Significance
The evolutionary transformation from a blood-feeding to an obligate nonbiting lifestyle is occurring uniquely within the genetic background of a single species of mosquito, Wyeomyia smithii, as a product of selection in nature. Associated genetic changes in metabolic pathways indicate a high anticipatory metabolic investment prior to consuming blood, presumably balanced by the reproductive benefits from an imminent blood meal. This evolutionary transformation provides a starting point for determining pivotal upstream genetic changes between biters and nonbiters and for identifying universal nonbiting genes or pathways in mosquitoes. If there is no bite, there is no transmission of pathogens; hence W. smithii offers a different approach to investigate control of blood-feeding vectors of human diseases.
Keywords: genetic background, directional selection, malaria, KEGG metabolic pathways, genomics
Abstract
The spread of blood-borne pathogens by mosquitoes relies on their taking a blood meal; if there is no bite, there is no disease transmission. Although many species of mosquitoes never take a blood meal, identifying genes that distinguish blood feeding from obligate nonbiting is hampered by the fact that these different lifestyles occur in separate, genetically incompatible species. There is, however, one unique extant species with populations that share a common genetic background but blood feed in one region and are obligate nonbiters in the rest of their range: Wyeomyia smithii. Contemporary blood-feeding and obligate nonbiting populations represent end points of divergence between fully interfertile southern and northern populations. This divergence has undoubtedly resulted in genetic changes that are unrelated to blood feeding, and the challenge is to winnow out the unrelated genetic factors to identify those related specifically to the evolutionary transition from blood feeding to obligate nonbiting. Herein, we determine differential gene expression resulting from directional selection on blood feeding within a polymorphic population to isolate genetic differences between blood feeding and obligate nonbiting. We show that the evolution of nonbiting has resulted in a greatly reduced metabolic investment compared with biting populations, a greater reliance on opportunistic metabolic pathways, and greater reliance on visual rather than olfactory sensory input. W. smithii provides a unique starting point to determine if there are universal nonbiting genes in mosquitoes that could be manipulated as a means to control vector-borne disease.
The word “mosquito” immediately conjures thoughts of being bitten and then, likely, the myriad of human and livestock diseases that these insects transmit. A third of Earth’s human population remains at risk for dengue, with an annual incidence of 390 million infections/y (1), and public concern over the spread of malaria and West Nile, Zika, and Chikungunya viruses continues to increase. Hundreds of laboratories have been employing heroic means to eradicate pathogens of humans transmitted by mosquitoes, and all these efforts assume that a bite will occur. However, what if there is no bite? If there is no bite, there is no disease transmission. In actuality, blood feeding is not universal among mosquitoes. Three genera (Toxorhynchites, Malaya, and Topomyia) and several species in otherwise biting genera never bite (2–7). Individuals of many species may or may not take a blood meal (bite) for the first ovarian cycle, but all these species require a blood meal for their second and subsequent ovarian cycles (8–10). In all cases in which biting and nonbiting species are compared, the results are not robust because differences in allelic composition or gene expression are confounded by the independent evolution of multiple, unrelated traits in genetically incompatible taxa. To address this concern, we use an approach that separates genes associated with avid blood feeding from those associated with nonbiting within a single polymorphic population. We then use differential gene expression (DGE) as a direct consequence of selection on blood feeding within the polymorphic population to identify genes involved in the evolutionary transformation between blood-feeding and obligate nonbiting populations within a single species in nature.
Among the known contemporary species of mosquitoes, only one species blood feeds (bites) in one part of its range but is an obligate nonbiter in the remainder of its range: the pitcher plant mosquito Wyeomyia smithii (11–17). All populations of W. smithii are fully interfertile regardless of geographic origin or propensity to bite, and northern, obligate nonbiting populations are derived from more southern, biting ancestors (18–23). Hence, W. smithii provides a unique opportunity to use selection to concentrate or dilute biting genes within a polymorphic population and then compare DGE of selected avid biters with obligate nonbiters.
We started with a Florida (FL) population that has a low propensity to bite and imposed direct selection for blood feeding. After selection, we had a line of avid biters (FLavid) whose gene-expression profile we could compare with that of disinterested nonbiters (FLdis) isolated from the same polymorphic Florida population. We then were able to compare DGE between avid biters and disinterested nonbiters derived from a common genetic background. We continued by comparing DGE between the avid biters (FLavid) with an obligate nonbiting population from Maine (MEonb). Selection on blood feeding, isolation of disinterested nonbiters, assays of propensity to bite, and experimental sampling of avid biters, disinterested nonbiters, and obligate nonbiters all occurred between 1,200 and 1,400 h subjective mosquito time to factor out variation due to diurnal or circadian rhythmicity. Biters were defined as females that broke the skin with their proboscis, but they were sampled before they actually consumed blood to factor out any DGE due to the presence of blood. Thus, our protocol was specifically designed to detect alterations in gene expression that occur before the intake of blood, i.e., switches associated with the anticipatory costs and metabolic adjustments of a blood-feeding lifestyle.
