Significance
Neurons and nonneuronal cells in the developing brain dynamically regulate gene expression as neural connectivity is established. However, the specific gene programs activated in distinct cell populations during the assembly and refinement of many intact neuronal circuits have not been thoroughly characterized. In this study, we take advantage of recent advances in transcriptomic profiling techniques to characterize gene expression in the postnatal developing lateral geniculate nucleus (LGN) at single-cell resolution. Our data reveal that genes involved in brain development are dynamically regulated in all major cell types of the LGN, suggesting that the establishment of neural connectivity depends upon functional collaboration between multiple neuronal and nonneuronal cell types in this brain region.
Keywords: lateral geniculate nucleus, LGN, retinogeniculate, transcriptomics, refinement
Abstract
Coordinated changes in gene expression underlie the early patterning and cell-type specification of the central nervous system. However, much less is known about how such changes contribute to later stages of circuit assembly and refinement. In this study, we employ single-cell RNA sequencing to develop a detailed, whole-transcriptome resource of gene expression across four time points in the developing dorsal lateral geniculate nucleus (LGN), a visual structure in the brain that undergoes a well-characterized program of postnatal circuit development. This approach identifies markers defining the major LGN cell types, including excitatory relay neurons, oligodendrocytes, astrocytes, microglia, and endothelial cells. Most cell types exhibit significant transcriptional changes across development, dynamically expressing genes involved in distinct processes including retinotopic mapping, synaptogenesis, myelination, and synaptic refinement. Our data suggest that genes associated with synapse and circuit development are expressed in a larger proportion of nonneuronal cell types than previously appreciated. Furthermore, we used this single-cell expression atlas to identify the Prkcd-Cre mouse line as a tool for selective manipulation of relay neurons during a late stage of sensory-driven synaptic refinement. This transcriptomic resource provides a cellular map of gene expression across several cell types of the LGN, and offers insight into the molecular mechanisms of circuit development in the postnatal brain.
The transcriptome of the developing brain is highly dynamic. Early embryonic stages of neural tissue induction, brain region specification, and neuronal differentiation are all regulated, in part, by the expression of specific transcription factors that coordinate unique gene programs within each cell type and tissue (1–3). However, the cellular heterogeneity and anatomical complexity of the developing brain continue to present a major challenge to the study of how transcriptional changes contribute to the assembly and refinement of neural circuits. Recent advances in single-cell RNA sequencing have enabled the profiling of gene expression patterns across thousands of individual cells in the mature brain, revealing a remarkable degree of previously underappreciated taxonomic diversity (4–11). However, the contribution of coordinated, cell type-specific gene expression programs to postnatal brain development remains poorly understood.
The establishment and refinement of neural connectivity are fundamental aspects of neurodevelopment. Circuit assembly is initially specified by a molecular code of complementary guidance and adhesion molecules. During this multistage process, potential synaptic partners make direct contact through molecular interactions between axon guidance cues, such as Ephrins and Eph receptors (12–14). Synapses are then specified and further differentiated through physical contact between synaptogenic adhesion molecules, such as Neurexins and Neuroligins, that are embedded in pre- and postsynaptic membranes and bind across the cleft to organize synaptic architecture and composition (15–17). The process of synaptogenesis is accompanied by myelination, which accrues as the circuit matures, ensuring the efficiency of action potential propagation within both local and long-range circuits (18, 19). Finally, synapses are strengthened or eliminated based upon a process of activity-dependent synaptic refinement that is in part mediated through the tagging of synapses with complement proteins such as C1q and C3 (20–22). Thus far, the molecular mechanisms of circuit assembly and refinement have primarily been examined on a gene-by-gene basis and have for the most part not yet been studied at a genome-wide level. In addition, how nonneuronal cell types in the developing brain contribute to the development and plasticity of synapses requires further investigation.
The temporal, spatial, and cell-type specificities of the molecular code that defines synapse formation and refinement are dependent upon the active regulation of gene transcription (22). To date, two types of experiments have primarily contributed to our understanding of the transcriptional dynamics underlying postnatal brain development: whole-tissue sequencing, and sequencing of specific cell populations isolated through the use of mouse genetics or by mechanical techniques such as FACS and immunopanning. The application of whole-tissue sequencing to heterogeneous brain regions such as the prefrontal cortex has shown that the transcriptome of the embryonic brain is highly dynamic and becomes more stable as the brain matures (23–26). However, this approach has been technically limited by an inability to disentangle cell type-specific gene expression from population-level changes. Furthermore, most studies have focused on transcriptional changes between embryonic development and adulthood, with few studies examining the critical first few weeks of postnatal brain development during which neuronal connectivity is established.
On the other hand, strategies of transcriptional analysis following the mechanical isolation of tissues are limited to investigator-selected cell types and may not allow for the profiling of a large number of cell types within a given sample, which can be useful for providing biological context and for comparing levels of a gene of interest across multiple cell types (27–29). Due to technical limitations, these studies have not fully addressed the transcriptional heterogeneity within distinct cell types during development, nor have they completely characterized transcriptional dynamics in nonneuronal cells.
To address these gaps in knowledge, we took advantage of recent advances in droplet-based single-cell transcriptomics to profile gene expression in the lateral geniculate nucleus (LGN) of the thalamus across postnatal development (30–32). We chose the LGN for single-cell RNA sequencing (scRNA-seq) because the mouse LGN undergoes a well-characterized process of circuit assembly, remodeling, and refinement during the first month of postnatal life (33, 34). We analyzed the transcriptomes of over 35,000 cells across four time points: postnatal days 5, 10, 16, and 21. These time points were selected because they coincide with distinct but somewhat overlapping developmental events that occur across this period, including axonal targeting and retinotopic mapping, synaptogenesis, oligodendrocyte differentiation and myelination, and activity- and experience-dependent synaptic refinement (33). We identified and characterized changes in gene expression across these stages of postnatal development in excitatory relay neurons, oligodendrocytes, astrocytes, microglia, and endothelial cells. Remarkably, genes that were coregulated with similar temporal kinetics but in different cell types were found to share biological functions, consistent with precise, cell type-specific transcriptional control of subsets of functionally related genes during postnatal development. We used this dataset to predict candidate regulators of distinct aspects of retinogeniculate development, such as retinotopic mapping, and to identify and characterize the Prkcd-Cre transgenic mouse line as a tool for manipulating relay neurons specifically during late-stage postnatal refinement.
The resource presented here provides a detailed, transcriptome-wide map of gene expression within neurons, glia, and vascular cells at four time points during the postnatal development of the mouse LGN. These data reveal that genes involved in synapse and circuit development are highly expressed and dynamically regulated in both neurons and nonneuronal cells, providing further support for the conclusion that the assembly and refinement of neural connectivity during postnatal development depend upon collaboration between the majority of cell types in the LGN.
Results
Single-Cell Capture and Identification of Cell Types.
