Significance
Insulin-like growth factor 1 (IGF1) plays an important role in the regulation of metabolism and growth. Epidemiological studies revealed that IGF1 deficiency is associated with cancer protection. Genomic analyses conducted on patients with Laron syndrome (LS), a rare form of dwarfism linked to mutation of the growth hormone receptor, identified thioredoxin-interacting protein (TXNIP) as a downstream target for IGF1 action. TXNIP is a metabolic gene involved in redox regulation that can also function as a tumor suppressor. Data identify a regulatory link between the IGF1 signaling pathway and TXNIP. Elevated TXNIP levels in patients with LS may account for cancer protection in this pathology. Furthermore, TXNIP may constitute a biomarker to predict and/or monitor responsiveness to anti-IGF1R therapy.
Keywords: insulin-like growth factor 1, IGF1 receptor, TXNIP, Laron syndrome
Abstract
Laron syndrome (LS), or primary growth hormone (GH) insensitivity, is the best-characterized entity among the congenital insulin-like growth factor 1 (IGF1) deficiencies. Life-long exposure to minute endogenous IGF1 levels is linked to low stature as well as a number of endocrine and metabolic abnormalities. While elevated IGF1 is correlated with increased cancer incidence, epidemiological studies revealed that patients with LS do not develop tumors. The mechanisms associated with cancer protection in LS are yet to be discovered. Recent genomic analyses identified a series of metabolic genes that are overrepresented in patients with LS. Given the augmented expression of these genes in a low IGF1 milieu, we hypothesized that they may constitute targets for IGF1 action. Thioredoxin-interacting protein (TXNIP) plays a critical role in cellular redox control by thioredoxin. TXNIP serves as a glucose and oxidative stress sensor, being commonly silenced by genetic or epigenetic events in cancer cells. Consistent with its enhanced expression in LS, we provide evidence that TXNIP gene expression is negatively regulated by IGF1. These results were corroborated in animal studies. In addition, we show that oxidative and glucose stresses led to marked increases in TXNIP expression. Supplementation of IGF1 attenuated TXNIP levels, suggesting that IGF1 exerts its antiapoptotic effect via inhibition of TXNIP. Augmented TXNIP expression in LS may account for cancer protection in this condition. Finally, TXNIP levels could be potentially useful in the clinic as a predictive or diagnostic biomarker for IGF1R-targeted therapies.
Insulin-like growth factor 1 (IGF1) plays an important role in the regulation of metabolism, nutrition, and growth (1–3). Endocrine and tissue IGF1 levels are tightly controlled throughout development, with peak values typically reached at puberty (4) and reduced during adulthood. Elevated IGF1 is associated with increased risk for cancer development (5–7). Likewise, overexpression of the IGF1 receptor (IGF1R) has been linked to malignant transformation (8, 9). Enhanced expression of components of the IGF system in cancer cells reflects the prosurvival activities of this growth factor axis.
Laron syndrome (LS), or primary growth hormone (GH) insensitivity, is a rare form of dwarfism caused by deletion or mutation of the GH receptor (GHR) gene, leading to congenital IGF1 deficiency (10–13). The typical features of LS are short stature, typical face, obesity, and high GH and low IGF1 serum levels (10, 14). Epidemiological analyses provide evidence that homozygous GH-insensitive individuals are protected from cancer (15–17). This finding is consistent with the concept that congenital IGF1 deficiency confers protection against future development of a tumor. Hence, epidemiological studies highlight the key role of IGF1 in cancer. The mechanisms responsible for cancer protection in LS are yet to be identified.
Genome-wide association studies conducted on patients with LS identified genes and signaling pathways that are differentially regulated in LS and may provide a biochemical and genetic basis for cancer evasion (18). Bioinformatics analyses allowed us to cluster differentially expressed genes according to their biological functions. Of interest, about 15% of the genes are involved in metabolic pathways. Given the key role of IGF1 in regulation of cancer cell metabolism, it is reasonable to assume that alteration in the expression of IGF1-dependent metabolic genes may constitute an important mechanism that contributes to establishment of a malignant phenotype.
Thioredoxin-interacting protein (TXNIP) was initially discovered as a vitamin D3-induced gene in leukemia (19). Yeast two-hybrid analyses identified TXNIP as a thioredoxin (TRX)-binding protein (20). TXNIP binds to the catalytic active center of reduced TRX and inhibits its expression and activity, highlighting the critical participation of TXNIP in redox regulation (21–25). TXNIP belongs to the α-arrestin family (21) and inhibits proliferation via activation of apoptosis signal regulating kinase 1 (ASK1) (26–29). TXNIP functions as a tumor suppressor, being commonly silenced by genetic or epigenetic events in cancer cells (30–32).
Given the important role of the IGF1 pathways in the regulation of metabolic processes and in view of emerging information regarding the physiological and pathological activities of TXNIP, we examined the hypothesis that TXNIP constitutes a target for IGF1 action. In addition, we investigated the involvement of TXNIP in IGF1R-mediated glucose metabolism and oxidative stress. Our results identify a regulatory link between the IGF1 pathway and TXNIP. Specifically, we provide evidence that TXNIP gene expression is negatively regulated by IGF1. The ability of IGF1 to down-regulate TXNIP under a number of stress conditions suggests that the antiapoptotic effect of IGF1 is mediated via inhibition of TXNIP. Elevated TXNIP levels in patients with LS may account for cancer protection in this pathology. Finally, we report a mechanism for TXNIP autoregulation.
Results
Identification of Metabolic Genes Differentially Expressed in LS.
