Significance
Short interspersed elements (SINEs) are retrotransposons that have accumulated in genomes through a “copy-and-paste” mechanism. Once inserted, these sequences have the potential to gain novel functions, including regulating gene expression. Here, we demonstrate the unexpected finding that independently evolved SINEs have accumulated in the 3′-untranslated regions of orthologous human and mouse genes. We demonstrate that, in some cases, this has resulted in their regulating gene expression by Staufen-mediated mRNA decay (SMD) in human and mouse myoblasts. Thus, SINE-directed SMD has convergently evolved to regulate myoblast metabolism. This represents an example of the convergent evolution of posttranscriptional gene regulation using SINEs.
Keywords: Staufen1, UPF1, Staufen-mediated mRNA decay, SINEs, convergent evolution
Abstract
Primate-specific Alu short interspersed elements (SINEs) as well as rodent-specific B and ID (B/ID) SINEs can promote Staufen-mediated decay (SMD) when present in mRNA 3′-untranslated regions (3′-UTRs). The transposable nature of SINEs, their presence in long noncoding RNAs, their interactions with Staufen, and their rapid divergence in different evolutionary lineages suggest they could have generated substantial modification of posttranscriptional gene-control networks during mammalian evolution. Some of the variation in SMD regulation produced by SINE insertion might have had a similar regulatory effect in separate mammalian lineages, leading to parallel evolution of the Staufen network by independent expansion of lineage-specific SINEs. To explore this possibility, we searched for orthologous gene pairs, each carrying a species-specific 3′-UTR SINE and each regulated by SMD, by measuring changes in mRNA abundance after individual depletion of two SMD factors, Staufen1 (STAU1) and UPF1, in both human and mouse myoblasts. We identified and confirmed orthologous gene pairs with 3′-UTR SINEs that independently function in SMD control of myoblast metabolism. Expanding to other species, we demonstrated that SINE-directed SMD likely emerged in both primate and rodent lineages >20–25 million years ago. Our work reveals a mechanism for the convergent evolution of posttranscriptional gene regulatory networks in mammals by species-specific SINE transposition and SMD.
Short interspersed elements (SINEs) are retrotransposable DNA sequences of ∼100–600 base pairs (bp). Lineage-specific SINEs have expanded to now account for ∼8% of mouse and ∼13% of human genomic sequences (1, 2). SINEs that have integrated into RNA polymerase (Pol) II transcription units have adopted an array of roles that include generating pre-mRNA splice sites or polyadenylation sites, and regulating mRNA localization or mRNA translation (3).
The large number of SINEs in mRNAs provides the opportunity for intermolecular base pairing to form a partially complementary duplex between SINEs in, e.g., a long noncoding RNA (lncRNA) or another mRNA. Such duplexes can contain binding sites for the double-stranded (ds) RNA-binding proteins (RBPs) Staufen (STAU)1 and/or STAU2 (4–6), which, when present in an mRNA 3′-UTR, can trigger STAU-mediated mRNA decay (SMD), a process that additionally requires the ATP-dependent RNA helicase UPF1 (4, 5, 7–9). SMD has been implicated in the regulation of myogenesis (10, 11), adipogenesis (12), cell mobility and invasion (4, 5), and stress-induced apoptosis (13).
In primates, SMD can be triggered by base pairing between Alu elements (4, 5). Alu elements emerged from a dimer of 7SL RNA and expanded in the primate lineage to ∼1.4 million copies per human genome (2, 14). In rodents, which lack Alu elements, homoduplexes can form between B1, B2, B4, or identifier (ID) elements (11). Like Alu elements, B1 elements were derived from 7SL RNA, whereas B2 elements were derived from tRNA, B4 elements from the fusion of a tRNAAla-derived ID element at the 5′-end and a B1 element at the 3′-end, and ID elements from tRNAAla or a neuronally expressed brain cytoplasmic (BC1) RNA (1, 15–17). Alu and B or ID (B/ID) SINE families emerged after primates and rodents diverged ∼90 million years ago (MYA), and have continuously amplified independently in the two mammalian lineages (1, 18, 19) (Fig. 1A).
