Significance
There is a lack of a general method to noninvasively and remotely manipulate cells with high spatiotemporal precisions. We developed an ultrasound-based mechanogenetics system to achieve this goal. Cells were engineered with the mechanosensor Piezo1 and genetic transducing modules to perceive the mechanical perturbation generated by the ultrasound wave and transduce it into genetic activities. Mechanosensitive and ultrasound-controllable T cells were further engineered to target and eradicate tumor cells with inducible chimeric antigen receptors. This mechanogenetics approach can be extended to remotely control, in principle, any gene activity in live cells for the reprogramming of cellular functions. This method should also provide a general approach to remotely control molecular functions for biological studies and clinical applications, particularly cell-based cancer immunotherapy.
Keywords: ultrasound, mechanogenetics, remote control, cancer immunotherapy, synthetic biology
Abstract
While cell-based immunotherapy, especially chimeric antigen receptor (CAR)-expressing T cells, is becoming a paradigm-shifting therapeutic approach for cancer treatment, there is a lack of general methods to remotely and noninvasively regulate genetics in live mammalian cells and animals for cancer immunotherapy within confined local tissue space. To address this limitation, we have identified a mechanically sensitive Piezo1 ion channel (mechanosensor) that is activatable by ultrasound stimulation and integrated it with engineered genetic circuits (genetic transducer) in live HEK293T cells to convert the ultrasound-activated Piezo1 into transcriptional activities. We have further engineered the Jurkat T-cell line and primary T cells (peripheral blood mononuclear cells) to remotely sense the ultrasound wave and transduce it into transcriptional activation for the CAR expression to recognize and eradicate target tumor cells. This approach is modular and can be extended for remote-controlled activation of different cell types with high spatiotemporal precision for therapeutic applications.
With the use of central memory T cells capable of suppressing cancer relapse for several years (1), chimeric antigen receptor (CAR) T-cell immunotherapy is transforming cancer therapy (2). However, major challenges remain for CAR-based immunotherapy against solid tumors. For example, the nonspecific targeting of the CAR T cells against normal/nonmalignant tissues (on-target but off-tumor toxicities) can be life-threatening (3). There is hence an urgent need for the control of CAR T-cell activation with high spatiotemporal precision.
Existing methods to control genetic activities typically rely on chemical inducers (e.g., dimerizers), radio waves, and magnetic or light activation (e.g., optogenetics). Chemical inducers, radio waves, and magnetic fields typically lack spatial resolution, with radio and magnetic waves mostly requiring the signal amplification and transmission by nanoparticles (or particle aggregates) to stimulate genetically engineered cells (4–7). Although light activation or optogenetics can enable a precise control of genetic regulation in space and time, it cannot reach deep tissues (8). In contrast, ultrasound and its associated energy can be safely and noninvasively delivered into small volumes of tissue deep inside the body with high spatiotemporal resolution (9, 10). In fact, high frequency ultrasound (HFU) can be focused for mechanical stimulation of single cells at a subcellular region of less than 10 μm (11). Microbubbles, which have been well established as ultrasound imaging contrast agents and approved by the Food and Drug Administration for clinical use (12), can further amplify the impact of low-frequency ultrasound stimulation with the capability of long-distance penetration of cells physically coupled to microbubbles, as a result of a large difference in acoustic impedance between the surrounding media and the air inside the bubbles (13–15).
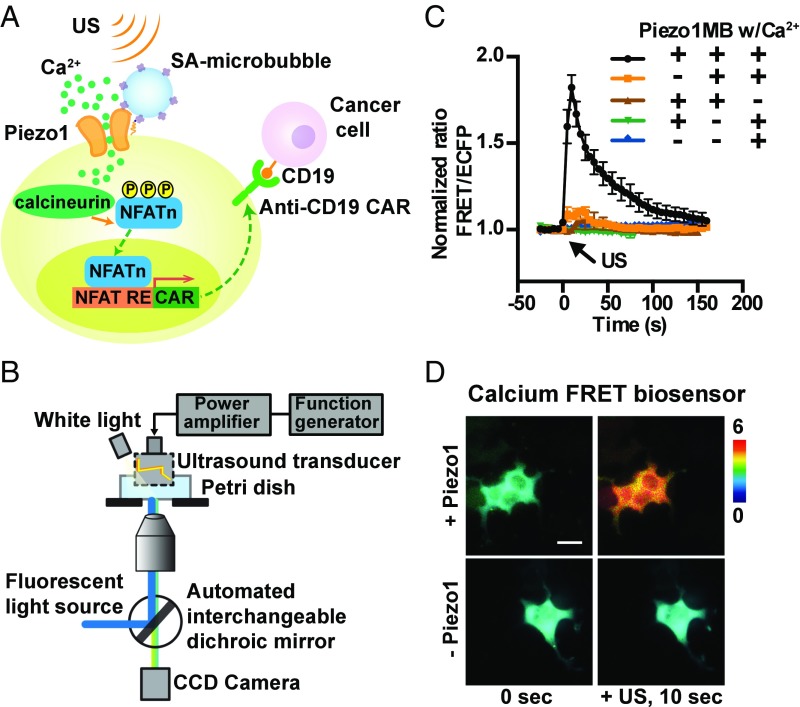
We have developed a modular method to engineer cells that can sense ultrasound stimulation applied remotely and noninvasively to control genetics and CAR protein expression for recognizing antigens to kill the target tumor cells. This method utilizes the Piezo1 ion channel as a mechanical sensor (Fig. 1A), which has been identified as a component of mechanically activated (MA) cation channels (16) and is inherently mechanosensitive (17). The ultrasound stimulation of Piezo1 and the consequent calcium influx can activate calcium-sensitive phosphatase calcineurin to dephosphorylate a transcription factor, the nuclear factor of activated T-cells (NFAT), which can then translocate to the nucleus for the activation of a NFAT response element (RE) to drive the expression of designed target genes (18).