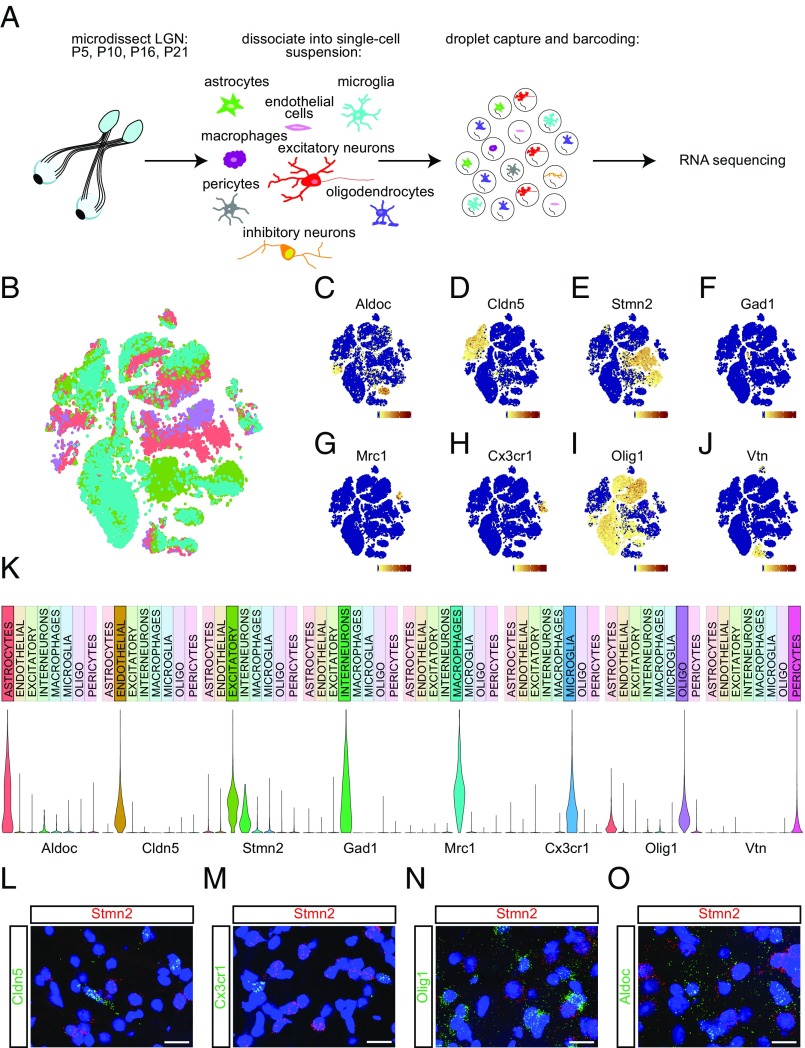
To map gene expression in the developing LGN, we prepared single-cell suspensions of microdissected dorsal LGN from mice at postnatal day (P)5, P10, P16, and P21. Dissociated single cells were captured and their mRNA was barcoded using a droplet-based strategy termed “inDrops” (30) (Fig. 1A). Following library preparation and whole-transcriptome sequencing, we performed a quality control (QC) assessment to remove cells that may represent doublets instead of individual cells. Specifically, we removed from the dataset cells that expressed aberrantly high or low levels of genes, or cells expressing a particularly high percentage of mitochondrial genes (SI Experimental Procedures). After removing these, we proceeded to analyze the remaining 35,326 cells that passed QC (7,499 cells from P5, 7,596 cells from P10, 13,091 cells from P16, and 7,140 cells from P21).
Fig. 1.
Capture, sequencing, and classification of cell types. (A) Schematic of the experimental design. Dorsal LGNs were microdissected from mice at four developmental ages: P5, P10, P16, or P21. Tissue was dissociated into a single-cell suspension, and then cells were captured and barcoded using the inDrops platform. Libraries were prepared and whole-transcriptome RNA sequencing was performed. (B) t-SNE plot of all cell types clustered by principal component analysis. Purple, P5; pink, P10; green, P16; teal, P21. (C) Expression pattern of the astrocyte marker Aldoc across cell clusters. (Scale, 0 to 150 transcripts per cell.) (D) Expression pattern of the endothelial cell marker Cldn5 across cell clusters. (Scale, 0 to 200 transcripts per cell.) (E) Expression pattern of the excitatory neuron marker Stmn2 across cell clusters. (Scale, 0 to 120 transcripts per cell.) (F) Expression pattern of the inhibitory neuron marker Gad1 across cell clusters. (Scale, 0 to 70 transcripts per cell.) (G) Expression pattern of the macrophage marker Mrc1 across cell clusters. (Scale, 0 to 225 transcripts per cell.) (H) Expression pattern of the microglial marker Cx3cr1 across cell clusters. (Scale, 0 to 120 transcripts per cell.) (I) Expression pattern of the oligodendrocyte marker Olig1 across cell clusters. (Scale, 0 to 320 transcripts per cell.) (J) Expression pattern of the pericyte marker Vtn across cell clusters. (Scale, 0 to 200 transcripts per cell.) (K) Violin plots displaying normalized expression levels of each cell type-specific marker across all cell types. (L–O) Confocal images of coronal LGN sections following FISH, probed for the excitatory neuron marker Stmn2 (red) and endothelial cell marker Cldn5 (green; L); microglial marker Cx3cr1 (green; M); oligodendrocyte marker Olig1 (green; N); and astrocyte marker Aldoc (green; O). (Scale bars, 10 μm.)
Using the Seurat software package for R, we next identified highly variable genes by calculating the average expression and distribution of each gene across all cells (31). Genes with high cell-to-cell variability were used to perform principal component analysis (PCA) and linear dimension reduction. We conducted a semisupervised clustering analysis that included an unsupervised approach followed by manual filtering. Given the multidimensional nature of the data, we used t-distributed stochastic neighbor embedding (t-SNE) to visualize cell clustering. Through analysis of all cells across all time points, we identified 29 cell clusters with distinct gene expression signatures with greater than 100 cells per cluster (Fig. 1B).
Based on the expression of genes encoding the neuronal markers Stmn2 and Snap25 (a cytoskeletal regulator and synaptic protein, respectively), the 29 cell clusters were divided into 8 neuronal (Stmn2/Snap25-positive) and 19 nonneuronal (Stmn2/Snap25-negative) clusters; 2 clusters were excluded due to the presence of two or more cell-type markers, suggesting that the cluster may not represent a single cell type. As expected from single-cell expression data from other brain regions, more cells were classified as nonneuronal than neuronal (10). This is a technical feature of the dissociation protocol and does not reflect the cellular composition of the dorsal LGN, which we found to be composed of roughly 65% excitatory neurons by in situ hybridization. The eight neuronal clusters were nearly uniformly glutamatergic, as evidenced by expression of Slc17a6 (vesicular glutamate transporter 2; VGLUT2). There were rare GABAergic neurons, identified by expression of Slc32a1 (vesicular inhibitory amino acid transporter; VIAAT) and Gad1. Nonneuronal clusters were grouped into oligodendrocytes, microglia, astrocytes, endothelial cells, pericytes, and macrophages using a variety of established marker genes including Olig1, Cx3cr1, Aqp4, Cldn5, Vtn, and Mrc1, respectively (Fig. 1 C–K and SI Experimental Procedures). We confirmed that nonoverlapping populations of cells in the LGN expressed these markers by performing single-molecule fluorescence in situ hybridization (FISH; Fig. 1 L–O). We then proceeded to analyze each cell population separately.
Analysis of Neuronal Diversity.
