Significance
GDF8 is a signaling protein that inhibits muscle mass. Inhibitors of GDF8 are highly sought as therapeutics for the treatment of muscle-wasting diseases. During synthesis, GDF8 is made as a precursor where the signaling segment is cleaved from the N-terminal prodomain, which remains associated and inhibits signaling. Activation involves an additional cleavage of the prodomain. We demonstrate GDF8 signaling could be gained through a conformational change where the prodomain remains associated with the signaling segment. Alteration of the prodomain can weaken the interactions causing GDF8 to signal, thus alleviating inhibition by the prodomain. This study illuminates how GDF8 transitions from an inhibited state to an active state—information that will help to understand the mechanism of GDF8 signaling.
Keywords: GDF8, latent activation, GDF8 prodomain, myostatin activation, myostatin prodomain
Abstract
Growth/differentiation factor 8 (GDF8), or myostatin, negatively regulates muscle mass. GDF8 is held in a latent state through interactions with its N-terminal prodomain, much like TGF-β. Using a combination of small-angle X-ray scattering and mutagenesis, we characterized the interactions of GDF8 with its prodomain. Our results show that the prodomain:GDF8 complex can exist in a fully latent state and an activated or “triggered” state where the prodomain remains in complex with the mature domain. However, these states are not reversible, indicating the latent GDF8 is “spring-loaded.” Structural analysis shows that the prodomain:GDF8 complex adopts an “open” configuration, distinct from the latency state of TGF-β and more similar to the open state of Activin A and BMP9 (nonlatent complexes). We determined that GDF8 maintains similar features for latency, including the alpha-1 helix and fastener elements, and identified a series of mutations in the prodomain of GDF8 that alleviate latency, including I56E, which does not require activation by the protease Tolloid. In vivo, active GDF8 variants were potent negative regulators of muscle mass, compared with WT GDF8. Collectively, these results help characterize the latency and activation mechanisms of GDF8.
One of the most thoroughly described negative regulators of skeletal muscle mass is the TGF-β superfamily ligand growth/differentiation factor 8 (GDF8), also known as myostatin (1, 2). Genetic disruption of Gdf8 results in substantial skeletal muscle growth (1, 2). Further, a significant increase in muscle fiber size is also observed when adult animals are treated with agents that bioneutralize GDF8 (reviewed in ref. 3). As such, targeted inhibition of GDF8 is currently being pursued for the treatment of skeletal muscle-related disorders and associated symptoms (4, 5).
GDF8, like numerous TGF-β family members, is a disulfide-linked dimer that is synthesized as a precursor protein which requires cleavage by a furin-like protease to yield an N-terminal prodomain and a C-terminal mature, signaling domain (6). Interestingly, for a number of TGF-β ligands the role of the prodomain extends beyond ligand maturation and folding support (7, 8), remaining noncovalently associated with the mature ligand following secretion in either a low-affinity, noninhibitory or high-affinity, inhibitory fashion (reviewed in ref. 9). For example, the prodomains of TGF-β1, TGF-β2, TGF-β3, GDF11, and GDF8 hold the mature ligand in a latent or inactive state mediated by a noncovalent, yet high-affinity, ligand-specific interaction (6, 10–13), whereas mature Activin A and BMP9 remain associated with, but are not inhibited by, their prodomain (14, 15). Activation of TGF-β1 and TGF-β3 requires covalent interactions with the extracellular matrix and cellular contractile forces to release the mature ligand (16–18). In fact, resolution of the latent TGF-β1 crystal structure provided a molecular explanation for how latency is exerted by the prodomain via a coordinated interaction between the N-terminal alpha helix (alpha-1), latency lasso, and fastener of the prodomain with type I and type II receptor epitopes of the mature domain (18). However, GDF8 activation requires a second cleavage event within the prodomain via proteases from the BMP1/Tolloid (TLD) family of metalloproteases (13). However, the molecular and structural details of the GDF8 latent state have yet to be determined.
Based on sequence conservation and prior biochemical data describing the N-terminal portion of the GDF8 prodomain (10), it is plausible that the molecular interactions and overall structure of the GDF8 latent complex may be similar to that of TGF-β1. However, the prodomains of a number of TGF-β family members share similar sequence conservation, yet they do not regulate the mature ligand in the same fashion and also exhibit significant structural diversity (14, 15). Therefore, while one might expect that GDF8 and TGF-β1 would share certain elements for how the prodomain binds and confers latency, it is possible that significant structural and molecular differences in these interactions occur as they exhibit profoundly different mechanisms of activation. However, this comparison is hindered by a lack of understanding of the GDF8 latent complex at the molecular level.
In this study, we utilized small-angle X-ray scattering (SAXS) and mutagenesis to characterize the GDF8 latent complex. Interestingly, SAXS analysis reveals that the GDF8 latent complex adopts a more “open” conformation, similar to the overall structure of the BMP9 and Activin A prodomain complexes, which are not latent. The open conformation of the GDF8 latent complex is in stark contrast to the “closed” conformation adopted by the TGF-β1 latent complex. Furthermore, we identify key residues in the GDF8 prodomain that are responsible for promoting latency, indicating that GDF8 and TGF-β1 share similar features for latency including a latency lasso. We further show that certain mutations in the prodomain of GDF8 can reduce latency, producing a more active ligand both in vitro and in vivo. Overall, our data provide insight toward the molecular mechanisms of GDF8 latency and activation.
Results
Prodomain–GDF8 Can Exist in a Latent and Active Complex.
Initial characterization in adult mice showed that GDF8 is secreted into the systemic circulation as a latent protein complex that requires activation to trigger GDF8 signaling (11). While the biological mechanism for activation remained unknown, it was shown that a GDF8-specific signal derived from the serum of a WT mouse, but not a Gdf8−/− mouse, could be detected following exposure to acidic conditions, referred to here as “acid activation” (11). The premise for acid activation stemmed from a similar observation that was made during the characterization of TGF-β, which is similarly regulated by its prodomain (19, 20), and provided the initial basis that latent GDF8 and latent TGF-β are likely to be very similar in terms of activation and prodomain release.
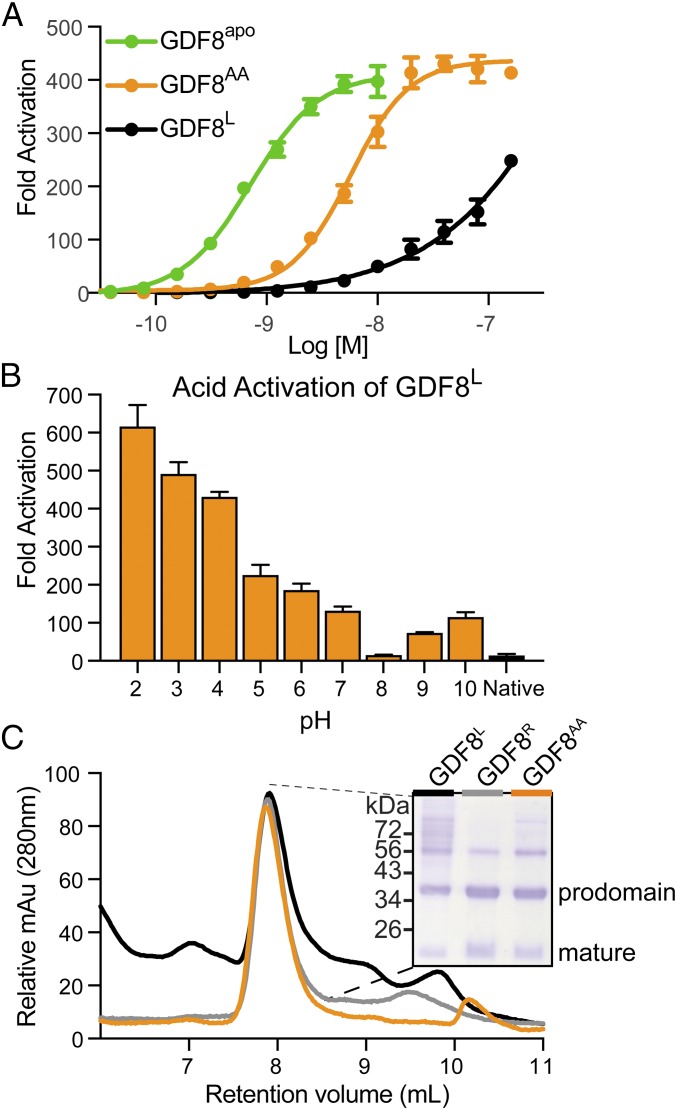
While a molecular basis to describe how acid activation alleviates ligand latency remains unknown, it is thought that the acidic conditions simply dissociate the prodomain from the mature domain, thereby freeing the ligand from inhibition (11). However, our initial attempts to purify the mature domain from the prodomain after acid activation using an affinity column to the high-affinity antagonist, follistatin, failed, even though the complex exhibited significant activity. This observation suggested that perhaps the prodomain remained bound to the mature domain but was not in a fully inhibitory state. To extend these initial observations, we isolated the mammalian-derived latent proGDF8 complex (GDF8L) and compared its signaling activity to both the acid-activated state (GDF8AA) and to the mature, unbound GDF8 (GDF8apo) using a SMAD3-responsive (CAGA)12 luciferase-reporter HEK293 cell line (21–25). As expected and consistent with our previous report, GDF8apo readily signaled with a calculated half-maximal effective concentration (EC50) of 0.72 nM (25), whereas media containing GDF8L did not readily signal and required nearly 10,000-fold more protein to achieve a similar response compared with GDF8apo (Fig. 1A). In contrast, acid activation of media containing GDF8L at pH 2 to generate GDF8AA resulted in a significant gain in activity compared with non-acid-activated latent GDF8 (Fig. 1A). Interestingly, the calculated EC50 for GDF8AA (5.7 nM) still did not reach the EC50 of GDF8apo, suggesting that under these conditions we were unable to observe the full signaling potential of mature GDF8. Since GDF8apo is stable and stored in 10 mM HCl, we do not expect this difference in activity to be caused by subjecting GDF8L to extreme conditions. We next evaluated the activation of GDF8L as a function of pH by subjecting the complex to various pH ranges (pH 2–10) for 1 h, followed by neutralization and (CAGA)12 activation (Fig. 1B). We determined that at the concentration tested (40 nM) the level of activation increases with a decrease in pH (Fig. 1B); however, substantial activation was observed throughout the pH range examined. The shape of the titration experiment suggests that multiple ionizable groups could be involved in the latency mechanism or that shifts in the pH cause disruption in the structure of the prodomain that affects its ability to inhibit GDF8.