Fig. 1.
Design of synthetic genetic circuits remotely activatable by ultrasound. (A) Schematic drawing of ultrasound-induced cell activation and gene expression. Microbubbles can be coupled to the surface of a cell, where mechanosensitive Piezo1 channels are expressed. Upon exposure to ultrasound waves, the mechanical stimulation can activate the Piezo1 ion channels. The subsequent calcium entry triggers the downstream pathways, including calcineurin activation, NFAT dephosphorylation, and translocation into the nucleus. The nucleus-translocated NFAT can bind to upstream response elements to initiate gene expression through one-stage or two-stage genetic transducing modules. (B) Diagram of an integrated system of ultrasound stimulation and FRET imaging. The CCD camera captures fluorescent images of FRET biosensors with two emission filters controlled by a filter changer (480DF40 for CFP and 535DF35 for YFP). The ultrasonic transducer was driven by a function generator and a power amplifier. (C) The time courses of the normalized FRET/ECFP (enhanced cyan fluorescent protein) ratio (mean ± SEM) of a D3cpv calcium biosensor in HEK293T cells before and after 5 s of ultrasound stimulation under different conditions with or without the presence of the three components (Piezo1, microbubbles, and Ca2+) in the medium: with all three components (black, n = 32), without Piezo1 (orange, n = 37), without microbubbles (green, n = 10), without Ca2+ (brown, n = 14), or without both Piezo1 and microbubbles (blue, n = 11). (D) Representative FRET/ECFP ratio images of the D3cpv calcium biosensor in HEK293T cells expressed with (Top) or without (Bottom) Piezo1 before (Left) and after (Right) ultrasound stimulation. (Scale bar: 20 μm.)
Results
We first examined whether ultrasound-induced mechanical stimulation can activate the Piezo1 channel to elicit calcium influx. HEK293T cells cotransfected with Piezo1-tdTomato and the D3cpv FRET calcium biosensor were examined by an ultrasound-FRET imaging system (Fig. 1B and Fig. S1) (19). Upon a focused HFU (150 MHz) stimulation with a distance of 1 mm, a Piezo1-dependent calcium influx was clearly detected in the target cell (Fig. S2B), suggesting that Piezo1 can serve as a mechanosensor to sense the ultrasound stimulation. For this 150-MHz focused transducer, the focused acoustic pressure applied on the cell membrane can directly activate the Piezo1 ion channel to mediate calcium influx (Fig. S2B). However, this high-frequency 150-MHz transducer has a limited working distance (1 mm) that is much shorter than the effective working distance of centimeters with low-frequency transducers. To develop a technology for the remote control of gene activities in live cells, we reasoned that microbubbles mechanically coupled to Piezo1 can be applied to amplify the mechanical waves of low-frequency 2 MHz ultrasound at a distance in centimeters (4–8). Indeed, microbubbles with a 2-μm diameter can be stimulated at a depth of 5 cm by a low-frequency ultrasound transducer (Figs. S3 and S4). Hence, microbubbles were coated with streptavidin and coupled to biotinylated RGD peptides, which were engaged with the membrane receptor integrins and connected to Piezo1 via cytoskeleton and membrane tension (20, 21). Upon a 2-MHz ultrasound stimulation at a distance of ∼1 cm, calcium influx into the cells was immediately detected in the presence of calcium-containing HBSS media (Fig. 1C and Movie S1), but not in HBSS media without calcium. In control HEK293T cells transfected only with a calcium biosensor but without Piezo1, there was no significant calcium influx in HBSS media with or without calcium (Fig. 1C). This 2-MHz transducer activates the Piezo1 channel differently from the 150-MHz transducer. For the 2-MHz transducer, the activation depends on several factors, e.g., the oscillation of microbubbles and possibly the resulting fluid microstreaming/shear stress associated with the ultrasound excitation. Indeed, the Piezo1-expressing cells without microbubble coupling did not respond to ultrasound to trigger calcium signals (Fig. 1C). Ultrasound power was robustly controlled in a range between 22.1 and 31.6 V to create a sufficient ultrasound wave to mechanically stimulate the cells without causing damage of the target cells (Fig. S5). Indeed, intermittent ultrasound waves could be applied to elicit multiple calcium waves (Fig. S5B), suggesting minimal ultrasound-induced alteration of the target cells.