One advantage of single-cell RNA-seq over whole-tissue transcriptome analysis is the ability to perform de novo discovery of cellular subtypes with distinct transcriptional signatures (31). Because mice rely less heavily on vision than higher-order mammals and because their LGNs lack the typical multilayered structure common to other species, excitatory relay neuron diversity is thought to be less complex in the mouse LGN (35, 36). However, this has not been sufficiently investigated at the level of gene expression. We investigated the possibility that the scRNA-seq analysis of excitatory neurons at each developmental time point might reveal stage-specific subpopulations. Therefore, we separated the neuronal cells by developmental time point, identified genes whose expression varied significantly between excitatory neurons at each different time point, and then applied PCA and the cell clustering algorithm. We were able to identify distinct neuronal subpopulations with characteristic transcriptional signatures (Fig. S1A). We then used a differential gene expression approach to identify genes that represent these unique subclusters of neurons.
Interestingly, we found several genes expressed only in a subset of excitatory neurons, suggesting that small groups of excitatory relay neurons may execute unique genetic programs across development. These genes were identified through differential gene expression analysis between clusters using the software packages Seurat and Monocle 2 (37). As an example, at P5 and P10, our clustering analysis identified a subset of neurons strongly expressing insulin-growth factor 2 (Igf2; Fig. S1B), which we validated by FISH (Fig. S1C). Expression of Igf2 was correlated with that of insulin-growth factor-binding protein 7, suggesting that the same subpopulation of excitatory neurons expresses both of these related signaling proteins (Igfbp7; Fig. S1D; Pearson r = 0.18, P = 2.5 × 10−18 at P5; Pearson r = 0.15, P = 1.8 × 10−9 at P10). While other populations of cells also express Igf2, most prominently pericytes, the expression of Igf2 in only a small subset of excitatory neurons suggests that it may play an important role in excitatory neurons in a spatially restricted manner. Indeed, in other systems, Igf2 has been shown to promote synapse formation and maturation via NF-κB activation following binding to cell-surface Igf receptors (38–40). On the other hand, Igfbp7 is thought to attenuate these biological functions by binding to insulin-growth factor receptors extracellularly (41, 42). Our data show that endothelial cells are the highest expressers of the Igf2 receptor Igf1R in the LGN, consistent with roles for Igf signaling in blood–brain barrier permeability (43). This raises the possibility that subpopulations of Igf2-positive relay neurons may remodel distinct parts of the LGN vasculature as it develops.
Consistent with the possibility that Igf2 may regulate synaptic remodeling near its point of neuronal secretion, we found that the expression of Igf2 in individual excitatory neurons was also strongly correlated with the expression of several collagen isoforms (Col1a1, Col3a1, Col4a1, and Col18a1), important components of the extracellular matrix (ECM) (Pearson r = 0.31, 0.47, 0.33, and 0.26, respectively; P = 3.6 × 10−57, 1.2 × 10−143, 3.1 × 10−63, and 9.2 × 10−40, respectively) (44). Expression of these isoforms suggests that this subpopulation of neurons may contribute to the formation and maintenance of synapses by modulating the ECM. Furthermore, these ECM components expressed by Igf2-positive relay neurons may serve to restrict the spread of secreted Igf2 so that it acts more locally.
Consistent with modification of the ECM as a general developmental mechanism, we found that distinct subsets of relay neurons expressed the zinc-dependent metalloproteinase neprilysin at P5 and P10. Neprilysin is an enzyme that is released into the ECM and cleaves peptides including amyloid beta (45). Interestingly, previous work has shown that zinc is highly localized to the ipsilateral region of the immature LGN and may therefore influence axon targeting as the circuit matures (46). More broadly, the modulation of the ECM by subsets of relay neurons is consistent with evidence that remodeling of the ECM is an important feature of circuit development (47).
In higher-order mammals (including primates), the LGN is composed of layers of magnocellular and parvocellular cells, which have distinct functional and transcriptional features (48). The mouse LGN, by contrast, lacks this stereotypical layered structure and magnocellular/parvocellular division (49). Utilizing a previously published microarray dataset from the macaque, we probed the scRNA-seq data from our excitatory cell clusters to determine whether genes differentially enriched in primate parvocellular or magnocellular cells demonstrate differential expression in the mouse (50). Many of these genes, including Tcf7l2—a marker for parvocellular cells in primates—and Eef1a2—a marker for magnocellular cells in primates—demonstrated diffuse expression across relay neurons (Fig. S1 E and F). However, we observed that the magnocellular marker Ppp2r2c demonstrated particularly high expression in a subset of neurons, which was most prominent earlier in development (Fig. S1 G and H).
Collectively, these results suggest that mouse relay neurons may be more transcriptionally heterogeneous than previously appreciated. Additional work is necessary to define the biological significance of this heterogeneity, and to determine whether it represents the range of developmental maturity within a given sample or functionally distinct cell states.
Analysis of Nonneuronal Diversity.
Among nonneuronal cell populations, oligodendrocytes were the most abundant class profiled. We characterized progenitor, intermediate, and mature oligodendrocyte populations, as well as their corresponding gene expression profiles, at each developmental stage. The relative abundance of these populations varied across development (Fig. S2 A–D). At P5, we found that 81% of Olig1-positive cells expressed Pdgfra, a marker of immature oligodendrocyte progenitor cells (Fig. S2E). Pdgfra-positive oligodendrocytes were enriched for chondroitin sulfate proteoglycan 5 (Cspg5), a negative axonal guidance cue, as well as matrix metalloproteinase 15 (Mmp15), an enzyme that actively remodels the ECM. A subset of Pdgfra-positive cells expressed genes associated with cell-cycle regulation, including cyclin B1 and Top2a, a DNA topoisomerase (6, 51). This population likely represents the actively proliferating subset of oligodendrocyte progenitor cells.
Oligodendrocyte maturation progressed across development, as evidenced by a decrease in the proportion of cells expressing Pdgfra (Fig. S2 E–H) and an increase in the proportion of cells that expressed the mature oligodendrocyte markers myelin basic protein (Mbp; Fig. S2 I–L), myelin-associated glycoprotein (Mag), myelin oligodendrocyte glycoprotein (Mog), and/or tubulin polymerization protein (Tpp). Markers of late stages of myelination, including Mog and Ermin, a cytoskeletal protein involved in myelin wrapping and compaction, emerged as a subset of Mbp-positive cells at P10 (52, 53). The population of myelinating oligodendrocytes was enriched for the genes encoding proteolipid protein 1 (Plp1), a component of the myelin sheath, Sirt2, a positive regulator of oligodendroglial differentiation, and neurofascin (Nfasc), a cell-adhesion molecule necessary for the assembly of nodal and paranodal domains of myelinated axons (54–56).
Interestingly, we found that a small subset of Olig1-positive cells at each developmental stage was enriched for Bmp4, a member of the transforming growth factor superfamily that is a negative regulator of oligodendrogenesis (Fig. S2 M–P) (57). Bmp4 increases astroglial and decreases oligodendroglial lineage commitment (58). This suggests that we may have captured a population of Olig1-positive cells that are shifting toward astroglial commitment.
