Significance
The molecular basis of immunodeficiency–centromeric instability–facial anomalies (ICF) syndrome is poorly understood. ICF is caused by mutations in HELLS, CDCA7, or the DNA methyltransferase DNMT3b. While these mutations all cause DNA methylation defects, little is known about the molecular function of CDCA7. It has been speculated that HELLS, which belongs to the SNF2 family ATPase, facilitates DNA methylation by DNMT3b through nucleosome remodeling, but HELLS on its own fails to exhibit such an activity. Here, starting from a comparative proteomic analysis of chromatin proteins in frog egg extracts, we demonstrated that CDCA7 is required for HELLS to associate with chromatin and exert nucleosome remodeling activity. Our finding delineates the molecular pathway involving CDCA7, HELLS, and DNMT3b.
Keywords: nucleosome remodeling, ICF, HELLS, Xenopus, chromatin proteomics
Abstract
Mutations in CDCA7, the SNF2 family protein HELLS (LSH), or the DNA methyltransferase DNMT3b cause immunodeficiency–centromeric instability–facial anomalies (ICF) syndrome. While it has been speculated that DNA methylation defects cause this disease, little is known about the molecular function of CDCA7 and its functional relationship to HELLS and DNMT3b. Systematic analysis of how the cell cycle, H3K9 methylation, and the mitotic kinase Aurora B affect proteomic profiles of chromatin in Xenopus egg extracts revealed that HELLS and CDCA7 form a stoichiometric complex on chromatin, in a manner sensitive to Aurora B. Although HELLS alone fails to remodel nucleosomes, we demonstrate that the HELLS–CDCA7 complex possesses nucleosome remodeling activity. Furthermore, CDCA7 is essential for loading HELLS onto chromatin, and CDCA7 harboring patient ICF mutations fails to recruit the complex to chromatin. Together, our study identifies a unique bipartite nucleosome remodeling complex where the functional remodeling activity is split between two proteins and thus delineates the defective pathway in ICF syndrome.
Immunodeficiency–centromeric instability–facial anomalies (ICF) syndrome is a rare recessive autosomal disorder, causing severe infections (1). One of the diagnostic characteristics of patients with ICF is a unique metaphase chromosome morphology in lymphocytes stimulated by mitogens, where the juxtacentromeric heterochromatin of chromosomes 1 and 16 enriched with repetitive satellites 2 and 3, becomes stretched and fragile (2). While the molecular basis of ICF is unknown, it is thought to be caused by defective DNA methylation, as the majority of patients have mutations in the de novo DNA methyltransferase DNMT3b with hypomethylated satellites 2 and 3 DNA (3). More recently, mutations in a transcription regulator ZBTB24, a putative nucleosome remodeler helicase, lymphoid specific (HELLS) (also known as LSH and SMARCA6), and CDCA7 have been found to also cause reduced satellite DNA methylation and ICF (4, 5). ZBTB24 promotes CDCA7 transcription (6), but how HELLS and CDCA7 mutations contribute to ICF syndrome is unknown.
HELLS is a SNF2 ATPase family protein, required for de novo DNA methylation of repetitive elements and developmentally programmed genes through interaction with DNMT3b (7). Among six groups of SNF2 family proteins, HELLS belongs to a SNF2-like subfamily, including SMARCA2/4 (BRM/BRG1), CHD3–5 (Mi-2), CHD1/2, CHD6–9, SMARCA1/5 (SNF2L/SNF2h, ISWI) and ALC1 (8). While the core ATPase domain of all other SNF2-like subfamily proteins exhibit nucleosome remodeling activity (9–15), mouse HELLS does not possess ATP-dependent nucleosome remodeling activity in vitro (16). Furthermore, while many SNF2-like ATPases form large multisubunit complexes, it has been suggested that HELLS does not (17). However, ATPase-defective point mutations in HELLS are unable to support de novo DNA methylation during embryonic stem cell differentiation (18) and in fibroblasts (19). Additionally, the Arabidopsis thaliana homolog of HELLS, DDM1, exhibits nucleosome remodeling activity (20). Therefore, it remains a mystery why HELLS lacks nucleosome remodeling activity. CDCA7, containing a highly conserved 4CXXC zinc finger domain, has been suspected to be a transcription factor under the control of c-Myc (21–24), but little is known about its molecular function or its relationship to HELLS and DNMT3b.
In this study, we determine how the proteomic composition of chromatin is affected by the combinatorial effects of the cell cycle, the heterochromatin mark H3K9me3, and the chromosomal passenger complex (CPC), which contains the protein kinase Aurora B and plays a number of critical roles in mitotic chromosome structure and function (25). The comparative proteomic analysis demonstrates that most known protein complex subunits associate with chromatin in equivalent amounts, and their stoichiometry is preserved even as the absolute levels of chromatin binding drastically change between conditions. Using this principle, we identify a chromatin-associated complex comprising ICF proteins HELLS and CDCA7, and demonstrate that HELLS–CDCA7 is a nucleosome remodeling complex, providing insight into the molecular basis of the ICF disorder.
Results
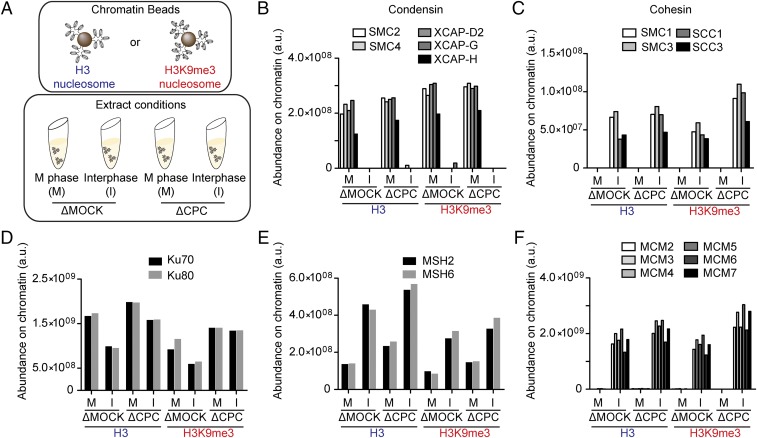
To assess the impact of the cell cycle, the heterochromatic mark H3K9me3, and Aurora B on the proteomic composition of chromatin, we used Xenopus egg extracts arrested in meiotic metaphase II by the cytostatic factor (CSF extract). The extract was subject to immunodepletion using control IgG (∆MOCK) or anti-INCENP (∆CPC), which can effectively deplete the CPC, including Aurora B (26). Magnetic beads coated with nucleosome arrays reconstituted with recombinant Xenopus core histones, with or without H3K9me3, were added to these extracts (27, 28). Extracts were then either left in metaphase or released into interphase by adding calcium before purification of beads and liquid chromatography tandem mass spectrometry (LC-MS/MS) of chromatin-bound proteins. This enabled a direct quantitative comparison of chromatin proteomics under eight different conditions harboring a combination of three different perturbations (cell cycle, ±CPC, and ±H3K9me3) (Fig. 1A and Dataset S1).
Fig. 1.
Proteomic profiling of chromatin regulation by the cell cycle, the CPC, and H3K9me3. (A) Schematic of the procedure to identify chromatin-bound proteins in Xenopus egg extracts. Beads coated with unmodified or H3K9me3 nucleosome arrays were incubated in M phase or interphase Xenopus egg extracts subject to CPC or mock depletion. Beads were recovered, washed, and bound proteins were analyzed by LC-MS/MS. (B–F) Abundance of condensin (B), cohesin (C), Ku70–Ku80 (D), MSH2–6 (E), and MCM2–7 (F), identified by LC-MS/MS on the indicated chromatin beads. Abundance (arbitrary units) was calculated by integrating LC-MS signals for each peptide, as described in Methods.
The MS analysis revealed that known chromatin-associated proteins largely exhibited the expected binding patterns across the eight conditions. For example, all condensin and cohesin subunits exclusively associated with chromatin in M phase and in interphase, respectively (Fig. 1 B and C). HP1 and Suv39 exclusively interacted with H3K9me3-chromatin in a manner sensitive to the CPC, consistent with the notion that Aurora B-dependent H3S10ph dissociates these chromodomain-containing proteins (Fig. S1A), while suppression of HP1 binding to H3K9me3 nucleosomes in interphase is likely due to delayed dephosphorylation of H3S10ph (29, 30). In contrast, ATRX (and its binding partner DAXX) was recruited to H3K9me3 in a manner independent of the CPC, consistent with its H3S10ph-insensitive binding to H3K9me3 (Fig. S1B) (31, 32). Notably, many known stoichiometric chromatin-associated complexes, differing in abundance by orders of magnitude, including Ku70–Ku80, MSH2–6, MCM, FACT, FANCD2–FANCI, and astrin–kinastrin showed remarkable regularity across different conditions (Fig. 1 D–F and Fig. S1 C–F).