The objectives of this study were first to determine whether the evolutionary transformation from blood feeding to obligate nonbiting has taken place through a process of selection or through a process of drift and correlated response to selection on other traits. We pursued this objective by testing for a positive association, or lack thereof, between DGE due to known directional selection on blood feeding in the Florida population and DGE between the avid Florida biters and the obligate nonbiting Maine mosquitoes as end points of evolution. The second objective was to determine if there are anticipatory costs identifiable from Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways that differ within and between populations representing diverse biting lifestyles. Third, we used an ongoing evolutionary process in nature as a template, the ultimate goal being to interrupt the spread of mosquito blood-borne disease by turning off the biting phenotype itself. Such a transformation is already taking place within a single species in nature; we just have to discover how. Hence, this third objective is an initial investigation into the “how” for moving forward.
Results
We compare patterns of DGE among selected avid biters (FLavid), isolated disinterested nonbiters (FLdis), and obligate nonbiters (MEonb) and relate these results to functional metabolic pathways.
Patterns of DEG.
As indicated above, none of the mosquitoes used to determine DGE was allowed to consume blood. Selected avid biters (FLavid) were scored as biting after fully inserting their proboscis into the skin of the host but before they took up blood. Disinterested nonbiters (FLdis) were individuals that showed no inclination to insert their proboscis into a host after being given eight opportunities to do so over a 17-d period. During this period, any females biting or attempting to bite were immediately discarded. Obligate nonbiters (MEonb) were observed throughout the same sample time in the presence of a rat; in no case did any of those mosquitoes attempt to insert their proboscis.
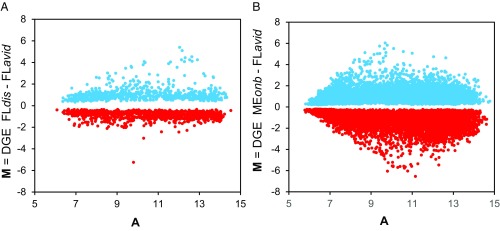
After seven generations of selection, the incidence of biting in the selected line doubled from 19 to 40%. We first determined DGE between the avid biters in the selected line (FLavid) with the disinterested nonbiting isolates (FLdis) (Fig. 1A). This comparison maximized the genetic differences between avid and disinterested nonbiters in the same polymorphic population. We also determined DGE between the avid biters (FLavid) and the obligate nonbiters (MEonb) (Fig. 1B). This comparison maximized the genetic differences between avid biters as a specific response to direct selection on blood feeding and obligate nonbiting as an end point of evolution in nature. The fundamental question then is: What is the association between differentially expressed genes due to known selection on blood feeding (Fig. 1A) and the differential expression of those same individual genes between selected avid biters and obligate nonbiters from a natural nonbiting population (Fig. 1B)? Specifically, are the M-values in Fig. 1B associated with the M-values in Fig. 1A? (M-values are defined in Fig. 1.) A positive association is evidence for the evolution of obligate nonbiting being due to selection in nature; the lack of a significant association or a negative association is evidence for the evolution of nonbiting being due to drift or to a correlated response to selection on other traits.
Fig. 1.
M/A plots of DGE of contigs and singletons from two comparisons. (A) DGE between individuals selected for avid biting (FLavid) and isolated disinterested nonbiters (FLdis) from the polymorphic FL population. (B) DGE between the same individuals from the avid biting line (FLavid) and individuals from the obligate nonbiting population (MEonb). Each M/A plot (SI Microarray Platform) represents two replicates of two dye swaps. On a two-dye microarray, one treatment is labeled with a green dye and the other with a red dye so that the difference in fluorescence between the two dyes represents the difference in gene expression between the two treatments; hence, “M” (for “minus”) = log2(red) − log2(green). To account for differences in red vs. green dye fluorescence, the dyes are “swapped” between treatments in a separate comparison. “A” symbolizes the average log intensities of the two dyes: 1/2 [log2(red) + log2(green)].
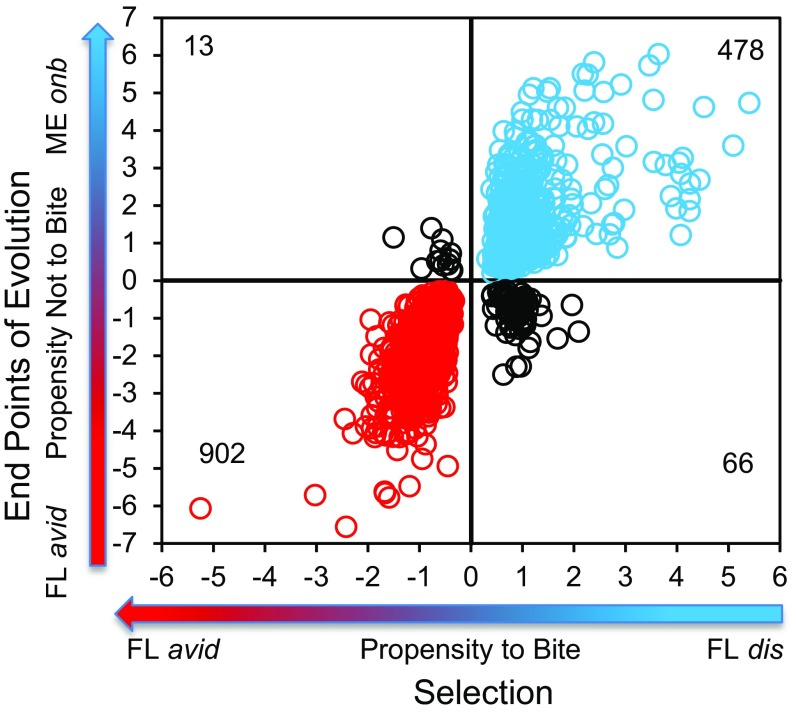
To illustrate the correlation between DGE due to selection for avid biters and due to obligate nonbiting, we developed “Quad” plots (Fig. 2) that show gene-by-gene M-values from the two M/A plots in Fig. 1. In Fig. 2, the horizontal axis shows the M-values from DGE between avid biters (FLavid) vs. disinterested nonbiters (FLdis) (Fig. 1A); the vertical axis shows the M-values from DGE between avid biters (FLavid) vs. obligate nonbiters (MEonb) (Fig. 1B), but subject to the restriction that the false-discovery rate (24) must be q < 0.01 for both M-values. Each gene is represented only once in the Quad plot: The x axis constitutes the M-value of a gene predicted from direct selection on blood feeding, and the y axis constitutes the M-value of the same gene observed between the end points of evolution between avid and obligate nonbiters. The DGE on the lower left-to-upper right diagonal indicates a positive association between direct selection on blood feeding and the evolution of obligate nonbiting. The DGE on the lower right-to-upper left diagonal indicates a negative or no correlation between DGE and the evolution of obligate nonbiting. Of the 21,618 genes identified by the differentially expressed sequence exploration tool (DEET), a broadly useful discriminating algorithm developed in our laboratory (SI DEET: Refinement of the W. smithii Transcriptome), 1,459 met the criteria for inclusion in the Quad plot (SI Collection and Rearing), and 95% of the genes in the Quad plot fell along the axis of positive association.