To identify differentially expressed genes that may account for cancer protection in LS, a genome-wide profiling was conducted using Epstein-Barr virus (EBV)-immortalized lymphoblastoid cells derived from four female patients with LS and four healthy controls of the same ethnic group (Irak, Iran, and Yemen) and age range (LS, 44.25 ± 6.08 y; controls, 51.75 ± 11.3 y; mean ± SD; P value = 0.29) (18). Thirty-nine annotated genes that were differentially expressed in LS compared with controls were identified (with a P value of <0.05 and fold-change difference cutoff >|2|). Functional analyses identified a number of metabolic genes that are highly expressed in LS, in comparison with control, cells. These genes include: (i) UDP-glucuronosyltransferase 2 (UGT2B15; fold change = 7.68); (ii) ZYG-11 homolog A (ZYG11A; fold change = 3.06); (iii) ribosomal modification protein RimK-like family member B (RIMKLB; fold change = 3.04); and (iv) thioredoxin-interacting protein (TXNIP; fold change = 2.04). These genes have not been previously linked to the IGF1-insulin signaling pathway (33). Given the role of TXNIP as a candidate tumor suppressor, and in view of its enhanced expression in patients with LS, further experiments were aimed at evaluating the hypothesis that TXNIP constitutes a target for inhibitory regulation by IGF1.
Real-Time Quantitative Polymerase Chain Reaction Validation of TXNIP mRNA Expression in LS.
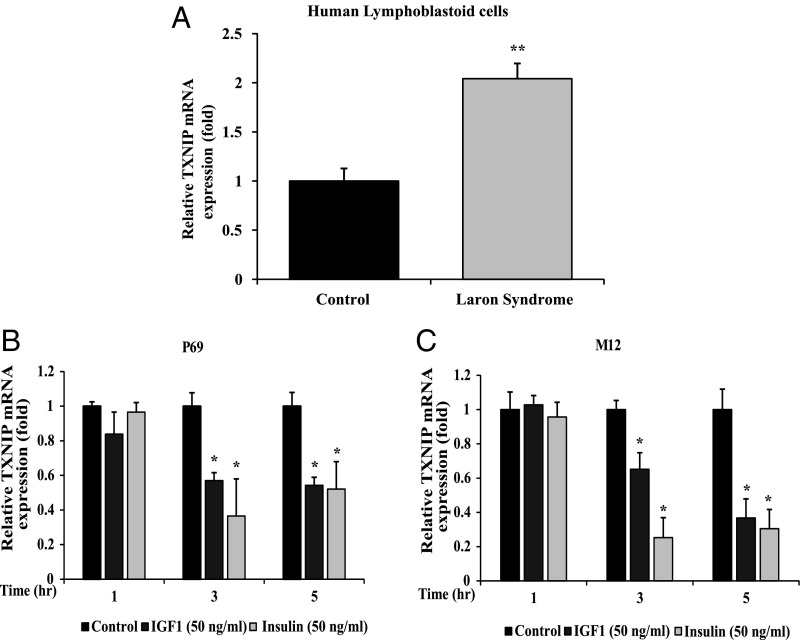
To validate the differences in gene expression between patients with LS and controls identified in gene arrays, real-time quantitative polymerase chain reaction (RT-QPCR) was performed. Levels of TXNIP mRNA were markedly elevated in LS-derived lymphoblastoid cells compared with gender-, age-, and ethnic origin-matched controls (2.04-fold) (Fig. 1A). These measurements confirm gene array data, suggesting that the TXNIP gene is highly expressed in patients with LS.
Fig. 1.
Effect of IGF1 and insulin on TXNIP gene expression. (A) Total RNA was obtained from individual LS and control lymphoblastoid cell lines and TXNIP mRNA levels were measured by RT-QPCR. The bars represent the mean ± SEM of three individual cell lines. **P < 0.01, significantly different versus controls. P69 (B) and M12 (C) prostate cells were deprived of serum for 24 h and then exposed to IGF1 (50 ng/mL, gray bars) or insulin (clear bars) for 1, 3, or 5 h. At the end of the incubation, cells were harvested and levels of TXNIP mRNA were measured by RT-QPCR. The bars represent the mean ± SEM of three independent experiments, each performed in triplicate. *P < 0.01 relative to serum-deprived conditions. A value of 1 was given to TXNIP mRNA levels in control cells (solid bars).
In Vivo TXNIP Expression in GHRKO and Hepatic IGF1 Transgenic Mice.
Expression of TXNIP was measured in livers of 4-mo-old GHRKO (“Laron”) mice, with undetectable serum and tissue IGF1 levels, and hepatic IGF1 transgenic (HIT) mice, with an approximately twofold increase in serum IGF1 (Fig. S1 A and B). As expected from our in vitro data in LS-derived lymphoblastoid cells, TXNIP expression increased approximately twofold in GHRKO mice, while it decreased by ∼65% in HIT mice (Fig. S1C). These data support the notion that the TXNIP gene is a physiological target for IGF1 action.
Effect of IGF1 and Insulin on TXNIP Gene Expression.
To investigate the effect of IGF1 and insulin on TXNIP expression, the P69 and M12 prostate cancer cell lines were grown in serum-free medium for 24 h, after which they were treated with IGF1 or insulin (50 ng/mL) for different periods. TXNIP mRNA expression was quantified by RT-QPCR and normalized to GAPDH mRNA. Short-term (3 and 5 h) treatment with IGF1 or insulin down-regulated TXNIP mRNA expression in both cells in comparison with controls (Fig. 1 B and C). At 3 h, IGF1 inhibited TXNIP mRNA levels in M12 cells by 34.8%, whereas insulin reduced mRNA levels by 74.7%. At the same time, IGF1 inhibited TXNIP mRNA levels in P69 cells by 43.0%, whereas insulin reduced expression by 63.5%. At 5 h, insulin and IGF1 exhibited a similar inhibitory effect in both prostate cells (∼57% reduction). Hence, results are consistent with an inhibitory role of both IGF1 and insulin on TXNIP gene expression.