Identification of the HELLS–CDCA7 Complex.
As subunits of known chromatin-associated complexes behaved similarly on chromatin, we wondered whether we could identify novel chromatin-associated complexes based on similar proteomic profiles, which have successfully been used to identify soluble complexes (33). However, an initial hierarchical clustering attempt failed to cluster several known protein complex subunits, due to the presence of proteins that are highly abundant in the extracts and likely stick nonspecifically to chromatin beads. To circumvent this issue, we assessed relative chromatin enrichment values by normalizing the chromatin-associated amount by the reported protein concentrations in eggs (34). Removing proteins whose enrichment values do not surpass the 70th percentile at any condition, effectively reduced this confounding effect by nonspecific binders. Using this refined dataset, hierarchical clustering broadly partitioned the proteins into M phase or interphase-specific interactors. Further branching clustered stoichiometric subunits of many known protein complexes (Fig. 2A and Fig. S1 G–J).
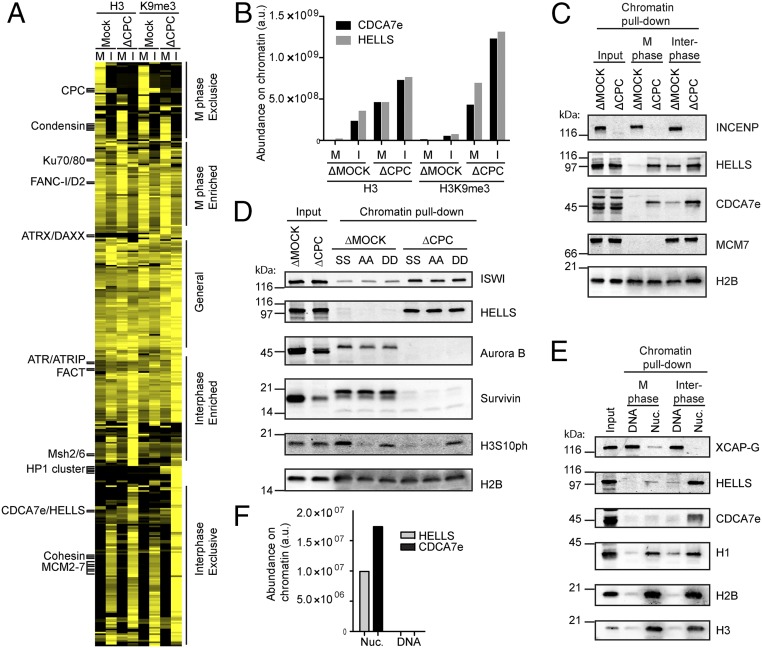
Fig. 2.
Chromatin binding of HELLS–CDCA7e is coregulated by the CPC and nucleosomes. (A) Unsupervised clustering of chromatin-associated proteins. A heatmap of each protein’s normalized profile is displayed, with proteins enriched on a given chromatin sample colored yellow. Examples of known stoichiometric chromatin-associated complexes cluster together and are indicated. Fine-grained protein complex clusters (Left) and coarse-grained cell-cycle clusters (Right) are labeled. (B) LC-MS/MS analysis HELLS and CDCA7e copurified with unmodified or H3K9me3 nucleosome beads recovered from M phase or interphase mock or CPC-depleted extracts. (C) Western blot analyses of HELLS and CDCA7e copurified with nucleosome beads recovered from M phase or interphase mock or CPC-depleted extracts. Representative of n = 2 independent experiments. (D) Western blot analyses of proteins copurified with nucleosome beads recovered from M phase mock or CPC-depleted extracts. Nucleosomes containing wild-type H3 (SS), H3S10AS28A (AA), or H3S10DS28D (DD) were used. (E) Western blot analyses of proteins copurified with nucleosome or DNA beads recovered from M phase or interphase H3–H4-depleted extracts. (F) Abundance of HELLS and CDCA7e on nucleosome or DNA beads recovered from M phase H3–H4-depleted extracts quantified by LC-MS/MS. Data from ref. 28 (originally missing CDCA7e) was reanalyzed using an mRNA reference database containing CDCA7e (34).
The hierarchical clustering successfully sensed subtle differences in the proteomic profiles so that the subunits of known protein complexes clustered together, including complexes such as FACT and ATR/ATRIP along with ATR activators, ETAA1 and DNA2 (35, 36), which were only subtly affected by the experimental conditions (Fig. 2A and Fig. S1H). Remarkably, the method successfully clustered Ku70–80 with its F-box containing promoter of DNA binding-dependent ubiquitylation, FBXL12 (Fig. S1I) (37), and RPA1–3, with known binding partners SMARCAL1 and HELB (38–43). Interestingly, TOPBP1, a known activator of ATR (44), clustered with its binding partners, FANCJ (45) and RAD17 (46, 47), particularly on M phase chromatin, but not with ATR/ATRIP, perhaps reflecting its reported mitotic chromosome localization (48).
Encouraged by these examples of successful clustering, we sought novel protein interactions. Among proteins that are not previously known to form a complex, CDCA7e, the embryonic variant of CDCA7, and HELLS clustered in our unsupervised method (Fig. 2A and Fig. S1K) and showed strikingly similar profiles, where equivalent amounts of these two proteins copurified with chromatin beads across all eight conditions (Fig. 2B). Quantitative Western blotting using antibodies against HELLS and CDCA7e confirmed this stoichiometric relationship between HELLS and CDCA7 (Fig. S2).
One of the most apparent characteristics of the HELLS–CDCA7e behavior is its eviction from M phase chromatin in a manner dependent on the CPC (Fig. 2 B and C). As Aurora B is responsible for mitotic phosphorylation of H3S10 and H3S28, we wondered whether phosphorylation of these H3 residues was responsible for HELLS/CDCA7e eviction. Reconstituting chromatin beads harboring alanine or aspartate mutations at H3S10 and H3S28 showed that HELLS is regulated by the CPC independently of phosphorylation at these residues (Fig. 2D). ISWI, another remodeling complex shown to be evicted from M phase chromatin by Aurora B (49), was similarly unaffected by H3S10 and H3S28 mutations (Fig. 2D).
As with many nucleosome remodeling complexes, including ISWI and FACT (28), HELLS and CDCA7e are dependent on nucleosomes for chromatin association in egg extracts (Fig. 2 E and F). Many chromatin proteins that fail to associate with nucleosome-free DNA in physiological cytoplasmic extract (28) possess DNA binding activity in isolation. These include H1 (50), RCC1 (51), topoisomerase II (28), and FACT (52). Similarly, purified HELLS–calmodulin-binding peptide (CBP) and maltose-binding protein (MBP)–CDCA7e also have DNA-binding activity in vitro (Fig. S3 A and B), despite their nucleosome-dependent chromatin binding in egg extracts. This suggests that under physiological conditions, HELLS–CDCA7e either cannot compete with endogenous DNA binding proteins for binding to naked DNA, or that proteins exist in egg extract that remove HELLS–CDCA7e from DNA but not from nucleosomes.
As the amounts of HELLS and CDCA7e on chromatin are equivalent through a variety of conditions, we hypothesized that they form a complex. Indeed, coimmunoprecipitation experiments from chromatin-free Xenopus egg extracts showed that HELLS and CDCA7e interact in soluble cytoplasmic extract (Fig. 3 A and B). Furthermore, purified HELLS–CBP coimmunoprecipitated with MBP–CDCA7e in isolation, indicating a direct interaction between HELLS and CDCA7e (Fig. 3C). However, immunodepletion of either component from Xenopus egg extract did not codeplete the other (Fig. 3D), despite their equimolar presence in eggs (∼100 nM) (34). This suggests that only a fraction of HELLS and CDCA7e interacts in the cytoplasm, consistent with a previous report that HELLS exists as a monomer in mouse and human cells (17).