Fig. 1.
Activity and analysis of the latent GDF8 prodomain complex. (A) HEK293 (CAGA)12 cells treated with latent GDF8 prodomain complex (GDF8L; black), acid-activated (GDF8AA; orange), and free mature (GDF8apo; green) ligand. Experiments were performed at least twice with each data point measured in triplicate. Shown is a representative experiment. Data were fit by nonlinear regression to a variable slope to determine the EC50. (B) Activity measurement following 1-h incubation of purified GDF8L (40 nM) at the indicated pH, followed by neutralization before exogenous administration to HEK293 (CAGA)12 cells. Data are shown as the mean ± SEM. (C) SEC analysis of GDF8L, GDF8AA, and reformed (GDF8R) prodomain:ligand complexes. (Inset) The protein composition of the peak visualized by SDS/PAGE under nonreducing conditions.
Given that titration of the GDF8 prodomain against mature GDF8 results in potent ligand inhibition (10), we hypothesized that we were unable to recover the full signal from acid-activated latent GDF8 due to the possibility that the prodomain may still be able to provide some level of antagonism, through a noncovalent interaction, despite being acid-activated. To test this hypothesis, we subjected GDF8L and GDF8AA to size-exclusion chromatography (SEC) followed by SDS/PAGE/Coomassie staining. In addition, we combined isolated GDF8 prodomain with GDF8apo, both components derived from mammalian cell expression, and applied the mixture to SEC. We determined that all three variations of GDF8 complexes had similar retention volumes with both components (coelution of the prodomain and GDF8apo), indicative of complex formation (Fig. 1C). This result supports the idea that during the acid activation the prodomain can reassociate with the mature domain and partially inhibit signaling. However, since the GDF8AA has significant activity, it also suggests that the latent interaction between the prodomain and mature domain is not completely reversible. Nevertheless, our finding that following exposure to acidic conditions mature GDF8 remains associated with the prodomain, but in an active state, provided the opportunity for further comparison with latent GDF8.
SAXS Analysis Reveals Conformational Differences Between GDF8 and Other TGF-β Prodomain–Ligand Complexes.
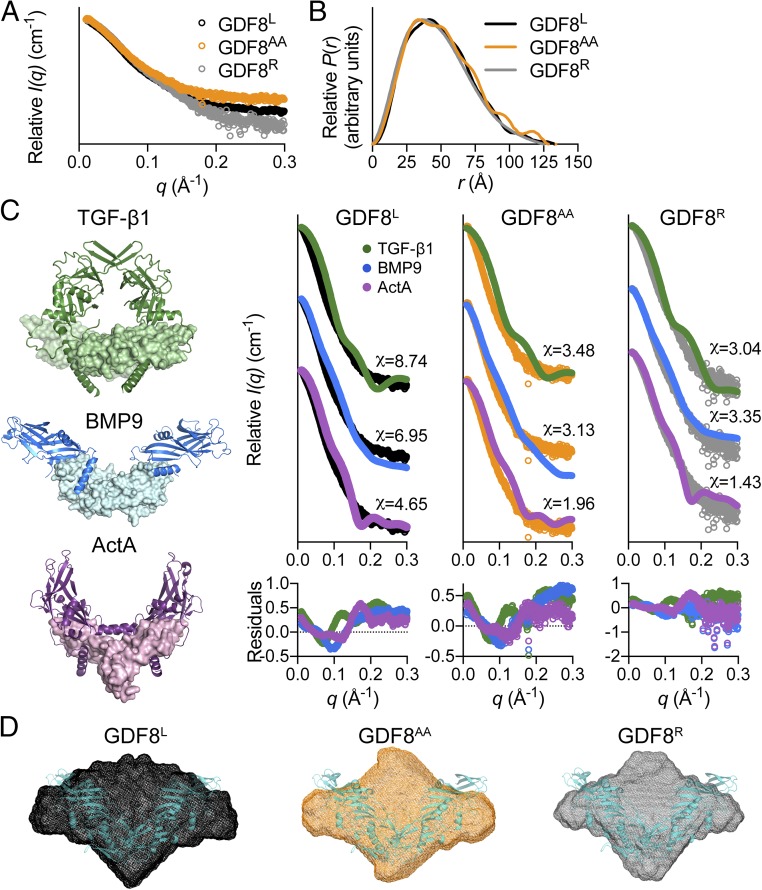
While the aforementioned data suggest that the latent and acid-activated forms of GDF8 could adopt different molecular states, limited structural information is available for the prodomain:GDF8 complex. Given that high-resolution structural information for prodomain:ligand complexes of latent TGF-β1 (18) and nonlatent ligands BMP9 (14) and Activin A (15) have demonstrated a series of configurations that range in compactness, we next wanted to determine how the prodomain:GDF8 complex compared with these other prodomain:ligand complexes. Therefore, we used the solution-based technique SAXS to analyze the purified prodomain:GDF8 complexes, including GDF8L, recombined and purified prodomain:GDF8 (GDF8R), and GDF8AA (Fig. 2 and Table S1). Samples were well-behaved in solution and did not show evidence of interparticle repulsion or aggregation over multiple protein concentrations (Fig. 2A and Table S1). From the Gunier analysis we determined that GDF8L has a lower radius of gyration (Rg) than GDF8AA, 41.1 ± 0.85 versus 46.8 ± 0.86 Å (Table S1), respectively, which suggests that acid activation of GDF8L altered the overall conformation of the complex. This is further supported by the appearance of a more “featured” pairwise distribution plot [P(r)] for the GDF8AA complex compared with the GDF8L complex (Fig. 2B). Additionally, we determined that the GDF8R complex had a scattering profile, pairwise distribution curve, and associated SAXS-derived values similar to those of the GDF8L complex (Fig. 2 A and B and Table S1).
Fig. 2.
SAXS analysis of latent, acid-activated, and reformed GDF8 prodomain complex. (A) SAXS scattering profile showing the intensity distribution and (B) the pairwise distribution function for the various GDF8 prodomain complexes. (C) The crystal structures of various prodomain:ligand complexes used to generate theoretical scattering profiles for comparison [TGF-β1, Protein Data Bank (PDB) ID code 3RJR (18); BMP9, PDB ID code 4YCG (14); ActA (Activin A), PDB ID code 5HLY (15)]. The chi (χ) value was determined using the FoXS webserver (26). Residuals for each comparison are shown below the scattering profiles. Note that the latent TGF-β1 structure exemplifies a closed conformation unlike the nonlatent, but prodomain:ligand-associated BMP9 and Activin A (ActA) structures are in an open conformation. (D) Ab initio SAXS envelope (DAMFILT model) of the GDF8L (black), GDF8AA (orange), and GDF8R (gray) complexes. Note that the reconstruction of the GDF8AA complex appears more elongated compared with the other GDF8 prodomain complexes. The recently resolved GDF8 prodomain complex crystal structure [PDB ID code 5NTU (37)], shown in teal, is superimposed on the various ab initio molecular envelopes.