Effect of Serum Deprivation on TXNIP Gene Expression.
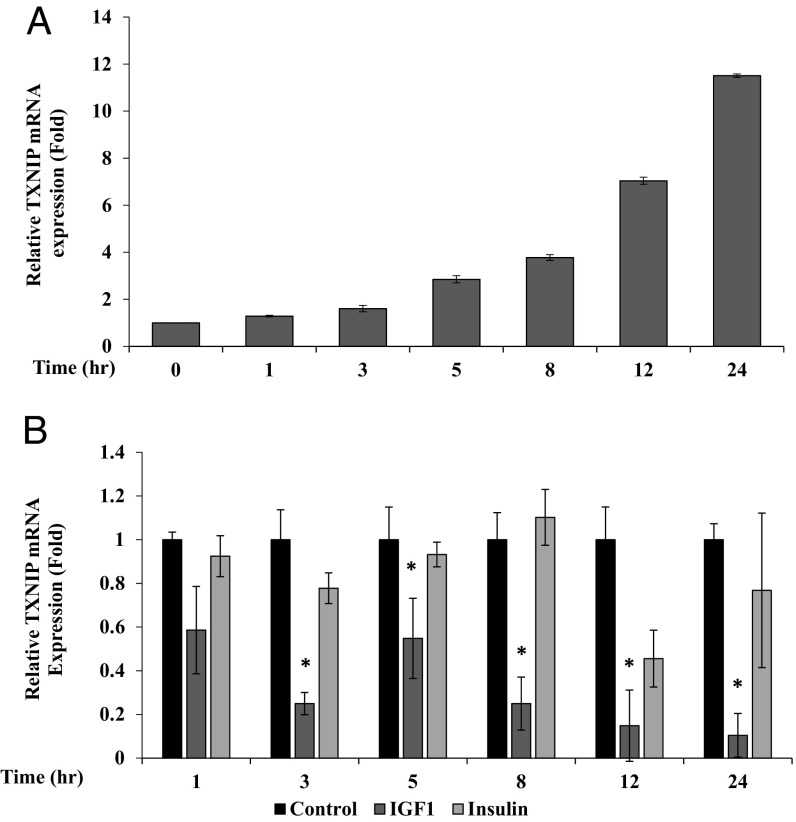
To evaluate the hypothesis that TXNIP expression is up-regulated upon exposure to stress, we assessed the impact of starvation on TXNIP mRNA. To this end, human embryonic kidney HEK293 cells were grown in serum-free conditions for 24 h, after which TXNIP mRNA levels were measured at different times using RT-QPCR. Results show that TXNIP expression increased in a time-dependent manner upon serum deprivation, reaching maximal expression levels at 24 h. Thus, after 24-h starvation, TXNIP mRNA levels were ∼11.5-fold higher than at time 0 (Fig. 2A).
Fig. 2.
Effect of serum deprivation on TXNIP gene expression. (A) Confluent human embryonic kidney HEK293 cells were serum starved for 24 h. Cells were collected at different times and TXNIP mRNA levels were measured by RT-QPCR. Bars represent the mean ± SEM of three independent experiments. A value of 1 was given to the TXNIP mRNA levels at time 0. (B) Effect of IGF1 and insulin on TXNIP gene expression. Serum-starved HEK293 cells were treated with IGF1 or insulin for different periods. Bars represent the mean ± SEM of three independent experiments. *P < 0.01 relative to serum-deprived conditions. A value of 1 was given to the TXNIP mRNA levels in untreated cells (solid bars).
Effect of IGF1 and Insulin on Starvation-Induced TXNIP Gene Expression.
To assess the effect of IGF1 and insulin on starvation-induced TXNIP expression, 24-h serum-starved HEK293 cells were treated with IGF1 or insulin and harvested for RNA preparation. RT-QPCR indicate that both hormones down-regulated TXNIP expression at all time points (Fig. 2B). Of interest, IGF1 was more potent than insulin in inhibiting TXNIP mRNA levels. Thus, at 12 h, IGF1 inhibited TXNIP mRNA by 85%, whereas insulin decreased expression by only 54%. At short times, insulin had a reduced effect. These results indicate that IGF1 and, to a lower extent, insulin abrogate the effect of starvation on TXNIP mRNA. The effect of both hormones on TXNIP protein is described in SI Text and Fig. S2.
Transcriptional Regulation of TXNIP Promoter Activity by IGF1.
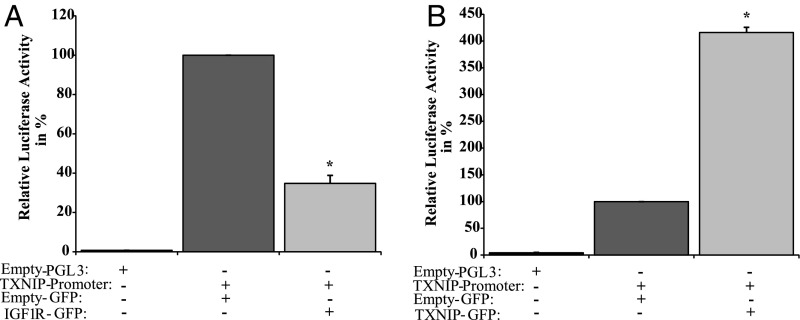
To assess whether the effect of IGF1 on TXNIP mRNA levels was mediated at the transcription level, cotransfections were performed in P69 cells using a TXNIP promoter-luciferase reporter, in the presence or absence of an IGF1R expression vector. Cells were harvested 48 h after transfection, and luciferase activity was measured. IGF1R overexpression led to an ∼65% reduction in TXNIP promoter activity compared with pcDNA3-transfected cells (Fig. 3A). Hence, the effect of IGF1 on TXNIP expression is mediated at the level of transcription.