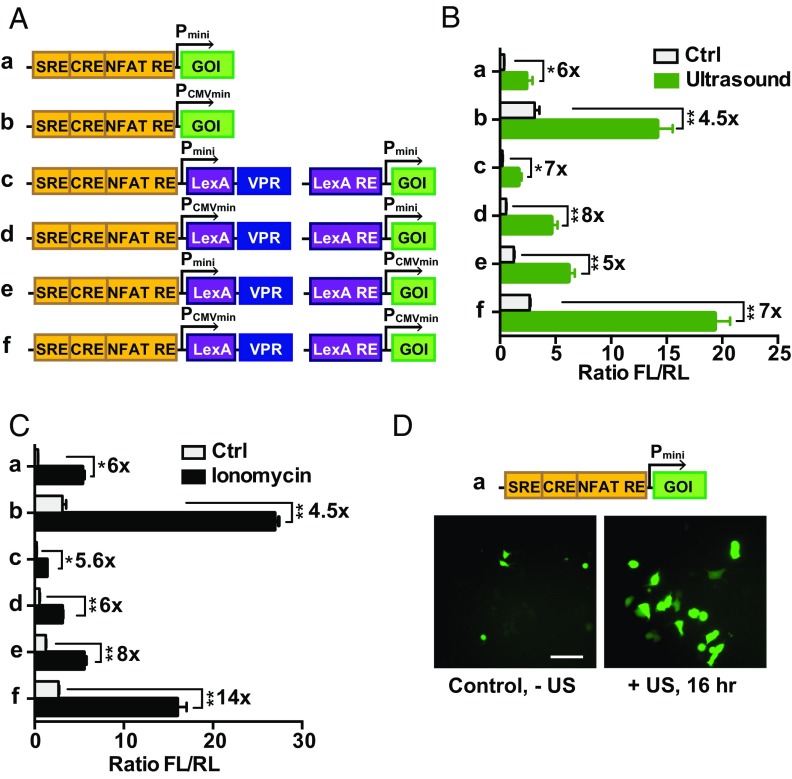
We then examined whether the ultrasound-induced calcium influx can be applied to control gene expression. Genetic transducing modules (GTMs) were engineered to transmit the calcium-mediated NFAT activation and other related signaling into genetic activities. A one-stage GTM design was developed consisting of three calcium responding elements in cis: a serum responding element (SRE), a cyclic adenosine monophosphate response element (CRE), and a NFAT response element (NFAT RE), placed in tandem upstream of the minimal promoters controlling target gene expression (5) (Fig. 2 A, a and b). To reduce the potentially leaky protein expression in cells and enhance the induction specificity upon stimulation, a two-stage GTM was designed using the potent tripartite transcriptional activator VPR (22) (Fig. 2 A, c–f). In these designs, the first induced product upon ultrasound stimulation and calcium influx is a DNA-binding domain LexA fused to VPR (LexA-VPR). Upon induction, this LexA-VPR activates a second gene response element (LexA RE) for the expression of target genes (Fig. 2 A, c–f). Two minimal promoters were compared in their efficiency and specificity; the one derived from the CMV promoter was more robust but exhibited a higher basal level noise than the other one derived from the commercial vector pGL4.30 (Promega). Collectively, six potential candidates were designed (Fig. 2A) with luciferase or fluorescent proteins as target reporter genes to characterize the activation potency. Indeed, 2-MHz ultrasound stimulation for 10 min (see detailed information in Materials and Methods) caused a clear induction of reporter genes measured by luciferase activities (Fig. 2B), similar to the chemical induction by ionomycin (Fig. 2C). We observed a significant difference in the basal and induced expression levels of target genes upon stimulation with these six different designs (Fig. 2 A and B). Among them, design a was chosen as the candidate for further applications due to its simplicity, low basal level, and more efficient activation potency. Indeed, the inducible promoter based on design a can be activated by ultrasound stimulation to clearly drive the expression of the GFP variant mNeonGreen (Fig. 2D). These results indicate that the mechanosensor Piezo1 and GTMs can be engineered and integrated to coordinate with the endogenous molecular network for the sensing of remote ultrasound stimulation to guide gene activations. We also showed that the RGD-tagged microbubble engagement of integrins on the cell surface alone was not sufficient to significantly activate the specifically designed gene expression controlled by genetic transducers (Fig. S6A), suggesting a high specificity of the ultrasound controllable gene activation in our system. While the 10-min application can induce relatively robust mechanogenetic effects, shorter durations of ultrasound stimulation can also induce gene expression, but with a weaker efficiency (Fig. S6B).
Fig. 2.
Characterization of GTMs remotely activatable by ultrasound. (A) The design of ReCoM GTMs. Calcium-responding elements consisting of SRE, CRE, and NFAT RE were followed by target genes of interest (GOI) controlled by minimal promoter (a, Pmin) or minimal CMV promoter (b, PCMVmin). For two-stage constructs, LexA-VPR was designed as the product of stage 1 gene cassette, which can bind to the LexA-responding element and activate stage 2 target genes (c, d, e, and f). (B) Ultrasound stimulation induced gene products of different GTMs. Microbubble-coupled HEK293T cells transfected with Piezo1 and GTMs showed a significant increase in luciferase gene expression upon ultrasound stimulation (n = 3). (C) Lonomycin stimulation induces gene expression. HEK293T cells transfected with Piezo1 and genetic cassettes showed a significant increase in luciferase gene expression 16 h after ionomycin stimulation (n = 3). (D) Fluorescence microscopy images of HEK293T cells transfected with Piezo1 and gene activation cassette a (mNeonGreen as the reporter) without (Left) and 16 h after (Right) ultrasound stimulation (for 10 min). (Scale bar: 40 μm.) Error bars indicate SEM, *P < 0.05, **P < 0.01 from two-tailed Student t test.