Our data suggest that the transcriptional program accompanying the developmental evolution of oligodendrocytes may contribute to the establishment of the extracellular milieu as retinal inputs are formed and remodeled. The transcriptional hallmarks of oligodendrocyte development in our data are consistent with other studies indicating that oligodendrocytes undergo a stereotyped process of differentiation across postnatal development resulting in the establishment of myelination as circuits remodel and stabilize. However, we find that a smaller subset of oligodendrocytes remains in an immature state, possibly serving as a reservoir in the mature brain for replacing mature oligodendrocytes that have become injured or have died.
Dynamic Regulation of Gene Expression Across Postnatal Development.
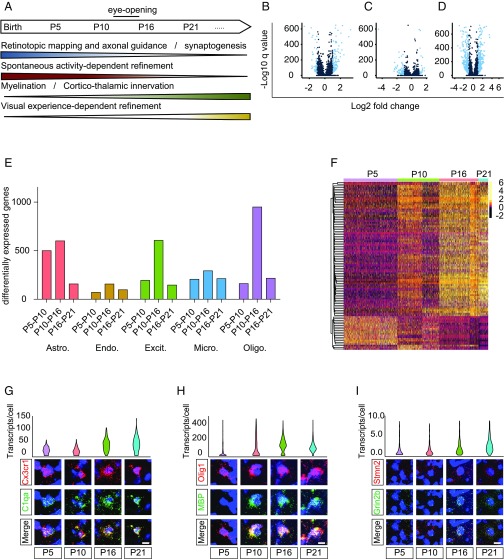
During brain development, cells can transition from one functional state to another in response to intrinsic gene programs and extrinsic factors such as neuronal activity, secreted neurotrophic proteins, and cell–cell interactions (59). Subsequently, gene transcription during cell-state changes drives the expression of unique protein repertoires that are critical for cell function (60). Such differentially expressed proteins may be important for distinct phases of development. In the LGN, postnatal development has been well-characterized and involves distinct but overlapping stages of circuit assembly and refinement (Fig. 2A). Early phases between P5 and P10 involve axonal targeting and a nuanced interplay of synaptogenesis and spontaneous activity-dependent synaptic pruning. Corticothalamic innervation emerges between P10 and P20, when oligodendrocyte differentiation and myelination are occurring at a high level. Subsequently, visual experience-dependent synaptic refinement largely occurs between P20 and P30.
Fig. 2.
Transcriptional dynamics across LGN development. (A) Timeline of postnatal LGN development, highlighting overarching developmental processes and their correlations with distinct phases. (B–D) Volcano plots indicating genes identified as developmentally regulated (light blue dots) in excitatory neurons based on an absolute log2-fold change >1 and false discovery rate <0.05 between P5 and P10 (B), P10 and P16 (C), and P16 and P21 (D). (E) Number of genes that change significantly (≥2-fold, FDR <0.05) across each temporal transition in astrocytes, endothelial cells, excitatory neurons, microglia, and oligodendrocytes (left to right). (F) Heatmap of gene transcript count (log-normalized to 10,000 transcripts per cell) for the top 100 genes that vary across development, clustered by expression pattern across development. Rows represent genes and columns represent excitatory cells, grouped by developmental time point. (G) Violin plot based on scRNA-seq data (Top) and confocal FISH images (Bottom) showing developmental regulation of C1qa (green) in microglia identified by expression of Cx3cr1 (red). Ages are listed below. (Scale bar, 5 μm.) (H) Violin plot based on scRNA-seq data (Top) and confocal FISH images (Bottom) showing developmental regulation of Mbp (green) in oligodendrocytes identified by expression of Olig1 (red). Ages are listed below. (Scale bar, 5 μm.) (I) Violin plot based on scRNA-seq data (Top) and confocal FISH images (Bottom) showing developmental regulation of Grin2b (green) in excitatory neurons identified by expression of Stmn2 (red). Ages are listed below. (Scale bar, 5 μm.)
We investigated the temporal features of transcription across early LGN development in each of the cell types, with the goal of identifying components of the transcriptional program underlying axonal targeting and synaptogenesis, synaptic refinement, and myelination. We leveraged the power of our single-cell expression resource to profile coordinated changes in gene expression within the five predominant cell types across postnatal development (excitatory relay neurons, oligodendrocytes, astrocytes, microglia, and endothelial cells). For each of the cell types, we performed a differential gene expression analysis using Monocle 2 to identify genes whose expression changed by ≥2-fold between two adjacent time points with a false discovery rate (FDR) <5% (37). This approach identified significant transcriptional changes in each cell type across development. For example, we found 199, 612, and 152 genes significantly changed between P5 and P10, P10 and P16, and P16 and P21 in excitatory neurons, respectively (Fig. 2 B–D). In addition to neurons, oligodendrocytes and astrocytes were very dynamic, differentially expressing a total of 1,148 and 1,142 genes, respectively. Microglia and endothelial cells also exhibited coordinated changes in gene expression, with microglia differentially expressing 648 genes and endothelial cells differentially expressing 308 genes between P5 and P21.
We found that different cell types exhibit diverse temporal patterns of transcriptional activity, likely correlating with unique functions of each cell type at different points of development (Fig. 2E). For example, the astrocyte transcriptome is highly dynamic between P5 and P10, during which time astrocytes robustly prune retinogeniculate synapses. Consistent with a relationship between transcription and function, we find that astrocytes up-regulate Mertk, a receptor that is required for astrocyte-driven synaptic pruning, between P5 and P10 (61). On the other hand, neurons were most transcriptionally dynamic between P10 and P16, two time points that flank the onset of visual experience at eye opening (Fig. 2 D and E). Consistently, we observe the up-regulation of visual activity-dependent genes such as the phosphatase Dusp1 and the chemokine Cx3cl1 at P16 in neurons. As a negative regulator of MAPK signaling, Dusp1 may serve to shut off this pathway following its activation at the onset of visual experience. Although Cx3cl1 has not been studied in the context of neural development, its enrichment in neurons and the fact that it is secreted are consistent with possible roles in the establishment of neuronal connectivity (62).
We augmented this temporal analysis with a weighted linear regression analysis to identify genes in excitatory neurons whose variation was most strongly associated with postnatal age. We then clustered the genes based on their temporal expression kinetics across the four developmental time points (Fig. 2F). This analysis affirms that modules of genes in excitatory neurons demonstrate distinct kinetics, and that the bulk of age-dependent genes demonstrate increased expression from P10 to P16.
Our data therefore show that each of the five cell types profiled exhibits coordinated changes in gene expression during postnatal development, and suggest that developmentally regulated genes are particularly important for cell type-specific functions across development.
Coregulated Genes Share Biological Functions.
To investigate whether genes regulated with similar temporal dynamics share biological functions, we performed unbiased gene ontology enrichment analyses of coregulated genes. Within each cell type, we found examples of enriched ontologies for genes with expected cell type-specific functions. For example, genes up-regulated in excitatory neurons at P16 included those involved in glutamatergic neurotransmission, such as the NR2B subunit of the NMDA receptor (Grin2b). On the other hand, microglia exhibited dynamic regulation of genes involved in chemokine and cytokine signaling, such as the complement protein C1qa, which facilitates synaptic pruning during the first postnatal week of retinogeniculate development. We confirmed by FISH the cell type-specific developmental regulation of these genes as well as other genes with previously described cell-type distributions, including the secreted growth factor Ndnf in endothelial cells, the synaptogenic molecule Hevin in astrocytes, and the myelin component Mbp in oligodendrocytes (Fig. 2 G and H and Fig. S3).