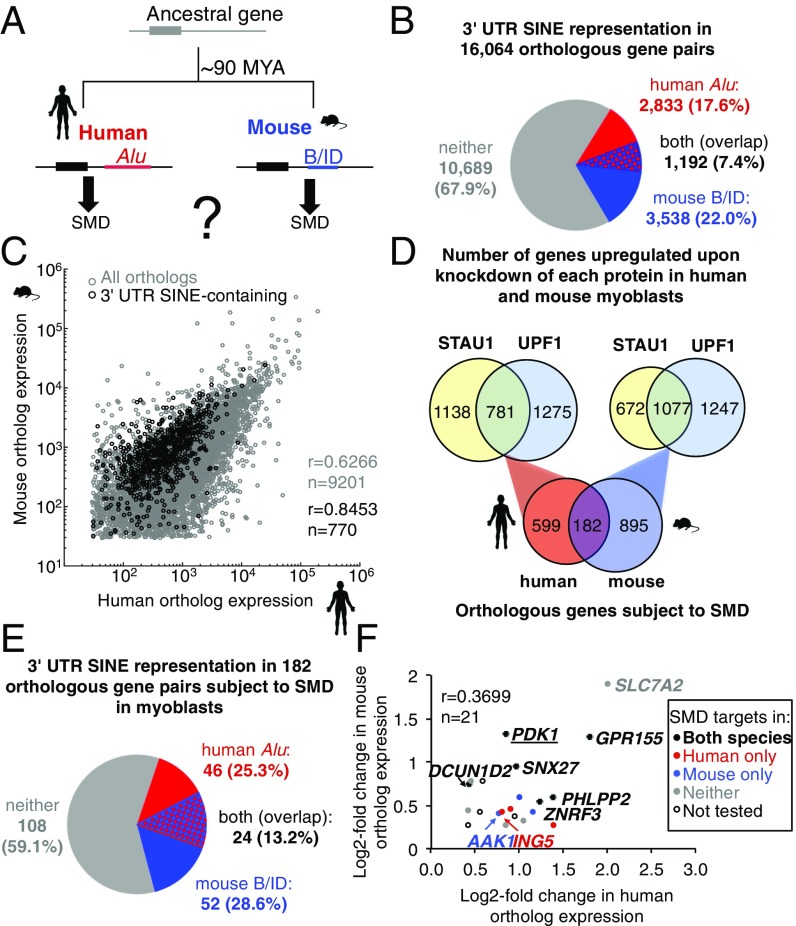
Fig. 1.
Identification of 3′-UTR SINE-containing orthopairs that are targets of Staufen-mediated mRNA decay (SMD). (A) Diagram of an archetypal orthopair that independently acquired a 3′-UTR Alu SINE in humans and a B or ID (B/ID) SINE in mouse. (B) Pie chart showing the proportion of human:mouse orthopairs that are 3′-UTR SINE-containing in human (red), mouse (blue), both (red and blue), or neither ortholog (gray). (C) Scatterplot showing the correlation in orthologous gene expression in control siRNA-treated human and mouse myoblasts (MBs); r, Pearson correlation coefficient; n, number of observations. (D) Venn diagrams demonstrate that hMBs and mMBs share 182 orthologs that are putative SMD targets, i.e., transcripts significantly up-regulated relative to their level in control siRNA-treated cells when STAU1 and, separately, UPF1 was depleted using siRNA in hMBs or mMBs. (E) Pie chart showing the proportion of the 182 orthopairs that produce putative SMD targets in both hMBs and mMBs and are 3′-UTR SINE-containing in the human ortholog (red), mouse ortholog (blue), or both orthologs (red and blue). (F) Scatterplot comparing the log2-fold change after hSTAU1 or mStau1 knockdown of each 3′-UTR SINE-containing human and mouse orthopair (3 of the 24 were excluded from this analysis as the major 3′-UTR isoform lacks the SINE). Color coding indicates SMD target validation using RT-qPCR (SI Appendix, Fig. S2).
Despite integrating independently in their respective genomes, the positions of extant Alu and B/ID elements correlate over large genomic regions (15), are predominately found in introns (20), and are enriched in upstream and intronic sequences of groups of genes with similar functions (21). Additionally, retrotransposons, including SINEs, are overrepresented in the 3′-UTRs of both human and mouse and correlate with decreased expression of their encoded transcripts (22). The large number, similar positioning in the human and mouse genomes, and the analogous function of Alu and B/ID SINEs in SMD led us to speculate that alterations to posttranscriptional regulation resulting from lineage-specific 3′-UTR SINE insertion may have resulted in parallel alterations to gene expression in the two lineages.
Here we identify 1,192 orthologous gene pairs (orthopairs) in human and mouse that are 3′-UTR SINE-containing. Using RNA sequencing (RNA-seq) and gene-specific assays, we identify orthopairs that are SMD targets in both human and mouse. We demonstrate that these independent networks have converged to fine tune the activity of the metabolic regulator PDK1 in both human and mouse myoblasts. Finally, we identify the 3′-UTR features that correlate with SINE presence and extend our observations to SINEs in additional species.
Results
Identifying 3′-UTR SINE-Containing Orthologs That Are Regulated by SMD.
Although 3′-UTR SINEs are known to trigger SMD in human and mouse (4, 5, 11), orthopairs in which distinct species-specific SINEs trigger SMD remain undiscovered (Fig. 1A). To search for such pairs, we analyzed the 16,064 human and mouse orthopairs annotated by the University of California, Santa Cruz (UCSC) and Mouse Genome Informatics (MGI) (22, 23) and found 1,192 (7.4%), wherein both orthologs are 3′-UTR SINE-containing (Fig. 1B). Similarly, 1,109 (7.0%) of the 15,736 1:1 orthopairs generated by the mouse ENCODE project (24) are likewise 3′-UTR SINE-containing (SI Appendix, Table S1). This overlap represents a statistically significant approximately twofold enrichment relative to a random insertion model (SI Appendix, Table S1). One possible source of this enrichment might be parallel evolution of SMD control; however, several correlated features of mouse and human 3′-UTRs with the potential to lead to biased SINE insertion also contribute to this enrichment (discussed below).