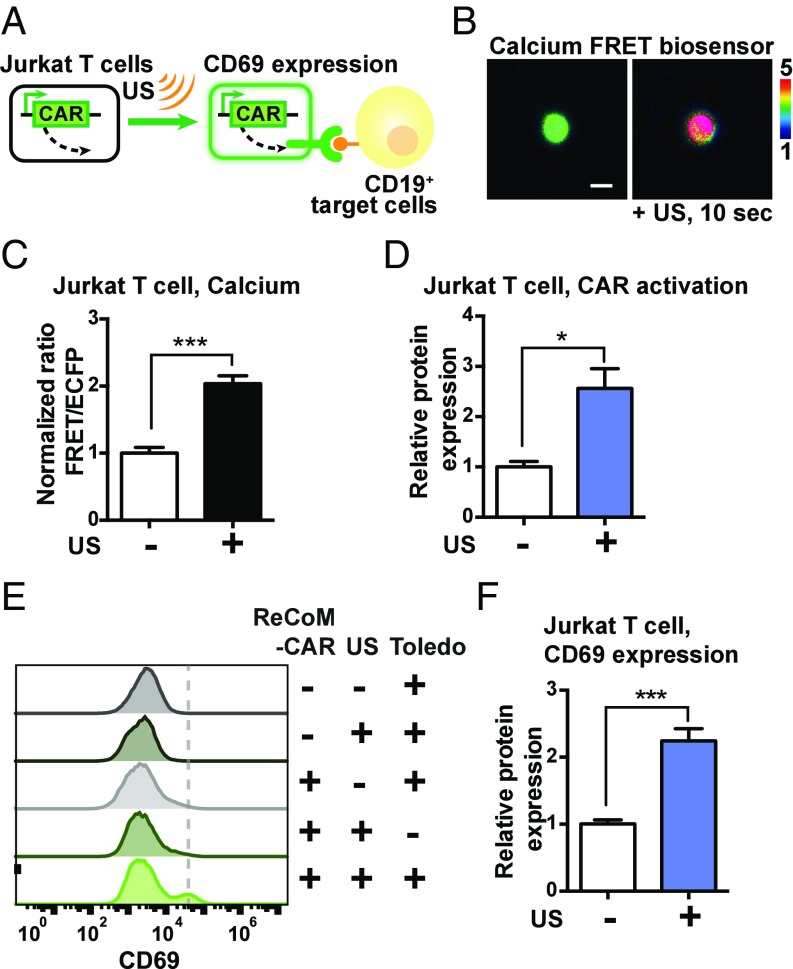
We next applied this system of remote-controlled mechanogenetics (ReCoM) based on design a to control the expression of anti-CD19 CAR in Jurkat T-cell lines and primary human T cells (peripheral blood mononuclear cells, or PBMCs) for cancer immunotherapy since anti-CD19 CAR and CD19 have been well established as an interaction pair for the immunotherapy of hematopoietic malignancies (2, 23). Jurkat and PBMC T cells express high levels of endogenous Piezo1 (24). Indeed, when microbubbles coated by streptavidin were coupled to the biotinylated surface membrane of Jurkat cells without the introduction of additional exogenous Piezo1 (Fig. 3A), calcium influx was observed upon ultrasound stimulation (Fig. 3 B and C). Further experiments validated ReCoM in controlling the production of the mNeonGreen reporter driven by different minimal promoters upon ultrasound stimulation in Jurkat T cells without the need for coexpression of exogenous Piezo1 (Fig. S7 A and B). We then applied ReCoM to control anti-CD19 CAR production by ultrasound in Jurkat cells. The encoding mRNA and expression percentage of anti-CD19 CAR in Jurkat cells was significantly increased after ultrasound stimulation (Fig. 3D and Fig. S7C). The Jurkat cells with the ultrasound-induced CAR expression were then incubated with CD19 antigen-expressing target tumor cells (Toledo lymphoma tumor cells that express high levels of CD19) (25). Upon the engagement of ultrasound-induced Jurkat and target Toledo cells for 24 h, the surface marker CD69 reflecting T-cell activation (26) was clearly up-regulated in the Jurkat cells (Fig. 3 E and F). These results indicate that the ultrasound-induced CAR production in Jurkat T cells can functionally mediate the engagement with antigens on the target tumor cells and activate Jurkat cells.
Fig. 3.