To determine if the GDF8L complex adopted a conformation similar to that of the other known prodomain:ligand structures (Fig. 2C) we compared our experimental scattering profile to the theoretical scattering profile using FoXS (Fig. 2D) (26). We first compared the experimental profile of GDF8L to the theoretical profiles based on the prodomain:ligand structures of TGF-β1, BMP9, and Activin A. This analysis showed that the overall structure of the GDF8L complex did not show substantial similarity to any structure as indicated by the calculated chi values (Fig. 2D), which is also consistent with a larger Rg value than the other prodomain:ligand structures. Nevertheless, the most similarity was found with Activin A (χ = 4.65), which has an open conformation, whereas the least similarity was found with TGF-β1 (χ = 8.74; Fig. 2D). Interestingly, both the GDF8AA and GDF8R complexes were more similar to Activin A (χ = 1.96 and χ = 1.43, respectively) while still a poor fit with BMP9 (χ = 3.13 and χ = 3.35, respectively) and TGF-β1 (χ = 3.48 and χ = 3.04, respectively), suggesting that GDF8AA and GDF8R are also likely in an open conformation and that there are likely additional differences in these complexes compared with the GDF8L complex. To extend these observations, we calculated the SAXS-derived ab initio molecular envelopes for each state. The overall shape of the envelopes for each state further supported our initial observation that structural differences likely exist between the activity states (Fig. 2E). However, following superposition of the prodomain:GDF8 complex crystal structure, which was resolved during the preparation of this paper (Discussion), we observed that there are poorly defined regions within the envelopes, which may be the result of structural flexibility inherent to the GDF8 prodomain complexes.
Specific Mutations Within the Prodomain Enhance GDF8 Activity.
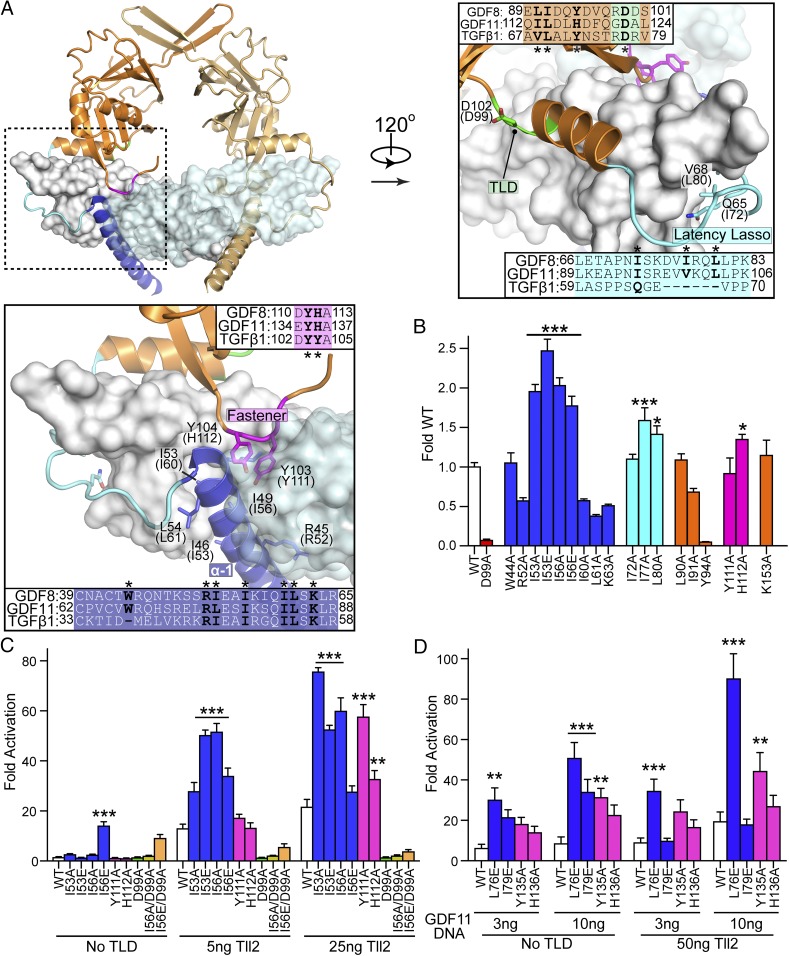
Our SAXS analysis revealed that the GDF8L complex likely adopts a different overall conformation compared with TGF-β1. Despite this, GDF8 and TGF-β1 share high sequence conservation in the N-terminal alpha-1 helix, latency lasso, alpha-2 helix, and fastener regions (Fig. 3A). Thus, we hypothesized that these regions could interact with the mature GDF8 ligand and are important for forming the noncovalent interactions required for latency, such that removing these interactions might generate a more active GDF8 ligand (i.e., remove latency). One might also expect that disruption of these interactions might disrupt folding, as observed for TGF-β1 (27). Therefore, to test our hypothesis, we utilized the TGF-β1 structure as a guide to systematically mutate specific residues in regions of the GDF8 prodomain and compared their activity to WT GDF8L. For this evaluation we developed a robust cell-based (CAGA)12 luciferase-reporter assay where we could assess the variants through transient transfection. Our first goal was to determine which TLD family protease member [e.g., BMP1/mTLD, tolloid-like 1 (Tll1) or tolloid-like 2 (Tll2)] yielded the most optimal activation of WT GDF8L (13, 28). Using an assay format similar to one previously described (25), we compared the activity of WT GDF8 following transient cotransfection of WT GDF8, furin, and either BMP1, Tll1, or Tll2 using HEK293 (CAGA)12 luciferase cells (Fig. S1A). As predicted, we observed little to no signal when the TLDs were not included in the assay, indicating that little to no basal TLD is present and incapable of activating GDF8L (Fig. S1A). However, when cells were cotransfected with DNA from one of the three TLDs, we observed a dose-dependent increase in signal with increasing concentrations of WT GDF8 DNA (Fig. S1A). As predicted, we observed differences in the fold activation of WT GDF8 when cotransfected with the various TLDs, where the highest activation resulted from Tll2 (Tll2 > Tll1 > BMP1; Fig. S1A). Although this result is consistent with previous reports (13, 28) indicating that differences in the magnitude of activation by TLDs, we cannot rule out the possibility this increase in activity is due to differences in TLD protein expression levels or differential regulation of TLD maturation needed for activation (29–31). Regardless, since Tll2 was the most effective activator of WT GDF8, with increases ranging from 20- to 60-fold activation, it was used in the remaining assays unless otherwise noted.
Fig. 3.
Mutations within the GDF8 prodomain and activation by Tolloid. (A) Latent TGF-β1 structure highlighting inhibitory elements of the prodomain: alpha-1 helix (blue), latency lasso (cyan), and the fastener (magenta). Sequence alignment between TGF-β1, GDF8, and GDF11 are shown with residues investigated marked with an asterisk. Labels correspond to TGF-β1 with human GDF8 in parentheses. (B) Transfection assay to determine GDF8 mutant activity. HEK293 (CAGA)12 cells were cotransfected with 25 ng of each GDF8 mutant, 50 ng of furin (human), and 25 ng of Tll2 (human) DNA. Fold activation was determined by dividing the signal from no GDF8 plasmid (25 ng of empty vector, 50 ng of furin, and 25 ng of Tll2). (C) HEK293 (CAGA)12 cells were cotransfected with 25 ng of GDF8 mutant DNA, 50 ng of furin, 25 ng of empty psF-IRES, and 0, 5, or 25 ng of Tll2. The luciferase signal was normalized to Renilla. Fold activation was calculated similar to B. (D) Transfection assay to determine GDF11 mutant activity. HEK293 (CAGA)12 were cotransfected with 3 or 10 ng of the GDF11 mutant DNA and 50 ng of furin, with or without 50 ng of Tll2. Transfection, normalization, and fold activation were calculated as they were in C. All mentioned experiments were performed at least twice where individual points were measured in triplicate. Error is shown as mean ± SEM. Bar graphs were compared using one-way (B) or two-way (C and D) ANOVA with Bonferroni correction against WT (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
The panel of mutations is shown in Fig. 3B and is categorized based on the anticipated location in the prodomain. Within these regions we primarily focused our attention on mutation of hydrophobic residues, since hydrophobic interactions commonly drive known inhibitory interactions within the TGF-β family (reviewed in ref. 9). For example, GDF8 maintains a number of hydrophobic resides that are predicted to align to one side of the alpha-1 helix, similar to the register of TGF-β1. Of particular interest, we identified two hydrophobic residues in the alpha-1 helix, I53 and I56, which showed more than twofold higher activity compared with WT GDF8 when mutated to either an alanine (I53A, I56A) or glutamate (I53E and I56E; Fig. 3B). Additionally, mutation of residues outside of the alpha-1 helix, I77A within the latency lasso, and H112A within the fastener showed an increase in activity compared with WT, whereas the mutants generated in the alpha-2 helix did not show any significant gain in activity compared with WT (Fig. 3B). In contrast, Y94A resulted in little to no activity. As a control, we tested the activity of D99A, which has previously been shown to eliminate activation by TLD (13). As expected, introduction of D99A abolished activity, supporting that the assay is specific to the plasmid carrying the GDF8 gene. In addition, we tested the K153R mutant, which was previously shown to enhance furin processing but not to influence activation by TLD. K153R had activity similar to WT, indicating that TLD processing is optimal (32).