To search for the existence of 3′-UTR SINE-containing orthopairs where both orthologs are under SMD control, we sought a common biological context within which a large number of orthopairs would be similarly expressed, including a minimum of distinct mRNA isoforms with widely variable 3′-UTRs that often typify different cell types and differentiation states (25). A common context is also expected to depend on SMD in similar ways, as the efficiency of SMD can vary among different cell types and can change during the process of differentiation (10–12). We chose human hSkMc skeletal muscle myoblasts (hMBs) and mouse C2C12 skeletal muscle myoblasts (mMBs), knowing that SMD is efficient in mMBs (4, 10, 11), and expecting it would also be efficient in hMBs. Triplicate cultures of MBs from each species were treated with control or SMD factor-targeting siRNA, either STAU1 siRNA or UPF1 siRNA, and poly(A)+ RNA from each condition was sequenced and analyzed (see below) using DESeq (26). Mean expression values for >9,000 orthopairs from the three control replicates for mouse and human were compared (Fig. 1C), and a Pearson correlation coefficient of 0.6266 was calculated, similar to a previous report (r = 0.76) comparing orthopairs in human and mouse muscle tissue (1, 27). The lower value calculated here likely reflects expression differences between hMB and mMB cell lines that are not present in muscle tissue. Expression values for 3′-UTR SINE-containing orthopairs are also highly correlated (Fig. 1C). These data suggest that the gene expression characteristics of these two MB cell lines from different species are sufficiently similar to support comparison of the regulation of orthopairs by SMD.
To identify which 3′-UTR SINE-containing orthologous mRNA pairs are SMD targets in human and mouse MBs, we identified transcripts that are under SMD control in human and mouse MBs based on their up-regulation (≥1.2-fold, P < 0.05) upon siRNA-mediated depletion of STAU1 and, separately, UPF1 using DESeq (SI Appendix, Fig. S1 A and B and Dataset S1) (7, 26). Although depletion of UPF1 inhibits both nonsense-mediated mRNA decay (NMD) and SMD (10), and STAU1 plays other roles in gene expression (9, 28), a proportion of the genes up-regulated upon depletion of either protein is also up-regulated in response to depletion of the other protein in human (781 genes) and mouse (1,077 genes) MBs (Fig. 1D). Of these, 163 (20.9%) and 373 (34.6%), respectively, are 3′-UTR SINE-containing, a significant enrichment relative to all orthopairs (Fig. 1B) [P = 0.0238 (human), P value < 0.0001 (mouse), χ2 test]. Nevertheless, we noted that the majority of human or mouse MB mRNAs with a 3′-UTR SINE are not detectably SMD targets, likely for a number of reasons. First, not all 3′-UTR SINEs trigger SMD as not all SINEs bind STAU1 (see, e.g., ref. 29); however, the features that define which SINEs are competent for STAU1 binding remain unknown. Second, steady-state measurements, including standard RNA-seq, can be insensitive to subtle changes in mRNA stability when transcription rates are high. Third, alternative polyadenylation that excludes the SINE from the major mRNA isoform expressed in human or mouse MBs may obscure SMD-target detection for the minor mRNA isoform. In this regard, it is notable that for 10 of the 57 orthologous genes having a 3′-UTR SINE whose transcripts detectably underwent SMD in hMBs only, the SINE is present in only a minor mRNA isoform in mMBs.
Despite the limited sensitivity inherent in this approach, both mRNAs from 182 orthopairs are among those up-regulated after separate depletion of both UPF1 and STAU1 (Fig. 1D and Dataset S1), and 24 of these (13.2%) carry a 3′-UTR SINE in both members of the orthopair (Fig. 1E and SI Appendix, Tables S1 and S2). This is a significant enrichment relative to the 7.4% of all orthopairs that are 3′-UTR SINE-containing (Fig. 1B) (P = 0.0058, χ2 test). We did not observe a similar enrichment when we examined the orthopairs with a SINE in only one ortholog (P = 0.48, Alu only; P = 0.85, B/ID only; χ2 test), suggesting that SINE acquisition may have contributed to the parallel evolution of Staufen regulatory networks since the last common euarchontal ancestor of humans and mice.
