Significance
The human airway tree is a filter of noxious particulate matter, the primary cause of chronic obstructive pulmonary disease (COPD). We demonstrate that variation in central airway tree branching occurs in over one-quarter of the general population and increases COPD susceptibility, particularly among smokers. We show that these central airway branch variants are biomarkers of altered distal lung structure, the primary site of COPD pathobiology. Finally, we demonstrate the heritability of central airway branch variants within families and identify and replicate an association with FGF10. These findings suggest that central airway branch variants, easily detectable by computed tomography, represent heritable biomarkers of widely altered lung structure and a COPD susceptibility factor.
Keywords: airway branching, fibroblast growth factor, chronic obstructive pulmonary disease, computed tomography
Abstract
Susceptibility to chronic obstructive pulmonary disease (COPD) beyond cigarette smoking is incompletely understood, although several genetic variants associated with COPD are known to regulate airway branch development. We demonstrate that in vivo central airway branch variants are present in 26.5% of the general population, are unchanged over 10 y, and exhibit strong familial aggregation. The most common airway branch variant is associated with COPD in two cohorts (n = 5,054), with greater central airway bifurcation density, and with emphysema throughout the lung. The second most common airway branch variant is associated with COPD among smokers, with narrower airway lumens in all lobes, and with genetic polymorphisms within the FGF10 gene. We conclude that central airway branch variation, readily detected by computed tomography, is a biomarker of widely altered lung structure with a genetic basis and represents a COPD susceptibility factor.
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death and a major cause of disability globally (1, 2). Cigarette smoking is the major COPD risk factor (3), but COPD is not rare among those who have never smoked cigarettes, and many smokers do not develop COPD (4). Furthermore, approximately half of older adults with COPD exhibit low lung function early in life (5). These observations suggest that host factors beyond smoking may contribute to COPD risk and may create opportunities for personalized disease prevention and treatment.
The tracheobronchial tree serves as the conduit for gas exchange and acts as a filter of inhaled particulate matter. Airway bifurcations are major sites of inhaled particulate matter deposition (6), and alterations in these sites may contribute to differential deposition patterns of harmful particulate matter (e.g., cigarette smoke) and beneficial particles (e.g., inhaled bronchodilators) that could, respectively, affect disease risk and therapeutic response. Airway lumen caliber directly determines airflow resistance (7), which contributes to airflow limitation and respiratory symptoms (8). Hence, image-based anatomic markers of widely altered airway tree structure may facilitate personalization of COPD risk and treatment.
Genome-wide association studies (GWAS) for lung function and COPD have identified several genes (9, 10), including HHIP, LTBP4, SOX5, and TGFBR3, that regulate tracheobronchial tree formation in utero (11–14) and thus may have consequences for airway structure and postnatal disease susceptibility. Small autopsy studies showed frequent variation in the branching of the segmental airways in the lower, but not upper, lobes (15, 16). Comparative physiologists believe this variation may exist because the human species is not ventilation limited, thus reducing the selection pressure for a single conserved ventilation apparatus (17). In our modern environment, however, the airway tree must interact with noxious particulate matter (e.g., cigarette smoke, air pollution), the primary cause of COPD (8). Therefore, in this modern context, airway branch variation may be implicated in COPD pathogenesis.
In the present study we demonstrate that lower-lobe segmental airway branch variants are common in a large multiethnic population-based sample and among smokers in a COPD cohort and that these airway branch variants are associated with COPD and respiratory symptoms. In addition, we demonstrate that these common airway branch variants, easily identifiable on computed tomography (CT), indicate more generalized alterations in airway lumen caliber and branching and emphysema throughout the lung. Finally, we demonstrate the heritability of these airway branch variants and evaluate candidate genes known to regulate airway tree morphogenesis. Taken together, our findings suggest that a simple CT-based measure of central airway anatomy reflects genetically determined alterations in structure throughout the lung that predispose to COPD risk and provides a tool to facilitate personalized strategies for prevention and treatment.
Results
Prevalence of Central Airway Branch Variation in the General Population.
We first sought to determine the prevalence of airway branch variants in a large human cohort sampled from the general population. The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study performed chest CT scans for 3,169 participants (SI Appendix, Fig. S1). Their mean age was 69 ± 9 y, 48% were male, and the race/ethnic distribution was 39% white, 27% African-American, 21% Hispanic, and 13% Asian-American. Fifty-four percent were current or former smokers, with a median exposure of 15 pack-years (calculated as the number of years of smoking × the number of cigarette packs smoked per day ÷ 20) (SI Appendix, Table S1).
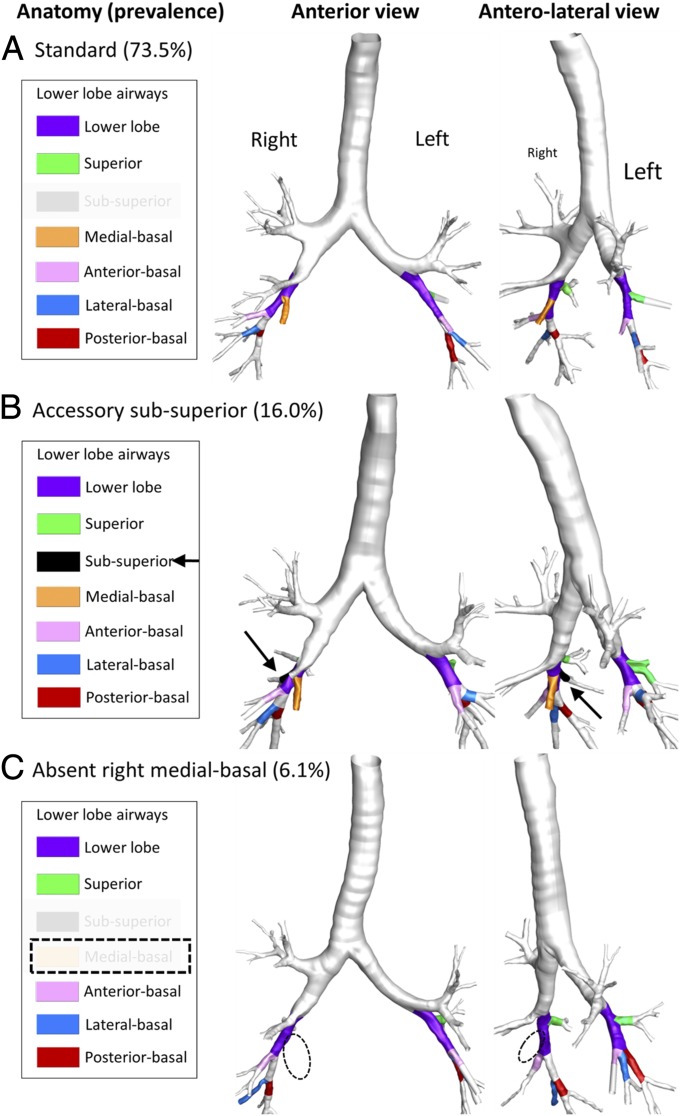
Airway branch variation was observed in 26.5% (95% CI: 24.9–27.8%) of participants. The most common airway branch variant was an accessory subsuperior airway (16.0%; 95% CI: 14.6–17.2%), followed by an absent right medial-basal airway (6.1%; 95% CI: 5.3–6.9%). These common airway branch variants are shown in Fig. 1. Four percent of participants had rare airway branch variants, such as an accessory left medial-basal airway, tracheal or carinal airway, or combinations of the above-mentioned variants. The remaining 73.5% had no airway branch variants, that is, standard airway anatomy.
Fig. 1.