In addition to the observation that each cell type coregulates genes associated with known functions, we also observed several unexpected patterns that point toward previously unappreciated developmental roles for each cell type. One unexpected finding was the enrichment of genes involved in oligodendrocyte differentiation and myelination in cell types other than oligodendrocytes, predominantly astrocytes, microglia, and neurons. Although expressed most highly in oligodendrocytes, each of the other cell types exhibited developmental regulation of Plp1 and Mobp, two predominant components of the myelin sheath, at P16, a time at which myelination accelerates as retinogeniculate axons remodel and corticothalamic axons newly innervate the LGN (63, 64). Additionally, both astrocytes and microglia induce other candidate regulators of myelination around P16. For example, astrocytes up-regulate the expression of the gene encoding the secreted signaling molecule Fgf1, which has been implicated in a variety of functions including the regulation of myelination in vivo (65, 66). This result supports multicellular cooperation in the establishment of myelination, perhaps due to the need for a large amount of myelin as both retinogeniculate and corticothalamic axons remodel.
Perhaps still more unexpected is the finding that microglia may also regulate aspects of myelination in the LGN, in addition to their known roles in synaptic pruning. Although there is still relatively little functional evidence that microglia regulate myelination in vivo, supplementation of oligodendrocyte-neuron coculture media with microglia-conditioned media enhanced myelin protein synthesis (67, 68). Consistent with unexplored roles for microglia in oligodendrocyte differentiation and myelination, we found that one of the most highly up-regulated genes in microglia at P16 encodes the secreted enzyme Autotaxin, which converts lysophosphatidylcholine into the lipid signaling molecule lysophosphatidic acid (LPA) (69). LPA is critical for myelination in the CNS and functions through binding to LPA receptor 1 (LPAR1) in oligodendrocytes, a receptor that is required for myelination in the cerebral cortex (70, 71). Our analysis revealed that oligodendrocytes up-regulate LPAR1 at the same time that microglia up-regulate Autotaxin, indicating that this pathway might represent a novel mechanism whereby microglia can regulate myelination in the LGN.
Another aspect of retinogeniculate development that is supported by multicellular collaboration is synaptogenesis. Of particular interest are the synaptogenic protein Hevin (Sparcl1) and its antagonist, Sparc. Hevin and Sparc were identified as astrocyte-derived secreted signaling molecules that positively and negatively regulate synaptogenesis, respectively, in retinal ganglion cell (RGC) cultures and in the superior colliculus, another visual structure in the brain that receives input from RGCs (72–74). Roles for these molecules in synaptic development have been primarily attributed to their secretion by astrocytes, but our data indicate a more complex regulation of reciprocal Hevin/Sparc signaling in the LGN. Unlike in the superior colliculus, we found that microglia, neurons, and oligodendrocytes in the LGN, but not astrocytes, up-regulate Hevin early in postnatal development. In fact, we found that neuronal Hevin expression at P5 is restricted to a subpopulation of excitatory neurons, and that the proportion of neurons expressing Hevin increases at P10 and P16, with near-uniform expression at P21. Subsequently, we find that astrocytes join neurons and oligodendrocytes in up-regulating Sparc expression as late as P16, suggesting that multicellular up-regulation of this antisynaptogenic molecule may be required to provide a “brake” on Hevin-dependent synaptogenesis and/or a transition to synaptic refinement. Together, our data suggest that the integration of cell type-specific, dynamic gene expression in multiple populations underlies critical aspects of retinogeniculate development, including synaptogenesis and myelination.
Identification of Candidate Pathways Underlying Axonal Targeting and Synaptic Refinement.
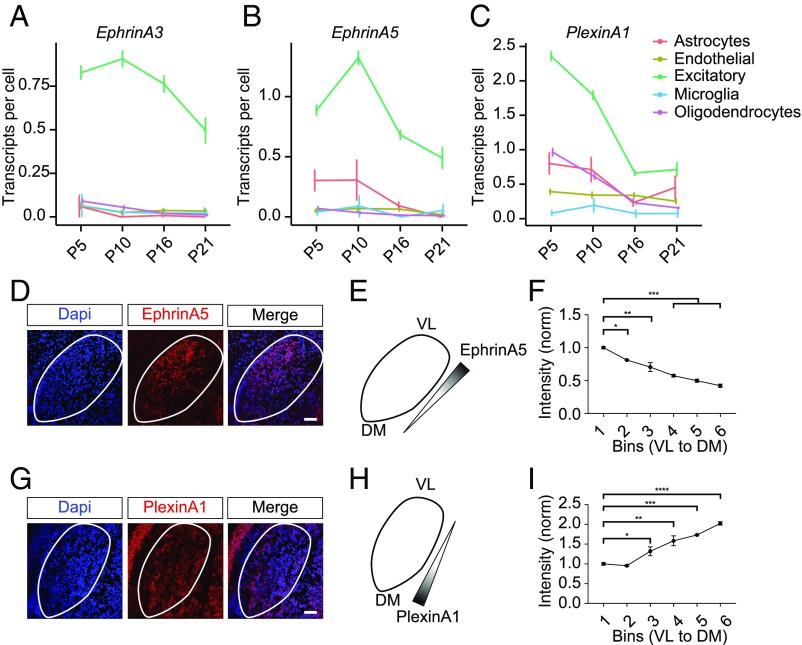
Ontological analysis of our scRNA-seq data revealed that genes regulated with similar temporal expression patterns share biological functions, suggesting that this database may be useful in identifying candidate regulators of retinogeniculate development. We tested this possibility by identifying genes whose expression patterns are highly similar to those of known regulators of distinct aspects of retinogeniculate development, first focusing on axonal targeting and retinotopic mapping, processes regulated by EphrinA/EphA signaling (75–77).
In the LGN of the mouse, the axon guidance cues EphrinA2, -A3, and -A5 are expressed according to a ventral–lateral–anterior>dorsal–medial–posterior spatial gradient, providing a molecular code that directs the appropriate targeting of retinal axons to distinct domains (75–77). However, despite some mapping deficits, major features of retinal axon targeting are still intact in mice lacking EphrinA2, -A3, and -A5, indicating that additional molecules must be involved in this process. Consistent with their known functions, our data indicate that EphrinA3 and -A5 are highly enriched in relay neurons during early postnatal development at P5 and P10, and then decrease as the circuit refines (Fig. 3 A and B). Further, we confirmed by FISH that EphrinA5 is expressed in a gradient pattern, as previously reported (Fig. 3 D–F).
Fig. 3.