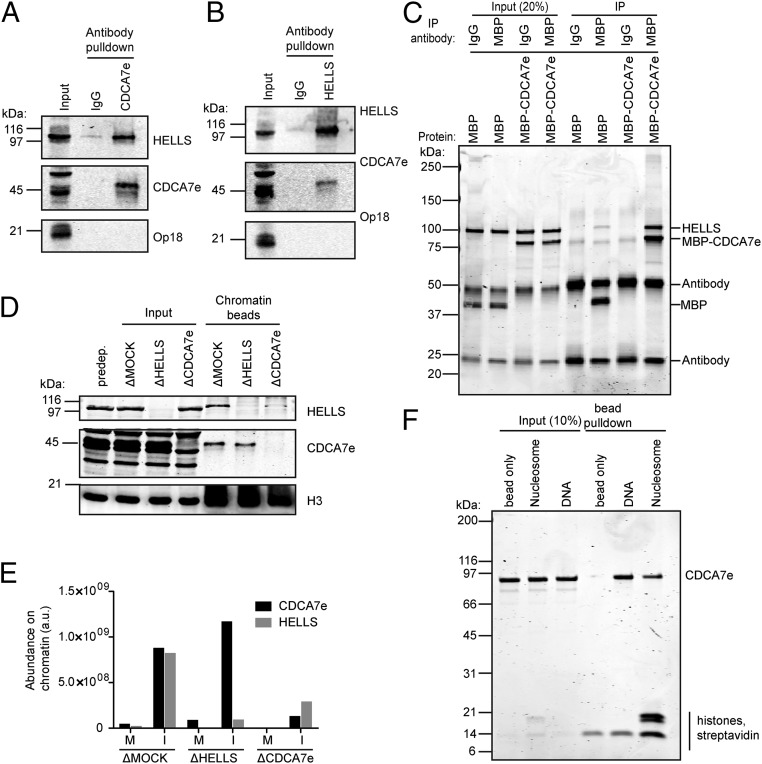
Fig. 3.
CDCA7e directly recruits HELLS to chromatin. (A and B) Western blot analysis of CDCA7e (A) and HELLS (B) immunoprecipitation from M phase Xenopus extracts. Preimmune rabbit IgG was used to control for nonspecific binding. Representative of n = 3 independent experiments. (C) Coomassie-stained gel of purified HELLS–CBP coimmunoprecipitation with MBP–CDCA7. Purified HELLS–CBP was incubated with MBP–CDCA7e or MBP alone and immunoisolation was performed using beads coupled with anti-MBP antibodies or control IgG. Representative of n = 2 independent experiments. (D) Western blot analyses of proteins copurified with nucleosome beads recovered from interphase extracts mock depleted or depleted of HELLS or CDCA7e. Representative of n = 4 independent experiments. (E) Abundance of HELLS and CDCA7e on nucleosome beads recovered from interphase or M phase extracts mock depleted or depleted of HELLS or CDCA7e, quantified by LC-MS/MS. (F) Coomassie-stained gel of a pulldown of nucleosome or DNA beads incubated with MBP–CDCA7e. Uncoupled beads were used to control for nonspecific binding. Representative of n = 2 independent experiments.
In Xenopus, many genes have an embryonic/egg specific paralog, such as histone H1 (53), dasra A (26), and survivin (54). In agreement with CDCA7e being the only detectable CDCA7 paralog in the egg (55), we only detected peptides for CDCA7e in our proteomics data. However, exogenously expressed somatic CDCA7 paralogs (CDCA7 and CDCA7L) also interacted with HELLS in egg extract (Fig. S3C), indicating that HELLS–CDCA7 interaction is conserved among different CDCA7 paralogs.
CDCA7e Is Required for Chromatin Loading of HELLS.
Presence of near identical amounts of HELLS and CDCA7e on chromatin, despite their unstable complex formation in egg extracts, can be explained by their dependency for chromatin binding. Indeed, while immunodepletion of HELLS from interphase extracts did not affect chromatin binding of CDCA7e, depleting CDCA7e greatly reduced HELLS recruitment to chromatin (Fig. 3D), indicating that CDCA7e is required for HELLS chromatin localization, but not vice versa. Although a majority of HELLS and CDCA7e dissociate from chromatin in M phase, MS-based analysis on chromatin beads depleted of either CDCA7 or HELLS indicated that binding of HELLS to chromatin also depends on CDCA7e in M phase (Fig. 3E). Additionally, purified MBP–CDCA7e can bind DNA, nucleosome arrays containing linker DNA, and mononucleosomes lacking linker DNA in vitro (Fig. 3F and Fig. S3D). Collectively, these data suggest that CDCA7e directly binds a nucleosome and recruits HELLS.
To test whether the CDCA7-dependent chromatin binding of HELLS is conserved in human somatic cells, CDCA7 was depleted from HeLa cells using siRNA. Quantitative Western blotting analysis showed that CDCA7 knockdown by RNAi reproducibly caused a decrease in chromatin-associated HELLS (Fig. S4 A and B), suggesting that CDCA7 indeed facilitates binding of HELLS to chromatin in somatic cells. In agreement with our data in Xenopus extracts, HELLS and CDCA7 chromatin binding was increased following treatment of mitotic cells with the Aurora B inhibitor, ZM447439 (Fig. S4C). Together with the stoichiometric association of CDCA7e and HELLS on chromatin that covaries across a variety of conditions (Fig. 2 B and F), its chromatin-independent interaction capacity (Fig. 3 A–C), and CDCA7e/CDCA7-dependent chromatin association of HELLS (Fig. 3 D and E and Fig. S4), we suggest that CDCA7e (and CDCA7) and HELLS form a complex on chromatin.
CDCA7e Stimulates Nucleosome Remodeling Activity of HELLS.
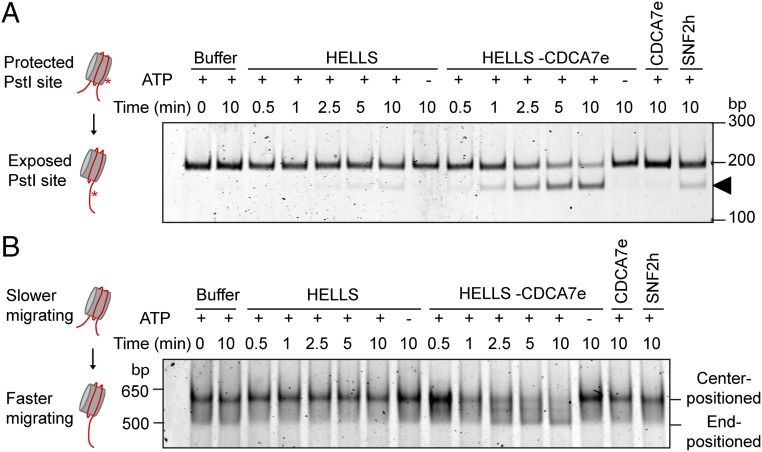
We next tested whether CDCA7e is required for HELLS remodeling activity. In agreement with a previous study (16), we saw little remodeling with purified HELLS–CBP in a restriction enzyme accessibility assay where productive remodeling exposes a restriction site within the nucleosome (Fig. 4A). However, adding MBP–CDCA7e to HELLS–CBP resulted in robust remodeling activity, while MBP–CDCA7e alone did not remodel the nucleosome. Using native gel electrophoresis, which can distinguish between end-positioned and center-positioned nucleosomes, we saw that coincubation of HELLS–CBP and MBP–CDCA7e shifts a center-positioned nucleosome to the end of the DNA fragment, while HELLS–CBP or MBP–CDCA7e alone did not mobilize nucleosomes (Fig. 4B). Free DNA was not generated during the course of the remodeling, indicating that HELLS–CDCA7e did not evict the nucleosome (Fig. S5A). Unlike SNF2h, HELLS–CDCA7e failed to generate center-positioned nucleosomes from end-positioned nucleosomes (Fig. S5B). However, HELLS–CDCA7e can still mobilize end-positioned nucleosomes in a restriction enzyme accessibility assay (Fig. S5C), suggesting that HELLS–CDCA7e does not permanently stall at DNA ends, but continuously mobilizes end-positioned nucleosomes. As previously reported (16), HELLS possesses intrinsic DNA and nucleosome-stimulated ATPase activity (16), which is enhanced by MBP–CDCA7e (Fig. S5 D and E), indicating that CDCA7 may couple HELLS ATPase activity to productive nucleosome sliding. Together, our data show that CDCA7 is required to activate HELLS remodeling activity and answer the longstanding question of whether HELLS is a bona fide nucleosome remodeler.
Fig. 4.