Representative 3D airway reconstructions from chest CT scans of participants with standard lower-lobe anatomy (A), an accessory subsuperior airway branch variant (B), and absent right medial-basal airway branch variant (C). Standard lower-lobe segmental airway anatomy was defined as the presence of right and left superior, anterior-basal, lateral-basal, and posterior-basal segmental airways; the presence of the right medial-basal airway; absence of the left medial-basal airway; and absence of subsuperior segmental airway. (See SI Appendix, Fig. S2 for details.) A variant lower-lobe segmental airway, defined as any deviation from standard anatomy, with a prevalence above 5% in the MESA Lung Study was considered common. The remaining 4% of participants in the MESA Lung Study had rare or combinations of variants.
An accessory subsuperior segmental airway was more common among whites and less common among Asian-Americans, whereas an absent right medial-basal airway was more common among Asian-Americans and less common among African-Americans (SI Appendix, Table S2). There were minor differences in gender, body size, lung volume, and smoking status with airway branch variants, whereas the prevalence of a childhood diagnosis of asthma and living with a smoker during childhood were similar (P ≥ 0.125) (SI Appendix, Table S2).
Common Airway Branch Variation and COPD.
We next determined whether the common airway branch variants were associated with COPD defined by standard postbronchodilator spirometry (8) in the MESA Lung Study and in the Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS), a large case-control study of COPD that used the identical CT protocol. There were 243 cases of COPD and 2,065 controls without airflow limitation in the MESA Lung Study (SI Appendix, Table S3) and 1,823 cases of COPD and 923 controls in the SPIROMICS. The characteristics of MESA Lung and SPIROMICS participants by COPD status are summarized in SI Appendix, Table S4.
The presence of an accessory subsuperior segmental airway was associated with 1.64 higher odds of COPD (95% CI: 1.18–2.29; P = 0.004) compared with standard anatomy in the MESA Lung Study. This airway branch variant was also significantly associated with COPD in SPIROMICS (Table 1), and the pooled odds ratio (OR) for COPD in the two cohorts was 1.40 (95% CI: 1.19–1.64; P < 0.001). This association changed little with multivariable adjustment for potential confounders (Table 1) and did not differ between smokers and nonsmokers (P interaction = 0.222).
Table 1.
ORs for COPD by common airway branch variants in the MESA Lung Study, SPIROMICS, and both studies pooled
| Study | Standard anatomy | Accessory subsuperior airway | Absent right medial-basal airway |
| MESA Lung Study | |||
| Participants with COPD | 165 | 53 | 16 |
| Participants without COPD | 1,548 | 303 | 120 |
| OR for COPD | |||
| Unadjusted | Reference group | 1.64 (1.18–2.29) | 1.25 (0.73–2.16) |
| P = 0.004 | P = 0.42 | ||
| Adjusted for age, gender, race/ethnicity, height, weight, smoking status, pack-years | Reference group | 1.65 (1.14–2.39) | 1.47 (0.81–2.68) |
| P = 0.008 | P = 0.20 | ||
| SPIROMICS | |||
| Participants with COPD | 1,047 | 556 | 122 |
| Participants without COPD | 603 | 240 | 41 |
| OR for COPD | |||
| Unadjusted | Reference group | 1.33 (1.11–1.60) | 1.71 (1.19–2.47) |
| P = 0.002 | P = 0.004 | ||
| Adjusted for age, gender, race/ethnicity, height, weight, smoking status, pack-years | Reference group | 1.23 (1.02–1.50) | 1.59 (1.08–2.34) |
| P = 0.033 | P = 0.018 | ||
| Pooled* | |||
| Participants with COPD | 1,212 | 609 | 138 |
| Participants without COPD | 2,151 | 543 | 161 |
| OR for COPD | |||
| Unadjusted | Reference group | 1.40 (1.19–1.64) | 1.55 (1.15–2.08) |
| P < 0.001 | P = 0.004 | ||
| Adjusted for age, gender, race/ethnicity, height, weight, smoking status, pack-years | Reference group | 1.31 (1.10–1.55) | 1.57 (1.14–2.17) |
| P = 0.002 | P = 0.006 |
ORs for COPD were calculated using logistic regression with standard anatomy as the reference group.
Pooled analysis models include a term for study cohort. Participants with rare airway branch variants consisting of presence of the left medial-basal airway, or combinations of the above-mentioned variants are not shown (MESA: n = 103; SPIROMICS: n = 137).
Absence of the right medial-basal segmental airway was not significantly associated with COPD in the MESA Lung Study (Table 1); however, there was evidence for an interaction by smoking status (P interaction = 0.006), and among smokers the OR for COPD was 2.04 (95% CI: 1.13–3.71; P = 0.019). Among smokers in SPIROMICS, this variant was associated with 1.71 higher odds of COPD (95% CI: 1.19–2.47; P = 0.004). The pooled OR for COPD was 1.55 (95% CI: 1.15–2.08; P = 0.004) overall and 1.78 (95% CI: 1.27–2.49; P < 0.001) among smokers.
The associations between these common airway branch variants and COPD did not differ by gender (P interaction ≥ 0.161), race/ethnicity (P interaction ≥ 0.112), presence of childhood asthma (P interaction ≥ 0.222), or living with a smoker during childhood (P interaction ≥ 0.247) in study-specific or pooled analyses. Maternal smoking during pregnancy, assessed in SPIROMICS only, did not differ by common airway branch variant status (P = 0.348) and did not modify the association between these airway branch variants and COPD (P interaction ≥ 0.586). Defining COPD by the lower limit of normal (18) or by the combination of spirometric criteria and symptoms (8) yielded similar results (SI Appendix, Tables S5 and S6).
The presence of a tracheal or carinal airway was uncommon (0.8% in both studies), and there was no statistical association with COPD in study-specific or adjusted pooled analyses, although CIs were wide (SI Appendix, Table S7).
Common Airway Branch Variation and Respiratory Symptoms.
An accessory subsuperior airway was associated with chronic bronchitis, which is defined by chronic productive cough (19), in SPIROMICS and in the pooled analysis (SI Appendix, Table S8). It was also associated with worse scores on the COPD Assessment Test (CAT) (multivariate mean difference 0.9, 95% CI: 0.2–1.6; P = 0.008) and the St. George Respiratory Questionnaire for COPD (SGRG-C) (multivariate mean difference 2.4, 95% CI: 0.8–4.0; P = 0.004) (SI Appendix, Table S9). These differences were of greater magnitude and approached thresholds of clinically significant differences among patients with COPD (multivariate mean difference in CAT and SGRQ-C: 1.2 95% CI: 0.4–2.0 and 3.6 95% CI: 1.6–5.5, respectively; P ≤ 0.004) (SI Appendix, Table S10). This airway branch variant was not significantly associated with dyspnea (SI Appendix, Table S8).
In contrast, the absence of a right medial-basal airway was significantly associated with dyspnea in SPIROMICS and pooled analyses (SI Appendix, Table S8) but was not significantly associated with symptoms of chronic bronchitis or with CAT or SGRQ-C scores (SI Appendix, Tables S8 and S9).
Common Airway Branch Variation and Lung Structure.
Since it would be surprising that central airway branch variation alone would contribute to disease and symptoms, we examined if branch variation represented more widespread changes in lung morphology, particularly in nonaffected lobes of the lung.
In the MESA Lung Study and SPIROMICS, we found that CT-resolved airway segment lengths were 3.7% shorter (95% CI: 2.5–4.9%; P < 0.001) among participants with an accessory subsuperior airway than among those with standard anatomy, independent of lung volume. Shorter central airways were present not just in the variant-containing lobe but also in all other lobes in the lung (SI Appendix, Fig. S4), suggesting a generalized developmental process.