Of the 24 orthopairs that were putative SMD targets, three lacked a SINE in the 3′-UTR of the major isoform and were excluded from our analysis. Of the remaining 21, we used RT-qPCR to quantify mRNA and pre-mRNA of 17 orthopairs in control, UPF1-depleted, and STAU1-depleted cells. From this, we confirmed that 6 orthopairs that contain a SINE in the 3′-UTR of the expressed isoform in MBs were significantly up-regulated (t test, P < 0.05) upon depletion of each SMD factor (Fig. 1F and SI Appendix, Fig. S2 E and F), indicating that they are bona fide SMD targets in MBs of both species. We further confirmed that 5 of the defined putative SMD targets copurify with mouse STAU1 in MBs using a monoclonal antibody that we developed to the C-terminal 73 amino acids of human STAU1 (SI Appendix, Fig. S1). Notably, the up-regulation of SLC7A2 mRNA detected in RNA-seq experiments was due to changes in transcription, rather than mRNA stability, because both human and mouse SLC7A2 mRNA (i) did not increase more than its pre-mRNA upon STAU1 or UPF1 depletion and (ii) did not detectably copurify with mouse or human STAU1 by α-STAU1 immunoprecipitation (SI Appendix, Fig. S1 E–H). This highlights the specificity of our approach to identify bona fide SMD targets. As further evidence that the mRNAs we identify are SMD targets by virtue of retrotransposons that are lineage specific, rather than conserved, we failed to find conserved mammalian-wide interspersed repeats (MIRs) or long interspersed elements (LINEs) in the 3′-UTRs of the 5 orthopairs that we demonstrate are SMD targets in human and mouse. The small number of the ∼1,100 SINE-containing orthopairs identified in these experiments is only a subset of those for which convergent evolution may have taken place across both genomes, given the technical sensitivity limits (described above) and the fact that MBs represent only one of a large number of expression states that human and mouse cells can occupy.
Independently Evolved 3′-UTR SINEs Direct SMD of the Metabolic Regulator PDK1.
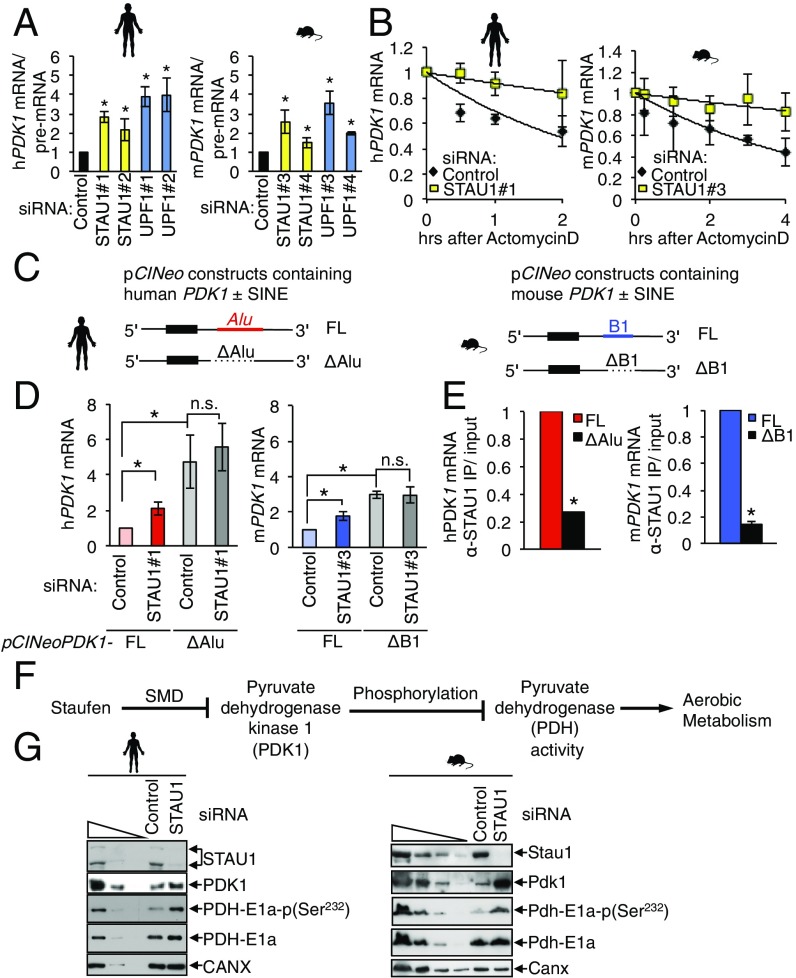
To provide direct evidence that the SINE of orthologous transcripts is required for SMD, we chose to analyze the orthopair encoding pyruvate dehydrogenase kinase 1 (PDK1), because (i) each PDK1 mRNA harbors only a single SINE in the major 3′-UTR isoform expressed in MBs (SI Appendix, Table S2); (ii) the SINE is sufficiently downstream of the termination codon to prevent interruption of the ORF, allowing for a simple deletion experiment; and (iii) the full-length mRNA is short enough to be readily clonable. Each mRNA was up-regulated in response to each of two nonoverlapping STAU1 and UPF1 siRNAs (Fig. 2A) and stabilized in response to STAU1 siRNAs (Fig. 2B). To interrogate the role of each SINE in SMD, we generated PDK1 cDNA expression constructs containing either the native 3′-UTR or a SINE-deletion (ΔSINE) variant (Fig. 2C). Consistent with a role for each SINE in promoting SMD, each ΔSINE mRNA was expressed at a significantly higher level than its corresponding SINE-containing mRNA and was unresponsive to STAU1 depletion, whereas the corresponding SINE-containing mRNAs responded to STAU1 depletion as did mRNA from the native gene (Fig. 2D). Deletion of the SINE results in more up-regulation than depletion of STAU1 protein, likely because residual SMD remains, due to both incomplete depletion of STAU1 and the contribution of STAU2 to SMD (6) (SI Appendix, Fig. S3). We further demonstrated that the interaction of both human and mouse PDK1 mRNA with STAU1 depends on the SINE, as deleting the SINE decreased the coimmunoprecipitation with STAU1 by ∼5–10 fold (Fig. 2E). Although the human MBs were able to take up siRNAs, they were inefficiently transfected with DNA constructs. Thus, we employed human HeLa cells to demonstrate the general dependence of the SMD of PDK1 mRNA on the Alu element (Fig. 2 D and E). We conclude that the primate-specific Alu and rodent-specific B1 SINEs regulate both human and mouse orthologs of PDK1 by SMD (Fig. 2 A–E) and that neither element was present in the common ancestor. While both PDK1 3′-UTR SINEs are 7SL derived, it is likely that tRNA-derived SINEs would also function analogously, considering their demonstrated function in SMD (11).