Fig. 3.
Transcriptional regulation of TXNIP promoter activity. (A) P69 cells were transiently transfected with a luciferase reporter under the control of the proximal TXNIP promoter (or empty pGL3 luciferase vector), along with a β-galactosidase vector. After 24 h, cells were cotransfected with an IGF1R expression vector (IGF1R-GFP), or empty vector (GFP), for an additional 24 h. At the end of the incubation, cells were harvested and luciferase and β-galactosidase values were measured. Promoter activities are expressed as luciferase normalized to β-galactosidase. (B) Autoregulation of the TXNIP gene. 3T3-L1 cells were transfected with a TXNIP promoter-luciferase gene (or empty pGL3), together with a β-galactosidase vector. Twenty-four hours after transfection, cells were cotransfected with a TXNIP expression vector (or empty GFP), and processed as described above. The results represent the mean ± SEM of three independent experiments. *P < 0.01 versus GFP-transfected cells.
Autoregulation of the TXNIP Gene.
To evaluate the hypothesis that TXNIP can autoregulate its own promoter at the level of transcription, transient transfections were performed in 3T3-L1 fibroblasts using a TXNIP promoter-luciferase reporter, in the presence or absence of a TXNIP expression vector. Luciferase measurements indicate that TXNIP overexpression led to an ∼415% increase in TXNIP promoter activity compared with pcDNA3-transfected cells (Fig. 3B). These data provide evidence for a mechanism for TXNIP gene autoregulation.
Effect of Oxidative Stress on TXNIP Expression.
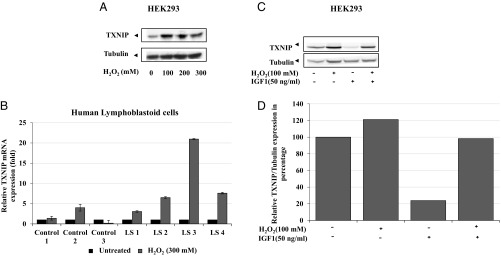
To investigate the effect of oxidative stress on TXNIP expression, HEK293 cells were serum starved for 24 h and then exposed to increasing concentrations of hydrogen peroxide (H2O2) for an additional 2 h. All doses of H2O2 increased TXNIP protein expression compared with untreated cells (Fig. 4A). Next, the impact of oxidative stress was examined in individual LS and control lymphoblastoid cell lines. To this end, cells were treated with 300 mM of H2O2 for 2 h and TXNIP mRNA levels were quantified. There was little change in TXNIP mRNA levels in three healthy lymphoblastoid lines upon H2O2 treatment (Fig. 4B). On the other hand, H2O2 increased TXNIP mRNA levels in LS cells. Thus, in three of four patient-derived lines, H2O2 increased TXNIP mRNA by ∼7- to 20-fold. Data indicate that TXNIP expression is increased in LS, but not healthy, cells upon oxidative insult. To examine the effect of IGF1 on oxidation-induced TXNIP levels, HEK293 cells were serum starved for 24 h, followed by addition of H2O2 for 2 h in the presence or absence of IGF1. Results indicate that the H2O2-induced TXNIP expression was attenuated by IGF1 (Fig. 4 C and D). The effect of oxidative stress on TXNIP expression in breast cancer cells is detailed in SI Text and Fig. S3.
Fig. 4.
Effect of oxidative stress on TXNIP levels. (A) Serum-starved HEK293 cells were treated with H2O2 (100, 200, 300 mM) or left unstimulated. Lysates (100 μg) were analyzed by Western blotting for TXNIP and tubulin levels. (B) Effect of oxidative stress on TXNIP levels in LS-derived and control lymphoblastoids. Four individual LS and three control lymphoblastoid cell lines were treated with 300 mM of H2O2 or left unstimulated. Cells were harvested after 2 h and levels of TXNIP mRNA were measured by RT-QPCR. A value of 1 was given to TXNIP mRNA levels in untreated cells (solid bars). (C) Serum-starved HEK293 cells were treated with H2O2 (100 mM) or IGF1 (50 ng/mL) or both for 2 h. Lysates (100 μg) were analyzed by Western blotting. (D) Densitometric analysis of Western blot shown in Fig. 4C.
Effect of IGF1 on High Glucose-Induced TXNIP Expression.