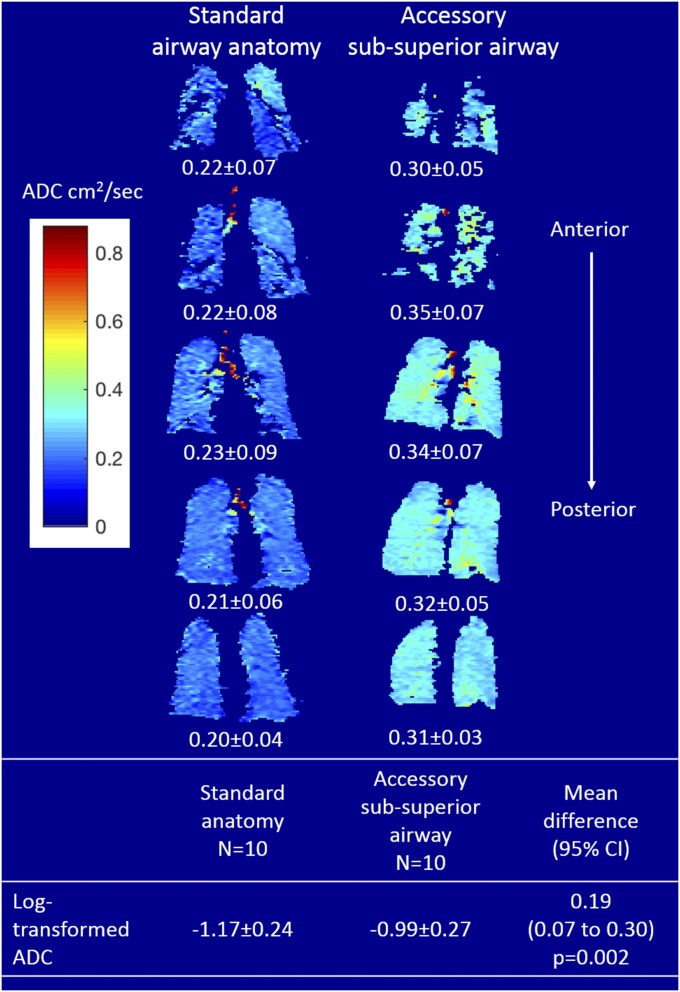
We therefore examined the peripheral lung structure of this airway branch variant. The presence of an accessory subsuperior airway demonstrated a higher percentage of emphysema-like lung in all lobes in SPIROMICS and in the pooled analysis (P < 0.001; SI Appendix, Table S11). Further examination using inhaled hyperpolarized helium-3 (3He) apparent diffusion coefficients (ADCs), a measure of alveolar size, among 20 smokers matched for gender, height, and COPD status (SI Appendix, Table S12) revealed that the presence of an accessory subsuperior airway was associated with higher ADC than seen with standard anatomy (P = 0.002) (Fig. 2). This difference remained significant when restricting the analysis to participants without emphysema on CT (n = 14; P = 0.001). The central airway lumen caliber (cross-sectional area) in the four nonaffected lobes of participants with an accessory subsuperior airway was similar to those in subjects with standard anatomy (SI Appendix, Fig. S4).
Fig. 2.
Inhaled 3He magnetic resonance-assessed ADCs among participants with an accessory subsuperior airway and standard anatomy. Shown are representative anterior-to-posterior coronal ADC heat maps from a participant with an accessory subsuperior airway (Right) and a participant with standard anatomy (Left). Participants with an accessory subsuperior airway demonstrated higher ADCs than participants with standard anatomy matched for gender, height, and COPD status. See SI Appendix, Table S11 for mean differences in CT-assessed percent emphysema.
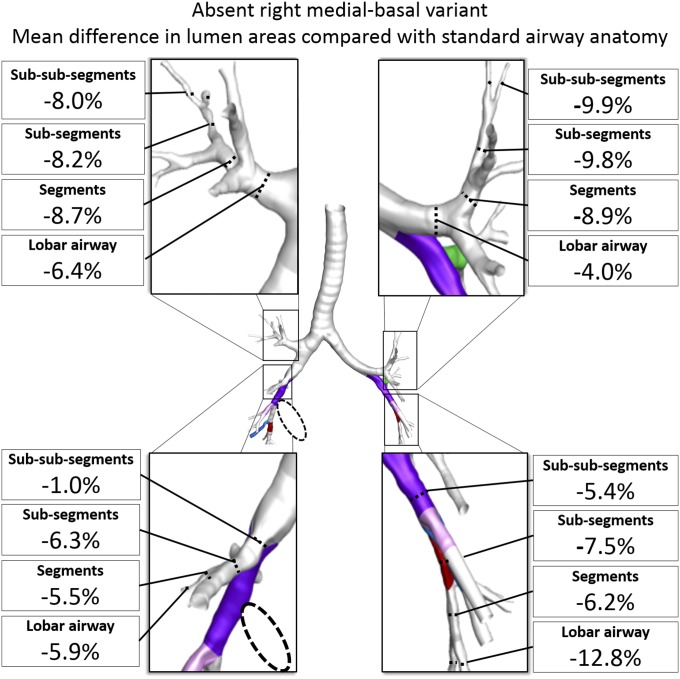
Airway morphology was different in participants with an absent right medial-basal airway. These individuals had significantly smaller airway lumen areas in all lobes of the lung, suggesting a generalized process (Fig. 3 and SI Appendix, Fig. S4). CT-resolved airway lengths in the branch variant-containing lobe were greater than in participants with standard anatomy (SI Appendix, Fig. S4) but were similar in the lobes not containing the branch variant, and there was no evidence in adjusted analyses that percent emphysema was increased in any lobe (SI Appendix, Table S11) or that ADC was increased (P = 0.841).
Fig. 3.
Mean differences in cross-sectional airway lumen areas by lobe and by anatomical level among participants with an absent right medial-basal airway branch variant (dashed circle) compared with standard anatomy in unaffected lobes. Airway lumen area comparisons are adjusted for lung volume. All mean differences were statistically significant (P < 0.01). See SI Appendix, Fig. S4 for standard airway anatomy lumen areas and mean differences with 95% CI.
Results stratified by cohort (MESA Lung Study and SPIROMICS) were largely similar (SI Appendix, Figs. S5 and S6), without evidence of effect modification by cohort (P interaction > 0.144).
Stability of Common Airway Branch Variants over 10 y.
Given the caliber and central position of these common airway branch variants in the airway tree, we hypothesized that their presence/absence did not change over time. Among the 300 MESA participants in whom anatomy was compared using images spanning a 10-y interval, there was no discordance in lower-lobe segmental airway anatomy (κ = 1.0), suggesting that these airway branch variants are not acquired in late adulthood.
Candidate Gene-Association Analysis of Common Airway Branch Variants.
Given the stability of central airway branch variants over a decade, we examined their heritability, hypothesizing a developmental origin. Sixteen unrelated participants in the MESA Family Study exhibiting a common airway branch variant were identified, and their sibling airway anatomy was assessed while masked to index case anatomy (sibship size range: 2–6; n = 64). The prevalence of an accessory subsuperior airway in siblings was 46% (95% CI: 30–62%), higher than the prevalence of 16% observed in the overall study population (P = 0.002). The prevalence of an absent right medial-basal airway in a sibling was 31% (95% CI: 8–69%), higher than the prevalence of 6% seen in the overall study population (P = 0.02). Given these results, we hypothesized that airway branch variants could reflect altered function of genes regulating airway morphogenesis.