To validate these observations and perform a more rigorous cross-comparison between WT GDF8 and these mutants we inserted the ligand DNA into the pSF-CMV-FMDV-Rluc vector, which allowed us to normalize our data for transfection efficiency. We focused on the I53A/E and I56A/E mutants within the alpha-1 as well as the Y111A and H112A mutants within the fastener region due to their apparent importance when examining the structure of TGF-β1. This approach was used because previous efforts to detect the secreted ligand in the conditioned medium in this assay format were unsuccessful, likely due to protein levels below the limit of detection. We determined that all mutants retain significantly higher activity than WT GDF8 in a dose-dependent fashion with respect to titration of ligand DNA and Tll2 DNA (Fig. S1B). Given that activation of WT GDF8 is differentially regulated by the various TLDs, we tested whether or not our mutants retained higher activity when activated by the other TLDs, Tll1 and BMP1. Overall, our results indicated that our mutants were more active than WT GDF8, although there were a few differences in the activation across the various TLDs (Fig. S1C). Except for the I53A and I56A variants of the Ile mutations cotransfected with Tll2, all mutants showed enhanced activity in the presence of either Tll2 or Tll1, while only the Y111A and H112A mutants showed enhanced activity when cotransfected with BMP1 (Fig. S1C). These results were unexpected and likely suggest that the enhanced activity of our mutants may occur because of multiple mechanisms, such as whether or not TLD is still required for activation.
To determine if the enhanced ligand activity was dependent on TLD activity (i.e., TLD-dependent), we compared the activity of these mutants transfected with and without Tll2 (Fig. 3C). Interestingly, of the mutants tested, the I56E mutant showed significant activity compared with WT GDF8 in the absence of Tll2 (Fig. 3C), whereas the other mutants (I53A/E, Y111A, and H112A) required the presence of TLD for enhanced activity (Fig. 3C). To confirm that GDF8 with the I56E mutation is not dependent on TLD we generated the double mutant I56E/D99A, which would eliminate the potential for Tll2 activation. Similar to I56E, transfection of the I56E/D99A mutant showed enhanced activity, thus demonstrating that the I56E mutation results in nonlatent and active GDF8 ligand (Fig. 3C). However, we did observe that cotransfection of Tll2 further enhanced the activity of the I56E mutant, suggesting that more activity from this mutant can still be gained, but not in the presence of D99A. Thus, I56E has activity without the requirement of TLD, but TLD can further potentiate I56E’s activity. The I56A mutant did not show the same Tll2 independence as I56E (Fig. 3C), suggesting that introduction of the charged residue may destabilize the interaction between the prodomain and the mature ligand, perhaps by disrupting a hydrophobic pocket or core.
GDF11 is a closely related ligand to GDF8 and is regulated in a similar fashion as GDF8 in terms of latency and the requirement of TLD processing to alleviate latency (33). Therefore, we tested if mutation of similar residues in GDF11 as GDF8 would also enhance ligand activity (Fig. 3 A and D). To test this hypothesis, we assessed ligand activity following cotransfection of the HEK293 (CAGA)12 cells with and without Tll2 (Fig. 3D). Similar to that of GDF8, WT GDF11 activity was increased when Tll2 was present. Furthermore, mutation of similar residues in the alpha-1 (L76E and I79E) and fastener (Y135A and H136A) regions in GDF11 significantly enhanced ligand activity. Specifically, GDF11 L76E showed significantly enhanced ligand activity compared with WT that was independent of Tll2 (Fig. 3D). Interestingly, unlike in GDF8 (Y111A), GDF11 Y135A also showed enhanced ligand activity compared with WT GDF11 in the absence of Tll2 (Fig. 3D), suggesting that there may be unique molecular contacts in this region, which may account for these differences. Overall, our results indicate that mutation of specific residues in the GDF11 prodomain can affect ligand latency and activity.
GDF8 Prodomain Mutations Exhibit Reduced Antagonism.
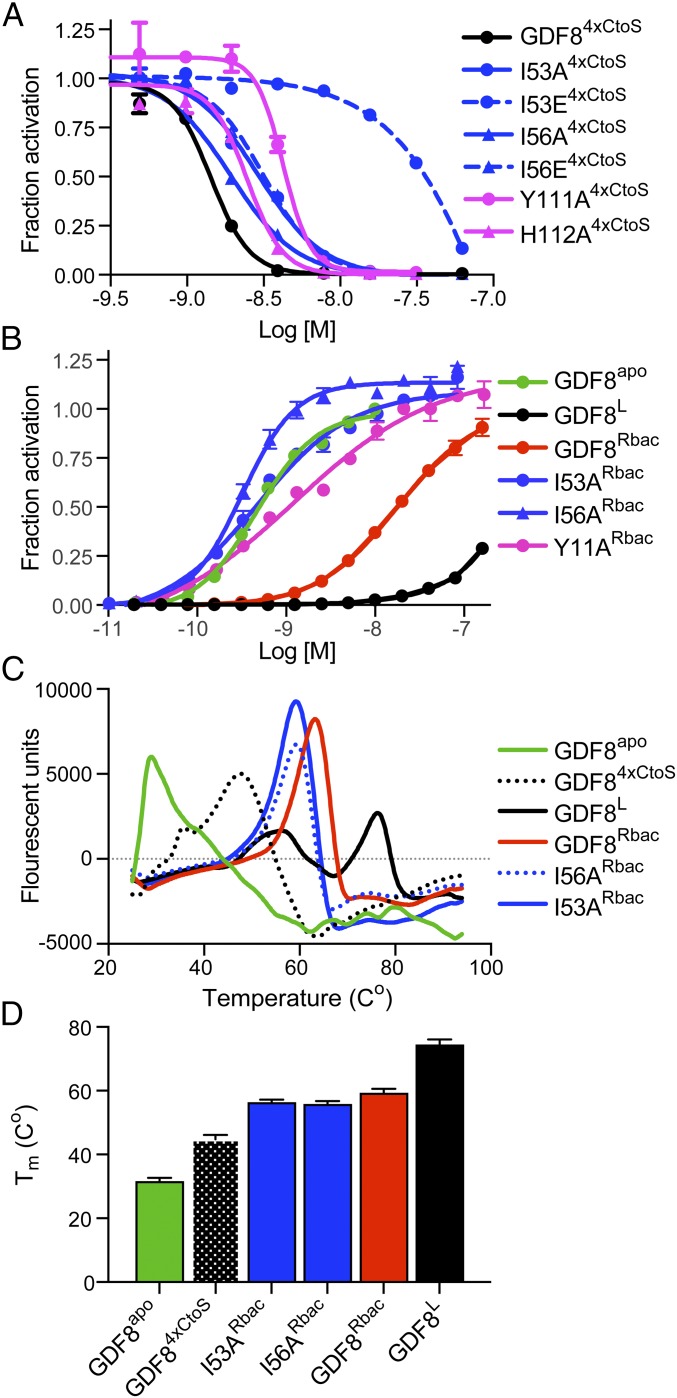
As mentioned earlier, GDF8 mature ligand signaling can be antagonized by titrating increasing amounts of purified GDF8 prodomain. Therefore, we next wanted to determine if the prodomains with activating mutations had an altered capacity to inhibit the mature GDF8 ligand. To accomplish this, we produced and purified the GDF8 prodomain mutants in bacteria (Fig. S2A) and determined their half-maximal inhibitory potential (IC50) against a constant concentration of mammalian-derived, mature GDF8 (Fig. 4A). To improve the production and solubility of the bacteria-derived GDF8 prodomain mutants, we mutated all four cysteines in the prodomain to serine (GDF84xCtoS; Materials and Methods). Using the SMAD3-responsive (CAGA)12 luciferase-reporter HEK293 cell line described above, we determined the IC50 for several of the activating prodomain mutations (Fig. 4A and Table S2). Results show that mutations in the fastener region, Y111A and H112A, had an IC50 similar to GDF84xCtoS, whereas mutations in the alpha-1 helix (I53A, I56A and I56E) were three- to fourfold less potent. However, the most dramatic effect was observed with I56E, which was ∼16-fold less potent than GDF84xCtoS.
Fig. 4.
Characterization of bacterially produced GDF8 prodomains and purified prodomain:ligand complexes. (A) Representative IC50 curve of serially diluted bacterially expressed prodomains mixed with exogenous, mammalian-derived GDF8apo (0.62 nM) and added to HEK293 (CAGA)12 cells. Fraction activation was calculated using the signal of GDF8apo treated with the prodomain divided by GDF8apo alone or maximum signal. Data were fit to a nonlinear regression with variable slope to determine the IC50. (B) EC50 curves of reformed GDF8 (mammalian-derived) prodomain (bacterial derived) complexes denoted with Rbac superscript. Data were fit by nonlinear regression to a variable slope to determine the EC50. (C) Representative derivative plot of melt curves from 24 to 100 °C generated by thermal shift and reported as fluorescent units. (D) Representative Tm (Co) for each mutant shown in C. All experiments were performed at least twice where individual points were measured in triplicate for A and B and duplicate for C and D. All data are shown as mean ± SEM.