CDCA7e is an essential stimulator of HELLS nucleosome remodeling activity. (A) Restriction enzyme accessibility nucleosome remodeling assay with HELLS–CBP and MBP–CDCA7e. The 601-positioned mononucleosomes with a PstI site engineered 15 bp into the nucleosome with 20 bp flanking DNA on each end were incubated with the indicated remodeling proteins and PstI. Productive nucleosome sliding exposes the PstI site, resulting in cleaved DNA (arrow). DNA was resolved on a 10% polyacrylamide gel and visualized with SYBR Gold. Representative of n = 2 independent experiments. (B) Native gel nucleosome remodeling assay with HELLS–CBP and MBP–CDCA7e. Center-positioned mononucleosomes (same as in A) were incubated with the indicated remodeling proteins. Reactions were stopped, resolved on a 5% polyacrylamide gel, and visualized with SYBR Gold. Sliding results in end-positioned nucleosomes, which migrate faster. Representative of n = 2 independent experiments.
CDCA7 Patient ICF Mutations Fail to Recruit HELLS to Chromatin.
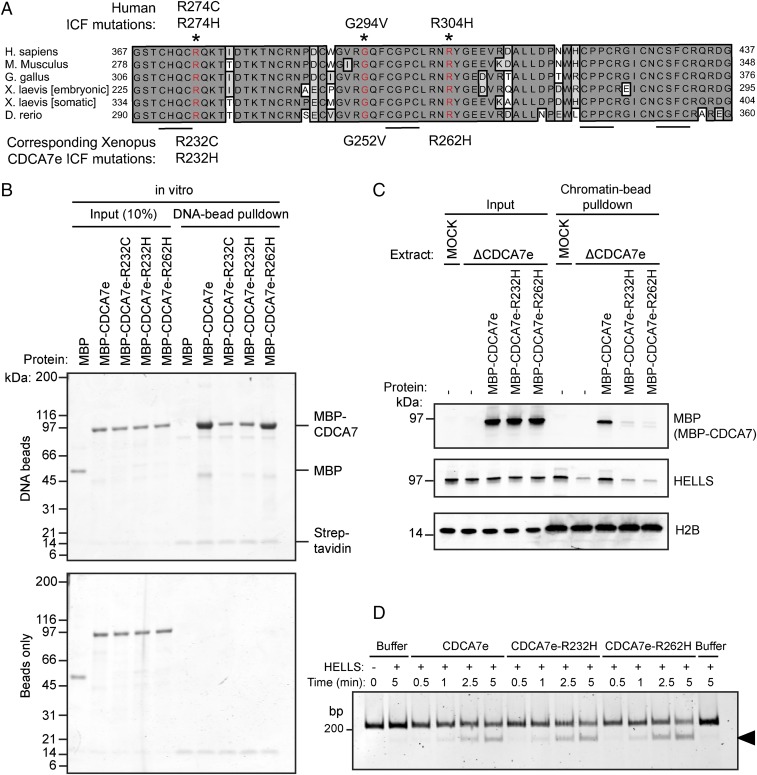
Thus, far, all identified CDCA7 mutations in patients with ICF map to the highly conserved zinc finger domain (Fig. 5A) (4). To determine the molecular defect of these CDCA7 mutations, we purified recombinant MBP–CDCA7e engineered with three corresponding patient mutations (R232C, R232H, or R262H). All three tested ICF mutants could interact with HELLS–GFP generated in reticulocyte lysate (Fig. S6A), although the possibility that R262H mutant has a slightly reduced affinity to HELLS cannot be ruled out (Fig. S6B). However, CDCA7e–R232C/H mutations displayed a strong DNA binding defect in vitro (Fig. 5B). Accordingly, while wild-type MBP–CDCA7e supported chromatin recruitment of HELLS in egg extracts depleted of endogenous CDCA7e, MBP–CDCA7e–R232C/H failed to associate with chromatin (Fig. 5C and Fig. S6C), and to recruit HELLS. Interestingly, CDCA7e–R262H maintained DNA-binding activity in vitro (Fig. 5B), but it failed to associate with chromatin in egg extract (Fig. 5C). Collectively, these data show that the primary defect of CDCA7 R232 and R262 ICF mutations is an inability to recruit the HELLS–CDCA7 complex to chromatin.
Fig. 5.
CDCA7 ICF mutants are defective in chromatin recruitment of the HELLS–CDCA7 complex. (A) Alignment of CDCA7 zinc finger domains from the indicated species. The 4CXXC motif is underlined, and residues with identified ICF mutations (*) are highlighted in red. (B) Coomassie-stained gel of a pulldown of DNA beads incubated with MBP–CDCA7e harboring the indicated ICF mutations. Uncoupled beads (Bottom) were used to control for nonspecific binding. Representative of n = 2 independent experiments. (C) Western blot analyses of proteins copurified with chromatin beads recovered from interphase extracts mock depleted or depleted of CDCA7e. Beads coated with 19 × 601 naked DNA were chromatinized in interphase extract for 90 min before addition of 1 μM recombinant MBP–CDCA7e harboring the indicated ICF mutations. Following an additional 60-min incubation, chromatin beads were recovered. Representative of n = 2 independent experiments. (D) Restriction enzyme accessibility nucleosome remodeling assay with HELLS–CBP and MBP–CDCA7e with the indicated ICF mutations. The 601-positioned mononucleosomes with a 34 and 15 bp flanking DNA on the 5′ and 3′ end, respectively, incubated with the indicated remodeling proteins and MspI endonuclease. Productive nucleosome sliding exposes an MspI site, resulting in cleaved DNA (arrow). DNA was resolved on a 10% polyacrylamide gel and visualized with SYBR Gold.
Because HELLS can bind DNA independently of CDCA7e in vitro (Fig. S3B), the DNA-binding activity of CDCA7e may not be required for nucleosome remodeling under purified conditions. We therefore wondered whether CDCA7e–R232C/H DNA-binding defective mutations could still activate HELLS remodeling activity. Strikingly, the CDCA7e–R232H ICF mutation was capable of activating HELLS remodeling activity under both saturating (Fig. 5D) and nonsaturating (Fig. S6 D and E) conditions, indicating that CDCA7e can stimulate HELLS remodeling independently of its DNA-binding activity. Additionally, the CDCA7e–R262H, which retains DNA-binding activity in vitro, can also stimulate remodeling activity (Fig. 5D and Fig. S6 D and E). These data suggest that in vivo CDCA7e serves two separable functions whereby CDCA7e is required to recruit HELLS to chromatin, and once there, CDCA7e activates HELLS remodeling activity.
Discussion
Comparative MS-based proteomic analysis on chromatin components in this study revealed a complex composed of HELLS and CDCA7e, which possesses nucleosome remodeling activity. Here, we name this complex the CDCA7–HELLS ICF-related nucleosome remodeling complex (CHIRRC).
HELLS Requires CDCA7 to Execute Nucleosome Remodeling.
Many well-studied nucleosome remodeling complexes contain a core ATPase subunit, which possesses intrinsic remodeling activity, such as BRM/BRG1, Mi-2, CHD1/2, CHD6–9, SNF2L/SNF2h, and ALC1 (9–15), though remodeling activity and directionality can be modulated by associated proteins. These core subunits contain an ATPase domain with flanking chromatin recognition domains such as the tandem chromodomains in CHD1, which couple its ATPase activity to productive nucleosome sliding. In this study, we identify a unique remodeling complex configuration where the nucleosome remodeling activity of the CHIRRC is split over two polypeptides, HELLS and CDCA7. The core ATPase subunit HELLS has no intrinsic remodeling activity, but requires CDCA7 to couple the ATPase activity to productive remodeling.
CDCA7 has two somatic paralogs, CDCA7 and CDCA7L, which likely activate HELLS in a similar manner. Somatic CDCA7 and CDCA7L contain the highly conserved zinc finger domain, which may facilitate their chromatin recruitment, while their divergent N termini may modulate their remodeling activity, or recruit HELLS to different genomic loci. By splitting the core remodeling protein into two polypeptides, the CHIRRC may be differentially regulated during development by CDCA7 paralogs. While DDM1, the Arabidopsis homolog of HELLS, can remodel nucleosomes without any additional protein in vitro (20), our observation that CDCA7 mutants defective in recruiting HELLS to chromatin can still support remodeling activity opens a possibility that DDM1 possesses CDCA7-like proteins that control chromatin targeting.
Our results indicate that there are two distinct mechanisms by which CDCA7 regulates HELLS. First, CDCA7 is required to recruit HELLS to chromatin. Second, CDCA7 stimulates the DNA-dependent ATPase activity and the nucleosome remodeling of HELLS independently of its capacity to recruit HELLS to chromatin (Fig. 6). Indeed, in vitro, all of the tested ICF mutations of CDCA7e that affect DNA or chromatin binding still retained remodeling activity. Although CDCA7 is critical for recruitment of HELLS to chromatin in physiological conditions, this functionality may be bypassed in the reconstituted system where HELLS can interact with DNA independently of CDCA7e (Fig. S3B). The mechanism by which HELLS interacts with DNA in isolation in a manner independently of CDCA7e is not clear, but it is likely that nonspecific DNA-binding activity of HELLS is suppressed by a chaperone or by other competing DNA-binding proteins. Whatever the mechanism, the fact that CDCA7e without chromatin-binding capacity stimulates remodeling activity strongly suggests that CDCA7e can activate HELLS by an allosteric mechanism in addition to its role in chromatin recruitment of HELLS.