Remote-controlled activation of Jurkat T cells with ReCoM. (A) Jurkat cells were transfected with inducible ReCoM-CAR. After ultrasound stimulation, they were mixed with antigen CD19-expressing target tumor cells and evaluated for their activation level (CD69 expression). (B) The representative FRET/ECFP ratio images of the D3cpv calcium biosensor in Jurkat cells before (Left) and after (Right) ultrasound stimulation. (Scale bar: 10 μm.) (C) The FRET/ECFP ratio percentage change of D3cpv calcium FRET biosensor averaged among multiple Jurkat cells before and 10 s after ultrasound stimulation (n = 15). (D) Normalized expression of anti–CD19-CAR in Jurkat cells 16 h after ultrasound stimulation (n = 3). (E) Representative histograms of T-cell activation in Jurkat cells by quantifying the expression of the cell-surface protein marker CD69. Jurkat and Toledo mixtures were stained with Alexa647-conjugated anti-CD69 antibody and analyzed by flow cytometry. (F) The bar graphs represent CD69 up-regulation (normalized percentage of CD69-positive cells) in ultrasound-induced Jurkat cells upon Toledo cell engagement (n = 8). Error bars indicate SEM, *P < 0.05, ***P < 0.001 from two-tailed Student t test.
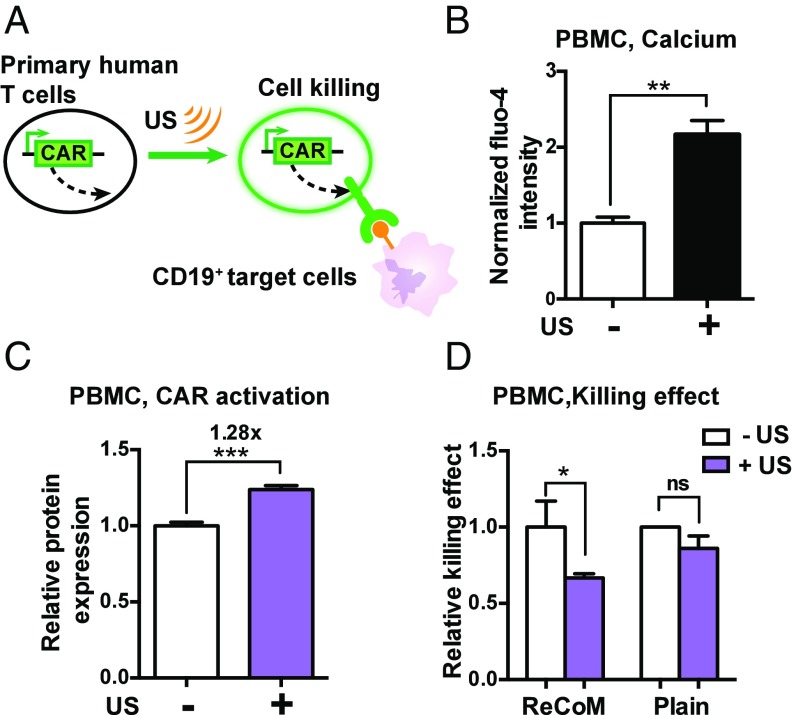
Then, we applied ReCoM to remotely control the CAR production in PBMCs and examine their efficacy in tumor cell killing (Fig. 4A). Calcium influx in PBMCs can be clearly observed upon ultrasound stimulation (Fig. 4B). We further measured the ReCoM-mediated expression of anti-CD19 CAR in PBMCs. The average expression level of anti-CD19 CAR was significantly increased after ultrasound stimulation (Fig. 4C). These ultrasound-induced PBMCs were incubated with a target B-cell leukemia cell line (Nalm6) expressing CD19-antigen and luciferase (27, 28), which allowed for convenient tracking of the target tumor cell growth by measuring the luciferase activities. The results of luciferase activity in surviving target cells revealed that the ultrasound-induced ReCoM PBMCs can cause significantly more toxicity of the target Nalm6 cells than the ReCoM PBMCs not exposed to ultrasound or the plain PBMCs exposed to ultrasound but without ReCoM GTMs (Fig. 4D).
Fig. 4.
Remote-controlled activation of PBMCs with ReCoM. (A) PBMCs were transfected with inducible ReCoM-CAR. After ultrasound stimulation, they were mixed with antigen CD19-expressing target tumor cells and evaluated for killing efficiency by functional assays. (B) The Fluo-4 calcium indicator intensity change in PBMCs before and after 5 s of ultrasound stimulation (n = 4). (C) Expression of ReCoM-CAR in PBMCs 16 h after ultrasound stimulation (n = 5). (D) Cytotoxicity of target Nalm6 tumor cells caused by CAR expression upon ultrasound stimulation in PBMC cells transfected with ReCoM-CAR GTMs, measured by luciferase-based killing assay (n = 4). Cytotoxicity of plain PBMCs upon ultrasound stimulation was also measured (n = 3). Error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001 from two-tailed Student t test.
Discussion
We report here the development of ReCoM technology to allow the remote and noninvasive control of gene expression in live cells with high spatiotemporal precision using ultrasound. Although there have been disparate trial-and-error approaches to apply ultrasound to mechanically perturb cells and Caenorhabditis elegans (29–31), there was no established method as the ReCoM presented here to convert the ultrasound mechanical signals into genetic controls of cells. Indeed, we have demonstrated that this ReCoM system can remotely control the expression of luciferase or mNeonGreen in various adherent or suspension cells. We further established that ReCoM is effective in controlling CAR expression in T cells to guide the recognition and eradication of tumor cells for controllable cancer immunotherapy. This system is highly modular as shown by multiple designs of genetic circuits with various basal levels and activation potencies, thus allowing continuous evolution and optimization to target multiple types of cancers and precancerous conditions. In summary, ReCoM will usher in an era in life science and medical technology by bringing the full power of remote control of gene and cell activation to scientific and clinical communities for precise control of therapeutics in space and time.