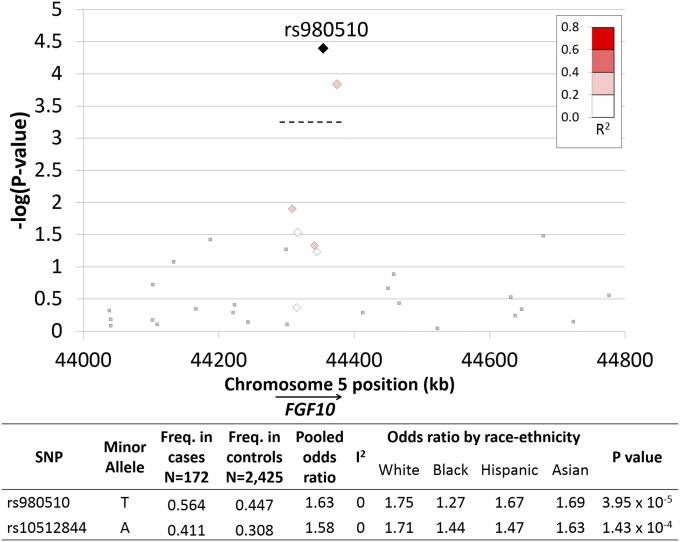
A targeted gene analysis involving 109 SNPs located within 11 candidate genes was conducted (SI Appendix, Table S13). The absence of the right medial-basal airway was associated with two SNPs on chromosome 5. SNPs rs980510 and rs10512844 are separated by 20.2 kb, are in low linkage disequilibrium (r2 = 0.38), and are located within the same intron of FGF10 (Fig. 4). Multiplicative and recessive allelic association models identified the same locus but with greater statistical significance (P = 1.33 × 10−5 and P = 2.94 × 10−6, respectively). There was no evidence of heterogeneity by race/ethnic subgroups (I2 = 0%). In contrast, the presence of an accessory subsuperior airway was not associated with the selected SNPs in the 11 candidate genes (SI Appendix, Table S13).
Fig. 4.
Regional association plot of candidate SNPs located within the FGF10 gene and the absent right medial-basal airway variant. Candidate SNPs were selected a priori based on their location within genes implicated in airway morphogenesis (11 genes, 109 SNPs). Genotyping: Affymetrix 6.0, excluding MAF <0.5, missingness per SNP >0.1, missingness per subject >0.1, and linkage disequilibrium >0.7. Candidate SNPs within FGF10 are indicated by diamond symbols. The degree of linkage disequilibrium with rs980510 is indicated by the R2 color scale. Square symbols indicate regional FGF10 SNPs not included in the candidate gene analysis. ORs and P values were calculated using an additive allelic model for each race/ethnic group, adjusted for gender and genetic ancestry principle components, and pooled by random effects meta-analysis. I2 is an index of effect measure heterogeneity by race/ethnicity. The Bonferroni P threshold of 4.59E−04 [−log(4.59E−04) = 3.34] is indicated by the dashed line.
The genetic association between FGF10 and absence of the right medial-basal airway was replicated in an independent cohort (SPIROMICS; rs980510; OR: 1.42, P = 0.008) (SI Appendix, Table S14).
Discussion
Variant branching of the segmental airways was observed in one-quarter of participants in a large population-based multiethnic sample. The most common airway branch variant, an accessory subsuperior airway, was associated with higher odds of COPD and chronic bronchitis, a higher number of central airway bifurcations, and larger airspaces in all lobes. The second most common airway branch variant, absence of the right medial-basal airway, was associated with higher odds of COPD among smokers and of dyspnea and with smaller airway lumens in all lobes. This airway branch variant exhibited familial aggregation and was associated with intronic SNPs in FGF10, which replicated in an independent cohort. These findings suggest that airway branch variation, which is common and easily identifiable, reflects widely altered lung structure and provides a genetically determined anatomical basis for COPD susceptibility.
This study represents by far the largest assessment of variant airway anatomy, but the observed prevalence was comparable to that in early surgical/autopsy series (15, 16). For example, Yamashita reported an accessory subsuperior airway prevalence of 14.0–14.3% and an absent right medial-basal airway prevalence of 7.8% in 180 specimens. The minor differences in prevalence estimates are likely due to the small numbers and overrepresentation of the diseased lungs in autopsy series and, potentially, differences by race/ethnicity.
This study assesses the association of common airway branch variation with COPD and symptoms. The magnitude of the observed associations with COPD was as large or larger than those reported for gene variants in GWAS (9), suggesting that personalized intervention strategies based on these airway variants might yield clinically important results. For example, these readily detected airway variants may facilitate the identification of people at increased risk of COPD or, conversely, identify the COPD subgroup of persons who exhibit low lung function in early life but who do not experience accelerated disease progression (5). Longitudinal research is needed to determine if such image-based phenotyping can facilitate the identification of those most likely to benefit from COPD prevention or treatment strategies and minimize testing of ineffective interventions in those at low risk of disease incidence or progression (20).
Airway tree structure is fractal in nature (patterning is similar across scales) (7), and study of murine lung development suggests that proximal variation in airway anatomy, arising from an altered genetic branching program, may iterate to produce an altered distal airway tree (21), the primary site of COPD pathophysiology (22, 23). The presence of an accessory subsuperior airway was associated with shorter central airways in all lobes, independent of lung volume. While these differences were modest over four airway generations, the cumulative deficit in airway length may require larger airspaces to fill the remaining lung volume. Indeed, the presence of an accessory subsuperior airway was associated with higher percent emphysema on CT and higher ADC for inhaled 3He, suggesting larger airspaces (24, 25). Alternatively, if airway development is programmed to fill the lung volume, then the observed 3.7% shorter central airways observed in the present analysis would require three to five additional peripheral airway generations [∼75,000–1,000,000 excess airway bifurcations, depending on the conducting airway model (26–28)] compared with standard anatomy. Airway bifurcations are major sites of inhaled particle deposition, including particle sizes typical of cigarette smoke and environmental air pollution (29, 30), and may contribute to the higher odds of chronic bronchitis observed among smokers with this airway branch variant.
The absence of the right medial-basal airway was associated with smaller airway lumens in all lobes for a given lung volume (dysanapsis) (31). Airway lumen caliber is a major structural determinant of airways resistance and airflow (32) and may contribute to the higher odds of COPD and dyspnea associated with this variant. The absence of the right medial-basal airway was also associated with FGF10 intronic SNPs, suggesting a genetically programmed airway structure–function endotype, either through changes in protein coding or through gene regulation. FGF10 is critical for airway tree morphogenesis (33–36), and FGF10 mutations are associated with a rare syndrome of aplasia of the lacrimal and salivary glands (ALSG) (37). The salivary and lacrimal glands, similar to the airway tree, are branched structures. A study of young patients with ALSG (mean age: 39 y) showed greater airflow obstruction than in their unaffected siblings, but an endotypic mechanism was not identified (38). Notably, the FGF10-associated airway branch variant was associated with COPD only among smokers. This observation suggests that absence of the right medial-basal airway may be a biomarker of FGF10-mediated airway lumen narrowing and increased airways resistance that are insufficient to cause COPD alone but may interact with environmental exposures, such as cigarette smoke, to augment susceptibility to clinically symptomatic COPD. Alternatively, airway branch variation and COPD may not be mechanistically related but instead may share a common cause (e.g., FGF10 may regulate airway morphogenesis, yielding branch variation, and, independently, regulate airway injury response that contributes to COPD) (39–41).