Fig. 2.
SMD regulates the expression of human and mouse PDK1 mRNAs in hMBs and mMBs by virtue of their 3′-UTR SINEs. (A) Histogram representing RT-qPCR quantifications of each mRNA normalized to its pre-mRNA and then to control siRNA treatment after treatment with each of two nonoverlapping siRNAs to STAU1 or UPF1. (B) RT-qPCR quantitation of human (Left) or mouse (Right) PDK1 mRNA in hMBs or mMBs at indicated time points after treatment with actinomycin D normalized to 18S rRNA and then to 0 h. (C) pCINeo expression vectors containing human (Left) or mouse (Right) PDK1 cDNA that does or does not (ΔSINE) harbor the 3′-UTR SINE. (D) RT-qPCR quantifications of human (Left) or mouse (Right) PDK1 transgene mRNA normalized to the level of MUP transgene mRNA and then to control siRNA treatment. (E) RT-qPCR quantitation of human or mouse PDK1 transgene mRNA that coimmunoprecipitated with STAU1 from human HeLa or mMB cells transfected with the plasmids described in C, normalized first to the level before immunoprecipitation (IP), then to the SINE-containing (FL) transcript. (F) Flowchart showing how Staufen1, via SMD, may inhibit human and mouse PDK1-mediated phosphorylation of pyruvate dehydrogenase subunit E1a (PDH-E1a) at serine 232 potentially leading to alterations in aerobic metabolism. (G) Western blotting of lysates of hMBs (Left) or mMBs (Right) collected 2 d after transfection with the specified siRNA. The level of calnexin (CANX) serves as a loading control. All results derive from three or more independent experiments. Histograms represent the average and SD. *P < 0.05.
Showing the adaptive value of a SINE insertion that brings an mRNA under SMD control in human and mouse would be very difficult. But if control of the PDK1 gene by the STAU1 regulatory network is physiologically important in the MBs of both species, metabolic consequences of SMD loss on the downstream events that depend on correct expression of PDK1 should be evident and similar in both species. In both human and mouse, PDK1 is one of a family of four kinases that phosphorylates pyruvate dehydrogenase (PDH) at one of three serines on the E1a subunit (PDH-E1a), thereby reducing pyruvate dehydrogenase activity so as to control the metabolic state of the cell (30) (Fig. 2F). We found for both human and mouse MBs that STAU1 depletion increased the level of PDK1 protein and concomitantly increased the level of PDH-E1a phosphorylation at serine 232, which is uniquely phosphorylated by PDK1 (30), while total PDH-E1a protein levels were not significantly changed in the MBs of either species (Fig. 2G). These data are consistent with a similar contribution of 3′-UTR SINEs to MB metabolism in human and mouse and demonstrate that species-specific SINE insertions can promote parallel evolution of posttranscriptional regulatory networks by employing STAU1 proteins in SMD.
The 3′-UTR SINEs Reside in Orthopairs with Long 3′-UTRs and High A+T Content.
Given that human Alu SINEs and mouse B/ID SINEs have expanded independently, and assuming that every 3′-UTR has an equal probability of containing a SINE, the expected number of orthopairs in which both members harbor a 3′-UTR SINE would be ∼624 (SI Appendix, Table S1). This is significantly fewer than the 1,192 genes we found (P value < 10−100, χ2 test), suggesting that SINEs have preferentially integrated and/or been maintained after random insertion in the 3′-UTRs of orthopairs after primates and rodents diverged. Although Alu, B, and ID SINEs all depend on endonuclease and reverse transcriptase activities encoded by the LINE L1 (31, 32), factors that influence SINE positions are not well understood.