Fig. 2.
W. smithii: DGE on the horizontal axis is associated with selection on blood feeding, and DGE on the vertical axis is associated with the evolution of nonbiting. The 902 biting genes are concentrated in the lower left; the 478 nonbiting genes are concentrated in the upper right. The orthogonal axis of 79 genes shows DGE not associated with selection on blood feeding. Note that each gene appears on this plot only once, factoring out any DGE between populations unrelated to direct selection on blood feeding.
qPCR Verification.
Only two of 15 genes (actin and eip71cd) deviated from the expected microarray quadrant and the observed quadrant determined by qPCR data (Table S2). In this study, we compared pathways and genes within those pathways only when two or more genes were differentially expressed in the same direction.
Functional Metabolic Pathways.
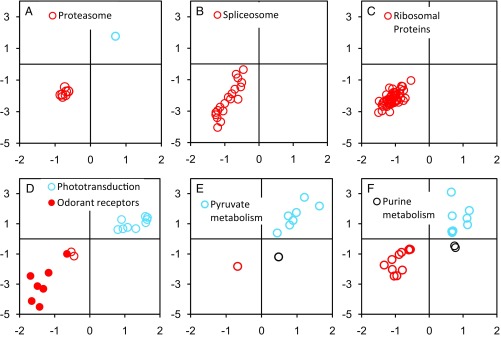
Among the differentially expressed genes in the Quad plot (Fig. 2), KEGG pathway enrichment analysis (25) identified seven metabolic pathways that differed significantly at P < 0.05 following P-value adjustment using the sequential Bonferroni correction (Fig. 3).
Fig. 3.
KEGG pathways: proteosome (A), spliceosome (B), ribosomal proteins (C), phototransduction and odorant receptors (D), pyruvate metabolism (E), and purine metabolism (F). Axes are as in Fig. 2: DGE on the horizontal axis is associated with direct selection on biting, and DGE on the vertical axis is associated with the evolutionary transition from blood feeding to obligate nonbiting. The lower left quadrant plots DGE associated with blood feeding; the upper right quadrant plots genes associated with nonbiting.
Ribosome, spliceosome, ribosome, and proteasome proteins.
DGE of eight of nine proteasome proteins, of all 19 spliceosome proteins, and of all 53 ribosome proteins was associated with blood feeding (Fig. 3 A–C). These results show that, relative to nonbiting, blood-feeding females are investing in protein degradation (proteasome), in posttranscriptional RNA editing (spliceosome), and in the translation of edited RNA into proteins (ribosome).
Phototransduction.
Of the 11 differentially expressed phototransduction proteins on the microarray (Fig. 3D), nine were associated with nonbiting; the seven furthest from the origin all belonged to the actin family of genes; the other two were calcium-binding proteins involved in lipid metabolism and oxidoreductase activity. Two were associated with blood feeding: Arrestin-2 and β-adrenergic-receptor kinase (Gprk1), whose joint action interferes with metarhodopsin, thereby attenuating sensitivity to light (26). The attenuation of light sensitivity in the blood-feeders prompted a query for odorant-related proteins. Of the 21 contigs/singletons associated with odorant reception in the W. smithii transcriptome, eight were differentially expressed, and all were associated with blood feeding (Table S1). These results show that there was an attenuation of light sensitivity coincident with an increase in odorant receptivity in blood-feeding females relative to nonbiting females.
Pyruvate metabolism.