The present study has limitations. First, the human cross-sectional analyses may have introduced some selection bias; although one study was population-based, the proportion included of those selected was high (90%), and variant segmental airway anatomy was unrelated to missing spirometry. Cross-sectional studies can also be subject to reverse causation; however, it is very unlikely that COPD caused such proximal changes in the airways, and we observed no changes in lower-lobe segmental airway anatomy over 10 y. Nevertheless, longitudinal data are required to determine whether central airway branch variants precede clinical lung disease or instead may indicate low lung function in early life (5). Furthermore, such longitudinal data should assess other early life exposures not accounted for in the present studies (e.g., prematurity).
Blinding of the anatomy readers may have been incomplete due to visualization of emphysema on CT. It is highly unlikely, however, that this affected results: Assessment of the central airway anatomy is straightforward (SI Appendix, Fig. S2), resulting in excellent reproducibility [better than the scoring of emphysema by expert radiologists (42, 43)]; the scans were not assessed for emphysema at the same time; and the association of emphysema and COPD is modest (44).
We performed a candidate gene analysis with focused SNP analysis rather than a hypothesis-free GWAS. This targeted approach identified an association with FGF10 polymorphisms. We believe this is unlikely to represent a false-positive result, as we replicated the association in an independent cohort. Nevertheless, experimental and functional data are needed, and it is likely that other genes are implicated in the development of these common airway branch variants.
In summary, central airway branch variation, readily detected by CT, was common in a large multiethnic population-based sample, was associated with COPD as well as with respiratory symptoms in two large cohorts, and reflected diffuse changes throughout the lungs. Further, we have identified at least one form of central airway branch variation associated with COPD that appears to be linked to a genetic etiology. Taken together, our findings suggest that certain central airway branch variants serve as a biomarker of more widely altered lung structure that has implications for airflow resistance and gas exchange and also may influence the pulmonary deposition of environmental toxicants (including cigarette smoke) and inhaled therapies. Additional studies are needed to determine if personalized intervention based upon these variants is warranted.
Methods
Prevalence of Central Airway Branch Variation in the General Population and Associations with COPD and Respiratory Symptoms.
Study participants.
The MESA is a multicenter, prospective cohort study of whites, African-Americans, Hispanics, and Chinese-Americans in the United States (45). MESA recruited 6,814 men and women 45–84 y of age in 2000–2002 from the general population in six communities. Exclusion criteria were clinical cardiovascular disease, weight over 136 kg, pregnancy, and impediments to long-term follow-up. The MESA Lung Study enrolled participants sampled from MESA who underwent measurements of endothelial function, consented to genetic analyses, and completed an examination in 2004–2006 (46) and all participants in the MESA Air Pollution Study, who were recruited under the same enrollment criteria as MESA (47). The present analysis included MESA Lung Study participants who underwent a chest CT scan in the fifth MESA examination in 2010–2012 (SI Appendix, Fig. S1) and used measures from this examination, except as noted.
The SPIROMICS recruited cases of COPD and controls, 40–80 y of age with at least 20 pack-years of smoking, as well as 200 nonsmoker controls, in 2010–2015 (48). Exclusion criteria included other chronic lung diseases except asthma, body mass index >40 kg/m2, prior lung resection, metal in the chest, and pregnancy. Nonsmoker controls were excluded from the current analysis.
Institutional review board approval was obtained at each of the clinical study sites for both studies [MESA: Columbia University, Johns Hopkins University, Northwestern University, University of California (UC) Los Angeles, University of Minnesota, Wake Forest University; SPIROMICS: Columbia University, Johns Hopkins University, National Jewish, Temple University, University of Alabama at Birmingham, University of Illinois, University of Iowa, (UC) Los Angeles, University of Michigan, (UC) San Francisco, University of Utah, Wake Forest University] (https://www.mesa-nhlbi.org/ and www.spiromics.org/spiromics/ParticipatingInstitutions). Written informed consent was obtained from all participants.
Chest CT acquisition.
Participants underwent full-lung chest inspiratory CT on 64-slice or 128-slice helical scanners (120 kVp, 0.625–0.75 mm slice thickness, 0.5 s rotation time) following the same highly standardized protocol in both studies (49).
MESA Lung Study participants also underwent cardiac CT scans in 2000–2002 (50), which provided complete imaging of the lower-lobe segmental airways.
Airway branch variation.
Standard multiplanar image reformatting software (OsiriX 64-bit v8.4; Pixmeo) was used to visualize segmental airways arising from the local long axis of the right and left lower-lobe airways. We focused on lower-lobe segmental airway variants based on the high prevalence described in early autopsy studies (15, 16). In contrast, these studies reported accessory or absent segmental airways in other lobes to be rare (<1%), and we confirmed this in an initial five-lobe reading of 318 MESA COPD CT scans, in which we observed no accessory or absent segmental airways in the upper lobes.
Airway branch variants were ascertained by readers unaware of other participant information. The presence or absence of superior, subsuperior, medial-basal, anterior-basal, lateral-basal, and posterior-basal segmental airways was determined according to the descriptions of Yamashita (SI Appendix, Fig. S2) (15). Examples of the multiplanar images are shown in SI Appendix, Fig. S3. Interrater reproducibility of airway branch variants by masked assessment of 50 CT scans by two raters was excellent (interrater κ 0.94; 95% CI: 0.86–1.00; P < 0.0001).
The superior segmental airway was defined as the first (i.e., most proximal) airway originating from the lower-lobe stem. The subsuperior segmental airway was defined as an airway originating from the lower-lobe stem, inferior (i.e., distal) to the superior segmental airway but not extending to the basal region. The medial-basal airway was defined as an airway originating from the lower-lobe stem, oriented medially, extending to the basal regions of the lobe, and passing anterior, posterior, or both anterior and posterior to the basal vein (15). The remaining basal segmental airways (anterior-basal, lateral-basal, and posterior-basal) were defined as airways originating from the lower-lobe stem and extending to their respective basal regions. Airways arising distal to these basal segmental branches were not evaluated.
Standard lower-lobe segmental airway anatomy was defined as the presence of right and left superior, anterior-basal, lateral-basal, and posterior-basal segmental airways; presence of the right medial-basal segmental airway; absence of the left medial-basal airway; and absence of the subsuperior segmental airway (15). Variant lower-lobe segmental airway anatomy was defined as any deviation from standard anatomy.
We also assessed CT scan images for the presence of an airway originating from the trachea or main carina (16).
COPD and respiratory symptoms.
Spirometry was performed following American Thoracic Society recommendations (51) on a dry-rolling-seal spirometer in the MESA Lung Study and a pneumotachograph in SPIROMICS. Predicted values and limits of normal were calculated using reference equations (52). COPD was defined as a postbronchodilator forced expiratory volume in 1 s (FEV1) to forced vital capacity (FVC) ratio <0.7 (8). COPD defined by an FEV1/FVC ratio below the lower limit of normal and by an FEV1/FVC ratio <0.7 with dyspnea or chronic bronchitis symptoms was used in sensitivity analyses (8).
Dyspnea was assessed using the modified Medical Research Council breathlessness scale (53), with scores above 0 corresponding to increasing levels of dyspnea-associated disability. Chronic bronchitis was defined by affirmative responses to questions about cough and phlegm production for ≥3 mo each year for at least two consecutive years (19). In SPIROMICS, respiratory health status was assessed using the CAT (54) and the SGRQ-C, with higher scores indicating greater impairment (55). The CAT test consists of eight questions and yields a score from 0 (no impact) to 40 (very high impact). The SGRQ-C consists of 40 questions and yields a score from 0 (no impairment) to 100 (worst possible health). The minimum clinically important differences for the CAT and SGRQ-C scores are 2 and 4, respectively (56, 57).