Reformed Complexes Using the GDF8 Prodomain Mutants Are More Active and Exhibit Decreased Thermal Stability.
We next wanted to determine if we could reform the prodomain:ligand complex using the various mutant prodomain constructs and subsequently assess their signaling activity. Therefore, we combined the mutated prodomains with the mature ligand and isolated the complex by SEC. We calculated the EC50 of the complexes using the HEK293 (CAGA)12 luciferase-reporter cells and compared these results to GDF8L complex, GDF84xCtoS complex (referred to as GDF8Rbac), and GDF8apo (Fig. 4B and Table S3). We were unable to isolate a stable complex using the prodomain mutations of I53E and I56E, presumably due to a loss in affinity for the mature GDF8 (Fig. 4A and Table S2). Interestingly, all reformed mutant complexes showed significant activity with EC50 values similar to that of the mature GDF8, indicating that the prodomain:ligand inhibitory complex was less stable during the assay and could not function to inhibit GDF8 signaling. This is in contrast to the GDF8Rbac complex, which had significantly less activity but still had more activity than GDF8L complex (Fig. 4B and Table S3).
To further determine if the enhanced activity shown by the mutants, specifically the alpha-1 mutants (I53 and I56), may be explained in part by destabilization of the prodomain:mature ligand complex we performed a thermal shift assay. In this assay the binding of the hydrophobic dye Rox was measured as a function of temperature (Fig. 4 C and D). We determined that the mammalian-derived GDF8L complex had the highest melting temperature (Tm) whereas the reformed GDF84xCtoS complex (GDF8Rbac) and GDF8 I53A and I56A mutant complexes showed a lower Tm suggestive of diminished stability or differences in the binding mode compared with the GDF8L complex (Fig. 4 C and D). Both the GDF8L and mutant complexes showed increased stability compared with GDF8apo and the unbound GDF84xCtoS prodomain, indicating that the difference in Tm is not due to excess GDF8apo ligand within the sample or as a result of dissociated, unbound prodomain (Fig. 4 C and D). In addition to the higher Tm maxima for the GDF8L complex, we detected a second maxima at a lower temperature, not observed in the reformed complexes, suggesting that a complex destabilization event occurred for the GDF8L complex (Fig. 4C). Taken together, these data suggest that mutation of the residues within the alpha-1 helix alleviates GDF8 latency through disruption of the interaction between the prodomain and mature domain.
GDF8 Mutants Enhance Muscle Atrophy Compared with WT GDF8.
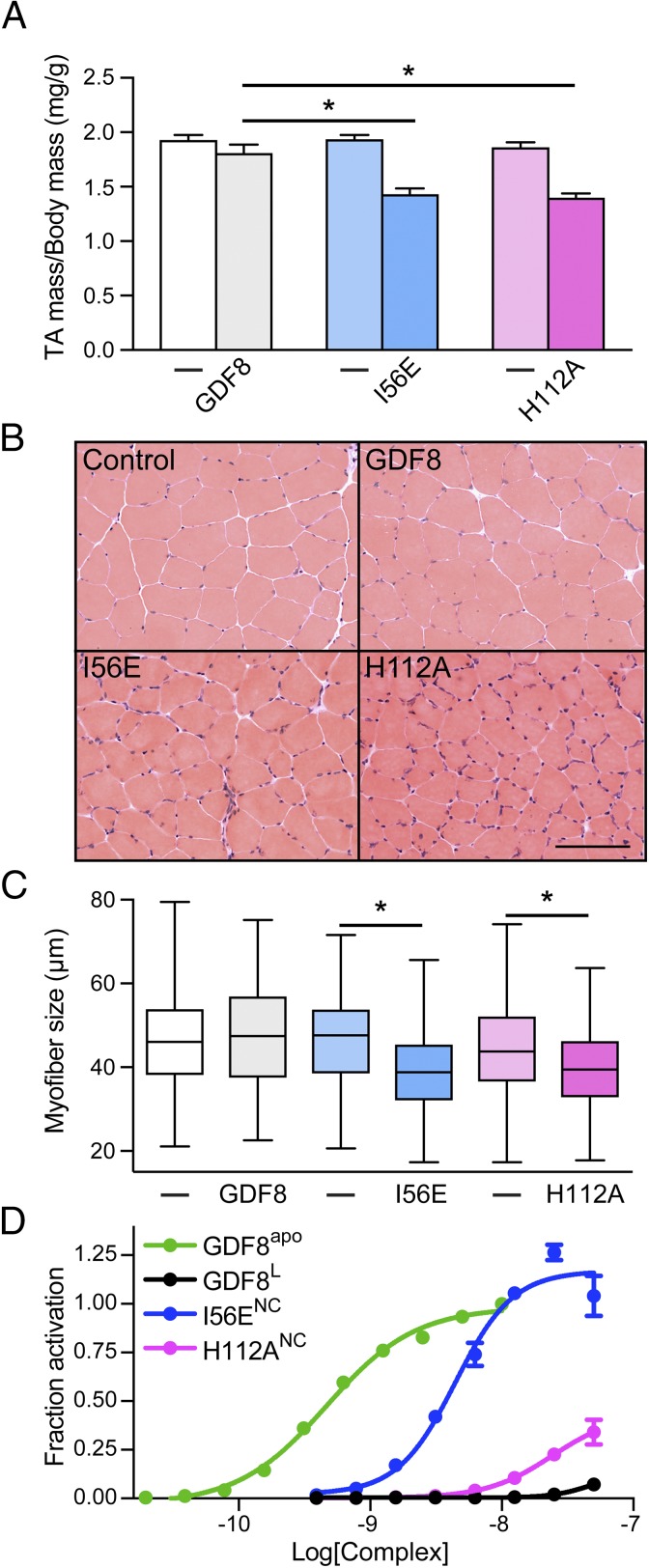
Having demonstrated that mutation of specific residues within the GDF8 prodomain result in a more active or less latent ligand in vitro, we next wanted to determine if the enhanced activity would be recapitulated in vivo in a model of skeletal muscle atrophy. To test this, we generated AAV6 vectors encoding either WT GDF8 or the activating GDF8 mutants, I56E and H112A, and locally injected them in into the tibialis anterior (TA) muscles of 6- to 8-wk-old male C57BL/6 mice. Eight weeks after AAV injections, WT GDF8, which is secreted in a latent form, induced a modest (∼7%) decrease in TA mass (from 61.2 mg to 56.7 mg) (Fig. 5A). In contrast, the decrease in muscle mass induced by GDF8 I56E (from 53.2 mg to 40 mg) and GDF8 H112A (from 50.4 mg to 37.3 mg) was much greater (∼25%) (Fig. 5A), which is consistent with the in vitro finding that these mutations enhance the ligand activity (Fig. 3 and Fig. S1). Histological analysis, using hematoxylin and eosin staining of GDF8-treated TA muscles, revealed that the decreased muscle mass was a product of muscle fiber atrophy (Fig. 5B), as indicated by decreased fiber diameter (Fig. 5C). GDF8 I56E and GDF8 H112A also provoked a significant endomysial cellular infiltration, which was not evident in WT GDF8-treated muscles (Fig. 5B). As we have shown previously with Activin A, these cells are likely collagen-secreting myofibroblasts (34) and their presence is indicative of enhanced GDF8 activity. Collectively, these data indicate that activating mutations in GDF8 markedly increase in vivo activity of this TGFβ superfamily ligand.
Fig. 5.
Activating mutations in GDF8 increase in vivo activity. The right TA muscles of 6- to 8-wk-old male C57BL/6 mice were injected with AAV6 vectors encoding for GDF8, GDF8 (I56E), or GDF8 (H112A) (left TA muscles were injected with equivalent doses of an AAV6 vector lacking a transgene). (A) Eight weeks after AAV6 injection, the TA muscles were harvested and weighed (n = 4–6, paired Student’s t test, data groups with different letters achieved significance, P < 0.05; *significantly different from WT GDF8, P < 0.05). (B) Hematoxylin and eosin staining of TA muscles was performed on cryosections (scale bar, 100 µm) and (C) muscle fiber diameter quantified (n = 3, paired Student’s t test, data groups with different letters achieved significance of P < 0.05, at least 150 myofibers were counted per TA muscle). (D) Mammalian-derived (NC, native complex) and purified GDF8 prodomain complexes were serially titrated and exogenously added to HEK293 (CAGA)12 cells and reported as fraction activation compared with GDF8apo. Data shown are representative of one of three independent experiments that were performed with duplicate wells for each data point. Data were fit by nonlinear regression to a variable slope to determine the EC50. All data shown as mean ± SEM.