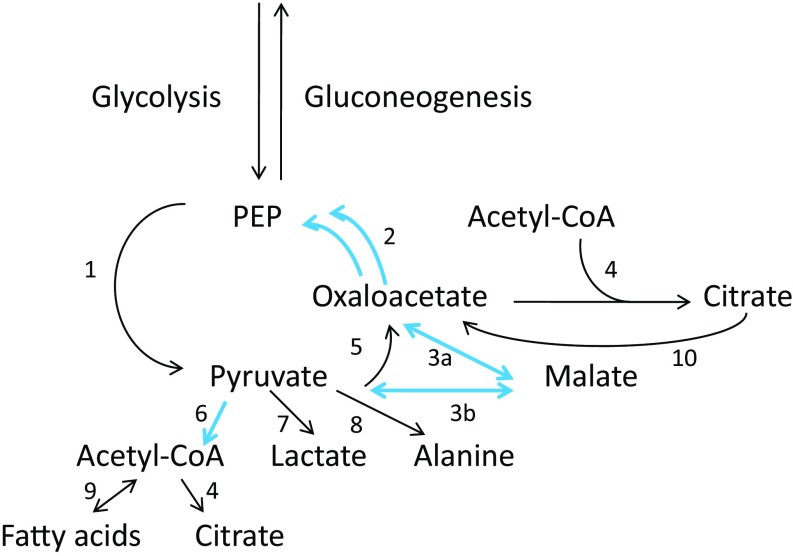
In the pyruvate pathway seven differentially expressed genes were associated with nonbiting. There was also one gene associated with blood feeding and one orthogonal to the bite/no-bite diagonal (Fig. 3E). The sole gene in the biting quadrant (lactoylgutathione lyase) and the off-diagonal gene (aldehyde dehydrogenase family seven member A1) were remotely located in the pyruvate pathway and were not connected with each other (Fig. S1). DGE in the pyruvate nonbiting quadrant (Fig. 4) clustered first around the conversion of pyruvate to acetyl-CoA. The generation of acetyl-CoA provides a basis for both the synthesis and degradation of fatty acids and entry into the citric acid cycle by combining with oxaloacetate (Fig. 4). However, neither enzymes linking acetyl-CoA to fatty acid metabolism nor enzymes linking acetyl-CoA to the citric acid cycle were differentially expressed. Second, pyruvate is potentially linked to hypoxia through lactate or alanine, but, again, neither of the linking enzymes was differentially expressed. Third, up-regulation of oxaloacetate from malate in nonbiters potentially signals gluconeogenesis through the action of phosphoenolpyruvate carboxykinase (PEPCK), which was also up-regulated but not from pyruvate directly, as indicated by the observation that the gene encoding pyruvate carboxylase was not up-regulated. Up-regulation of oxaloacetate also potentially enhances oxidative phosphorylation through the citric acid cycle, but the key linking enzyme, citrate synthetase, was not up-regulated. In combination, acetyl-CoA and oxaloacetate constitute gateways into fatty acid synthesis and degradation, oxidative phosphorylation, and gluconeogenesis, but the linking enzymes to these processes also were not up-regulated. Hence, the overall picture is one of increased preparedness to reallocate energy metabolism in diverse directions in nonbiting females but not to commit directly to any one of those directions.
Fig. 4.
Detail of KEGG pyruvate metabolism pathway 00620 showing genes up-regulated in nonbiters. Blue arrows indicate genes up-regulated in nonbiters, and black arrows indicate genes not significantly differentially expressed: 1, pyruvate kinase; 2, PEPCK (two paralogs); 3, malate dehydrogenase; 3a malate dehydrogenase; 3b malate dehydrogenase–oxaloacetate-decarboxylate (NADP+); 4, citrate synthase; 5, pyruvate carboxylase; 6, pyruvate dehydrogenase complex that includes the E1 component subunit alpha, the E1 component subunit beta, and the E2 component; 7, lactate dehydrogenase; 8, alanine transaminase; 9, pyruvate → acetyl-CoA carboxylase, or pyruvate ← acetyl-CoA C acetyl transferase, acetyl-CoA acetyl transferase 2; 10, citric acid cycle.
Purine and caffeine metabolism.
KEGG pathway analysis showed overlapping purine and caffeine pathways. DGE involving purine metabolism (Fig. 3F) includes three genes overlapping with caffeine: Uric acid oxidase is associated with nonbiting, xanthine dehydrogenase is associated with blood feeding, and a paralog of xanthine dehydrogenase is located in an off-diagonal quadrant from the nonbiting/blood-feeding axis. In the caffeine pathway, uric acid oxidase catalyzes the generation of dimethylurea (2 N per molecule), and xanthine dehydrogenase catalyzes the generation of methyluric acid (4 N per molecule). In sum, the overlap in DGE between purine and caffeine pathways involves the excretion of nitrogen.
DGE involving purine metabolism was divided between nonbiting (eight genes) and blood feeding (11 genes) with two genes in a quadrant off the blood-feeding/nonbiting diagonal (Fig. 3F and Fig. S2). Among nonbiters, differentially expressed genes involved inosine monophosphate (IMP) biosynthesis. IMP forms a branch point leading either to RNA and DNA or to ATP synthesis. Hence, DGE in nonbiting females indicates increased preparedness, relative to biting females, to allocate resources to alternate pathways, rather than directing metabolism to one in particular.
Among blood feeders, differentially expressed genes were directed toward the regulation of protein phosphorylation and proteolysis, cell cycle, nucleotide biosynthesis, DNA and RNA polymerases, and female germline ring canal formation (Table S3). These results show a preparation to break down proteins, presumably those ingested from the blood meal, and to initiate cell proliferation and ovarian development.