Candidate regulators of retinotopic mapping in the LGN. (A) EphrinA3 expression in transcripts per cell across development in all cell types. Error bars, SEM. Color coding (Right). (B) EphrinA5 expression in transcripts per cell across development in all cell types. Error bars, SEM. (C) PlexinA1 expression in transcripts per cell across development in all cell types. Error bars, SEM. (D) Confocal FISH images of P10 LGN stained with DAPI and probed for EphrinA5 (red). (Scale bar, 100 μm.) (E) Schematic of LGN and EphrinA5 gradient decreasing from ventrolateral (VL) to dorsomedial (DM). (F) Quantification of EphrinA5 mRNA-normalized fluorescence intensity across the LGN from VL to DM, measured in six bins. (G) Confocal FISH images of P10 LGN stained with DAPI and probed for PlexinA1 (red). (Scale bar, 100 μm.) (H) Schematic of LGN and PlexinA1 gradient increasing from DM to VL. (I) Quantification of PlexinA1 mRNA-normalized fluorescence intensity across the LGN from VL to DM, measured in six bins. For F and I, statistical significance was determined by a one-way ANOVA followed by a post hoc Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars, SEM.
One family of potential regulators of axonal targeting and retinotopic mapping in the LGN are Plexins, which bind Semaphorins to mediate axonal guidance and repulsion during brain development but whose expression and functions in the LGN are not known (78, 79). Remarkably, we found that most Plexins (8 out of 11) are dynamically regulated across LGN development in astrocytes, neurons, or oligodendrocytes. A particularly appealing candidate regulator of mapping is PlexinA1, the expression pattern of which is highly similar to that of EphrinA3 and -A5 (Fig. 3C). We found that PlexinA1 is expressed in a dorsomedial-to-ventrolateral gradient pattern counter to that of EphrinA5, consistent with a complementary role in establishing the synaptic architecture of the LGN (Fig. 3 G–I).
To identify novel candidate regulators of retinotopic mapping, we performed a correlation analysis of gene expression in individual cells to identify genes whose expression patterns were significantly correlated with EphrinA3 and -A5 at P10, when the expression of these genes in relay neurons peaks. Intriguingly, we identified many genes whose expression is highly correlated with either EphrinA3 or -A5. Intersecting the two datasets revealed that both EphrinA3 and -A5 expression are highly correlated with expression of an ECM-interacting α-Integrin (Itga11) at P10 (Pearson r = 0.14, P = 2 × 10−7 for EphrinA3; Pearson r = 0.12, P = 2.8 × 10−4 for EphrinA5), an interesting finding in light of our observation that the ECM is regulated dynamically across LGN development (80, 81). This raises the possibility that Itga11 may stabilize Ephrin-initiated axon–dendrite interactions by creating adhesions with the ECM. Another gene that was correlated with EphrinA3 and -A5 at P10 is the class I MHC molecule H2-Bl (Pearson r = 0.12, P = 3.8 × 10−5 for EphrinA3; Pearson r = 0.11, P = 3.6 × 10−4 for EphrinA5). Since class I MHC molecules regulate spontaneous activity-dependent synaptic pruning in the LGN before eye opening (82), H2-Bl may act as a real-time editor, pruning inappropriate synapses in a cell-autonomous manner as retinotopic mapping proceeds.
We applied the same correlation method to identify potential coregulators of other developmental processes in the LGN, including synaptic refinement. For example, we found that the expression of the complement molecule C1qa through which microglia engulf synapses early in postnatal development was highly correlated with its family members C1qb and C1qc (Pearson r = 0.51, P = 1 × 10−7 for C1qb; Pearson r = 0.4, P = 5.4 × 10−4 for C1qc). C1qa was also strongly correlated with the expression of Sparc (Pearson r = 0.44, P = 3.1 × 10−5), the secreted matricellular protein implicated in synapse elimination (83). Therefore, C1qa and Sparc may be collaborating as part of a transcriptional program to drive the refinement of neuronal connectivity.
Neural circuit development relies on endothelial guidance cues and activity-dependent changes in endothelial cells and vascular structures. Mfsd2a is a membrane transport protein that is an essential component of the blood–brain barrier, regulating blood–brain permeability and vascular remodeling (84, 85). We found that the expression of Mfsd2a in endothelial cells correlates strongly with proteins that are part of the ECM, including the proteoglycan Spock2 (Pearson r = 0.33, P = 1.1 × 10−16 at P5) and the von Willebrand A domain-related protein (Pearson r = 0.26, P = 1.1 × 10−9 at P5), which stabilizes the ECM and supports nerve development (86–89). Endothelial cells may therefore contribute to the organization of the ECM as the LGN remodels. This is particularly interesting within the context of Mfsd2a function, which mediates the establishment of the vasculature in the cortex and may perform similar functions in the LGN (83, 84).
The Prkcd-Cre Mouse Line Targets Relay Neurons During Late Postnatal Development.
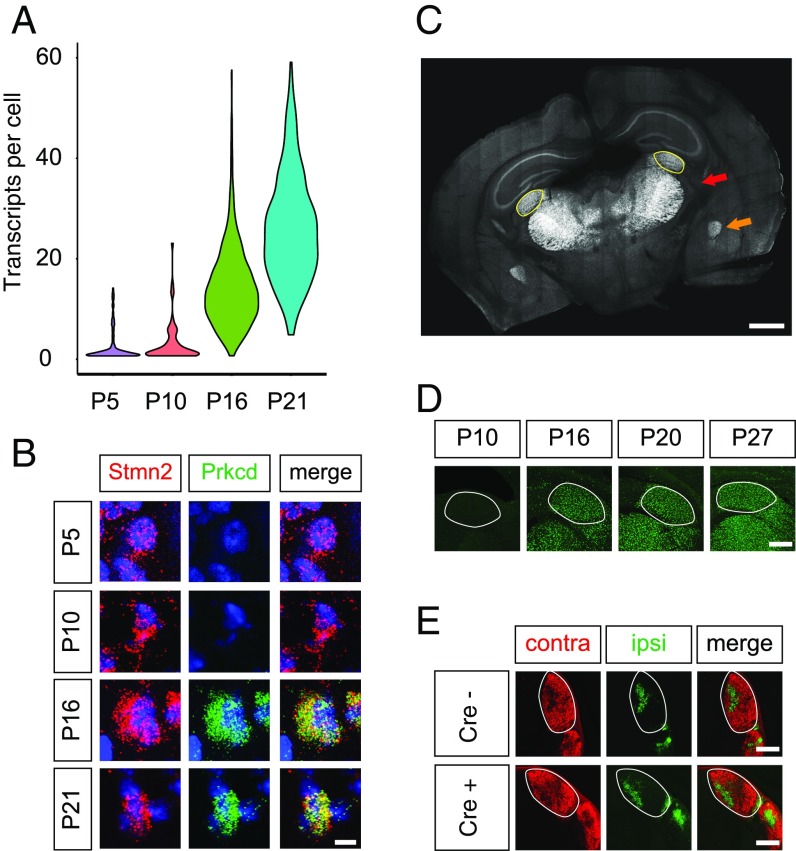
Sensory experience-dependent synaptic refinement in the LGN occurs largely between P20 and P30. Although this phase is critical for the proper refinement of synaptic connectivity, it has been much less studied than earlier phases of refinement, which are driven by spontaneous activity. One challenge has been a lack of mouse genetic tools for manipulating excitatory relay neurons during late-stage refinement. To manipulate relay neurons during the vision-sensitive period between P20 and P30, we used our resource of LGN gene expression to identify relay neuron-specific genes that are significantly up-regulated by the start of this phase, focusing on genes that increase in neurons between P10 and P16. We identified the gene encoding the intracellular signaling molecule Prkcd (Protein kinase C delta-type) as the single most highly up-regulated relay neuron-specific gene between P10 and P16, achieving a nearly 70-fold induction at the later time point (Fig. 4 A and B). A literature search identified a BAC transgenic Prkcd-Cre mouse line termed Tg(Prkcd-glc-1/CFP,-Cre) (hereafter Prkcd-Cre) that was initially generated and used to drive Cre expression in the amygdala for the dissection of striatal circuitry (90). Outside of the amygdala, the highest expression of Prkcd across the brain at P16 was seen in excitatory thalamic neurons, including relay neurons of the LGN (Fig. 4C).