Materials and Methods
Ultrasonic Transducer and Stimulation System.
Ultrasonic transducers were fabricated with standard procedures (32). The center frequency of the ultrasonic transducers is 2 MHz, and the aperture size is 5 mm. The transducer is unfocused with the natural focus at ∼8.3 mm. The 150-MHz high-frequency focused transducer has the focus at ∼1 mm. The developed 2 or 150 MHz ultrasonic transducers were integrated with a Nikon FRET microscope to stimulate cells using ultrasound as well as to monitor molecular signals using FRET imaging. The ultrasonic transducer was connected to a 3D mechanical stage to control the position of the transducer. A pulser/receiver (Olympus), and an oscilloscope (Lecroy) were used to place the natural focus of the ultrasonic transducer at the target cell as shown in Fig. 1B. To stimulate HEK293T cells for calcium imaging, the 150-MHz ultrasonic transducer was positioned perpendicularly to the dish bottom surface. The transducer was submerged in the medium 1 mm away from the cells. (Fig. S2A). The resultant peak-to-peak voltage (Vpp) was 22.12–31.6 V. The acoustic pressure was ∼0.6 MPa for the 2-MHz transducer at the site of the bubbles and ∼3 MPa at the focus site for the 150-MHz high-frequency transducer. The duration of 150-MHz ultrasound stimulation was 5–10 μs. The 2-MHz ultrasonic transducer was positioned at a 45° angle relative to the dish-bottom surface to avoid standing ultrasound wave in a dish. The transducer was submerged in the medium 8.3 mm away from the cells (Fig. S1A). The electrical signals were generated by a function generator and a power amplifier to drive the ultrasonic transducer. The resultant peak-to-peak voltage (Vpp) was 22.12–31.6 V. For the 2-MHz transducer, pulse repetition frequency (PRF) was 5 Hz, and the duty factor was 10%. The duration of ultrasound stimulation was 5 s for calcium imaging and 10 min for gene induction of cells. During the 10-min ultrasound application, cells on the glass-bottom dish were continuously scanned by ultrasound. Therefore, there were multiple calcium transients generated during this time period. In fact, results revealed varying strengths of mechanogenetic effects with different durations of ultrasound stimulation (Fig. S6B). The expectation for microbubbles to be attached to biotinylated surface of suspended cells is high. When the number ratio of microbubble/cell is at 40, there is a significant binding (>50%). When the number ratio of microbubble/cell is at 400, virtually 100% of the cells are bound to microbubbles. In our experiments, we maintained the number ratio of microbubble/cell at around 100, and on average, three to four microbubbles were attached to each suspended T cell.
A glass tube was used to contain Jurkat cells and PBMCs, and the ultrasonic transducer was positioned with a 45° angle with respect to the glass surface for ultrasound stimulation. The transducer was placed at 8.3 mm away from the cell–microbubble mixture inside the glass tube as shown in Fig. S1B. For the glass tube experiments, Vpp was 22.12–31.6 V. The PRF and duty factor were 100 Hz and 1%, respectively. The duration of ultrasound stimulation was 10 min for these suspension cells.
Image Acquisition.
The ultrasound stimulation/FRET imaging system was integrated with a Nikon fluorescence microscope for FRET imaging. Images were collected by a Nikon and a cooled charge-coupled device (CCD) camera using MetaFluor 6.2 and MetaMorph software (Universal Imaging) with a 420DF40 excitation filter, a 455DCXRU dichroic mirror, and two emission filters controlled by a filter changer (480DF40 for CFP and 535DF35 for YFP). A neutral density filter was used to control the intensity of the excitation light. The fluorescence intensity of nontransfected cells was quantified as the background signals and subtracted from the CFP and YFP signals on transfected cells. The pixel-by-pixel ratio images of YFP/CFP were calculated based on the background-subtracted fluorescence intensity images of CFP and YFP by the Metafluor program to allow quantification and statistical analysis of FRET responses. The emission ratio images were shown in the intensity modified display mode (33).
Constructs and Plasmids.
The construct of the mPiezo1-tdTomato was obtained from Ardem Patapoutian’s laboratory, Scripps Research Institute, La Jolla, CA (34). The D3cpv calcium FRET biosensor was previously described (35). The nucleotide sequence encoding the CD19-CAR, including the anti-human CD19 scFv, the human CD8α hinge, the CD28 transmembrane and costimulation domain, the human 4–1BB costimulation domain, and the CD3ζ ITAM-signaling domain were synthesized by Integrated DNA Technologies (1). Sequences encoding the VPR were obtained from Addgene (plasmids #63798) (22). The plasmid-containing sequences encoding the calcium-dependent response elements and minimal promoter were a gift from Jeffrey Friedman’s laboratory, Rockefeller University, New York. Standard molecular cloning techniques (PCR, restriction digestion, ligation, etc.) were applied to construct CAR expression plasmids and produce lentivirus using the second-generation self-inactivating lentiviral vector pHR’SIN:CSW.
Cell Culture and Reagents.