Protein Analysis Through Evolutionary Relationships (PANTHER) Overrepresentation Tests.
To interpret the gene-expression results in terms of known and phylogenetically conserved gene functions, plus genetic regulatory and metabolic pathways, we analyzed gene lists for enrichment of biological and molecular gene functions (Table S4). Biological processes included genes involved in translation and organonitrogen processes; molecular processes included genes structurally involved in ribosomes and molecular activity; proteins included genes involved in ribosomal and RNA-binding proteins. All these enriched functions were overrepresented in blood-feeding mosquitoes compared with their nonbiting counterparts, thus reinforcing the results from the KEGG pathway analysis that indicated an increased preparedness in blood feeders to deal with the exigencies of altered nitrogen balance and to advance cell-cycle activities through enhanced translation and ribosomal structure and function.
Discussion
Directional selection on blood feeding in a low-biting polymorphic population resulted in a direct response to selection, doubling the propensity to bite within seven generations and confirming that biting is a highly heritable trait. Response to selection also resulted in DGE between selected avid biters (FLavid) and isolated nonbiters (FLdis) in the same southern population. Of the DGE between FLavid and FLdis, 95% of the genes also showed DGE in the same direction between FLavid and an obligate nonbiting population (MEonb) (Fig. 2). The close association of DGE between direct response to selection and the evolutionary transition to obligate nonbiting in nature leads us to conclude that the evolution of the northern obligate nonbiting lifestyle from blood-feeding ancestors has been the consequence of selection through evolutionary time in nature within this single, fully interfertile species.
There are contemporary taxa of mosquitoes that never bite but persist and even thrive. The selective forces leading from a blood-feeding to a nonbiting lifestyle among different species and genera of mosquitoes are lost in evolutionary time, but blood feeding is not a free lunch. The added nutritional benefits of blood feeding are balanced by both extrinsic and intrinsic costs. There are known extrinsic costs of blood feeding that are incurred in finding and surviving on a host (27–29). Consuming a blood meal also involves intrinsic costs. First, there is a thermal shock due to imbibing a hot blood meal at ∼40 °C (30, 31). Second, the breakdown of hemoglobin specifically liberates toxic heme and iron that are excreted or bound and sequestered in the peritrophic matrix (32–37). DGE within and between populations of W. smithii, in which we can see a tight association between direct selection on blood feeding and the evolutionary transition to a nonbiting lifestyle in a single interfertile species, contrasts two broad patterns of gene expression: direct anticipatory costs and flexible metabolic opportunism.
First, the direct costs of consuming a blood meal in biting females of W. smithii are evidenced by the preparation for protein degradation by up-regulation of protein phosphorylation, the proteasome, and proteolysis (Fig. 3 A–C and Table S3), by a targeted increase in odorant receptors and decreased visual response (Fig. 3D), by investment in cell-cycle activity including nucleotide synthesis and DNA and RNA polymerases (Fig. S2 and Table S3), and by incipient ovarian maturation as indicated by up-regulating female germline ring canal formation (Table S3). In sum, these costs have the common theme of preparing to digest the expected blood meal and to initiate subsequent ovarian maturation. It is important to note that all these costs are incurred before blood is consumed.
Second, flexible metabolic opportunism in nonbiting females is evidenced by the up-regulation of visual over odorant reception (Fig. 3D) and by the up-regulation of metabolism leading to IMP (Fig. S2 and Table S3), which provides a branch point in purine metabolism that can lead, among other products, to RNA and DNA synthesis, nitrogen metabolism, or to the cytosolic second messengers cAMP and cGMP (38). Flexible metabolic opportunism is also represented by pyruvate-related enzymes that serve as gateways to diverse functional downstream processes (Fig. 4). Nonbiting is associated with the conversion of pyruvate to acetyl-CoA but not with the potential connection between acetyl-CoA and either fatty acid metabolism or entry into the citric acid cycle. At the same time, pyruvate in nonbiters is not directly linked to potential responses to hypoxia (lactate or alanine cycles) or to the direct generation of gluconeogenesis via the conversion of pyruvate carboxylase to oxaloacetate. Nonetheless, the potential for gluconeogenesis remains, because nonbiting is associated with the up-regulation of PEPCK, the crucial step in converting oxaloacetate to phosphoenolpyruvate in gluconeogenesis. In sum, nonbiting is associated with numerous “dogs that don’t bark”—the opportunistic potential for generating diverse downstream functional processes in response to appropriate environmental conditions.
Conclusion
The evolutionary transformation from a blood-feeding to an obligate nonbiting lifestyle in W. smithii is the product of selection in nature that has resulted in reduced anticipatory costs and the adoption of a more opportunistic lifestyle. The major remaining question is what specific upstream genes regulate the alteration of the associated metabolic processes that control this transformation. Mosquitoes have been called the “world’s most dangerous animal” (39) due to their ability to transmit heinous pathogens causing debilitating or lethal diseases in humans and livestock (1, 40–43). Identification of key genes responsible for the evolution of an obligate nonbiting lifestyle provides the potential to mitigate mosquito-borne diseases, because, if there is no bite, there is no disease transmission. At the genetic level, we have to determine the means by which this transformation has taken place and whether it leads to the identification of universal nonbiting genes or to universal target genetic pathways in mosquitoes. Is this determination possible? It should be: The genetic and genomic tools are available. We know that Mother Nature has done it, not only in the remote evolutionary past among different genera or species, but also today, as seen among the fully interfertile populations of a single species of mosquito.