Given the apparent activity differences between GDF8L, GDF8R, and GDF8Rbac prodomain complexes, it is plausible that the natively produced mutant prodomain:GDF8 complexes, such as in the AAV experiments above, may show differences in their TLD dependence and signaling potential. Therefore, we next wanted to determine if the activity of mammalian-produced mutant prodomain:GDF8 complexes were more active than the WT prodomain:GDF8 complex. Following expression and purification of the prodomain:ligand complexes we determined the EC50 of the complexes using the HEK293 (CAGA)12 luciferase-reporter cells and compared these results to GDF8L and GDF8apo (Fig. 5D, Fig. S2B, and Table S3). While not as active as GDF8apo, titration of the I56E complex showed significantly enhanced activity compared with both the H112A and WT GDF8L complexes and did not require the presence of TLD for enhanced activity (Fig. 5D). The H112A complex showed minimal elevation in activity compared with WT GDF8L (Fig. 5D). Together, these results support the notion that the muscle atrophy observed in our in vivo studies is likely the result of enhanced ligand activity.
Discussion
The goal of this study was to elucidate mechanisms of latent GDF8 activation and identify the residues within the prodomain that contribute to latency. Although many ligands have been shown to loosely associate with their prodomains, only the prodomains of TGF-β ligands, GDF8, and the highly related GDF11, have been shown to potently inhibit their respective ligands (reviewed in ref. 9). Through sequence alignment and structural modeling, we hypothesized that, despite the different modes of activation, the GDF8 prodomain confers latency through a binding mechanism similar to that observed in the latent TGF-β1 crystal structure (18). Using the low-resolution solution-based technique SAXS we demonstrated that the GDF8L complex exhibits an open conformation, unlike the closed conformation adopted by the latent TGF-β1 structure. This difference is not unexpected given the mechanistic differences required for their respective activation. It is possible that an open conformation is required for TLD activation of GDF8L to improve accessibility of the TLD-cleavage site or TLD-recognition motif, whereas a closed conformation may impede access. However, through site-directed mutagenesis of the GDF8 prodomain, based on sequence alignment to TGF-β1, we identified important residues within either the alpha-1 helix or fastener region which when mutated significantly enhance ligand signaling activity in vitro and in vivo. Together, our data support the conclusion that the GDF8 and TGF-β1 prodomains both utilize similar residues to confer latency, yet we have identified that significant overall structural differences exist between the two complexes.
Apart from biological mechanisms of activation, it has been shown that exposure of latent TGF-β (19, 20) and GDF8L (11, 25) to acidic conditions results in activation of the latent complexes. A molecular explanation for this mode of activation has yet to be determined, but it has been postulated that acid activation causes the prodomain and mature domain to dissociate, thus explaining the gain in ligand activity (11). Interestingly, our biophysical data strongly support that acid activation of the GDF8L does not dissociate the complex but rather the pro- and mature domains remain associated, yet in a different molecular state, referred to as a ‘triggered’ state. Interestingly, the triggered state is not as active as GDF8apo, suggesting that the prodomain needs to be dissociated for full activity. This might be through partially interfering with receptor binding and is consistent with exogenous addition of prodomain to inhibit GDF8apo. Moreover, we determined that reconstitution of the GDF8 prodomain:ligand complex (GDF8R) from individual components did not result in a fully latent complex as the GDF8R complex shows significant activity compared with the GDF8L complex, suggesting that the latent state and triggered state are not fully reversible. The notion that the GDF8 prodomain:ligand complex may exist in multiple activity states may explain, in part, why bacterially derived and refolded GDF8 prodomain:ligand complex has been shown to have significant ligand activity (35) and, therefore, may better represent the acid-activated or triggered state. Nonetheless, our findings raise the possibility that mature GDF8 may be held in a locked or spring-loaded state by its prodomain following biosynthesis, which can be triggered when exposed to changes in pH.
To extend our understanding of the molecular interactions that drive GDF8 latency we performed a targeted mutagenesis on the GDF8 prodomain, based on the latent TGF-β1 structure (18) and corresponding sequence alignment. Consistent with our hypothesis, we identified specific residues in the alpha-1 helix and the fastener region that when mutated resulted in a more active ligand compared with WT, whereas mutation of hydrophobic residues in the latency lasso region did not increase activity. Importantly, our data suggest that the increase in activity was not due to increased protein expression (Fig. S3). In fact, our most active mutant, I56E, showed the least detectable expression, perhaps due to rapid turnover of the mature ligand following receptor binding. Nonetheless, this observation is consistent with other groups that observed a reduction in ligand detection when corresponding residues were mutated in other TGF-β growth factors, although the effect of these mutations on TGF-β latency was not tested (27, 36).
Due to the overall complexity of GDF8 biosynthesis, latency, and activation we are unable to define the molecular mechanisms to describe or explain why these mutations enhance GDF8 activity. Surprisingly, all activating GDF8 mutants required the presence of TLD except the I56E mutant, which remained significantly active despite the incorporation of the TLD cleavage-resistant mutation D99A (GDF8 I56E/D99A; ref. 13). It is possible that incorporation of the I56E mutation disrupts the interaction between the alpha-1 helix and the mature ligand, which allows competition with GDF8 receptors. However, mutation of these regions may prevent GDF8 from fully entering the latent or spring-loaded state during biosynthesis. Instead, this variant may be secreted in a form similar to the triggered state that we have identified. This idea is supported by our data showing that the recombined mutant GDF8 prodomain:ligand complexes had signaling activity similar to that of GDF8apo. It is clear that further characterization of the mutant GDF8 prodomain:ligand complexes is necessary to pinpoint the molecular mechanism responsible for enhanced activity. However, we have identified specific residues within the GDF8 prodomain that can be modified to alleviate ligand latency without disrupting the function of the prodomain in folding and biosynthesis of the mature ligand (7, 8).
We extended our analysis of the GDF8 prodomain activating mutants in vivo using a model of skeletal muscle atrophy to determine if these mutants recapitulated our in vitro experiments. We focused our efforts on I56E, which showed the greatest activity independent of TLD and H112A, where increased activity is completely dependent on TLD. In both cases, AAV delivery of I56E or H112A decreased the size of the muscle fibers relative to control mice and mice that received AAV encoding WT GDF8. Similar to our in vitro experiments, we were unable to detect evidence of mature GDF8 in the muscle of mice that received the AAV encoding the I56E mutant, whereas we could reliably detect the mature ligand in the muscle from mice that received either the AAV encoding the H112A or WT proteins. As mentioned above, we speculate that loss of latency independent of TLD may enhance ligand turnover rate, thus making it challenging to detect the mature ligand. Together, these results are consistent with our previous observation that mutation of these residues results in a more active ligand.
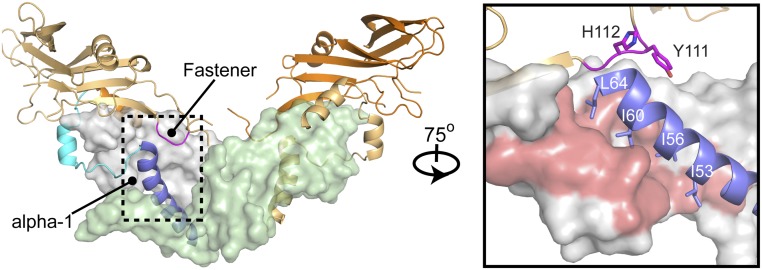
While this paper was in preparation, it became apparent that an X-ray crystal structure had been determined in the laboratory of Marko Hyvönen (Fig. 6 and Fig. S4) (37). Therefore, we wanted to compare how our low-resolution SAXS data compared with the overall shape of the GDF8 prodomain:ligand complex. Consistent with our initial SAXS-based observations, the crystal structure of the GDF8 prodomain:ligand adopts a more open conformation that is drastically different from that of TGF-β1 and more similar to that of Activin A or BMP9. In agreement with our hypothesis that GDF8 prodomain contains similar inhibitory elements comparable to TGF-β1, the alpha-1 helix, latency lasso, and fastener features are all present in the GDF8 prodomain:ligand complex. However, the conformation of the GDF8 prodomains in relation to their mature domain with which monomer they interact is significantly different from that of TGF-β1. For instance, the prodomain of one TGF-β1 monomer sits atop the other monomer of the homodimer, with all inhibitory elements imposed by one prodomain. However, the prodomain of one GDF8 monomer crosses over to interact with both mature domains of the dimer (Fig. 6). Notably, the alpha-1 helix and latency lasso inhibit the GDF8 monomer from the same chain while the fastener interacts with the adjacent monomer. The significance of this binding strategy on inhibition is unknown. However, this “fastener swap” may play a role to ensure homodimer formation and/or aid in exposure of the TLD protease site. Nevertheless, I53 and I56 in the alpha-1 helix are shown to interact directly with the GDF8 ligand. It is possible that mutation of I53 or I56 would destabilize the alpha-1 helix and disrupt binding of the prodomain to GDF8. One would also expect that mutation of I60 would show a similar, if not more, exaggerated phenotype compared with the I53 or I56 mutants. However, mutation of I60 did not result in enhanced activity, but rather even lower activity than WT GDF8. It is possible that I60 may be important for protein folding and loss of this residue is detrimental to this process. Furthermore, the GDF8 prodomain:ligand crystal structure supports our finding that mutation of the fastener residues, Y111 and H112, would destabilize the fastener interaction with the alpha-1 helix. This is similar to TGF-β1 where mutation of the fastener residues created a more active TGF-β1 ligand (18). Taken together, our mutational analysis of the GDF8 prodomain is highly consistent with the structure of the prodomain:GDF8 complex and also consistent with previous truncation analysis (35, 38–40). Our results are also consistent with results from the laboratory of Tim Springer, who performed a rigorous hydrogen-deuterium exchange followed by MS to map the interactions of the prodomain with the mature in solution (41).