HEK293T, Jurkat T cells, and Toledo cells were purchased from the American Type Culture Collection. HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium with 10% FBS (Gibco), 2 mM l-glutamine, 1 unit/mL penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate. Jurkat cells and Toledo cells were maintained in RPMI-1640 medium supplemented with 10% FBS, penicillin, and streptomycin. Primary human T cells were isolated from an anonymous healthy donor’s blood after apheresis (San Diego Blood Bank). Purified PBMCs were cryopreserved in culture RPMI-1640 medium supplemented with 10% DMSO until use. During experiments, PBMCs were maintained in RPMI-1640 medium supplemented with 10% FBS and 30 units/mL IL-2 (PeproTech). All cells were cultured at 37 °C with 5% CO2. Ionomycin was purchased from Sigma.
Cell Preparation for Ultrasound Stimulation.
For HEK293T cells, Targestar-SA lipid microbubbles (Targeson) (1 × 109/mL) were mixed with biotinylated Arg-Gly-Asp (RGD) peptides (0.01 mg/mL) (Peptide International) for 20 min (36). Immediately after removing the culture media in the dish, 5 μL of the microbubble–RGD mixture was added into the dish. The dish was then flipped upside down for 5 min to allow microbubbles to float up and attach to the cell membrane.
For Jurkat cells and PBMCs, membrane proteins on the cell surface were biotinylated by EZ-Link Sulfo-NHS-Biotin (2 mM) (ThermoFisher Scientific) for 15 min and washed with PBS before the cells were incubated with and coupled to Targestar microbubbles.
Gene Activation Assays.
For luciferase-based gene expression assay, the pRL-TK plasmid (Promega) was cotransfected with the ReCoM luciferase reporters using FUGENE 6 to normalize for transfection efficiency. Luciferase activity was assayed 36–48 h after transfection, using a dual-luciferase reporter assay system (Promega). In brief, cells were pelleted by centrifugation at 300 × g for 3 min, resuspended in lysis buffer for 15 min, and then processed with a plate reader (Tecan Infinite M1000 Pro) for luciferase assay and reading quantifications.
For flow cytometry assay of fluorescent reporter expression, cells were resuspended in FACS wash buffer (PBS + 0.5% BSA) and processed with a BD Accuri C6 cytometer. FlowJo software (TreeStar) was used to quantify fluorescence percentages and intensities.
For qPCR assay of CD19-CAR gene expression, total RNA from cells was extracted by TRIzol. cDNA was synthesized with 500 ng of total RNA using M-MLV transcriptase. Quantification of RNA levels was carried out using real-time PCR (KAPA SYBR FAST qPCR kit) (KAPA) and normalized to the level ACTB gene expression. Sequences of PCR primers are as follows: ReCoM-CAR, 5′-AAGTCCAGCTCCAGCAATCC-3′; and 5′-GGAAGGCTGACCCCACTAAC-3′.
Lentiviral Infection of Primary T Cells.
Pantropic VSV-G pseudotyped lentivirus was produced from Lenti-X 293T cells (#632180; Clontech Laboratories) cotransfected with a pHR′SIN:CSW transgene expression vector and the viral packaging plasmids pCMVdR8.91 and pCMV-VSV-G using Lipofectamine 2000 (Life Technologies). Viral medium/supernatant was collected 48 h after transfection for infection. Three days before viral infection, primary human T cells were thawed and activated using 2 μg/mL phytohemagglutinin (PHA) (ThermoFisher Scientific) in RPMI-1640 medium supplemented with 10% FBS, penicillin, and streptomycin for 72 h. PHA was then removed before cells were infected. Infected primary T cells (PBMCs) were maintained at ∼106/mL in culture RPMI medium supplemented with 30 units/mL IL2 for 5 d before experiments were conducted.
Verification of CAR Expression in T Cells.
Jurkat or primary human T cells were resuspended in FACS wash buffer (PBS + 0.5% BSA) and stained with Alexa Fluor 647 AffiniPure F(ab′)2 Fragment Goat Anti-Mouse IgG antibody (JacksonImmunoResearch). Stained cells were washed three times in washing buffer PBS + 0.5% BSA and processed with a BD Accuri C6 cytometer. FlowJo software (TreeStar) was used to quantify Alexa dye and/or mCherry fluorescence intensities and percentages.
Quantitation of CD69 Surface Expression.
Jurkat T cells after ultrasound activation were mixed with target Toledo cells at a 1:1 T-cell/target cell ratio in a 24-well plate. After 24 h of incubation, cells were pelleted by centrifugation at 300 × g for 5 min. Cells were resuspended in FACS wash buffer (PBS + 0.5% BSA) and stained with APC anti-human CD69 antibody (#310910; BioLegend). Stained cells were washed three times in FACS wash buffer and processed with a BD Accuri C6 cytometer. FlowJo software (TreeStar) was applied to compare Alexa fluorescence intensities of gated T cells (unique forward/side scatters) in samples. Data plots were generated using Prism software (GraphPad).
Luciferase-Based Cell Killing Assay.
PBMCs after ultrasound activation were mixed with NALM-6 target cells at a 10:1 T-cell/target cell ratio in a 96-well plate. After 24 h of incubation, cells were pelleted by centrifugation at 300 × g for 5 min. Cells were resuspended in 50 μL of lysis buffer (Promega) and processed with a plate reader (Tecan Infinite M1000 Pro) for luciferase assay.