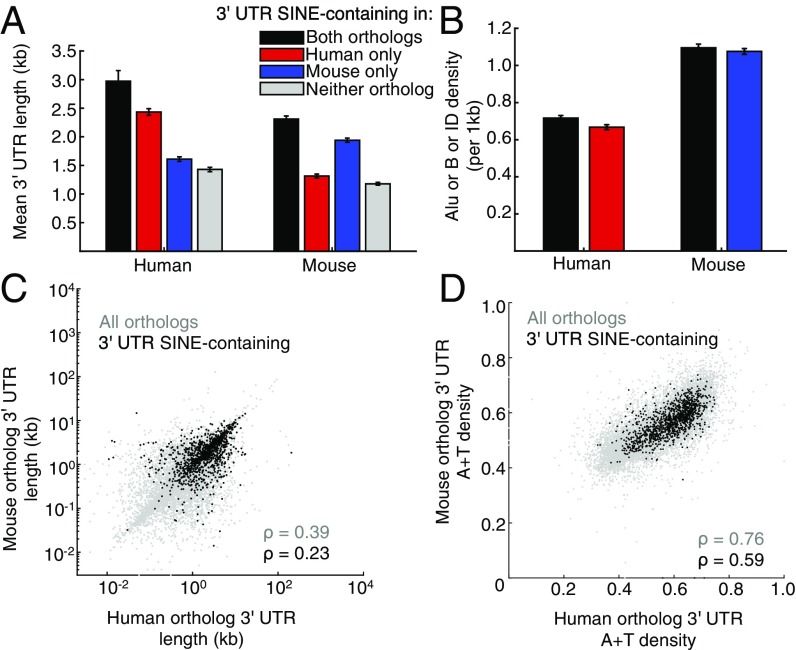
To understand more about the factors that influence SINE distributions, we systematically compared the sequence features of SINE-containing 3′-UTRs to SINE-lacking 3′-UTRs. We found that the enrichment observed is mostly accounted for by preferential insertion and/or retention in long 3′-UTRs because (i) SINE-containing 3′-UTRs are significantly longer than SINE-lacking 3′-UTRs (Fig. 3A and SI Appendix, Table S4); (ii) SINE density is comparable between 3′-UTRs in the same species (human P = 0.76072; mouse P = 0.44629) (Fig. 3B); (iii) 3′-UTR lengths of orthologous genes correlate (Fig. 3C); and (iv) statistical modeling to simulate SINE insertion based on 3′-UTR length predicts 932 ± 16 3′-UTR SINE-containing orthopairs, close to the observed number of 1,192 (SI Appendix, Table S3). To further analyze 3′-UTR lengths, we separated the 3′-UTRs by their SINE subtype, since subtype can provide a rough estimate of SINE insertion time. We found that all except the least-abundant SINE families (AluY in human and B4 in mouse) are present in significantly longer 3′-UTRs (SI Appendix, Table S5). This is consistent with SINEs being inserted and/or retained in longer 3′-UTRs, rather than 3′-UTR sequence accumulation after SINE insertion.
Fig. 3.
The 3′-UTR SINEs reside in orthopairs encoding mRNAs with long 3′-UTRs. (A) Histograms showing the mean 3′-UTR length of the longest mRNA isoform from RefSeq, excluding the SINE sequence, calculated for each orthopair that is 3′-UTR SINE-containing in both human and mouse (black), only human (red), only mouse (blue), or neither (gray). Error bars denote SEMs. (B) Histograms showing the number of Alu or B/ID SINEs per 1 kb in the 3′-UTRs of human or mouse transcripts from orthologous genes that either have or lack a 3′-UTR SINE. (C) Dot plot demonstrating the correlation between 3′-UTR length of the longest isoform of each human and mouse orthopair that lacks (light gray, ρ = 0.39, P < 10−100) or contains (black, ρ = 0.23, P = 4·10−15) 3′-UTR SINEs in both orthologs. (D) Same as in C but comparing 3′-UTR A+T content.
In addition to 3′-UTR length, we found that both human and mouse SINEs reside preferentially in 3′-UTRs that are conducive to L1 cleavage, i.e., (i) are enriched in TT dinucleotides, while human 3′-UTRs are additionally enriched for AA dinucleotides, increasing the probability of containing the TTAAAA L1 cleavage site (SI Appendix, Fig. S4 and Table S6); (ii) depleted of CC dinucleotides, increasing the rigidity of the DNA sequence (SI Appendix, Fig. S4); and (iii) contain nucleotides flanking the SINE insertion that display evidence of L1 cleavage and insertion (SI Appendix, Fig. S5). Consistent with these sequence features playing a role in the observed enrichment, the A+T content of orthologous 3′-UTRs is also correlated (ρ = 0.59, P = 3.5·10−111) (Fig. 3D).
Evidence of sequence conservation would provide support for functional retention of 3′-UTR SINEs. However, it is unlikely that STAU1-binding sites represent a specific sequence motif because (i) both sense Alus and antisense Alus (i.e., Alus that are the reverse complement of the sense Alu sequence) can trigger SMD to a similar degree (SI Appendix, Fig. S6); and (ii) we observe no enrichment of any particular Alu subtype in human or SINE type in mouse within those mRNAs that are up-regulated when STAU1 and, independently, UPF1 is knocked down (SI Appendix, Fig. S7). As anecdotal support for the hypothesis that 3′-UTR Alus in putative SMD targets may have experienced a selective pressure to maintain their sequence, we found that the percentage of the Alu sequence deleted relative to the consensus Alu sequence is significantly less in Alus residing in putative SMD targets relative to all 3′-UTR Alus (SI Appendix, Fig. S8).