Fig. 6.
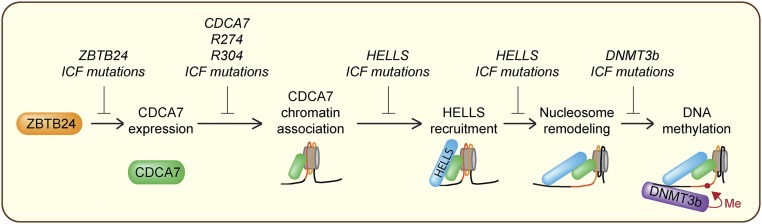
Functional mapping of proteins related to ICF syndrome. ZBTB24 stimulates transcription of CDCA7. CDCA7 associates with chromatin, recruits HELLS, and remodels nucleosomes to allow DNMT3b-mediated DNA methylation. CDCA7 ICF mutations inhibit chromatin binding and recruitment of HELLS to chromatin.
Interestingly, a majority of the CHIRCC dissociates from chromatin by Aurora B in M phase, reminiscent of the mitotic regulation of the ISWI remodeler (49). As the major histone targets of Aurora B (H3S10 and H3S28) were not responsible for their dissociation, uncovering the molecular determinants of this regulation will require further investigation. We speculate that eviction of these nucleosome remodeling proteins may modulate functionality of mitotic chromatin regulators, such as condensin, which preferentially interacts with nucleosome-free DNA (28).
CHIRCC, DNA Methylation, and ICF Syndrome.
ICF syndrome has been thought to be caused by defective de novo DNA methylation, since a majority of patients have mutations in DNMT3b and reduced DNA methylation (3). While the role of HELLS in DNA methylation through interaction with DNMT3b (17), and the importance of ZBTB24 in transcriptional induction of CDCA7 have been reported (6), how the ZBTB24–CDCA7 pathway contributes to the HELLS–DNMT3b DNA methylation previously remained unclear. Our data provide a mechanistic explanation of how all known ICF proteins converge on defects in DNA methylation (Fig. 6). DNMT3b has little activity on nucleosomes in vitro and in vivo, which require remodeling for complete de novo DNA methylation (56, 57). Therefore, we propose that the CHIRRC, a bona fide nucleosome remodeler, facilitates DNMT3b access to DNA. As HELLS deletion or ATPase mutations in mouse lead to reduced DNA methylation at repetitive elements (58), CDCA7 mutations in patients with ICF are likely defective in DNA methylation due to failure in chromatin recruitment and remodeling activity of HELLS.
Since the absence of HELLS causes ICF, but only hypomorphic CDCA7 mutants have been identified in CDCA7 (4), it is conceivable that CDCA7 possesses essential roles independent of HELLS regulation. It is also noteworthy that no DNMT3a/b was detected on chromatin in this study or in eggs (34), suggesting a role for the CHIRRC beyond DNA methylation control. However, in the context of ICF syndrome, DNMT3b, ZBTB24, CDCA7, and HELLS likely act in the same pathway. Further functional studies of the CHIRRC will help understand the molecular mechanisms behind the development of ICF phenotypes.
Methods
Xenopus Egg Extract Preparation and Immunodepletion.
The Rockefeller University Institutional Animal Care and Use Committee approved the protocol for the Xenopus laevis work, and all ethical regulations were followed. CSF-arrested X. laevis egg crude extracts were prepared as described (59). Extracts were kept on ice, and all extract experiments were performed at 20 °C unless specified. For experiments in interphase extract, CaCl2 was added to 0.3 mM. To immunodeplete HELLS, CDCA7e, or the CPC in 50-μL extract, 25 μg anti-HELLS, anti-CDCA7e, or anti-INCENP antibody was coupled to 100 μL Protein A Dynabeads (Thermo Fisher Scientific). INCENP antibody was crosslinked to the beads with BS3 (Thermo Fisher Scientific), following the manufacturer’s protocol. Antibody beads were washed extensively in sperm dilution buffer (5 mM Hepes, 100 mM KCl, 150 mM sucrose, 1 mM MgCl2, pH 8.0), split in half, and extract was depleted in two rounds at 4 °C, each for 45 min. Beads were removed on a magnet. Mock depletion was performed using purified preimmune rabbit IgG (Sigma-Aldrich). Histones H3–H4 were depleted as described (28). To deplete 50-μL extract, 130 μg anti-H4K12ac antibody was coupled to 12.5 μL rProtein A Sepharose (GE Healthcare). Extract was rotated with beads for 60 min at 4 °C and recovered. Depleted extract was incubated with 8.5 μL fresh rProtein A Sepharose to recover leached antibody for 35 min at 4 °C and recovered.
Protein Purification.
MBP–CDCA7–HIS was expressed in Escherichia coli at 16 °C overnight. Cells were centrifuged, resuspended in lysis buffer (1× PBS, 150 mM NaCl, 4 mM 2-mercaptoethanol, 20 mM imidazole, 0.1% Triton X-100, 100 μM ZnSO4, 1 mM PMSF, 0.25 mg/mL lysozyme, 5 μg/mL DNaseI, 10 μg/mL leupeptin, 10 g/mL pepstatin, and 10 μg/mL chymostatin, pH 7.8), and lysed by sonication. Lysate was centrifuged (Sorvall SS34 rotor, 14,000 rpm) for 30 min at 4 °C, and supernatant was incubated with 2 mL Ni-NTA agarose (Qiagen) for 1 h at 4 °C. Resin was washed three times with 20 mL wash buffer (1× PBS, 150 mM NaCl, 4 mM 2-mercaptoethanol, 100 μM ZnSO4, pH 7.8) supplemented with 20 mM imidazole, 10 mM ATP, and 2.5 mM MgCl2. Protein was eluted with wash buffer containing 400 mM imidazole. Eluate was incubated with 5 mL amylose resin [New England BioLabs (NEB)] for 1 h at 4 °C. Amylose resin was washed three times with 20 mL wash buffer, and MBP–CDCA7e was eluted with wash containing 20 mM maltose. Purifed MBP–CDCA7e was dialyzed in buffer (1× PBS, 150 mM NaCl, 4 mM 2-mercaptoethanol, 100 μM ZnSO4, pH 7.8) and concentrated via spin column (Amicon).
To purify HELLS–CBP, Saccharomyces cerevisiae containing pRS306G (60), containing HELLS–CBP were grown in YP raffinose at 30 °C to 1.3 × 107 cells per milliliter. Protein expression was induced by adding galactose (2% final) and incubating for 4 h. Cells were centrifuged, washed once with buffer A (25 mM Hepes, 1 M sorbitol, pH 7.6], and once with buffer B [45 mM Hepes, 0.02% Triton X-100, 10% glycerol, 5 mM Mg (OAc)2, 0.1 M K-glutamate, pH 7.6). Cells were resuspended in 0.5 volumes buffer B supplemented with 1 mM DTT, 10 μg/mL leupeptin, 10 μg/mL pepstatin, and 10 μg/mL chymostatin. Cells were frozen dropwise in liquid N2 and ground in a cryogenic grinding mill (SPEX). Cell powder was resuspended in buffer C (10 mM Hepes, 300 mM KCl, 10 mM 2-mercaptoethanol, pH 8.0) containing 2 mM CaCl2, and centrifuged (Sorvall SS34 rotor, 19,000 rpm) for 30 min at 4 °C. Supernatant was incubated with 5 mL calmodulin resin for 1 h at 4 °C. Resin was washed five times with 20 mL buffer C containing 2 mM CaCl2, and protein was eluted with buffer C, supplemented with 10 mM EDTA. Eluate was loaded on a heparin column (GE Healthcare), washed with buffer C, and eluted with buffer C containing 500 mM KCl. Purified HELLS–CBP was concentrated by covering HELLS-containing dialysis membrane with sucrose for 1 h at 4 °C.
Histone Purification.