Supplementary Material
Acknowledgments
We thank Professor Ardem Patapoutian (Scripps Research Institute) for the Piezo1 construct. This work was supported by National Institutes of Health Grants HL121365 and GM125379 (to S.C. and Y. Wang) and CA204704 and CA209629 (to Y. Wang); National Science Foundation Grants CBET1360341 and DMS1361421 (to Y. Wang); and the Beckman Laser Institute Foundation. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714900115/-/DCSupplemental.
References
- 1.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fesnak AD, June CH, Levine BL. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan RA, et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo D, et al. A mechanogenetic toolkit for interrogating cell signaling in space and time. Cell. 2016;165:1507–1518. doi: 10.1016/j.cell.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanley SA, et al. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science. 2012;336:604–608. doi: 10.1126/science.1216753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley SA, et al. Bidirectional electromagnetic control of the hypothalamus regulates feeding and metabolism. Nature. 2016;531:647–650. doi: 10.1038/nature17183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley SA, Sauer J, Kane RS, Dordick JS, Friedman JM. Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat Med. 2015;21:92–98. doi: 10.1038/nm.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 9.Ibsen S, Schutt CE, Esener S. Microbubble-mediated ultrasound therapy: A review of its potential in cancer treatment. Drug Des Devel Ther. 2013;7:375–388. doi: 10.2147/DDDT.S31564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon S, Aglyamov S, Karpiouk A, Emelianov S. The mechanical properties of ex vivo bovine and porcine crystalline lenses: Age-related changes and location-dependent variations. Ultrasound Med Biol. 2013;39:1120–1127. doi: 10.1016/j.ultrasmedbio.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon S, et al. Direct and sustained intracellular delivery of exogenous molecules using acoustic-transfection with high frequency ultrasound. Sci Rep. 2016;6:20477. doi: 10.1038/srep20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindner JR. Microbubbles in medical imaging: Current applications and future directions. Nat Rev Drug Discov. 2004;3:527–532. doi: 10.1038/nrd1417. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, et al. Two-bubble acoustic tweezing cytometry for biomechanical probing and stimulation of cells. Biophys J. 2015;108:32–42. doi: 10.1016/j.bpj.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon S, Aglyamov SR, Karpiouk AB, Kim S, Emelianov SY. Estimation of mechanical properties of a viscoelastic medium using a laser-induced microbubble interrogated by an acoustic radiation force. J Acoust Soc Am. 2011;130:2241–2248. doi: 10.1121/1.3628344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Z, et al. Acoustic tweezing cytometry for live-cell subcellular modulation of intracellular cytoskeleton contractility. Sci Rep. 2013;3:2176. doi: 10.1038/srep02176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syeda R, et al. Piezo1 channels are inherently mechanosensitive. Cell Rep. 2016;17:1739–1746. doi: 10.1016/j.celrep.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogan PG. Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium. 2017;63:66–69. doi: 10.1016/j.ceca.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TJ, et al. Distinct mechanisms regulating mechanical force-induced Ca2+ signals at the plasma membrane and the ER in human MSCs. Elife. 2015;4:e04876. doi: 10.7554/eLife.04876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox CD, et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat Commun. 2016;7:10366. doi: 10.1038/ncomms10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WL, Tan JW, Tan CN, Loo SC. Modulating drug release from gastric-floating microcapsules through spray-coating layers. PLoS One. 2014;9:e114284. doi: 10.1371/journal.pone.0114284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maus MV, Grupp SA, Porter DL, June CH. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petryszak R, et al. Expression Atlas update: An integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kochenderfer JN, et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother. 2009;32:689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol. 1996;3:301–304. doi: 10.1128/cdli.3.3.301-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brentjens RJ, et al. Genetically targeted T cells eradicate systemic acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2007;13:5426–5435. doi: 10.1158/1078-0432.CCR-07-0674. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Z, et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell. 2015;28:415–428. doi: 10.1016/j.ccell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legon W, et al. Transcranial focused ultrasound modulates the activity of primary somatosensory cortex in humans. Nat Neurosci. 2014;17:322–329. doi: 10.1038/nn.3620. [DOI] [PubMed] [Google Scholar]
- 30.Tufail Y, et al. Transcranial pulsed ultrasound stimulates intact brain circuits. Neuron. 2010;66:681–694. doi: 10.1016/j.neuron.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Ibsen S, Tong A, Schutt C, Esener S, Chalasani SH. Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat Commun. 2015;6:8264. doi: 10.1038/ncomms9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon S, et al. Dual-element needle transducer for intravascular ultrasound imaging. J Med Imaging (Bellingham) 2015;2:027001. doi: 10.1117/1.JMI.2.2.027001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 34.Cahalan SM, et al. Piezo1 links mechanical forces to red blood cell volume. Elife. 2015;4:e07370. doi: 10.7554/eLife.07370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 36.Heureaux J, Chen D, Murray VL, Deng CX, Liu AP. Activation of a bacterial mechanosensitive channel in mammalian cells by cytoskeletal stress. Cell Mol Bioeng. 2014;7:307–319. doi: 10.1007/s12195-014-0337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.