Fig. 6.
Crystal structure of the GDF8 prodomain complex. The GDF8 prodomain complex containing the mature dimer (gray and pale green), alpha-1 helix (blue), latency lasso (cyan), and fastener (magenta). (Middle Inset) Depiction of the alpha-1 helix and fastener regions following a 75° rotation about the y axis (Middle) Note the location of residues I53, I56, I60, and I64 within the alpha-1 helix and Y111 and H112 within the fastener regions. See also Fig. S4.
In summary, we determined that the latent GDF8 prodomain:ligand complex adopts a more open structural conformation unlike that of the TGF-β1 latent complex (reviewed in ref. 9). Interestingly, both ligands share commonality with respect to the alpha-1 and latency lasso inhibitory elements but show significant divergence with respect to the coordination of their respective fastener regions to confer latency. While it is unknown how this binding mode impacts or confers latency to GDF8 compared with TGF- β1, our data strongly support the notion that the GDF8 prodomain:ligand complex can exist in multiple conformational states which ultimately dictate ligand activity and that the interactions between the prodomain and mature domain can be modified to generate a less latent and more active signaling ligand. It is plausible that GDF8 circulates within serum (12) in these various conformational “activity” states, thus making it tempting to speculate that GDF8 biological regulation may include shifts in the balance of these activity states depending on the physiological context.
Materials and Methods
HEK293-(CAGA)12 Luciferase-Reporter Assay.
Luciferase-reporter assays for activation and inhibition were performed as previously described (21–25). Briefly, HEK293 (CAGA)12 cells stably transfected with plasmid containing Firefly luciferase-reporter gene under the control of SMAD3-responsive promoter were seeded in growth media at 20,000 cells per well in a 96-well plate. For transient transfection experiments, 200 ng total DNA (25–75 ng ligand DNA, 50 ng full length human furin, 5–50 ng of appropriate TLD DNA, filled to 200 ng with empty vector) per well was added directly to the growth media, incubated for 6 h, exchanged into serum-free media, and lysed 30 h posttransfection. Firefly luminescence was recorded followed by Renilla luminescence. To determine EC50 and IC50 values, the appropriate dilutions of either ligand alone or with antagonist, respectively, were serially titrated and added to the cells. Luminescence was recorded 18–24 h after ligand or antagonist addition. Data were fit to nonlinear regression with variable slope using GraphPad Prism 5 software. The EC50 and IC50 mean and SE was calculated for each experiment and the mean weighted to the SE as previously described (42).
Production and Purification of GDF8 Prodomain from Escherichia coli.
The prodomain of human GDF8 (residues 24–262) was cloned into a modified pET28a expression vector that contains an N-terminal 6x histidine tag, maltose binding protein (MBP), and a HRV-3C protease cleavage site. The cysteine residues in the human GDF8 prodomain (C39/C41/C137/C138) were mutated to serine to improve expression and solubility. E. coli Rosetta (DE3) strain carrying the appropriate prodomain construct was cultured until an OD of 0.8 at 600 nm was achieved and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for culture at 20 °C overnight. Cells were lysed and soluble 6xHis-MBP-GDF8 prodomain was applied to nickel affinity column (GE Lifesciences) followed by elution using a linear imidazole gradient. HRV-3C protease was added to the eluted protein and following cleavage the protein was dialyzed into 10 mM HCl and applied to a C4 reverse phase column (Sepax) and eluted with a linear gradient to 0.1% TFA, 95% acetonitrile over 30 column volumes to yield fractions containing the purified protein.
Mammalian-Derived Latent GDF8 Complex (GDF8L) and Mutant Complexes.
CHO cells stably producing GDF8 were used and protein was purified and quantified as previously described using SDS/PAGE/Coomassie staining and the quantified GDF8 mature as a standard (21, 24, 25, 43, 44). For the expression of mutant prodomain:GDF8 complexes, expi293 cells (Life Technologies) were transiently cotransfected with the mutant DNA and furin DNA. Protein was purified from conditioned medium and quantified as previously described (25).
Acid Activation.
Acid activation of GDF8 complex was performed as previously described (11, 25). In short, GDF8 complex was acidified to pH 2–7 using 1 M HCl and incubating for 1 h followed by neutralization with 1 M NaOH back to pH 8. Conversely, when a pH >8 was required 1 M NaOH was used which was neutralized accordingly with 1 M HCl. This material was then used in luciferase and SAXS analysis.
SAXS.
SAXS data were collected using SIBYLS mail-in SAXS service as previously described on purified GDF8L, GDF8AA, and GDF8R (24, 25). ScÅtter (SIBYLS) and the ATSAS program suite (EMBL) were used for data analysis. Comparison of the experimental scattering profiles to known crystal structures was performed using the FoXS webserver (26).
Western Analysis.
HEK293 (CAGA)12 cells were cultured and transfected with ligand DNA using conditions similar to those mentioned above. Thirty hours after transfection media was removed, concentrated ∼25×, and subjected to SDS/PAGE. Standard western protocols were utilized and the anti-GDF8 antibody from RnD Biosystems (AF788) was used as described by the manufacturer. Western blot signal was captured using the C-DiGit blot scanner (LI-COR).
Protein Thermal Shift.
Protein thermal shift assays were conducted using an OneStep real-time PCR system (Applied Biosystems), run by the StepOne Software v2.3, as described by the manufacturer. In short, 1 μg of protein was placed in 20 μL of 20 mM Hepes, pH 7.4, and 500 mM NaCl in the presence of 1× ROX reagent from the Protein Thermal Shift Dye Kit (Applied Biosystems). The melting temperature and Tm of each protein was conducted on a 1% gradient from 25 °C–100 °C, taking ∼40 min. Data were analyzed using Protein Thermal Shift Software v1.3, and curves were plotted from triplicate measurements.
Production of AAV Vectors.
The cDNA constructs encoding for WT GDF8, GDF8 I56E, and GDF8 H112A were cloned into an AAV expression plasmid consisting of a CMV promoter/enhancer and SV40 poly-A region flanked by AAV2 terminal repeats. These AAV plasmids were cotransfected with pDGM6 packaging plasmid into HEK293 cells to generate type-6 pseudotyped viral vectors. Briefly, HEK293 cells were seeded onto culture and were transfected with a vector-genome-containing plasmid and the helper plasmid pDGM6 by calcium phosphate precipitation. After 72 h, the media and cells were collected and subjected to three cycles of freeze–thaw followed by 0.22-µm clarification (Millipore). Vectors were purified from the clarified lysate by affinity chromatography using heparin columns (HiTrap; GE Healthcare); the eluent was ultracentrifuged overnight, and the vector-enriched pellet was resuspended in sterile physiological Ringer’s solution and quantified with a customized sequence-specific quantitative PCR-based reaction (Life Technologies).
Administration of AAV6 Vectors to Mice.
All experiments were conducted in accordance with the relevant code of practice for the care and use of animals for scientific purposes (National Health & Medical Council of Australia, 2016). Vectors carrying transgenes of GDF8 mutants were injected into the right TA muscle of 6- to 8-wk-old male C57BL/6 mice under isoflurane anesthesia at 1010 vector genomes (vg). As controls, the left TA muscles were injected with AAVs carrying an empty vector at equivalent doses. At the experimental endpoint, mice were humanely killed via cervical dislocation, and TA muscles were excised rapidly and weighed before subsequent processing.
Histological Analysis.
Harvested muscles were placed in optimal cutting temperature (OCT) cryoprotectant and frozen in liquid nitrogen-cooled isopentane. The frozen samples were cryosectioned through the middle of the muscle at 10-μm thickness and stained with hematoxylin and eosin. Tissue sections were imaged using a U-TV1X-2 camera mounted to an IX71 microscope and an Olympus PlanC 10×/0.25 objective lens. DP2-BSW acquisition software (Olympus) was used to acquire images. Images were separated into eight fields covering the whole of the TA muscle (designated A1–A4 and B1–B4). The minimum Feret’s diameter of muscle fibers in fields A2, B2, and B3 were determined using ImageJ software (NIH) by measuring at least 300 fibers per mouse muscle. The same fields were compared for each TA muscle examined.