Histones were purified as described (28). H2B, H2A, H3.2, and H4 were individually expressed in E. coli for 4 h at 37 °C. Cells were centrifuged, washed once with lysis buffer (50 mM Tris-Cl, 100 mM NaCl, 10 mM imidazole, 10 mM 2-mercaptoethanol, 0.25 mg/mL lysozyme, 5 μg/mL DNaseI, 10 μg/mL leupeptin, 10 μg/mL pepstatin, and 10 μg/mL chymostatin, pH 8.0), and resuspended in 30 mL lysis buffer. Cells were lysed by sonication and centrifuged (Sorvall SS34 rotor, 20,000 rpm) for 15 min at 4 °C. Inclusion bodies were rinsed with wash buffer (50 mM Tris-Cl, 100 mM NaCl, 10 mM imidazole, 10 mM 2-mercaptoethanol, pH 8.0) and resuspended in 15 mL wash buffer with 1% Triton X-100. Inclusion bodies were centrifuged (Sorvall SS34 rotor, 13,000 rpm), washed twice with wash buffer, and centrifuged again. Pellets were resuspended in 1 mL DMSO, diluted in 25 mL D500 buffer (6 M guanidine HCl, 500 mM NaCl, 50 mM Tric-Cl, 5 mM 2-mercaptoethanol, 7.5 mM imidazole, pH 8.0), and rotated overnight at room temperature. Mixture was centrifuged (Sorvall SS34 rotor, 30,000 rpm) for 20 min at 4 °C. H2B, H3.2, and H4 were further purified by incubation with 1.5 mL Ni-NTA agarose resin for 90 min at room temperature. Resin was washed five times with 20 mL D500 and three times with 20 mL D1000 (6 M guanidine HCl, 1 M NaCl, 50 mM Tric-Cl, 5 mM 2-mercaptoethanol, 7.5 mM imidazole, pH 8.0). Proteins were eluted with 5 mL elution buffer (6 M guanidine HCl, 1 M NaCl, 50 mM Tric-Cl, 5 mM 2-mercaptoethanol, 300 mM imidazole, pH 7.5). H3 harboring K9 trimethylation was generated by protein semisynthesis as described (61).
H3–H4 tetramers and H2A–H2B dimers were refolded as described (28). Each histone (45 μM) was added to 4.5 mL final volume D500. Histones were dialyzed stepwise in dialysis buffer (20 mM Mops, 500 mM EDTA, 1 mM EDTA, 4 mM DTT, pH 7.0) containing 10%, 5%, and 2.5% glycerol each for 4 h. Refolded histones were centrifuged, and the supernatant was incubated with 0.02 mg/mL TEV protease overnight at 16 °C. Refolded histones were purified by size exclusion chromatography (GE Healthcare HiLoad 16/60 Superdex 75).
Nucleosome Purification.
The 19 × 601 nucleosome arrays and mononucleosomes were prepared as described (28). Nineteen 601-positioning sequences separated by 53-bp linking DNA were digested from pAS696 with HaeII, DraI, EcoRI, and XbaI. The array was purified by PEG precipitation, and both ends were biotinylated by Klenow (NEB) fill-in with biotin-16-dUTP (ChemCyte). The 19 × 601 arrays were purified from free nucleotides by Illustra Nick columns (GE Healthcare). The 19 × 601 nucleosome arrays and mononucleosomes were prepared by gradient salt dialysis of H3–H4 tetramers, H2A–H2B dimers onto the DNA. The quality of the array was verified by AvaI digest, which cuts between 601 monomers, resulting in pure mononucleosomes when resolved on a native polyacrylamide gel. Nucleosome arrays (900 ng) were coupled to 3 μL M-280 streptavidin Dynabeads (Thermo Fisher Scientific) in bead coupling buffer (2.5% polyvinyl alcohol, 150 mM NaCl, 50 mM Tris-Cl, 0.25 mM EDTA, 0.05% Triton X-100, pH 8.0) at room temperature for 3 h. When coupling naked 19 × 601 DNA to Dynabeads, bead-coupling buffer contained 1.5 M NaCl. To prepare mononucleosomes lacking linker DNA, single-stranded 5′-biotinylated oligos of the 601 positioning sequence containing 5 bp of 3′-flanking DNA were ordered (IDT Technologies) and assembled into nucleosomes by salt dialysis as previously described.
Mass Spectrometry and Western Blots of Chromatin-Associated Proteins.
Analysis of chromatin-associated proteins was carried out as described (28). Nucleosome beads (0.15 μL/μL extract) were incubated in Xenopus egg extract for 2 h at 16 °C for MS experiments or 80 min at 20 °C for all other experiments, with flicking every 20 min. The extract was diluted with 10 volumes CSF-XB (10 mM Hepes, 100 mM KCl, 1 mM MgCl2, 50 mM sucrose, 5 mM EGTA, pH 8) and recovered on a magnet for 5 min at 4 °C. Beads were washed and recovered three times with 150 μL CSF-XB with 0.05% Triton X-100. Beads were resuspended in SDS/PAGE buffer and boiled. The beads were collected on a magnet and the supernatant resolved by gel electrophoresis. For mass spectrometry experiments, samples were run 1 cm into the gel.
Mass Spectrometry Analysis.
For MS analysis, standard trypsin digestion was performed followed by LC-MS/MS on a Finnigan Orbitrap XL (Thermo Fisher Scientific) mass spectrometer. MS/MS data were extracted with Proteome Discoverer (Thermo Fisher Scientific) and queried against an mRNA-derived X. laevis reference database (34) with Mascot (Matrix Science). This database was crucial, since the previously used National Center for Biotechnology Information (NCBI) database did not contain embryonic CDCA7e at the time. Mass tolerance of 20-ppm peptide precursor and 0.5-Da peptide fragments were used. Oxidized methionine, N-terminal acetylation, and up to three missed cleavage sites were allowed. Proteome Discoverer (Thermo Fisher Scientific) was used to quantify protein abundance on chromatin. LC-MS peaks of each identified peptide were integrated, and isotope peaks for each peptide were summed to give the total peptide area. Protein abundance was calculated by averaging of the three greatest peptide signals for each protein. Experimental reproducibility of this procedure has been reported (28).
Unbiased clustering was performed on the top 70th percentile of chromatin-associated proteins enriched over their extract concentration. Clustering was performed using the HOPACH algorithm (62) using cosine angle as the distance metric.
Coimmunoprecipitations from Xenopus Egg Extracts.
For coimmunoprecipitation from Xenopus egg extracts, anti-HELLS and anti-CDCA7e antibodies (25 μg) were coupled to 100 μL Protein A Dynabeads for 1 h at room temperature. Antibodies were crosslinked to the beads with BS3, following the manufacturer’s protocol. Antibody beads were washed extensively in sperm dilution buffer (5 mM Hepes, 100 mM KCl, 150 mM sucrose, 1 mM MgCl2, pH 8.0). A 50-μL extract was added to the beads and incubated on ice for 1 h with flicking every 20 min. The extract was diluted with 10 volumes CSF-XB (100 mM KCl, 1 mM MgCl2, 50 mM sucrose, 5 mM EGTA, and 10 mM Hepes, pH 8) and recovered on a magnet for 5 min. Beads were washed and recovered three times with 150 μL CSF-XB with 0.05% Triton X-100. Beads were resuspended in SDS/PAGE buffer and boiled. Control immunoprecipitation was performed using purified preimmune rabbit IgG (Sigma-Aldrich).
To test somatic CDCA7 and CDCA7L binding to HELLS, MYC–CDCA7, MYC–CDCA7L, and MYC–CDCA7e, mRNA were generated with an mMESSAGE mMACHINE SP6 kit (Life Technologies) according to the manufacturer’s instructions. mRNA was added to interphase Xenopus egg extract (100 μg/mL final) and incubated for 90 min before immunoprecipitation experiment, as described previously.
In Vitro Coimmunoprecipitations.
For in vitro coimmunoprecipitation experiments, 2.5 μg anti-MBP (NEB E8032) or preimmune mouse IgG (Sigma-Aldrich) were coupled to 10 μL Protein A Dynabeads for 1 h at room temperature. Beads were recovered on a magnet and washed extensively in sperm dilution buffer. Beads were resuspended in sperm dilution buffer with 0.05% Triton X-100 and 200 nM indicated proteins. Samples were agitated for 30 min at 20 °C. Beads were recovered on a magnet and washed three times with sperm dilution buffer with 0.05% Triton X-100. Beads were resuspended in SDS/PAGE buffer, resolved by gel electrophoresis, and stained with GelCode Blue (Thermo Fisher Scientific).