Materials and Methods
Larvae of a low-biting population of W. smithii were collected from Florida (30°N), and larvae of an obligate nonbiting population were collected from Maine (46°N). Stock populations of both the Florida (FL) and Maine (ME) populations were maintained under standard conditions (SI Collection and Rearing). From the wild-caught FL individuals, 14,000 larvae were reared to adulthood and selected for avid biting for seven generations (SI Directional Selection on Blood Feeding). The line selected for avid biting (FLavid), the low-biting FL stock population, and the obligate nonbiting ME stock population were synchronized in diapause. After 1 mo in diapause, four replicates of 1,200 larvae each of FLavid, FL, and ME were reared to adulthood in 12 separate adult cages (four replicates of three treatments). Starting 5 d after first adult female emergence, each of the 12 cages was offered a female Sprague–Dawley rat (Harlan Laboratories). Rats were anesthetized with a ketamine/xylazine mixture and offered as a blood source according to University of Oregon Institutional Animal Care and Use Committee protocols 10, 11, and 13–15. Rats were offered to mosquitoes three times a week for 15 min between 1,200 and 1,400 h subjective mosquito time to minimize diurnal or circadian rhythmicity on transcriptional profiles. The order in which rats were introduced into cages was rotated among the 12 cages during each sampling episode. Each cage was observed individually and continually during the 15 min that the rat was present.
FLavid.
From each replicate of the FLavid-selected line, any female that inserted her proboscis into the skin of the rat was scored as an avid biter (FLavid).
FLdis.
From each replicate of the FL stock population, any female that inserted her proboscis into the rat was scored as a biter, removed from the cage, and discarded. This process continued for 17 d, allowing females eight separate opportunities to bite. At the end of 17 d, 300 of the remaining females from each cage were scored as disinterested biters (FLdis). None of the disinterested females had made any attempt to bite.
MEonb.
In no cage did any of the females from the ME stock population attempt to bite. Samples of a few females were removed from the cage three times a week over a 17-d period and scored as obligate nonbiters (MEonb).
Once scored as an avid biter (FLavid), a disinterested biter (FLdis), or an obligate nonbiter (MEonb), females were flash frozen in 95% ethanol on dry ice and decapitated, and their heads were homogenized in cold TRI Reagent (TR-118; Zymo Research) and stored at −80 °C. Three hundred heads from each of the four replicates of each of the three treatments were sampled and processed as described in SI Tissue Collection and RNA Isolation.
Gene expression was measured using a custom W. smithii microarray (SI Microarray Platform) with two replicated dye swaps (SI Microarray Hybridization). Comparison among treatments used the limma package of Bioconductor (SI Microarray Analysis). Orthologs and paralogs/splice variants were scored using the DEET pipeline developed in our laboratory (SI DEET: Refinement of the W. smithii Transcriptome and SI Distribution of Paralogs and Splice Variants). DEG was verified with qPCR (SI qPCR Verification). Finally, differentially expressed pathways were assessed using KEGG and PANTHER analyses (SI KEGG and PANTHER).
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grants IOS-1255628 (to W.E.B.) and DEB-1455506 (OPUS) (to W.E.B. and C.M.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Transcriptome sequence, assembly, and annotation of Wyeomyia smithii are available through the National Center for Biotechnical Information (NCBI), https://www.ncbi.nlm.nih.gov/bioproject/?term=259209. The microarray data are available in the NCBI Gene Expression Omnibus repository (accession no. GSE100766). The source code for DEET can be browsed and is available for download at https://sourceforge.net/p/deet/code/ci/master/tree/.
See Commentary on page 836.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717502115/-/DCSupplemental.