Fig. 4.
Prkcd-Cre mouse as a tool for interrogating late-stage experience-dependent synaptic refinement. (A) Violin plot displaying expression of Prkcd in excitatory neurons across development. (B) Confocal FISH images validating up-regulation of Prkcd in excitatory neurons of the LGN between P10 and P16. (Scale bar, 5 μm.) (C) Coronal section of P16 mouse brain demonstrating CFP fluorescence in Cre-positive cells. Note that excitatory neurons across the thalamus are labeled with CFP. The dorsal LGN is outlined in yellow. Red arrow indicates thalamus, and orange arrow indicates amygdala. (Scale bar, 500 μm.) (D) Sun1-GFP expression in Cre-positive relay neurons of the thalamus in Prkcd-Cre/LSL-Sun1-GFP mice. The LGN is outlined. (Scale bar, 200 μm.) (E) Images of choleratoxin B conjugated to Alexa Fluor 647 (ipsilateral; pseudocolored green) and 555 (contralateral) in Prkcd-Cre–positive mice and Cre-negative littermates, demonstrating normal segregation of eye-specific inputs in Prkcd-Cre mice. The LGN is outlined. (Scale bars, 200 μm.)
We sought to determine whether this line could be used to drive Cre expression specifically in relay neurons of the LGN in a temporally controlled manner by crossing Prkcd-Cre mice to Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J mice, which express a GFP-tagged nuclear protein in the presence of Cre (29). As predicted, upon generating Sun1/Prkcd-Cre mice, we observed GFP-positive nuclear membranes in thalamic relay neurons of mice at P16 and later but not at P10, consistent with the expression pattern of Prkcd in our dataset (Fig. 4D).
Finally, we assessed retinogeniculate organization in Sun1/Prkcd-Cre mice compared with littermate Sun1 mice that lack Prkcd-Cre and therefore do not express Cre in relay neurons. We tested whether the expression of Cre from the Prkcd promoter affects baseline aspects of LGN development. RGC axon tracing in P21 mice showed similar eye-specific input organization in Cre-positive and Cre-negative littermate mice, suggesting that early stages of axonal targeting and synaptic pruning occur normally in the Prkcd-Cre, Sun1-positive mouse line (Fig. 4E). We therefore suggest that the Prkcd-Cre mouse line may be a valuable tool for achieving Cre-mediated recombination in relay neurons specifically during late postnatal development. These findings suggest that the gene expression resource provided here may be useful in identifying other points of genetic access for cell types in the LGN.
Discussion
Brain development is characterized by tight regulation of spatiotemporal changes in cell identity and function that are driven by dynamic transcriptional programs. The ability to investigate cell type-specific gene expression in vivo has, until recently, been difficult due to cellular heterogeneity and the complexity of the developing brain. scRNA-seq now provides a means to examine cell-specific changes that might otherwise be obscured during bulk tissue profiling.
In this study, we applied scRNA-seq to the early postnatal mouse LGN—a region that undergoes well-characterized periods of circuit assembly and refinement during the first month of life (33, 91, 92). The transcriptional programs underlying this period had not been previously characterized in detail. This is likely in part due to the small size and anatomical position of the LGN, which make it difficult to access, isolate, and dissociate, particularly in young mice. However, due to the fact that it is a relatively simple brain structure that undergoes extensive activity-dependent synaptic changes, the LGN is an attractive paradigm in which to study developmental gene regulation in parallel with synapse maturation and refinement.
Our differential gene expression analysis identified hundreds of genes that are dynamically regulated between adjacent developmental time points within neurons, glia, and endothelial cells. We grouped genes that varied with comparable temporal dynamics according to biological function and found that each LGN cell type expresses unique genes related to cell type-specific processes, such as vascular development genes in endothelial cells, as well as shared genes involved in tissue remodeling, including myelination and modification of the ECM. In excitatory neurons, we mapped expression kinetics across development and clustered genes according to their temporal expression profiles, identifying a previously unappreciated degree of transcriptional heterogeneity in this population. We present this map of developmental gene expression as a resource for the study of molecular mechanisms underlying circuit development at the retinogeniculate synapse and within other systems.
In addition to sequencing at single-cell resolution, this study is unique in its focus on the developing brain. Recent work by the Allen Institute for Brain Science has described single-cell sequencing of the adult mouse LGN using Cre-driven labeling and fluorescence-guided cell selection by FACS (93). Our study complements and extends this prior work by focusing on the developing LGN, profiling multiple developmental conditions, and characterizing multiple nonneuronal cell types, including astrocytes, microglia, and endothelial cells (93, 94).
We utilized the newly developed MetaNeighbor approach to compare the transcriptional signatures of excitatory neurons, inhibitory neurons, and oligodendrocytes at each postnatal time point in our data with those of the Allen Institute (https://www.biorxiv.org/content/early/2017/06/16/150524). This approach assesses the transcriptional similarity of a given cell type between separate datasets. Across postnatal development, the transcriptional signatures of excitatory and inhibitory neurons in our data were increasingly similar to those in the adult mouse LGN as sequenced by the Allen Institute. Oligodendrocytes demonstrated increasing similarity from P5 to P16 but slightly less similarity at P21 (SI Experimental Procedures and Fig. S4).
In addition to the creation of this single-cell expression resource, our experiments have also yielded several important insights into LGN development. First, we have defined cell type-specific marker genes for all cell populations in the LGN with a particular focus on the five predominant cell types, which can now be applied to the molecular study of LGN development by other groups interested in studying brain development. Next, we have found that all major cell types dynamically regulate their transcriptional states across postnatal LGN development. Our analysis of temporal gene kinetics allowed for classification of functional gene clusters with similar expression profiles across development in excitatory relay neurons. We observe particularly robust gene expression changes between P10 and P16. The heightened transcriptional activity between these two time points may reflect an important role for visual experience in shifting the transcriptional program as eye opening occurs between P12 and P14. The relative contributions of spontaneous activity and experience in driving transcriptional changes across early LGN development, and the identification of specific molecular regulators of these processes, should be further explored in future studies.