Because of multiple correlating features, it is difficult to estimate what fraction of the SINE-containing orthopairs has acquired SMD regulation and to what extent these SINEs have persisted through selectively advantageous regulatory properties.
Orthologs with 3′-UTR SINEs in Other Species Are Also Responsive to STAU1 and UPF1.
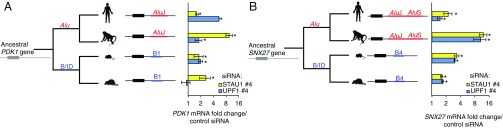
To ask when during mammalian evolution particular 3′-UTR SINEs became functional in SMD, we assayed PDK1 and SNX27 mRNAs in Chlorocebus sabaeus (African green monkey) COS-7 and Rattus norvegicus (rat) L6E9 cells, since each transcript is 3′-UTR SINE-containing. African green monkeys and humans last shared a common ancestor ∼28 MYA, and rats and mice last shared a common ancestor 18–23 MYA (33). Interestingly, African green monkey PDK1 and SNX27 and rat SNX27 mRNAs increased in response to STAU1 or UPF1 siRNA, consistent with regulation by SMD (Fig. 4). However, in the L6E9 myoblast cell line, rat PDK1 mRNA increased in response to STAU1 siRNA, but not UPF1 siRNA (Fig. 4A) and coimmunoprecipitated with STAU1 (SI Appendix, Fig. S9B), meeting two of the three criteria used to call an SMD target. While unclear, the failure of the rat ortholog of PDK1 to be up-regulated by UPF1 knockdown may reflect one of the many alternate functions of UPF1. Additionally, while rat L6E9 cells are, like mouse C2C12 cells, immortalized cultured myoblasts, important differences between the gene expression profiles of L6E9 cells and C2C12 cells have been observed, including a loss of myostatin expression in L6E9 cells and expression of myogenin before L6E9 cell differentiation (34). Since PDK1 is a key metabolic regulator, it may be sensitive to the differences between these model cell lines, possibly contributing to the different responses of mouse and rat PDK1 mRNA to UPF1 siRNA. Future studies may help to resolve whether PDK1 mRNA is an SMD target in rat tissues, which would better recapitulate the endogenous context than do L6E9 cells. Nevertheless, these findings support an emergence of SINE-directed SMD >20–25 million years ago.
Fig. 4.
Evidence that mRNAs containing independently evolved 3′-UTR SINEs are responsive to STAU1 and UPF1. (A, Left) Schematic phylogenetic tree indicating the approximate relative emergence of Alu elements or B/ID elements, and the type of 3′-UTR SINE in the indicated PDK1 ortholog. (Right) RT-qPCR quantitations of PDK1 mRNA in human MBs, African green monkey COS-7 cells, mouse MBs, and rat L6E9 MBs after treatment with the specified siRNA. Each mRNA is normalized to 18S rRNA, and the normalized level in control siRNA-treated cells is defined as 1. Data are shown on a log2 scale. (B) Same as in A, only analyzing SNX27 orthologs. All results derive from ≥3 independent experiments. Histograms represent the average and SD. *P < 0.05.
Discussion
Transposable elements (TEs) have contributed in diverse and significant ways to the evolution of human and mouse genomes with respect to the control of gene expression profiles (3, 35). In many cases, these events are specific to a particular lineage. However, there are examples whereby parallel exaptation of lineage-specific TEs in different organisms contributes to the regulation of specific genes by functioning as enhancers (36–38) or promoters (22) or by providing novel sites for splicing (20). Here, we show how the SMD regulatory network can combine with SINE exaptation to promote convergent evolution, bringing orthologous mammalian genes under parallel control regimes at the posttranscriptional level (Figs. 1 E and F, 2, and 4). The extent of the convergent evolution cannot simply be explained by the correlated length and A+T richness of human and mouse orthologous gene pairs (Fig. 3 and SI Appendix, Figs. S4 and S5 and Table S3) because, in each case, it also requires a trans-acting RNA (a lncRNA or other mRNA) to pair with the exapted SINE to form the double-stranded RNA-binding site for Staufen (4, 5, 11). Indeed, a number of studies have shown an enrichment of TE sequences in lncRNAs with evidence of sequence restraint (e.g., ref. 39). This predicts that there exist independently evolved, complex networks of interacting SINE-containing transcripts that have emerged to promote SMD of these orthologous pairs.
We found that approximately one-fifth of orthologous protein-coding genes in human and mouse encode at least one mRNA isoform that is 3′-UTR SINE-containing (Fig. 1B). Of these, approximately one-third are 3′-UTR SINE-containing in both orthologs (Fig. 1B). This suggests that SINEs have the potential to contribute to the evolution of posttranscriptional gene regulation of a large proportion of the human and mouse genomes. Indeed, our data are consistent with prior observations showing that retrotransposon inclusion in mRNA 3′-UTRs correlates with decreased mRNA levels in both human and mouse (22) and suggest that SMD of SINE-containing transcripts likely contributes to this effect. While here we show only a minority of these transcripts were detectably up-regulated in response to STAU1 and UPF1 depletion, our analysis likely underestimates the number of SMD targets due to the limits of detection in our assay (discussed above).