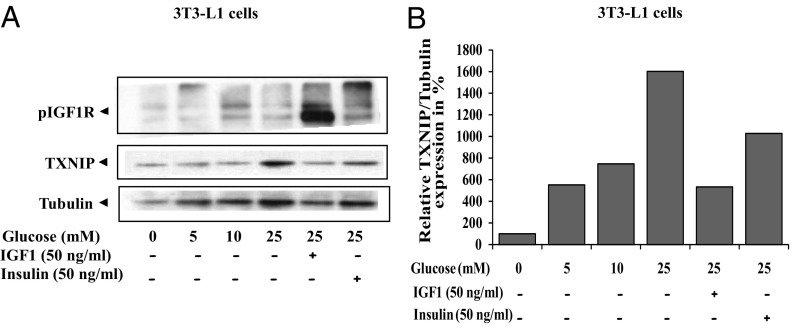
Evidence has been presented linking TXNIP action to glucose metabolism and diabetes (34, 35). The impact of increasing glucose concentrations on mouse embryonic 3T3-L1 fibroblasts and the added effect of IGF1 or insulin was next investigated. 3T3-L1 cells were serum starved for 24 h and then incubated with increasing concentrations of glucose for 6 h in the presence or absence of IGF1 or insulin. Control cells were grown in glucose-free media. Results of Western blots indicate that TXNIP expression was up-regulated in response to high glucose concentrations (160% at 25 mM glucose). Both IGF1 and insulin attenuated the glucose-stimulated TXNIP increase, albeit with different potencies. Thus, IGF1 down-regulated the glucose-induced TXNIP expression by ∼67%, whereas insulin decreased it by ∼36% (Fig. 5). A similar effect of IGF1 on glucose-stimulated TXNIP expression was seen in HEK293 cells (Fig. S4). The expression of TXNIP in benign and malignant prostate and breast cancer cells as well as the effect of TXNIP on migration is detailed in SI Text and Figs. S5 and S6.
Fig. 5.
Effect of IGF1 on glucose-induced TXNIP levels. (A) Serum-starved 3T3-L1 cells were maintained in medium with different concentrations of glucose, in the presence or absence of IGF1 or insulin for 6 h. Lysates were prepared and aliquots were analyzed by Western blots for TXNIP, phospho-IGF1R, and tubulin. (B) Densitometric analysis of blot shown in A.
Discussion
The role of the GH-IGF1 pathway in cancer biology has been the focus of clinical and basic research for several decades (1, 36, 37). While elevated endocrine IGF1 has been, for the most part, associated with enhanced cancer risk, the recent finding that patients with congenital IGF1 deficiency do not develop cancer reflects the fact that a reduction in IGF1 elicits protective pathways that might ultimately translate into tumor evasion (16, 17, 38). In genome-wide association studies conducted on patients with LS, we reported the identification of a series of genes that are differentially represented in patients compared with controls (18). Differences in gene expression occur in a number of pathways, including apoptosis, metabolism, Jak-STAT, and PI3K-AKT signaling, etc. Reductions in gene expression in patients with LS were noticed in a series of gene products usually involved in proliferation. These transcripts include cyclin D1, cyclin A1, serpin B2, versican, Sp1, etc. Decreases in these genes were correlated with corresponding reductions in proliferation, changes in cell cycle distribution, and enhanced apoptosis and autophagy. In addition, profiling analyses identified a series of metabolic genes whose expression was increased in patients with LS compared with healthy individuals (18, 33). It is reasonable to assume that enhanced expression of some of these genes may favor specific metabolic pathways that might eventually lead to protection from oxidative and genotoxic insults.
The present study identified TXNIP as a target for IGF1 and insulin action. Consistent with its elevated expression in LS, a prototypical case of congenital IGF1 deficiency, we provide evidence that IGF1 and insulin inhibit TXNIP mRNA and protein levels in a number of cultured cell lines. Animal studies using GHRKO (Laron) and HIT mice corroborated the in vitro data. The GHRKO mice are unique because of their resistance to GH action (39). This endocrine defect leads to growth retardation, delayed sexual maturation, reduced IGF1 levels, and elevated serum GH concentrations. Similar to patients with LS, GHRKO mice had reduced prostate and mammary tumor incidence in experimental models of cancer (40). With age, Laron mice become obese, hypoglycemic, and have an extended life span (39). The rationale for our measurements of TXNIP levels in liver of GHRKO and HIT mice is the physiological relevance of this organ in terms of TXNIP action. In accordance with its enhanced expression in LS cells, RT-QPCR revealed that TXNIP expression increased approximately twofold in livers of GHRKO mice (with reduced IGF1 levels), while it decreased in HIT mice (with increased serum IGF1).
Results of transfection assays indicate that the negative effect of IGF1 on TXNIP expression is mediated at the transcription level. Of interest, we provide evidence that TXNIP can act on its own promoter, leading to TXNIP autoregulation. Given the interplay between TRX and TXNIP in regulation of adipogenesis, we envision that this autoregulatory loop may be of importance in adipocyte control. The mechanisms responsible for TXNIP promoter regulation are unknown. TXNIP has been shown to function as a tumor suppressor that is often silenced in cancer cells (30, 31). Given the fact that patients with LS are subject to life-long low IGF1, it is conceivable that elevated TXNIP levels in LS result from relaxation of inhibitory control by insufficient IGF1.
Studies reported that TXNIP provides an integration point for both short- and long-term metabolic and signaling information, permitting the appropriate types and levels of cellular response to glucose availability and demand (41). We have previously provided evidence that LS cells display protection from oxidative stress (18). Consistently, we show here that TXNIP expression was up-regulated under oxidative stress. These results suggest that enhanced TXNIP levels may mediate the protection from oxidative stress. Since IGF1 is a major regulator of cell survival, we postulated that IGF1 could inhibit apoptosis by downregulating TXNIP. Indeed, our analysis revealed that IGF1 down-regulates the oxidative stress-induced TXNIP up-regulation in HEK293 cells. TXNIP acts as an oxidative stress mediator by inhibiting TRX activity or by limiting its bioactivity (21). The finding that TXNIP levels are increased in response to oxidation in patient-derived, but not control, lymphoblastoid cells is of major translational relevance.