To test MBP–CDCA7e ICF mutant binding to HELLS–GFP, HELLS–GFP or GFP alone was expressed in the TnT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions. TnT reaction was diluted 1:5 in binding buffer (10 mM Hepes, 100 mM NaCl, 0.025% Triton X-100, 0.25 mM TCEP, pH 7.8), containing 100 nM CDCA7e, and incubated for 20 min at 20 °C. To each sample, 10 μL anti-MBP coated protein A beads were added, and the sample was incubated for 20 min at 20 °C. Beads were recovered, washed, and resolved by gel electrophoresis as before. Gel was stained with GelCode Blue, dried, and exposed on a PhosphorImager screen.
In Vitro DNA and Nucleosome Binding.
To assay CDCA7e chromatin binding in vitro, nucleosome arrays or mononucleosomes were coupled to M280 streptavidin beads (Thermo Fisher Scientific) as described (28). For naked DNA samples, histones were washed off in 2 M NaCl. For binding experiments, 3 μL 19 × 601 DNA or nucleosome beads were incubated in 50 μL binding buffer (20 mM Hepes, 200 mM NaCl, 0.05% Triton X-100, 0.5 mM TCEP, pH 7.8) containing 1 μM of MBP–CDCA7e for 30 min at 20 °C. Beads were collected on a magnet and washed three times with binding buffer. Beads were resuspended in SDS/PAGE buffer, boiled, and collected on a magnet. The supernatant was resolved by gel electrophoresis and stained with GelCode Blue. To assay HELLS DNA binding, 3 μL DNA beads were incubated in 20 μL sperm dilution buffer with 0.05% Triton X-100 and 1 μM protein for 45 min at room temperature. Beads were collected on a magnet and washed three times with sperm dilution buffer with 0.05% Triton X-100. Beads were resuspended in SDS/PAGE buffer, boiled, and collected on a magnet. The supernatant was resolved by gel electrophoresis and stained with GelCode Blue.
Nucleosome Remodeling and ATPase Assays.
To assay nucleosome remodeling by restriction enzyme accessibility, mononucleosomes were positioned on a 601 sequence with a PstI site engineered 15 bp into the nucleosome and 20 bp flanking DNA on each end (a gift from N. Gamarra and G. Narlikar, University of California, San Francisco). Nucleosomes (15 nM) were added to remodeling buffer [6.5 mM Hepes, 2 mM ATP-Mg, 5 mM MgCl2, 70 mM KCl, 0.02% Triton X-100, 3 units/μL PstI (NEB), pH 7.5] containing 100 nM HELLS, CDCA7e, or SNF2h (a gift from N. Gamarra and G. Narlikar). Reactions were incubated at 25 °C. At the indicated times, 5 μL remodeling reaction was added to 5 μL stop buffer (20 mM Tris, 70 mM EDTA, 2% SDS, 20% glycerol, 0.2 mg/mL bromophenol blue, 3.3 mg/mL proteinase K, pH 7.5) and incubated at 50 °C for 20 min. A total of 5 μL of each sample was resolved on a 10% polyacrylamide gel in 1× TBE at 150 V for 3 h and stained with SYBR Gold for 30 min. Where indicated, a similar procedure was performed using a mononucleosome on a 601 sequence containing 34 bp and 23 bp flanking DNA on the 5′ and 3′ ends, respectively, and MspI restriction enzyme (NEB). To assay mobility of end-positioned nucleosomes (Fig. S5C), mononucleosomes were positioned on a 601 sequence containing 60 bp 3′ flanking DNA, and the protocol was repeated using HaeIII in place of PstI.
To assay nucleosome remodeling by native gel, aforementioned center-positioned or end-positioned mononucleosomes (20 nM) were added to remodeling buffer (12 mM Hepes, 2 mM ATP-Mg, 3 mM MgCl2, 0.02% Triton X-100, 70 mM KCl, 11% glycerol, pH 7.5) containing 100 nM HELLS–CBP, MBP–CDCA7e or SNF2h (100 nM), and incubated at room temperature. At the indicated times, 5 μL remodeling reaction was added to 5 μL stop solution (0.7 mg/mL plasmid DNA, 30 mM ADP, 20% glycerol). A total of 5 μL of each sample was resolved on a 5% polyacrylamide gel in 0.5× TBE at 80 V for 3 h and stained with SYBR Gold.
For ATPase assays, 100 nM protein was added to 10 μL ATPase buffer (25 mM Hepes, 60 mM KCl, 4% glycerol, 4 mM MgCl2, 1 mM cold ATP, 0.1 μCi/μL γ-33P ATP, pH 7.6) containing 40 ng/μL DNA or nucleosomes and incubated at 16 °C. At the indicated times, 1-μL reaction was spotted on a PEI cellulose TLC plate and dried. Plates were separated in 1.2 M KH2PO4, pH 3.8. Plates were dried and exposed on a PhosphorImager screen.
Antibody Production and Plasmids.
For antibody production, C-terminal peptides of Xenopus H1M (CGAPVKAGKKGKKVTN), HELLS (CQGVFKVVDSTEVTVS), and CDCA7e (CLNSLRNTKDEDSDGS) were synthesized (The Rockefeller University Proteomics Resource Center). Peptides were coupled to keyhole limpet hemocyanin according to the manufacturer’s protocol (Thermo Fisher Scientific) and used for rabbit immunization (Cocalico Biologicals). Antibodies were affinity purified from serum using the same peptides coupled to SulfoLink resin (Thermo Fisher Scientific) according to the manufacturer’s protocol.
To generate H3S10ph antibodies (MAB Institute, Inc), a hybridoma clone expressing H3S10ph-specific antibody CMA314 (3-7C4), using a synthetic peptide ARTKQTAR(trimethyl-K) (phospho-S)TGGKAPRKQC was conjugated with keyhole limpet hemocyanin (63). The specificity was evaluated by ELISA using various histone H3 peptides (Fig. S7) (64). For ELISA, microtiter plates were coated with the peptides conjugated with BSA and incubated with threefold dilutions of hybridoma supernatant, starting from 1:100 dilution with PBS, and then with peroxidase-conjugated anti-mouse IgG. After incubating with tetramethylbenzidine, the absorbance of 405 nm was measured using a plate reader. CMA314 reacted with H3S10ph regardless of the modification state of the neighboring K9. To purify IgG, hybridoma cells were grown in CD Hybridoma medium (Thermo Fisher Scientific) and the culture supernatant was passed through a protein A column (1 mL HiTrap protein A HP Sepharose; GE Healthcare). After eluting IgG with glycin-HCl (pH 2.8), the buffer was replaced with PBS using an Amicon ultrafilter (50-kDa cutoff; Millipore) (63).
Codon optimized Xenopus HELLS–TEV–CBP (Thermo GeneArt Strings) was Gibson assembled into pRS306G (60) yeast expression plasmid containing PGK1 3′ UTR. Xenopus embryonic CDCA7e (GE EXL1051-205982802) was Gibson assembled into pMAL (NEB). CDCA7 (GenBank accession no. BC130191), CDCA7L (GenBank accession no. BC126014), and CDCA7e were Gibson assembled with an N-terminal 6× MYC tag into pCS2. HELLS (NCBI Reference Sequence accession no. NM_001092973.1) was Gibson assembled with a C-terminal GFP tag into pCS2.
Western Blotting.
Anti-Op18 rabbit antibody (a gift from R. Heald, University of California, Berkeley, CA) was used as described (4.7 μg/mL) (65, 66). Xenopus HELLS, CDCA7e, and H1 were detected with aforementioned rabbit antibodies (1 μg/mL). Anti-human CDCA7 (15249-1-AP) was from Proteintech. Anti-human HELLS (ab3851), anti-RPA32 (ab16855), anti-H3 (Ab1791, 1 μg/mL), and anti-H2B (Ab1790, 1 μg/mL) antibodies were from Abcam. In Fig. S4, anti-H3 (05-499) was from EDM Millipore. Anti–phospho-Aurora B (2914, 1:200) rabbit antibodies were from Cell Signaling. Anti-MCM7 rabbit antibody was a gift from J. Walter (Harvard Medical School, Cambridge, MA) (1:9,000) (67). Anti-INCENP (1 μg/mL) (26), anti-Survivin (12 μg/mL) (68), anti-Aurora B (5 μg/mL) (66), anti-Dasra (1 μg/mL) (26), and anti–XCAP-G (28) (1 μg/ul) rabbit antibodies were used as described. Anti-ISWI rabbit antibody was a gift from T. Hirano, RIKEN, Wako, Saitama, Japan (49). Anti-H4K12ac antibody was a gift from H. Kimura (69). Anti-H3T3ph was from Millipore (07-424). H3S10ph was detected with a mouse monoclonal antibody (CMA314, 1 μg/mL), which allows H3K9 mono, di-, and trimethylation and H3K9 acetylation in Fig. 2, and with a mouse monoclonal antibody (7G1G7), which allows H3K9 mono- and dimethylation, and is excluded by H3K9 trimethylation and acetylation in Fig. S4 (63).