Supplementary Material
Acknowledgments
We thank Monash Micro Imaging staff, Alfred Medical Research and Education Precinct (AMREP) campus, for technical guidance and AMREP Precinct Animal Centre staff for animal husbandry; Dr. Hongwei Qian (Baker Heart and Diabetes Institute) for assistance with production of recombinant AAV vectors; and Georgia Goodchild for help with the histological analysis. This work was supported, in part, by NIH; a National Health and Medical Research Council (NHMRC) grant; the Muscular Dystrophy Association; the University of Cincinnati Graduate Dean Fellowship; American Heart Association Grants R01AG047131, R01AG040019, R03AG049657 (to R.T.L.), and 1078907 (to C.A.H.); App 1117835 for Gregorevic Senior Research Fellowship from NHMRC; Graduate Dean Fellowship and Grant 12PRE11790027 (to R.G.W.); and Grant R01GM114640 and Muscular Dystrophy Association Grant 240087 (to T.B.T.). This work was also supported by the Integrated Diffraction Analysis Technologies Program of the Department of Energy Office of Basic Energy Sciences awarded to the Advanced Light Source at Lawrence Berkeley National Laboratory. The Baker Heart and Diabetes Institute is supported in part by the Operational Infrastructure Support Program of the Victorian Government.
Footnotes
Conflict of interest statement: T.B.T. is a consultant for Acceleron Pharma. The University of Cincinnati and Monash University have filed for intellectual property on GDF8 and GDF11 listing R.G.W., T.B.T., and C.A.H. as inventors. Harvard University and Brigham and Women’s Hospital have filed for intellectual property on GDF11 listing R.T.L. as an inventor.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714622115/-/DCSupplemental.
References
- 1.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.McPherron AC, Lee S-J. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S-J. Extracellular regulation of myostatin: A molecular rheostat for muscle mass. Immunol Endocr Metab Agents Med Chem. 2010;10:183–194. doi: 10.2174/187152210793663748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suragani RNVS, et al. Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat Med. 2014;20:408–414. doi: 10.1038/nm.3512. [DOI] [PubMed] [Google Scholar]
- 5.Lach-Trifilieff E, et al. An antibody blocking activin type II receptors induces strong skeletal muscle hypertrophy and protects from atrophy. Mol Cell Biol. 2014;34:606–618. doi: 10.1128/MCB.01307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S-J, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray AM, Mason AJ. Requirement for activin A and transforming growth factor–beta 1 pro-regions in homodimer assembly. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- 8.Harrison CA, Al-Musawi SL, Walton KL. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-β superfamily ligands. Growth Factors. 2011;29:174–186. doi: 10.3109/08977194.2011.608666. [DOI] [PubMed] [Google Scholar]
- 9.Hinck AP, Mueller TD, Springer TA. Structural biology and evolution of the TGF-β family. Cold Spring Harb Perspect Biol. 2016;8:a022103. doi: 10.1101/cshperspect.a022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thies RS, et al. GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors. 2001;18:251–259. doi: 10.3109/08977190109029114. [DOI] [PubMed] [Google Scholar]
- 11.Zimmers TA, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- 12.Hill JJ, et al. The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem. 2002;277:40735–40741. doi: 10.1074/jbc.M206379200. [DOI] [PubMed] [Google Scholar]
- 13.Wolfman NM, et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi L-Z, et al. Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci USA. 2015;112:3710–3715. doi: 10.1073/pnas.1501303112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Fischer G, Hyvönen M. Structure and activation of pro-activin A. Nat Commun. 2016;7:12052. doi: 10.1038/ncomms12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wipff P-J, Hinz B. Integrins and the activation of latent transforming growth factor β1–An intimate relationship. Eur J Cell Biol. 2008;87:601–615. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence DA, Pircher R, Jullien P. Conversion of a high molecular weight latent beta-TGF from chicken embryo fibroblasts into a low molecular weight active beta-TGF under acidic conditions. Biochem Biophys Res Commun. 1985;133:1026–1034. doi: 10.1016/0006-291x(85)91239-2. [DOI] [PubMed] [Google Scholar]
- 20.Schultz-Cherry S, Murphy-Ullrich JE. Thrombospondin causes activation of latent transforming growth factor-beta secreted by endothelial cells by a novel mechanism. J Cell Biol. 1993;122:923–932. doi: 10.1083/jcb.122.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cash JN, et al. Structure of myostatin·follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem. 2012;287:1043–1053. doi: 10.1074/jbc.M111.270801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cash JN, Angerman EB, Keutmann HT, Thompson TB. Characterization of follistatin-type domains and their contribution to myostatin and activin A antagonism. Mol Endocrinol. 2012;26:1167–1178. doi: 10.1210/me.2012-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cash JN, et al. Development of a small-molecule screening method for inhibitors of cellular response to myostatin and activin A. J Biomol Screen. 2013;18:837–844. doi: 10.1177/1087057113482585. [DOI] [PubMed] [Google Scholar]
- 24.Walker RG, et al. Alternative binding modes identified for growth and differentiation factor-associated serum protein (GASP) family antagonism of myostatin. J Biol Chem. 2015;290:7506–7516. doi: 10.1074/jbc.M114.624130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker RG, et al. Structural basis for potency differences between GDF8 and GDF11. BMC Biol. 2017;15:19. doi: 10.1186/s12915-017-0350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton KL, et al. A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor beta (TGFbeta) ligands. J Biol Chem. 2009;284:9311–9320. doi: 10.1074/jbc.M808763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S-J. Genetic analysis of the role of proteolysis in the activation of latent myostatin. PLoS One. 2008;3:e1628. doi: 10.1371/journal.pone.0001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Solow-Cordero DE, Kessler E, Takahara K, Greenspan DS. Transforming growth factor-beta regulation of bone morphogenetic protein-1/procollagen C-proteinase and related proteins in fibrogenic cells and keratinocytes. J Biol Chem. 1997;272:19059–19066. doi: 10.1074/jbc.272.30.19059. [DOI] [PubMed] [Google Scholar]
- 30.Leighton M, Kadler KE. Paired basic/furin-like proprotein convertase cleavage of Pro-BMP-1 in the trans-Golgi network. J Biol Chem. 2003;278:18478–18484. doi: 10.1074/jbc.M213021200. [DOI] [PubMed] [Google Scholar]
- 31.Jasuja R, et al. Bone morphogenetic protein 1 prodomain specifically binds and regulates signaling by bone morphogenetic proteins 2 and 4. J Biol Chem. 2007;282:9053–9062. doi: 10.1074/jbc.M610929200. [DOI] [PubMed] [Google Scholar]
- 32.Szláma G, Trexler M, Buday L, Patthy L. K153R polymorphism in myostatin gene increases the rate of promyostatin activation by furin. FEBS Lett. 2014;589:295–301. doi: 10.1016/j.febslet.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Ge G, Hopkins DR, Ho W-B, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol. 2005;25:5846–5858. doi: 10.1128/MCB.25.14.5846-5858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen JL, et al. Elevated expression of activins promotes muscle wasting and cachexia. FASEB J. 2014;28:1711–1723. doi: 10.1096/fj.13-245894. [DOI] [PubMed] [Google Scholar]
- 35.Szláma G, Trexler M, Patthy L. Latent myostatin has significant activity and this activity is controlled more efficiently by WFIKKN1 than by WFIKKN2. FEBS J. 2013;280:3822–3839. doi: 10.1111/febs.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young GD, Murphy-Ullrich JE. Molecular interactions that confer latency to transforming growth factor-beta. J Biol Chem. 2004;279:38032–38039. doi: 10.1074/jbc.M405658200. [DOI] [PubMed] [Google Scholar]
- 37.Cotton TR, et al. 2017. Structure of the human pro-myostatin precursor and determinants of growth factor latency. bioRxiv:153403.
- 38.Jiang M-S, et al. Characterization and identification of the inhibitory domain of GDF-8 propeptide. Biochem Biophys Res Commun. 2004;315:525–531. doi: 10.1016/j.bbrc.2004.01.085. [DOI] [PubMed] [Google Scholar]
- 39.Ohsawa Y, et al. The inhibitory core of the myostatin prodomain: Its interaction with both type I and II membrane receptors, and potential to treat muscle atrophy. PLoS One. 2015;10:e0133713. doi: 10.1371/journal.pone.0133713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama K, et al. Identification of the minimum peptide from mouse myostatin prodomain for human myostatin inhibition. J Med Chem. 2015;58:1544–1549. doi: 10.1021/jm501170d. [DOI] [PubMed] [Google Scholar]
- 41.Le VQ, et al. 2017. Tolloid cleavage activates latent GDF8 by priming the pro-complex for dissociation. bioRxiv:154823.
- 42.Jones DC, et al. Identification of a κ-opioid agonist as a potent and selective lead for drug development against human African trypanosomiasis. Biochem Pharmacol. 2010;80:1478–1486. doi: 10.1016/j.bcp.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: Insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y-S, Lee S-J. Regulation of GDF-11 and myostatin activity by GASP-1 and GASP-2. Proc Natl Acad Sci USA. 2013;110:E3713–E3722. doi: 10.1073/pnas.1309907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.