Genomics Laboratory at NIG for supporting genome sequencing. This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS) (Grants JP15J06985, JP17K15162, and JP17K19411).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data have been deposited in the Gene Expression Omnibus (GEO) database of NCBI, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98424). ATAC-seq data have been deposited in the DRA of DNA Data Bank of Japan (DDBJ), www.ddbj.nig.ac.jp/index-e.html (accession no. DRA006161).
See Commentary on page 839.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713339115/-/DCSupplemental.
References
- 1.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, et al. Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell. 2016;167:355–368.e10. doi: 10.1016/j.cell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinstein M, de Souza FS. Evolution of transcriptional enhancers and animal diversity. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130017. doi: 10.1098/rstb.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki R, et al. Functional roles of Aves class-specific cis-regulatory elements on macroevolution of bird-specific features. Nat Commun. 2017;8:14229. doi: 10.1038/ncomms14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 6.Santangelo AM, et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spielmann M, Mundlos S. Structural variations, the regulatory landscape of the genome and their alteration in human disease. BioEssays. 2013;35:533–543. doi: 10.1002/bies.201200178. [DOI] [PubMed] [Google Scholar]
- 8.Conrad DF, et al. Wellcome Trust Case Control Consortium Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.English AC, et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics. 2015;16:286. doi: 10.1186/s12864-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green MC. New mutation. Mouse News Lett. 1964;31:27. [Google Scholar]
- 11.Kimura S, Schaumann BA, Shiota K. Ectopic dermal ridge configurations on the interdigital webbings of Hammertoe mutant mice (Hm): Another possible role of programmed cell death in limb development. Birth Defects Res A Clin Mol Teratol. 2005;73:92–102. doi: 10.1002/bdra.20108. [DOI] [PubMed] [Google Scholar]
- 12.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 13.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 14.Sagai T, et al. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh) Mamm Genome. 2004;15:23–34. doi: 10.1007/s00335-033-2317-5. [DOI] [PubMed] [Google Scholar]
- 15.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 16.Lettice LA, Hill AE, Devenney PS, Hill RE. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum Mol Genet. 2008;17:978–985. doi: 10.1093/hmg/ddm370. [DOI] [PubMed] [Google Scholar]
- 17.Masuya H, et al. A series of ENU-induced single-base substitutions in a long-range cis-element altering Sonic hedgehog expression in the developing mouse limb bud. Genomics. 2007;89:207–214. doi: 10.1016/j.ygeno.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Sweet HO. Hm and Hx are not alleles. Mouse News Lett. 1982;66:66. [Google Scholar]
- 19.Clark RM, Marker PC, Kingsley DM. A novel candidate gene for mouse and human preaxial polydactyly with altered expression in limbs of Hemimelic extra-toes mutant mice. Genomics. 2000;67:19–27. doi: 10.1006/geno.2000.6225. [DOI] [PubMed] [Google Scholar]
- 20.Furuse T, Takano-Shimizu T, Moriwaki K, Shiroishi T, Koide T. QTL analyses of spontaneous activity by using mouse strains from Mishima battery. Mamm Genome. 2002;13:411–415. doi: 10.1007/s00335-002-2168-5. [DOI] [PubMed] [Google Scholar]
- 21.Sagai T, et al. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136:1665–1674. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- 22.Sagai T, et al. Evolution of Shh endoderm enhancers during morphological transition from ventral lungs to dorsal gas bladder. Nat Commun. 2017;8:14300. doi: 10.1038/ncomms14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuzarte-Luís V, Hurlé JM. Programmed cell death in the developing limb. Int J Dev Biol. 2002;46:871–876. [PubMed] [Google Scholar]
- 24.Chen Y, Zhao X. Shaping limbs by apoptosis. J Exp Zool. 1998;282:691–702. [PubMed] [Google Scholar]
- 25.Lallemand Y, et al. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005;132:3003–3014. doi: 10.1242/dev.01877. [DOI] [PubMed] [Google Scholar]
- 26.Lettice LA, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- 27.Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- 28.Nishihara H, et al. Coordinately co-opted multiple transposable elements constitute an enhancer for wnt5a expression in the mammalian secondary palate. PLoS Genet. 2016;12:e1006380. doi: 10.1371/journal.pgen.1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki T, et al. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cande JD, Chopra VS, Levine M. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development. 2009;136:3153–3160. doi: 10.1242/dev.038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci USA. 2011;108:10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katayama K, et al. Disorganized innervation and neuronal loss in the inner ear of Slitrk6-deficient mice. PLoS One. 2009;4:e7786. doi: 10.1371/journal.pone.0007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of dorsal, twist, and snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antosova B, et al. The gene regulatory network of lens induction is wired through Meis-dependent shadow enhancers of Pax6. PLoS Genet. 2016;12:e1006441. doi: 10.1371/journal.pgen.1006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagai T, et al. SHH signaling directed by two oral epithelium-specific enhancers controls tooth and oral development. Sci Rep. 2017;7:13004. doi: 10.1038/s41598-017-12532-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudmant PH, et al. 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupiáñez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amano T, et al. Chromosomal dynamics at the Shh locus: Limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Swamynathan SK, Piatigorsky J. Orientation-dependent influence of an intergenic enhancer on the promoter activity of the divergently transcribed mouse Shsp/alpha B-crystallin and Mkbp/HspB2 genes. J Biol Chem. 2002;277:49700–49706. doi: 10.1074/jbc.M209700200. [DOI] [PubMed] [Google Scholar]
- 41.Hozumi A, et al. Enhancer activity sensitive to the orientation of the gene it regulates in the chordate genome. Dev Biol. 2013;375:79–91. doi: 10.1016/j.ydbio.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandyopadhyay A, et al. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pajni-Underwood S, Wilson CP, Elder C, Mishina Y, Lewandoski M. BMP signals control limb bud interdigital programmed cell death by regulating FGF signaling. Development. 2007;134:2359–2368. doi: 10.1242/dev.001677. [DOI] [PubMed] [Google Scholar]
- 45.Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: Inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merino R, et al. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- 47.Guha U, Gomes WA, Kobayashi T, Pestell RG, Kessler JA. In vivo evidence that BMP signaling is necessary for apoptosis in the mouse limb. Dev Biol. 2002;249:108–120. doi: 10.1006/dbio.2002.0752. [DOI] [PubMed] [Google Scholar]
- 48.Sanz-Ezquerro JJ, Tickle C. Autoregulation of Shh expression and Shh induction of cell death suggest a mechanism for modulating polarising activity during chick limb development. Development. 2000;127:4811–4823. doi: 10.1242/dev.127.22.4811. [DOI] [PubMed] [Google Scholar]
- 49.Niedermaier M, et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J Clin Invest. 2005;115:900–909. doi: 10.1172/JCI200523675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zúñiga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 51.Scherz PJ, Harfe BD, McMahon AP, Tabin CJ. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- 52.Weatherbee SD, Behringer RR, Rasweiler JJ, 4th, Niswander LA. Interdigital webbing retention in bat wings illustrates genetic changes underlying amniote limb diversification. Proc Natl Acad Sci USA. 2006;103:15103–15107. doi: 10.1073/pnas.0604934103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.