Primary antibodies were detected with LI-COR IRDye secondary antibodies and subsequently imaged and quantified on an Odyssey Infrared Imaging System.
Protein Alignment.
Protein alignment was performed with MacVector. Proteins were aligned using ClustalW with an open gap penalty of 10 and an extended gap penalty of 0.2.
Cell Cultures.
HeLa cells were maintained at 37 °C in a 5% CO2 atmosphere with 21% oxygen and grown in Dulbecco-modified MEM (DMEM) supplemented with 10% FBS (Atlanta Biologicals) and 100 units/mL penicillin–streptomycin (Life Technologies). Cells were treated with 40 ng/mL nocodazole alone or in combination with 2 μM ZM447439 for 20 h followed by collection by shakeoff.
RNAi.
On-TARGETplus RNAi Smartpool from Dharmacon was used for the down-regulation of CDCA7 or nontargerting siCONTROL (D001810). Cells were transfected with the RNAi using Lipofectamine/RNAiMAX (Thermo Fisher Scientific) and harvested after 72 h posttransfection.
Chromatin and Whole Cell Extracts.
Chromatin extracts were performed as previously described (70). Briefly, cells were lysed into chromatin buffer (150 mM NaCl, 50 mM Tris pH 8.0, 5 mM EDTA, 0.5% Nonidet P-40) supplemented with protease inhibitors (Roche), phosphatase, Protein Phosphatase Inhibitor 2 (NEB), and trichostatin A HDAC inhibitor (Sigma). For the whole cell extract, cells were lysed in 2× Laemmli buffer (71). Samples were sonicated at 4 °C using a Bioruptor for 45 min. Concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and samples were prepared for Western blotting.
Supplementary Material
Acknowledgments
We thank N. Gamarra, R. Heald, T. Hirano, L. Langston (The Rockefeller University, New York, NY), G. Narlikar, M. O’Donnell (The Rockefeller University), D. Remus (Memorial Sloan Kettering Cancer Center, New York, NY), and J. Walter for reagents; G. Narlikar and N. Gamarra for advice on nucleosome remodeling assays; H. Molina and J. Fernandez (Proteomics Resource Center for MS Analysis, The Rockefeller University) and C. Zierhut for helpful discussions; S. Ahmed, P. Choppakatla, and C. Zierhut for critical reading of the manuscript; and Naohito Nozaki (MAB Institute, Inc) for generating hybridoma cells. This work was supported by Grant R01GM075249 from the National Institutes of Health (to H.F.), Grants R37GM086868 and R01 GM107047 (to T.W.M.), a postdoctoral fellowship from the Swiss National Science Foundation (to M.M.M.), and Grants JP25116005 and JP26291071 from the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (KAKENHI) (to H.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717509115/-/DCSupplemental.
References
- 1.Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) Orphanet J Rare Dis. 2006;1:2. doi: 10.1186/1750-1172-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagleitner MM, et al. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome) J Med Genet. 2008;45:93–99. doi: 10.1136/jmg.2007.053397. [DOI] [PubMed] [Google Scholar]
- 3.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 4.Thijssen PE, et al. Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat Commun. 2015;6:7870. doi: 10.1038/ncomms8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Greef JC, et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet. 2011;88:796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, et al. Converging disease genes in ICF syndrome: ZBTB24 controls expression of CDCA7 in mammals. Hum Mol Genet. 2016;25:4041–4051. doi: 10.1093/hmg/ddw243. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oppikofer M, et al. Expansion of the ISWI chromatin remodeler family with new active complexes. EMBO Rep. 2017;18:1697–1706. doi: 10.15252/embr.201744011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang HB, Zhang Y. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 2001;29:2517–2521. doi: 10.1093/nar/29.12.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 13.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmeister H, et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017;45:10534–10554. doi: 10.1093/nar/gkx711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burrage J, et al. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci. 2012;125:5524–5534. doi: 10.1242/jcs.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J, et al. The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences. Nucleic Acids Res. 2015;43:1444–1455. doi: 10.1093/nar/gku1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Termanis A, et al. The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells. Nucleic Acids Res. 2016;44:7592–7604. doi: 10.1093/nar/gkw424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 21.Prescott JE, et al. A novel c-Myc-responsive gene, JPO1, participates in neoplastic transformation. J Biol Chem. 2001;276:48276–48284. doi: 10.1074/jbc.M107357200. [DOI] [PubMed] [Google Scholar]
- 22.Osthus RC, et al. The Myc target gene JPO1/CDCA7 is frequently overexpressed in human tumors and has limited transforming activity in vivo. Cancer Res. 2005;65:5620–5627. doi: 10.1158/0008-5472.CAN-05-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto Y, et al. JPO1/CDCA7, a novel transcription factor E2F1-induced protein, possesses intrinsic transcriptional regulator activity. Biochim Biophys Acta. 2006;1759:60–68. doi: 10.1016/j.bbaexp.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Gill RM, Gabor TV, Couzens AL, Scheid MP. The MYC-associated protein CDCA7 is phosphorylated by AKT to regulate MYC-dependent apoptosis and transformation. Mol Cell Biol. 2013;33:498–513. doi: 10.1128/MCB.00276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampath SC, et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 28.Zierhut C, Jenness C, Kimura H, Funabiki H. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat Struct Mol Biol. 2014;21:617–625. doi: 10.1038/nsmb.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 30.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 31.Eustermann S, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 32.Noh KM, et al. ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc Natl Acad Sci USA. 2015;112:6820–6827. doi: 10.1073/pnas.1411258112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan C, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wühr M, et al. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–321. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haahr P, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol. 2016;18:1196–1207. doi: 10.1038/ncb3422. [DOI] [PubMed] [Google Scholar]
- 37.Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2013;12:587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009;284:35951–35961. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciccia A, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guler GD, et al. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J Biol Chem. 2012;287:6469–6481. doi: 10.1074/jbc.M111.324582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 45.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen RT, Kruse T, Nilsson J, Oestergaard VH, Lisby M. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J Cell Biol. 2015;210:565–582. doi: 10.1083/jcb.201502107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacCallum DE, Losada A, Kobayashi R, Hirano T. ISWI remodeling complexes in Xenopus egg extracts: Identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nightingale KP, Pruss D, Wolffe AP. A single high affinity binding site for histone H1 in a nucleosome containing the Xenopus borealis 5 S ribosomal RNA gene. J Biol Chem. 1996;271:7090–7094. doi: 10.1074/jbc.271.12.7090. [DOI] [PubMed] [Google Scholar]
- 51.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith RC, Dworkin-Rastl E, Dworkin MB. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988;2:1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- 54.Niedzialkowska E, et al. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell. 2012;23:1457–1466. doi: 10.1091/mbc.E11-11-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peshkin L, et al. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felle M, et al. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 2011;39:6956–6969. doi: 10.1093/nar/gkr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muegge K. Lsh, a guardian of heterochromatin at repeat elements. Biochem Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- 59.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 60.Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. Chromatin constrains the initiation and elongation of DNA replication. Mol Cell. 2017;65:131–141. doi: 10.1016/j.molcel.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Müller MM, Fierz B, Bittova L, Liszczak G, Muir TW. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat Chem Biol. 2016;12:188–193. doi: 10.1038/nchembio.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J Stat Plan Inference. 2003;117:275–303. [Google Scholar]
- 63.Hayashi-Takanaka Y, Yamagata K, Nozaki N, Kimura H. Visualizing histone modifications in living cells: Spatiotemporal dynamics of H3 phosphorylation during interphase. J Cell Biol. 2009;187:781–790. doi: 10.1083/jcb.200904137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 65.Budde PP, Kumagai A, Dunphy WG, Heald R. Regulation of Op18 during spindle assembly in Xenopus egg extracts. J Cell Biol. 2001;153:149–158. doi: 10.1083/jcb.153.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelly AE, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walter J, Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 68.Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayashi-Takanaka Y, et al. Distribution of histone H4 modifications as revealed by a panel of specific monoclonal antibodies. Chromosome Res. 2015;23:753–766. doi: 10.1007/s10577-015-9486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giunta S, Funabiki H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc Natl Acad Sci USA. 2017;114:1928–1933. doi: 10.1073/pnas.1615133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.