Once a SINE has been integrated into the genome, exact excision of the SINE is rare (40). As a result, alternative polyadenylation may play a major role in determining the presence of 3′-UTR SINEs in mature mRNAs. Indeed, it is interesting to note that human PDK1 mRNA has an alternate mRNA isoform that excludes the Alu element while mouse PDK1 mRNA does not. This might provide the opportunity for differential regulation of this gene in the two organisms. Since alternative polyadenylation can be cell-type specific, it will be interesting to determine whether the sensitivity of orthologous gene pairs to SMD is differentially regulated by alternative polyadenylation in comparable cell types.
While our studies are limited to SINEs that function in SMD in MBs, studies of other human and mouse cell types will undoubtedly uncover more examples, given that there are cell-type differences in gene expression that produce mRNA isoforms having different 3′-UTRs and thus SINE content.
Here, we establish that lineage-specific 3′-UTR SINEs can function to regulate the levels of mRNAs from orthologous genes in human, mouse, African green monkey, and rat by directing SMD. Our findings demonstrate a previously underappreciated role that lineage-specific SINEs have played in not only the divergence but also the convergence of gene expression profiles between species. Here we provide a demonstration of the convergent evolution of 3′-UTR SINEs for posttranscriptional gene regulation.
Materials and Methods
Please see SI Appendix, SI Materials and Methods for detailed description of computational and statistical analyses, cell culture and transient transfections, cell lysis and Western blotting, RNA purification and RT coupled to quantitative (q)PCR, RNA-seq mapping and analysis of RNA-seq data, immunoprecipitations, mRNA half-life assays, plasmid constructions, plasmid constructions to generate and screen monoclonal anti-STAU1 antibody, and generation of monoclonal anti-STAU1 antibody. siRNA and primer sequences can be found in SI Appendix, Tables S7–S10.
Supplementary Material
Acknowledgments
We thank Francisco Lopez for plasmid preparations; Osamu Ohara for cDNA expression plasmids; Jason D. Fernandes and Sofie Salama for comments on a draft of the manuscript; and Yinghan Fu, Anthony Almudevar, and Tom Eickbush for computational and statistical advice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE89588). Perl scripts used in this text are available on Figshare https://figshare.com under doi:10.6084/m9.figshare.4765015.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715531115/-/DCSupplemental.
References
- 1.Kramerov DA, Vassetzky NS. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 2011;107:487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandal PK, Kazazian HH., Jr SnapShot: Vertebrate transposons. Cell. 2008;135:192–192.e1. doi: 10.1016/j.cell.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 3.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong C, Tang Y, Maquat LE. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2013;20:1214–1220. doi: 10.1038/nsmb.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013;4:423–435. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho H, Han S, Park OH, Kim YK. SMG1 regulates adipogenesis via targeting of Staufen1-mediated mRNA decay. Biochim Biophys Acta. 2013;1829:1276–1287. doi: 10.1016/j.bbagrm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Sakurai M, et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2017;24:534–543. doi: 10.1038/nsmb.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deininger P. Alu elements: Know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterston RH, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 16.Farwick A, et al. Automated scanning for phylogenetically informative transposed elements in rodents. Syst Biol. 2006;55:936–948. doi: 10.1080/10635150601064806. [DOI] [PubMed] [Google Scholar]
- 17.DeChiara TM, Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci USA. 1987;84:2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quentin Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 1994;22:2222–2227. doi: 10.1093/nar/22.12.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Sela N, et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsirigos A, Rigoutsos I. Alu and B1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLOS Comput Biol. 2009;5:e1000610. doi: 10.1371/journal.pcbi.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 23.Blake JA, et al. Mouse Genome Database (MGD)-2017: Community knowledge resource for the laboratory mouse. Nucl Acids Res. 2017;45:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue F, et al. Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian B, Manley JL. Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heraud-Farlow JE, Kiebler MA. The multifunctional staufen proteins: Conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014;37:470–479. doi: 10.1016/j.tins.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong C, Popp MW-L, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012;58:88–93. doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rardin MJ, Wiley SE, Naviaux RK, Murphy AN, Dixon JE. Monitoring phosphorylation of the pyruvate dehydrogenase complex. Anal Biochem. 2009;389:157–164. doi: 10.1016/j.ab.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 32.Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 33.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi S, Stoppani E, Gobbo M, Caroli A, Fanzani A. L6E9 myoblasts are deficient of myostatin and additional TGF-β members are candidates to developmentally control their fiber formation. J Biomed Biotechnol. 2010;2010:326909. doi: 10.1155/2010/326909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: From conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emera D, et al. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- 38.de Souza FSJ, Franchini LF, Rubinstein M. Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol Biol Evol. 2013;30:1239–1251. doi: 10.1093/molbev/mst045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EZ, Wespiser AR, Caffrey DR. The domain structure and distribution of Alu elements in long noncoding RNAs and mRNAs. RNA. 2016;22:254–264. doi: 10.1261/rna.048280.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.