Statistical analysis.
The prevalence of airway branch variants was calculated and 95% CIs were estimated using SAS 9.3 (SAS Software).
The ORs for COPD were calculated using logistic regression to adjust for age, gender, race/ethnicity, height, weight, smoking status, and pack-years of cigarette smoking (58). Effect modification was assessed with interaction terms in the full models. Pooled analyses of MESA Lung Study and SPIROMICS participants included a term for study.
Dyspnea, presence of chronic bronchitis, CAT, and SGRQ-C scores were compared according to airway branch variants using logistic and linear regression, respectively, adjusted for the same covariates. Analyses including SPIROMICS participants were additionally adjusted for COPD status to account for the case-control design.
Statistical analyses were performed using SAS 9.3 (SAS Software), and statistical significance was defined by a two-tailed P < 0.05.
Common Airway Branch Variation and Lung Structure.
Quantitative assessment of airway dimensions and percent emphysema.
Lobar volumes, attenuation, and central airway tree anatomy were assessed on chest CT scans for the MESA Lung Study and SPIROMICS at a single reading center (VIDA Diagnostics, Inc.) without knowledge of other participant information (32, 49).
Airways were labeled and measured anatomically from trachea to sub-subsegmental bronchi along five prespecified paths (RB1, RB4, RB10, LB1, and LB10), and segmentation and labeling were visually verified by a dedicated image analyst (49). Cross-sectional airway lumen areas were measured within an image plane perpendicular to the local airway segment long axis using a subvoxel-resolution algorithm, within an image plane, and measurements were averaged along the middle third of each labeled airway. Airway length was measured as the distance between bifurcation points.
The percentage of emphysema-like lung (percent emphysema) was defined as the percentage of voxels within the lung field less than −950 Hounsfield units, based on pathological comparisons (59), and calculated for the whole lung as well as each lobe.
The reproducibility of quantitative CT measures assessed by replicate scans was excellent (SI Appendix, Table S15).
3HE apparent diffusion coefficients.
The MESA COPD Study recruited cases of COPD and controls between 2010 and 2011 predominantly from MESA and the Emphysema and Cancer Action Project, a nonoverlapping lung cancer screening study, and also from the outpatient community at Columbia University Medical Center (42). Included participants were 50–79 y of age with at least a 10 pack-year smoking history. Exclusion criteria were clinical cardiovascular disease, advanced kidney disease, asthma before age 45 y, prior lung resection, pregnancy, or contraindication to MRI.
The definitions of postbronchodilator spirometry and COPD status matched the MESA Lung Study protocol (described above), as did the full-lung CT protocol and percent emphysema quantification (described above).
Between 2015 and 2017, 3He MRI was performed in a subset of 60 MESA COPD Study participants at the Columbia University site. Hyperpolarized 3He MRI ADCs were measured using a Phillips 3T scanner with a flexible wrap-around 3He radiofrequency coil. The 3He was polarized to 29 ± 5.7% using a GE spin-exchange polarizer. After a proton survey scan, 3D ADC scans were acquired without and with diffusion-sensitizing gradients corresponding to b-values of 0 and 1.6 s/cm2. The reconstructed voxel size was 3.5 × 3.5 × 40.0 mm. The participant was asked to breathe in 1 L of 3He gas mixture (350 mL of 3He gas with 650 mL of nitrogen) from residual volume with instructions to continue to breathe in a room-air chaser to “top off” the lungs up to total lung capacity. ADCs were calculated using a semiautomated approach programmed in Matlab to segment ventilated lung. An ADC heat map was generated for each of five coronal slices.
Covariates.
Age, gender, and race/ethnicity were self-reported. Current smoking was defined as self-reported cigarette smoking in the prior 30 d and was confirmed by plasma or urinary cotinine levels. Nonsmoker status was defined as having smoked fewer than 100 cigarettes lifelong. A childhood diagnosis of asthma, living with a smoker during childhood, and maternal smoking during pregnancy were self-reported, and the last was assessed in SPIROMICS only. Height and weight were measured using standardized protocols.
Statistical analysis.
Airway lumen areas and lengths by lobe were compared according to airway branch variant status using linear regression to adjust for the lung lobe volume achieved at the time of CT, study cohort, and COPD status where indicated.
Percent emphysema was log-transformed to approximate a normal distribution and was compared according to airway branch variant status using linear regression adjusted for age, gender, height, weight, smoking status, and pack-years of cigarette smoking.
ADC was log-transformed to approximate a normal distribution. Ten participants with an accessory subsuperior airway were matched 1:1 with 10 participants with standard anatomy, and two participants with an absent right medial-basal airway were matched 1:4 with participants with standard anatomy, for gender, height within 5 cm, and COPD status. Mean differences in ADC were calculated using generalized estimating equation regression to account for five ADC measures (slices) per participant and to adjust for matching cluster. Sensitivity analyses additionally adjusted for log-transformed percent emphysema and restricted analysis to those with less than 5% emphysema on CT.
Statistical analyses were performed using SAS 9.3, and statistical significance was defined by a two-tailed P < 0.05.
Stability of Common Airway Branch Variants over 10 y.
The 10-y change in airway anatomy was evaluated among 100 MESA participants with standard anatomy and 100 participants with each airway branch variant, sampled at random, by assessment of prior MESA cardiac CT scans, masked to findings from full-lung scans.
Within-participant stability of airway anatomy was assessed by κ statistic.
Candidate Gene-Association Analysis of Common Airway Branch Variants.
Familial aggregation.
The MESA Family Study enrolled and performed cardiac CT on 1,595 African American and Hispanic family members in 2004–2007 using the same inclusion/exclusion criteria as MESA (60). We assessed airway anatomy for 100 unrelated participants selected at random using the protocol described above (see Airway branch variation). This yielded 11 index cases with an accessory subsuperior variant and five index cases with an absent right medial-basal variant. We then assessed airway anatomy for the siblings of index cases in random order and blinded to index case anatomy. The prevalence of each airway branch variant among siblings of index cases was computed and compared with the population-based prevalence of the airway branch variant.
Genotyping.
MESA Lung Study participants who consented to genetic analysis were genotyped using the Affymetrix Human SNP array 6.0, of which 2,597 had full-lung CT imaging that permitted airway anatomy assessment (accessory subsuperior airway: 398; absent right medial-basal airway: 172).
SNPs included in the analysis were required to have minor allele frequency (MAF) above 0.05, missingness per marker below 0.1, missingness per individual below 0.1, and to pass the Hardy–Weinberg equilibrium test (P < 0.001). Markers in linkage disequilibrium above 0.7 were also excluded (Plink v1.07; 50 SNP window, five SNP shift), leaving 246,539 markers.
Replication of the top FGF10 polymorphism association (rs980510) was performed in SPIROMICS. Genotype data for the SPIROMICS sample were derived from OmniExpressExome BeadChip (Illumina) (61).
Candidate SNP selection.
Of 20 candidate genes selected a priori based on their known role in airway tree morphogenesis (62), 11 contained SNPs that passed the filtering and linkage disequilibrium criteria listed above, for a total of 109 candidate SNPs.
Power to detect an airway branch variant–SNP association was estimated using the observed prevalence of airway branch variants in the population-based cohort, assuming a disease allele frequency of 0.5 and accepting one false positive for the total number of SNPs compared. Because the central airway tree phenotype is thought to form prenatally (63), and systematic review of genetic effect sizes for early-life phenotypes demonstrates significantly higher effect sizes (OR >1.5) (64), we assumed a genotype effect size of 1.5. Power for additive and multiplicative association models for the accessory subsuperior airway branch variant was above 99% and for the absent right medial-basal airway branch variant was 76% and 92%, respectively (65).