Across evolution, alpha-arrestin family proteins like TXNIP are involved in nutrient sensing (42). TXNIP was reported to function as a strong glucose sensor, because its expression increased upon high-glucose stress (43). Of interest, TXNIP has been identified as one of the most potently suppressed skeletal muscle mRNAs in healthy individuals subjected to a hyperinsulinemic-euglycemic clamp (44). Consistent with these findings, our data indicate that TXNIP levels increased upon elevation of glucose concentrations in 3T3-L1 and HEK293 cells, whereas IGF1 and insulin down-regulated the glucose-induced TXNIP increase. This regulatory loop may be important in terms of improving the energy balance of the cells. Up-regulated TXNIP initiates apoptosis by interacting with TRX and translocating to mitochondria (45). Finally, we show that oxidative stress led to TXNIP nuclear localization in breast cancer cells.
Knockout of TXNIP resulted in a variable phenotype characterized by a deregulated metabolic signature, including adipogenesis and reduced gluconeogenesis (46). Livers of TXNIP KO animals showed decreased glucose uptake (41). Of interest, txnip-deficient mice are also predisposed to renal and hepatocellular carcinomas (47). TXNIP has been identified as a tumor suppressor gene, the down-regulation of which is associated with tumorigenesis and metastasis (35, 48). Low TXNIP expression has been observed in breast, liver, and stomach cancer (32). Consistent with its tumor suppressor role, basal expression of TXNIP was reduced in M12 cells, a metastatic prostate cancer line, compared with its benign counterpart. Likewise, TXNIP levels were reduced in aggressive MCF7 breast cancer, compared with benign MCF10A, cells. Finally, the tumor suppressor role of TXNIP was corroborated by cell migration assays.
Finally, the IGF1R emerged in recent years as a promising therapeutic target in oncology. There is, however, an urgent need to identify biomarkers that can help select patients who may benefit from IGF1R-directed therapies. In view of our results showing a regulatory link between the IGF1 axis and TXNIP, we propose that TXNIP may constitute a biomarker capable of predicting and/or monitoring response to anti-IGF1R therapy. Furthermore, elevated TXNIP levels in patients with LS may account for cancer protection in this pathology. The potential involvement of epigenetic mechanisms, particularly DNA methylation and histone acetylation, in inhibitory regulation of TXNIP gene expression by IGF1 is currently under investigation.
Materials and Methods
Cell Lines.
EBV-immortalized lymphoblastoid cell lines from patients with LS and healthy controls were obtained from the National Laboratory for the Genetics of Israeli Populations, Tel Aviv, Israel. The protocol was approved by the Tel Aviv University Ethics Committee. The P69 line was derived by immortalization of primary prostate epithelial cells, while the M12 line was derived from P69 cells by selection for tumor formation (49). P69 and M12 cells were a gift from Joy Ware (Medical College of Virginia, Richmond, VA). The androgen-independent prostate PC3 line was established from a metastasis (50). PC3 and breast cancer MCF7 and MCF10A cells were obtained from the American Type Culture Collection. Human embryonic kidney HEK293 cells were grown in DMEM/F12 with high, low, or no glucose. Mouse embryonic 3T3-L1 fibroblasts were grown in DMEM with high, low, or no glucose.
Animal Studies.
The generation of the GHRKO and HIT mouse models was previously described (51, 52). All mice were in the C57BL/6J (B6) genetic background. Weaned mice were allocated randomly into cages separated according to their sex. Mice were housed two to five animals per cage in a facility with 12-h light:dark cycles and free access to food and water. The analyses were performed in 4-mo-old male mice. All animal procedures were approved by the Institutional Animal Care and Use Committee of New York University School of Medicine (assurance number A3435-01, USDA licensed no. 465), and conform to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines (www.nc3rs.org.uk/arrive-guidelines).
Genome-Wide Association Studies.
Total RNA was prepared from lymphoblastoid cell lines using the TRIzol reagent. Affymetrix GeneChip Human Gene 1.0 ST arrays were used. Data analysis was performed on CEL files using Partek Genomics Suite. Data were normalized and summarized with the robust multiaverage method followed by ANOVA. Genes of interest that were differentially expressed with P < 0.05 and fold-change difference cutoff >|2| were obtained.
RT-QPCR.
Two micrograms of total RNA was reverse transcribed using the Superscript First-Strand Synthesis System for RT-PCR. RT-QPCRs were performed using Faststart Universal SYBR Green Master. Sequences of primers used to detect mRNAs are available upon request. The number of cycles to reach the fluorescence threshold was the cycle threshold (Ct). Each sample was tested in triplicate and mean Ct values are reported. For each reaction, a “no template” sample was included. Fold differences were calculated by the 2ΔΔCT method (53).
Western Blot Analyses.
Cells were grown to confluence, pelleted, and incubated with a lysis buffer for 20 min. The suspension was centrifuged and protein concentration was determined. Aliquots were boiled, resolved through 10% SDS/PAGE, and blotted onto nitrocellulose membranes. After blocking with 5% milk, the blots were incubated with the indicated antibodies.
Promoter Assays.
A TXNIP promoter-luciferase reporter plasmid (pGL3B-1081) was obtained from Addgene (cat. no. 18758). P69 cells were seeded in six-well plates 24 h before transfection and transfected with 0.5 μg of the TXNIP reporter plasmid, or empty pGL3, along with 0.2 μg of a β-galactosidase plasmid (pCMV, Clontech), using jetPEI (Polyplus). Twenty-four hours after transfection, cells were cotransfected with an IGF1R expression vector (IGF1R-GFP), or empty GFP. Cells were harvested after 24 h, and luciferase and β-galactosidase activities were measured (54). Promoter activities are expressed as luciferase values normalized for β-galactosidase. For TXNIP autoregulation analyses, 3T3-L1 cells were cotransfected with the TXNIP promoter reporter along with a TXNIP expression vector.
Overexpression Studies.