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downes JA. The feeding habits of biting flies and their significance in classification. Annu Rev Entomol. 1958;3:249–266. [Google Scholar]
- 3.Foster WA. Mosquito sugar feeding and reproductive energetics. Annu Rev Entomol. 1995;40:443–474. doi: 10.1146/annurev.en.40.010195.002303. [DOI] [PubMed] [Google Scholar]
- 4.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE. Illustrated keys to the mosquitoes of Thailand V. Genera Orthopodomyia, Kimia, Malaya, Topomyia, Tripteroides, and Toxorhynchites. Southeast Asian J Trop Med Public Health. 2007;38:1–65. [PubMed] [Google Scholar]
- 5.Wahid I, Sunahara T, Mogi M. The hypopharynx of male and female mosquitoes. Open Entomol J. 2007;1:1–6. [Google Scholar]
- 6.Miyagi I, et al. Three new phytotelmata mosquitoes of the genus Topomyia (Diptera: Culicidae) from Katibas, Lanjak-Entimau, Sarawak, Malaysia. J Sci Technol Tropics. 2012;8:97–117. [Google Scholar]
- 7.Zhou X, Rinker DC, Pitts RJ, Rokas A, Zwiebel LJ. Divergent and conserved elements comprise the chemoreceptive repertoire of the nonblood-feeding mosquito Toxorhynchites amboinensis. Genome Biol Evol. 2014;6:2883–2896. doi: 10.1093/gbe/evu231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spielman A. Bionomics of autogenous mosquitoes. Annu Rev Entomol. 1971;16:231–248. doi: 10.1146/annurev.en.16.010171.001311. [DOI] [PubMed] [Google Scholar]
- 9.Rioux J-A, Croset H, PechPérières J, Guilvard E, Belmonte A. L’autogenese chez les Dipteries Culicides. Tableau synoptique des espèces autogènes [Autogenesis in the Diptera Culicides. Synoptic table of autogenic species] Ann Parasitol Hum Comp. 1975;50:134–140. French. [PubMed] [Google Scholar]
- 10.O’Meara GF. Ecology of autogeny in mosquitoes. In: Lounibos LP, Rey JR, Frank JH, editors. Ecology of Mosquitoes: Proceedings of a Workshop. Florida Medical Entomology Laboratory; Vero Beach, FL: 1985. pp. 459–471. [Google Scholar]
- 11.Smith SM, Brust RA. Photoperiodic control of the maintenance and termination of larval diapause in Wyeomyia smithii (Coq.) (Diptera: Culicidae) with notes on oogenesis in the adult female. Can J Zool. 1971;49:1065–1073. doi: 10.1139/z71-165. [DOI] [PubMed] [Google Scholar]
- 12.Bradshaw WE. Blood-feeding and capacity for increase in the pitcher-plant mosquito, Wyeomyia smithii. Environ Entomol. 1980;9:86–89. [Google Scholar]
- 13.Bradshaw WE. Variable iteroparity as a life-history tactic in the pitcher-plant mosquito Wyeomyia smithii. Evolution. 1986;40:471–478. doi: 10.1111/j.1558-5646.1986.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 14.O’Meara GF, Lounibos LP, Brust R. Repeated egg clutches without blood in the pitcher-plant mosquito. Ann Entomol Soc Am. 1981;74:68–72. [Google Scholar]
- 15.Bradshaw WE, Holzapfel CM. Life cycle strategies in Wyeomyia smithii: Seasonal and geographic adaptations. In: Brown VK, Hodek I, editors. Diapause and Life Cycle Strategies in Insects. Dr W. Junk; The Hague: 1983. pp. 167–185. [Google Scholar]
- 16.Lounibos LP, Van Dover C, O’Meara GF. Fecundity, autogeny, and the larval environment of the pitcher-plant mosquito, Wyeomyia smithii. Oecologia. 1982;55:160–164. doi: 10.1007/BF00384482. [DOI] [PubMed] [Google Scholar]
- 17.Bradshaw WE, Holzapfel CM. Life-historical consequences of density-dependent selection ini the pitcher-plant mosquito, Wyeomyia smithii. Am Nat. 1989;133:869–887. [Google Scholar]
- 18.Bradshaw WE, Lounibos LP. Evolution of dormancy and its photoperiodic control in pitcher-plant mosquitoes. Evolution. 1977;31:546–567. doi: 10.1111/j.1558-5646.1977.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 19.Armbruster P, Bradshaw WE, Holzapfel CM. Evolution of the genetic architecture underlying fitness in the pitcher-plant mosquito, Wyeomyia smithii. Evolution. 1997;51:451–458. doi: 10.1111/j.1558-5646.1997.tb02432.x. [DOI] [PubMed] [Google Scholar]
- 20.Armbruster P, Bradshaw WE, Holzapfel CM. Effects of postglacial range expansion on allozyme and quantitative genetic variation in the pitcher-plant mosquito, Wyeomyia smithii. Evolution. 1998;52:1697–1704. doi: 10.1111/j.1558-5646.1998.tb02249.x. [DOI] [PubMed] [Google Scholar]
- 21.Armbruster P, Bradshaw WE, Steiner AL, Holzapfel CM. Evolutionary responses to environmental stress by the pitcher-plant mosquito, Wyeomyia smithii. Heredity (Edinb) 1999;83:509–519. doi: 10.1038/sj.hdy.6886040. [DOI] [PubMed] [Google Scholar]
- 22.Holzapfel CM, Bradshaw WE. Protandry: The relationship between emergence time and male fitness in the pitcher-plant mosquito. Ecology. 2002;83:607–611. [Google Scholar]
- 23.Merz C, et al. Replicate phylogenies and post-glacial range expansion of the pitcher-plant mosquito, Wyeomyia smithii, in North America. PLoS One. 2013;8:e72262. doi: 10.1371/journal.pone.0072262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey JD. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498. [Google Scholar]
- 25.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Leming MT, Metoxen AJ, Whaley MA, O’Tousa JE. Light-mediated control of rhodopsin movement in mosquito photoreceptors. J Neurosci. 2012;32:13661–13667. doi: 10.1523/JNEUROSCI.1816-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edman JD, Scott TW. Host defensive behavior and the feeding success of mosquitoes. Int J Trop Insect Sci. 1987;8:617–622. [Google Scholar]
- 28.Darbro JM, Harrington LC. Avian defensive behavior and blood-feeding success of the West Nile vector mosquito, Culex pipiens. Behav Ecol. 2007;18:750–757. [Google Scholar]