Statistical analysis.
Common airway branch variant-SNP associations were tested under an additive model using logistic regression to adjust for gender and principal components of genetic ancestry (66). Analyses were stratified by race/ethnicity and were pooled by random effects meta-analysis. A conservative Bonferroni threshold was used to define a statistically significant association (p-Bonferroni 0.05/109 = 4.59 × 10−4).
The main replication analysis was restricted to white participants because the sample was predominantly white and was adjusted for gender and principal components of genetic ancestry. Secondary analysis included all race/ethnicities and an additional adjustment term for race.
Information on how to access the human cohort data is available at the study websites: https://www.mesa-nhlbi.org/ and www.spiromics.com. Genotype and airway phenotype data are available through the database of Genotypes and Phenotypes (dbGaP) (phs000420.v6.p3).
Supplementary Material
Acknowledgments
We thank the other investigators, the staff, and the participants of the MESA Lung Study and SPIROMICS for their valuable contributions (a full list of participating MESA investigators and institutions can be found at https://www.mesa-nhlbi.org; more information about the study and how to access SPIROMICS data is available at www.spiromics.org) and acknowledge the following current and former investigators of the SPIROMICS sites and reading centers: Neil E. Alexis, PhD; Wayne H. Anderson, PhD; Richard C. Boucher, MD; Russell P. Bowler, MD, PhD; Elizabeth E. Carretta, MPH; Stephanie A. Christenson, MD; Alejandro P. Comellas, MD; Gerard J. Criner, MD; Ronald G. Crystal, MD; Jeffrey L. Curtis, MD; Claire M. Doerschuk, MD; Mark T. Dransfield, MD; Christine M. Freeman, PhD; Annette T. Hastie, PhD; Jerry A. Krishnan, MD, PhD; Lisa M. LaVange, PhD; Stephen C. Lazarus, MD; Deborah A. Meyers, PhD; John D. Newell, Jr, MD; Elizabeth C. Oelsner, MD, MPH; Wanda K. O’Neal, PhD; Robert Paine, III, MD; Nirupama Putcha, MD, MHS; Donald P. Tashkin, MD; Mary Beth Scholand, MD; J. Michael Wells, MD; and Robert A. Wise, MD. The project officers from the Lung Division of the National Heart, Lung, and Blood Institute (NHLBI) were Lisa Postow, PhD, Thomas Croxton, PhD, MD, and Antonello Punturieri, MD, PhD. SPIROMICS was supported by NIH/NHLBI Contracts HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C HHSN268200900019C, and HHSN268200900020C, which were supplemented by contributions made through the Foundation for the NIH and COPD Foundation from AstraZeneca, Bellerophon Pharmaceuticals, Boehringer-Ingelheim Pharmaceuticals, Inc., Chiesi Farmaceutici SpA, Forest Research Institute, Inc., GlaxoSmithKline, Grifols Therapeutics, Inc., Ikaria, Inc., Nycomed GmbH, Takeda Pharmaceutical Company, Novartis Pharmaceuticals Corporation, Regeneron Pharmaceuticals, Inc., and Sanofi. Financial support was provided by NIH/NHLBI Grants R01-HL130506, R01-HL077612, R01-HL093081, R01-HL112986, RC1HL100543, RD831697, N01-HC-95159, N01-HC-95160, N01-HC-95169, N01-HC-95162, N01-HC-95164, U01-HL114494, N01-HC95159-HC95169, the NIH/NHLBI contracts named above, the McGill University Health Center Research Institute, and the Fonds de la recherche en santé Québec (Quebec Health Research Fund).
Footnotes
Conflict of interest statement: E.A.H. is co-founder and share-holder in VIDA Diagnostics, which commercialized the software used to assess lung volumes and percent emphysema (but not airway anatomy) in this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715564115/-/DCSupplemental.
References
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vos T, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha P, et al. 21st-century hazards of smoking and benefits of cessation in the United States. N Engl J Med. 2013;368:341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 4.Lamprecht B, et al. BOLD Collaborative Research Group COPD in never smokers: Results from the population-based burden of obstructive lung disease study. Chest. 2011;139:752–763. doi: 10.1378/chest.10-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange P, et al. Lung-function trajectories leading to chronic obstructive pulmonary disease. N Engl J Med. 2015;373:111–122. doi: 10.1056/NEJMoa1411532. [DOI] [PubMed] [Google Scholar]
- 6.Phillips CG, Kaye SR. On the asymmetry of bifurcations in the bronchial tree. Respir Physiol. 1997;107:85–98. doi: 10.1016/s0034-5687(96)02506-6. [DOI] [PubMed] [Google Scholar]
- 7.Mauroy B, Filoche M, Weibel ER, Sapoval B. An optimal bronchial tree may be dangerous. Nature. 2004;427:633–636. doi: 10.1038/nature02287. [DOI] [PubMed] [Google Scholar]
- 8.Vogelmeier CF, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195:557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 9.Hobbs BD, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soler Artigas M, et al. International Lung Cancer Consortium; GIANT consortium Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersh CP, et al. SOX5 is a candidate gene for chronic obstructive pulmonary disease susceptibility and is necessary for lung development. Am J Respir Crit Care Med. 2011;183:1482–1489. doi: 10.1164/rccm.201010-1751OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanford LP, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterner-Kock A, et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16:2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita H. Roentgenologic Anatomy of the Lung. Igaku-Shoin; Tokyo: 1978. [Google Scholar]
- 16.Boyden EA. Segmental Anatomy of the Lungs; a Study of the Patterns of the Segmental Bronchi and Related Pulmonary Vessels. Blakiston Division; New York: 1955. [Google Scholar]
- 17.Weibel ER. Symmorphosis: On Form and Function in Shaping Life. Harvard Univ Press; Cambridge, MA: 2000. [Google Scholar]
- 18.Pellegrino R, et al. Interpretative strategies for lung function tests. Eur Respir J. 2005;26:948–968. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- 19.Kim V, et al. COPDGene® Investigators Clinical and computed tomographic predictors of chronic bronchitis in COPD: A cross sectional analysis of the COPDGene study. Respir Res. 2014;15:52. doi: 10.1186/1465-9921-15-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agustí A, Celli B, Faner R. What does endotyping mean for treatment in chronic obstructive pulmonary disease? Lancet. 2017;390:980–987. doi: 10.1016/S0140-6736(17)32136-0. [DOI] [PubMed] [Google Scholar]
- 21.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogg JC, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 23.Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med. 1968;278:1355–1360. doi: 10.1056/NEJM196806202782501. [DOI] [PubMed] [Google Scholar]
- 24.Gevenois PA, et al. Comparison of computed density and microscopic morphometry in pulmonary emphysema. Am J Respir Crit Care Med. 1996;154:187–192. doi: 10.1164/ajrccm.154.1.8680679. [DOI] [PubMed] [Google Scholar]
- 25.Choy S, Wheatley A, McCormack DG, Parraga G. Hyperpolarized (3)He magnetic resonance imaging-derived pulmonary pressure-volume curves. J Appl Physiol (1985) 2010;109:574–585. doi: 10.1152/japplphysiol.01085.2009. [DOI] [PubMed] [Google Scholar]
- 26.Fredberg JJ, Hoenig A. Mechanical response of the lungs at high frequencies. J Biomech Eng. 1978;100:57–66. [Google Scholar]
- 27.Weibel ER. Morphometry of the Human Lung. Springer; Berlin: 1963. [Google Scholar]
- 28.Tawhai MH, et al. CT-based geometry analysis and finite element models of the human and ovine bronchial tree. J Appl Physiol (1985) 2004;97:2310–2321. doi: 10.1152/japplphysiol.00520.2004. [DOI] [PubMed] [Google Scholar]