A TXNIP expression vector was obtained from Addgene (cat. no. 18759). PC3 cells were transfected with either GFP-TXNIP or empty vector using jetPEI. After 24 h, the medium was replaced with fresh medium. Overexpression of GFP-TXNIP was confirmed by Western blots. GFP fluorescence in PC3 cells was visualized by fluorescence microscopy.
Oxidative Stress Assays.
Cells were grown to 80% confluence, after which they were serum starved for 24 h. Oxidative stress was induced by treating the cells with increasing concentrations of H2O2, in the absence or presence of IGF1 (50 ng/mL) for 2 h. Cells were then washed and total RNA and protein were prepared.
Response to Glucose Stress.
Cells were grown as indicated above, followed by 24-h serum starvation. Glucose stress was induced by incubating the cells with increasing concentrations of glucose in the absence or presence of IGF1. Cells were then washed and total RNA and protein were prepared.
Statistical Analyses.
Statistical package SPSS was used and graphs were produced using Prism software. Nonparametric statistical methods (Mann–Whitney u test and Kruskal–Wallis test) were applied to determine the statistical significance difference between means. Parametric tests (Student’s t test and ANOVA test) were also used.
Supplementary Material
Acknowledgments
We thank Dr. Jay Ware for providing cell lines. This work was performed in partial fulfillment of the requirements for a PhD degree by K.N. at the Sackler Faculty of Medicine, Tel Aviv University. This research was supported by Grant 1403/14 from the Israel Science Foundation and Grant 20170079 from the Bernard Lieberman Foundation (Israel Cancer Association). H.W. is the incumbent of the Lady Davis Chair in Biochemistry (Tel Aviv University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715930115/-/DCSupplemental.
References
- 1.LeRoith D, Yakar S. Mechanisms of disease: Metabolic effects of growth hormone and insulin-like growth factor 1. Nat Clin Pract Endocrinol Metab. 2007;3:302–310. doi: 10.1038/ncpendmet0427. [DOI] [PubMed] [Google Scholar]
- 2.Yakar S, Adamo ML. Insulin-like growth factor 1 physiology: Lessons from mouse models. Endocrinol Metab Clin North Am. 2012;41:231–247. doi: 10.1016/j.ecl.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkes CP, Grimberg A. Insulin-like growth factor-1 is a marker for the nutrition state. Pediatr Endocrinol Rev. 2015;13:499–511. [PMC free article] [PubMed] [Google Scholar]
- 4.Klammt J, Pfäffle R, Werner H, Kiess W. IGF signaling defects as causes of growth failure and IUGR. Trends Endocrinol Metab. 2008;19:197–205. doi: 10.1016/j.tem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Chan JM, et al. Plasma insulin-like growth factor-I and prostate cancer risk: A prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 6.Hankinson SE, et al. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet. 1998;351:1393–1396. doi: 10.1016/S0140-6736(97)10384-1. [DOI] [PubMed] [Google Scholar]
- 7.Renehan AG, et al. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: Systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 8.Werner H. For debate: The pathophysiological significance of IGF-I receptor overexpression: New insights. Pediatr Endocrinol Rev. 2009;7:2–5. [PubMed] [Google Scholar]
- 9.Werner H. Tumor suppressors govern insulin-like growth factor signaling pathways: Implications in metabolism and cancer. Oncogene. 2012;31:2703–2714. doi: 10.1038/onc.2011.447. [DOI] [PubMed] [Google Scholar]
- 10.Laron Z, Pertzelan A, Mannheimer S. Genetic pituitary dwarfism with high serum concentation of growth hormoneA new inborn error of metabolism? Isr J Med Sci. 1966;2:152–155. [PubMed] [Google Scholar]
- 11.Laron Z. Laron syndrome (primary growth hormone resistance or insensitivity): The personal experience 1958-2003. J Clin Endocrinol Metab. 2004;89:1031–1044. doi: 10.1210/jc.2003-031033. [DOI] [PubMed] [Google Scholar]
- 12.Amselem S, et al. Laron dwarfism and mutations of the growth hormone-receptor gene. N Engl J Med. 1989;321:989–995. doi: 10.1056/NEJM198910123211501. [DOI] [PubMed] [Google Scholar]
- 13.Godowski PJ, et al. Characterization of the human growth hormone receptor gene and demonstration of a partial gene deletion in two patients with Laron-type dwarfism. Proc Natl Acad Sci USA. 1989;86:8083–8087. doi: 10.1073/pnas.86.20.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laron Z, Pertzelan A, Karp M, Kowadlo-Silbergeld A, Daughaday WH. Administration of growth hormone to patients with familial dwarfism with high plasma immunoreactive growth hormone: Measurement of sulfation factor, metabolic and linear growth responses. J Clin Endocrinol Metab. 1971;33:332–342. doi: 10.1210/jcem-33-2-332. [DOI] [PubMed] [Google Scholar]
- 15.Shevah O, Laron Z. Patients with congenital deficiency of IGF-I seem protected from the development of malignancies: A preliminary report. Growth Horm IGF Res. 2007;17:54–57. doi: 10.1016/j.ghir.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Steuerman R, Shevah O, Laron Z. Congenital IGF1 deficiency tends to confer protection against post-natal development of malignancies. Eur J Endocrinol. 2011;164:485–489. doi: 10.1530/EJE-10-0859. [DOI] [PubMed] [Google Scholar]
- 17.Guevara-Aguirre J, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapkina-Gendler L, et al. Identification of signaling pathways associated with cancer protection in Laron syndrome. Endocr Relat Cancer. 2016;23:399–410. doi: 10.1530/ERC-16-0054. [DOI] [PubMed] [Google Scholar]
- 19.Chen KS, DeLuca HF. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim Biophys Acta. 1994;1219:26–32. doi: 10.1016/0167-4781(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama A, et al. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 21.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi J, et al. Thioredoxin inhibits tumor necrosis factor- or interleukin-1-induced NF-kappaB activation at a level upstream of NF-kappaB-inducing kinase. Antioxid Redox Signal. 2000;2:83–92. doi: 10.1089/ars.2000.2.1-83. [DOI] [PubMed] [Google Scholar]