- 29.de Silva P, Jaramillo C, Bernal XE. Feeding site selection by frog-biting midges (Diptera: Corethrellidae) on anuran hosts. J Insect Behav. 2014;27:302–316. [Google Scholar]
- 30.Benoit JB, et al. Drinking a hot blood meal elicits a protective heat shock response in mosquitoes. Proc Natl Acad Sci USA. 2011;108:8026–8029. doi: 10.1073/pnas.1105195108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahondère C, Lazzari CR. Mosquitoes cool down during blood feeding to avoid overheating. Curr Biol. 2012;22:40–45. doi: 10.1016/j.cub.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 32.Pascoa V, et al. Aedes aegypti peritrophic matrix and its interaction with heme during blood digestion. Insect Biochem Mol Biol. 2002;32:517–523. doi: 10.1016/s0965-1748(01)00130-8. [DOI] [PubMed] [Google Scholar]
- 33.Geiser DL, Chavez CA, Flores-Munguia R, Winzerling JJ, Pham DQ-D. Aedes aegypti ferritin. Eur J Biochem. 2003;270:3667–3674. doi: 10.1046/j.1432-1033.2003.03709.x. [DOI] [PubMed] [Google Scholar]
- 34.Graça-Souza AV, et al. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol. 2006;36:322–335. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Devenport M, et al. Identification of the Aedes aegypti peritrophic matrix protein AeIMUCI as a heme-binding protein. Biochemistry. 2006;45:9540–9549. doi: 10.1021/bi0605991. [DOI] [PubMed] [Google Scholar]
- 36.Esquivel CJ, Cassone BJ, Piermarini PM. Transcriptomic evidence for a dramatic functional transition of the malpighian tubules after a blood meal in the Asian tiger mosquito Aedes albopictus. PLoS Negl Trop Dis. 2014;8:e2929. doi: 10.1371/journal.pntd.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikbakhtzadeh MR, Buss GK, Leal WS. Toxic effect of blood feeding in male mosquitoes. Front Physiol. 2016;7:4. doi: 10.3389/fphys.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton AC, Bootman MD, Scott JD. Second messengers. Cold Spring Harb Perspect Biol. 2016;8:a005926. doi: 10.1101/cshperspect.a005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weetman D, Clarkson CS. Evolving the world’s most dangerous animal. Trends Parasitol. 2015;31:39–40. doi: 10.1016/j.pt.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Beaty BJ, Marquardt WC, editors. The Biology of Disease Vectors. Univ of Colorado Press; Niwot, CO: 1996. [Google Scholar]
- 41.Braks M, et al. Vector-borne disease intelligence: Strategies to deal with disease burden and threats. Front Public Health. 2014;2:280. doi: 10.3389/fpubh.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anyamba A, et al. Recent weather extremes and impacts on agricultural production and vector-borne disease outbreak patterns. PLoS One. 2014;9:e92538. doi: 10.1371/journal.pone.0092538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grace D, Bett B, Lindahl J, Robinson T. 2015. Climate and Livestock Diseases: Assessing the Vulnerability of Agricultural Systems to Livestock Pests Under Climate Change Scenarios. (Consultative Group on International Agricultural Research Research Program on Climate Change, Agriculture and Food Security, Copenhagen), Climate Change Agricultural and Food Security Working Paper no. 116.
- 44.Schroeder A, et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tormey D, et al. Evolutionary divergence of core and post-translational circadian clock genes in the pitcher-plant mosquito, Wyeomyia smithii. BMC Genomics. 2015;16:754. doi: 10.1186/s12864-015-1937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: A hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez J, Colborne JK. 2010 Dual-Labeled Expression Microarray Protocol for High-Throughput Genomic Investigations. CGB Technical Report 2010-01. Available at https://wiki.cgb.indiana.edu/download/attachments/22446090/cgb-tr-2010-01_v2.5.pdf. Accessed December 10, 2017.
- 48.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solution Using R and Bioconductor. Springer International Publishing AG; Cham, Switzerland: 2005. pp. 397–420. [Google Scholar]
- 49.R Core Team 2013 R: A Language and Environment for Statistical Computing, Version R.2.15.1 (R Found Stat Comput, Vienna). Available at https://www.r-project.org/. Accessed January 4, 2015.
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 51.Pruitt K, Brown G, Tatusova T, Maglott D. 2002 The reference sequence (RefSeq) database. The NCBI Handbook, eds McEntyre J, Ostel J (National Center for Biotechnology Information, Bethesda), Chap 18, pp 1–24. [updated 2012], Available at https://www.ncbi.nlm.nih.gov/books/NBK21101/. Accessed September 23, 2017.
- 52.Reynolds JA, Clark J, Diakoff SJ, Denlinger DL. Transcriptional evidence for small RNA regulation of pupal diapause in the flesh fly, Sarcophaga bullata. Insect Biochem Mol Biol. 2013;43:982–989. doi: 10.1016/j.ibmb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Bustin SA, et al. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber W, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tennenbaum D. 2016 KEGGREST: Client-Side REST Access to KEGG, R Package Version 1.16.1. Available at bioconductor.org/packages/release/bioc/html/KEGGREST.html. Accessed September 10, 2017.
- 56.Mi H, et al. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coskun A, et al. Proteomic analysis of kidney preservation solutions prior to renal transplantation. PLoS One. 2016;11:e0168755. doi: 10.1371/journal.pone.0168755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho RJ, Campbell MJ. Transcription, genomes, function. Trends Genet. 2000;16:409–415. doi: 10.1016/s0168-9525(00)02065-5. [DOI] [PubMed] [Google Scholar]
- 61.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.