- 29.Lambert AR, O’Shaughnessy P, Tawhai MH, Hoffman EA, Lin CL. Regional deposition of particles in an image-based airway model: Large-eddy simulation and left-right lung ventilation asymmetry. Aerosol Sci Technol. 2011;45:11–25. doi: 10.1080/02786826.2010.517578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H, Kleinstreuer C, Zhang Z, Kim CS. Nanoparticle transport and deposition in bifurcating tubes with different inlet conditions. Phys Fluids. 2004;16:2199–2213. [Google Scholar]
- 31.Green M, Mead J, Turner JM. Variability of maximum expiratory flow-volume curves. J Appl Physiol. 1974;37:67–74. doi: 10.1152/jappl.1974.37.1.67. [DOI] [PubMed] [Google Scholar]
- 32.Smith BM, et al. The Multi-Ethnic Study of Atherosclerosis (MESA) COPD Study and the Subpopulations and Intermediate Outcomes in COPD Study (SPIROMICS) Comparison of spatially matched airways reveals thinner airway walls in COPD. Thorax. 2014;69:987–996. doi: 10.1136/thoraxjnl-2014-205160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 34.Volckaert T, et al. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mailleux AA, et al. Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development. 2005;132:2157–2166. doi: 10.1242/dev.01795. [DOI] [PubMed] [Google Scholar]
- 36.Ramasamy SK, et al. Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol. 2007;307:237–247. doi: 10.1016/j.ydbio.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Entesarian M, et al. Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat Genet. 2005;37:125–127. doi: 10.1038/ng1507. [DOI] [PubMed] [Google Scholar]
- 38.Klar J, et al. Fibroblast growth factor 10 haploinsufficiency causes chronic obstructive pulmonary disease. J Med Genet. 2011;48:705–709. doi: 10.1136/jmedgenet-2011-100166. [DOI] [PubMed] [Google Scholar]
- 39.Volckaert T, et al. Fgf10-Hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev Cell. 2017;43:48–59.e45. doi: 10.1016/j.devcel.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volckaert T, Campbell A, De Langhe S. c-Myc regulates proliferation and Fgf10 expression in airway smooth muscle after airway epithelial injury in mouse. PLoS One. 2013;8:e71426. doi: 10.1371/journal.pone.0071426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh N. FGF10: A multifunctional mesenchymal-epithelial signaling growth factor in development, health, and disease. Cytokine Growth Factor Rev. 2016;28:63–69. doi: 10.1016/j.cytogfr.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Smith BM, et al. Pulmonary emphysema subtypes on computed tomography: The MESA COPD study. Am J Med. 2014;127:94.e97-23. doi: 10.1016/j.amjmed.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barr RG, et al. COPDGene CT Workshop Group A combined pulmonary-radiology workshop for visual evaluation of COPD: Study design, chest CT findings and concordance with quantitative evaluation. COPD. 2012;9:151–159. doi: 10.3109/15412555.2012.654923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mets OM, et al. Identification of chronic obstructive pulmonary disease in lung cancer screening computed tomographic scans. JAMA. 2011;306:1775–1781. doi: 10.1001/jama.2011.1531. [DOI] [PubMed] [Google Scholar]
- 45.Bild DE, et al. Multi-ethnic study of atherosclerosis: Objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez J, et al. The association of pipe and cigar use with cotinine levels, lung function, and airflow obstruction: A cross-sectional study. Ann Intern Med. 2010;152:201–210. doi: 10.1059/0003-4819-152-4-201002160-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufman JD, et al. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: The multi-ethnic study of atherosclerosis and air pollution (MESA air) Am J Epidemiol. 2012;176:825–837. doi: 10.1093/aje/kws169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couper D, et al. SPIROMICS Research Group Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS) Thorax. 2014;69:491–494. doi: 10.1136/thoraxjnl-2013-203897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieren JP, et al. SPIROMICS Research Group SPIROMICS protocol for multicenter quantitative computed tomography to phenotype the lungs. Am J Respir Crit Care Med. 2016;194:794–806. doi: 10.1164/rccm.201506-1208PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carr JJ, et al. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: Standardized protocol of multi-ethnic study of atherosclerosis (MESA) and coronary artery risk development in young adults (CARDIA) study. Radiology. 2005;234:35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 51.Miller MR, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 52.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general US population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 53.Bestall JC, et al. Usefulness of the Medical Research Council (MRC) dyspnoea scale as a measure of disability in patients with chronic obstructive pulmonary disease. Thorax. 1999;54:581–586. doi: 10.1136/thx.54.7.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones PW, et al. Development and first validation of the COPD assessment test. Eur Respir J. 2009;34:648–654. doi: 10.1183/09031936.00102509. [DOI] [PubMed] [Google Scholar]
- 55.Meguro M, Barley EA, Spencer S, Jones PW. Development and validation of an improved, COPD-specific version of the St. George respiratory questionnaire. Chest. 2007;132:456–463. doi: 10.1378/chest.06-0702. [DOI] [PubMed] [Google Scholar]
- 56.Kon SS, et al. Minimum clinically important difference for the COPD assessment test: A prospective analysis. Lancet Respir Med. 2014;2:195–203. doi: 10.1016/S2213-2600(14)70001-3. [DOI] [PubMed] [Google Scholar]
- 57.Jones PW. St. George’s respiratory questionnaire: MCID. COPD. 2005;2:75–79. doi: 10.1081/copd-200050513. [DOI] [PubMed] [Google Scholar]
- 58.Wacholder S. Binomial regression in GLIM: Estimating risk ratios and risk differences. Am J Epidemiol. 1986;123:174–184. doi: 10.1093/oxfordjournals.aje.a114212. [DOI] [PubMed] [Google Scholar]
- 59.Gevenois PA, de Maertelaer V, De Vuyst P, Zanen J, Yernault JC. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. Am J Respir Crit Care Med. 1995;152:653–657. doi: 10.1164/ajrccm.152.2.7633722. [DOI] [PubMed] [Google Scholar]
- 60.Manichaikul A, et al. Genome-wide study of percent emphysema on computed tomography in the general population. The multi-ethnic study of atherosclerosis lung/SNP health association resource study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun W, et al. SPIROMICS Research Group; COPDGene Investigators Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 2016;12:e1006011. doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cardoso WV, Lü J. Regulation of early lung morphogenesis: Questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 63.Jeffrey PK. The development of large and small airways. Am J Respir Crit Care Med. 1998;157:S174–S180. doi: 10.1164/ajrccm.157.5.rsaa-1. [DOI] [PubMed] [Google Scholar]
- 64.Agopian AJ, Eastcott LM, Mitchell LE. Age of onset and effect size in genome-wide association studies. Birth Defects Res A Clin Mol Teratol. 2012;94:908–911. doi: 10.1002/bdra.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson JL, Clark CC, Li KW, Caron S, Abecasis G. 2016 Genetic Association Study (GAS) Power Calculator. Available at csg.sph.umich.edu/abecasis/cats/gas_power_calculator/index.html. Accessed November 16, 2015.
- 66.Powell R, et al. Genetic ancestry and the relationship of cigarette smoking to lung function and per cent emphysema in four race/ethnic groups: A cross-sectional study. Thorax. 2013;68:634–642. doi: 10.1136/thoraxjnl-2012-202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.