- 23.Shah A, et al. Thioredoxin-interacting protein mediates high glucose-induced reactive oxygen species generation by mitochondria and the NADPH oxidase, Nox4, in mesangial cells. J Biol Chem. 2013;288:6835–6848. doi: 10.1074/jbc.M112.419101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junn E, et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig DL, et al. Cloning, genetic characterization, and chromosomal mapping of the mouse VDUP1 gene. Gene. 2001;269:103–112. doi: 10.1016/s0378-1119(01)00455-3. [DOI] [PubMed] [Google Scholar]
- 26.Aitken CJ, et al. Regulation of human osteoclast differentiation by thioredoxin binding protein-2 and redox-sensitive signaling. J Bone Miner Res. 2004;19:2057–2064. doi: 10.1359/JBMR.040913. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg SF, et al. Melanoma metastasis suppression by chromosome 6: Evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63:432–440. [PubMed] [Google Scholar]
- 28.Yoshihara E, Chen Z, Matsuo Y, Masutani H, Yodoi J. Thiol redox transitions by thioredoxin and thioredoxin-binding protein-2 in cell signaling. Methods Enzymol. 2010;474:67–82. doi: 10.1016/S0076-6879(10)74005-2. [DOI] [PubMed] [Google Scholar]
- 29.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 30.Han SH, et al. VDUP1 upregulated by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–4046. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- 31.Minn AH, et al. Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochem Biophys Res Commun. 2005;336:770–778. doi: 10.1016/j.bbrc.2005.08.161. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P, et al. A novel indication of thioredoxin-interacting protein as a tumor suppressor gene in malignant glioma. Oncol Lett. 2017;14:2053–2058. doi: 10.3892/ol.2017.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner H, et al. Genome-wide profiling of congenital IGF1 deficient patients: Translational implications in cancer prevention and metabolism. Transl Med Rep. 2017;1:6657. [Google Scholar]
- 34.Blouet C, Schwartz GJ. Nutrient-sensing hypothalamic TXNIP links nutrient excess to energy imbalance in mice. J Neurosci. 2011;31:6019–6027. doi: 10.1523/JNEUROSCI.6498-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Chng WJ. Roles of thioredoxin binding protein (TXNIP) in oxidative stress, apoptosis and cancer. Mitochondrion. 2013;13:163–169. doi: 10.1016/j.mito.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 37.Malaguarnera R, Belfiore A. The emerging role of insulin and insulin-like growth factor signaling in cancer stem cells. Front Endocrinol (Lausanne) 2014;5:10. doi: 10.3389/fendo.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renehan AG, Frystyk J, Flyvbjerg A. Obesity and cancer risk: The role of the insulin-IGF axis. Trends Endocrinol Metab. 2006;17:328–336. doi: 10.1016/j.tem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Disruption of growth hormone signaling retards early stages of prostate carcinogenesis in the C3(1)/T antigen mouse. Endocrinology. 2005;146:5188–5196. doi: 10.1210/en.2005-0607. [DOI] [PubMed] [Google Scholar]
- 41.Waldhart AN, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep. 2017;19:2005–2013. doi: 10.1016/j.celrep.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JL, et al. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 2010;6:e1001093. doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong SY, Hagen T. 2-Deoxyglucose induces the expression of thioredoxin interacting protein (TXNIP) by increasing O-GlcNAcylation - implications for targeting the Warburg effect in cancer cells. Biochem Biophys Res Commun. 2015;465:838–844. doi: 10.1016/j.bbrc.2015.08.097. [DOI] [PubMed] [Google Scholar]
- 44.Parikh H, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devi TS, Hosoya K, Terasaki T, Singh LP. Critical role of TXNIP in oxidative stress, DNA damage and retinal pericyte apoptosis under high glucose: Implications for diabetic retinopathy. Exp Cell Res. 2013;319:1001–1012. doi: 10.1016/j.yexcr.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chutkow WA, Patwari P, Yoshioka J, Lee RT. Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J Biol Chem. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- 47.Sheth SS, et al. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25:3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- 48.Yoshihara E, et al. Thioredoxin/txnip: Redoxisome, as a redox switch for the pathogenesis of diseases. Front Immunol. 2014;4:514. doi: 10.3389/fimmu.2013.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae VL, et al. Metastatic sublines of an SV40 large T antigen immortalized human prostate epithelial cell line. Prostate. 1998;34:275–282. doi: 10.1002/(sici)1097-0045(19980301)34:4<275::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 50.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 51.Wu Y, Wang C, Sun H, LeRoith D, Yakar S. High-efficient FLPo deleter mice in C57BL/6J background. PLoS One. 2009;4:e8054. doi: 10.1371/journal.pone.0008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, et al. Serum IGF-1 is insufficient to restore skeletal size in the total absence of the growth hormone receptor. J Bone Miner Res. 2013;28:1575–1586. doi: 10.1002/jbmr.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Maor SB, Abramovitch S, Erdos MR, Brody LC, Werner H. BRCA1 suppresses insulin-like growth factor-I receptor promoter activity: Potential interaction between BRCA1 and Sp1. Mol Genet Metab. 2000;69:130–136. doi: 10.1006/mgme.1999.2958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.