The droplet-based scRNA-seq technology used in this study is rapidly evolving but still relatively young. A technical consideration in the interpretation of our findings is that, due to low transcript capture efficiencies, our data underestimate the absolute transcript counts per cell for any given gene (30, 31). Additionally, scRNA-seq is limited by so-called dropout events, in which a gene is expressed but not detected through sequencing. This phenomenon is caused by low RNA input and the stochastic nature of gene expression at the single-cell level. The statistical power to perform differential gene expression is limited by cell number and sequencing depth. Therefore, it is important to be cautious in the interpretation of findings from scRNA-seq indicating that a given gene is differentially expressed and to validate genes of interest by other methods. Finally, the scRNA-seq dissociation protocol preferentially isolates and preserves some cell types better than others (8). For example, our approach did not capture enough inhibitory interneurons to analyze their developmental gene expression, a feature of our single-cell dissociation technique that we observe in the cortex as well. In the future, it will be important to optimize the dissociation technique to allow for the capture and sequencing of this critical population of neurons. Despite these caveats, evolving scRNA-seq techniques now allow for the development of an extensive catalog of cell type-specific gene expression across the entire central nervous system in both immature and mature animals. In the future, it will be important to compare our dataset with those obtained from other brain regions to distinguish transcriptional features that are specific to the LGN from those that are more broadly relevant. Such experiments have the potential to yield critical mechanistic insight into nervous system development and function.
Experimental Procedures
All experiments using animals were performed according to protocols approved by the Harvard Medical Area Institutional Animal Care and Use Committee. Each time point includes data from eight mice total processed as four independent samples of two mice each. Single cells were captured, barcoded, and sequenced according to the inDrops technique as previously described (30). In situ hybridizations were performed using the RNAscope platform (Advanced Cell Diagnostics). Additional details are contained in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. Chinfei Chen, Dr. Corey Harwell, and members of the M.E.G. laboratory for discussions and critical reading of the manuscript. We thank Vance Soares for graphic illustration of Fig. 1A. This work was funded by R37 NS028829 (to M.E.G.), the Pediatric Scientist Development Program and March of Dimes Birth Defects Foundation (B.T.K.), and 5T32AG000222-23 and a Goldenson Postdoctoral Fellowship (to L.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108761).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717871115/-/DCSupplemental.
References
- 1.Kelly OG, Melton DA. Induction and patterning of the vertebrate nervous system. Trends Genet. 1995;11:273–278. doi: 10.1016/s0168-9525(00)89074-5. [DOI] [PubMed] [Google Scholar]
- 2.O’Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- 3.Imamura T, Uesaka M, Nakashima K. Epigenetic setting and reprogramming for neural cell fate determination and differentiation. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130511. doi: 10.1098/rstb.2013.0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson MB, Walsh CA. Cerebral cortical neuron diversity and development at single-cell resolution. Curr Opin Neurobiol. 2017;42:9–16. doi: 10.1016/j.conb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romanov RA, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci. 2017;20:176–188. doi: 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Wu X, Jiang L, Zhang Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 2017;18:3227–3241. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacar B, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Manno G, et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 11.Hanchate NK, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350:1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer KS, Miko IJ. Eph-ephrin signaling in nervous system development. F1000 Res. 2016;5:413. doi: 10.12688/f1000research.7417.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reber M, Hindges R, Lemke G. Eph receptors and ephrin ligands in axon guidance. Adv Exp Med Biol. 2007;621:32–49. doi: 10.1007/978-0-387-76715-4_3. [DOI] [PubMed] [Google Scholar]
- 15.Jang S, Lee H, Kim E. Synaptic adhesion molecules and excitatory synaptic transmission. Curr Opin Neurobiol. 2017;45:45–50. doi: 10.1016/j.conb.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 18.Mount CW, Monje M. Wrapped to adapt: Experience-dependent myelination. Neuron. 2017;95:743–756. doi: 10.1016/j.neuron.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitew S, et al. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2014;276:29–47. doi: 10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 22.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroeze Y, et al. Transcriptome analysis identifies multifaceted regulatory mechanisms dictating a genetic switch from neuronal network establishment to maintenance during postnatal prefrontal cortex development. Cereb Cortex. January 19, 2017 doi: 10.1093/cercor/bhw407. [DOI] [PubMed] [Google Scholar]
- 24.Fertuzinhos S, et al. Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep. 2014;6:938–950. doi: 10.1016/j.celrep.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillman AA, Cookson MR. Transcriptomic changes in brain development. Int Rev Neurobiol. 2014;116:233–250. doi: 10.1016/B978-0-12-801105-8.00009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dillman AA, et al. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci. 2013;16:499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molyneaux BJ, et al. DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo A, et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shekhar K, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong YK, Chen C. Wiring and rewiring of the retinogeniculate synapse. Curr Opin Neurobiol. 2011;21:228–237. doi: 10.1016/j.conb.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guido W. Refinement of the retinogeniculate pathway. J Physiol. 2008;586:4357–4362. doi: 10.1113/jphysiol.2008.157115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denman DJ, Contreras D. On parallel streams through the mouse dorsal lateral geniculate nucleus. Front Neural Circuits. 2016;10:20. doi: 10.3389/fncir.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krahe TE, El-Danaf RN, Dilger EK, Henderson SC, Guido W. Morphologically distinct classes of relay cells exhibit regional preferences in the dorsal lateral geniculate nucleus of the mouse. J Neurosci. 2011;31:17437–17448. doi: 10.1523/JNEUROSCI.4370-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaltschmidt B, Kaltschmidt C. NF-kappaB in long-term memory and structural plasticity in the adult mammalian brain. Front Mol Neurosci. 2015;8:69. doi: 10.3389/fnmol.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmeisser MJ, et al. IκB kinase/nuclear factor κB-dependent insulin-like growth factor 2 (Igf2) expression regulates synapse formation and spine maturation via Igf2 receptor signaling. J Neurosci. 2012;32:5688–5703. doi: 10.1523/JNEUROSCI.0111-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terauchi A, Johnson-Venkatesh EM, Bullock B, Lehtinen MK, Umemori H. Retrograde fibroblast growth factor 22 (FGF22) signaling regulates insulin-like growth factor 2 (IGF2) expression for activity-dependent synapse stabilization in the mammalian brain. Elife. 2016;5:e12151. doi: 10.7554/eLife.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agis-Balboa RC, et al. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011;30:4071–4083. doi: 10.1038/emboj.2011.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evdokimova V, et al. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci Signal. 2012;5:ra92. doi: 10.1126/scisignal.2003184. [DOI] [PubMed] [Google Scholar]
- 43.Bake S, Okoreeh AK, Alaniz RC, Sohrabji F. Insulin-like growth factor (IGF)-I modulates endothelial blood-brain barrier function in ischemic middle-aged female rats. Endocrinology. 2016;157:61–69. doi: 10.1210/en.2015-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubert T, Grimal S, Carroll P, Fichard-Carroll A. Collagens in the developing and diseased nervous system. Cell Mol Life Sci. 2009;66:1223–1238. doi: 10.1007/s00018-008-8561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukami S, et al. Abeta-degrading endopeptidase, neprilysin, in mouse brain: Synaptic and axonal localization inversely correlating with Abeta pathology. Neurosci Res. 2002;43:39–56. doi: 10.1016/s0168-0102(02)00015-9. [DOI] [PubMed] [Google Scholar]
- 46.Land PW, Shamalla-Hannah L. Transient expression of synaptic zinc during development of uncrossed retinogeniculate projections. J Comp Neurol. 2001;433:515–525. doi: 10.1002/cne.1157. [DOI] [PubMed] [Google Scholar]
- 47.Kwok JC, Dick G, Wang D, Fawcett JW. Extracellular matrix and perineuronal nets in CNS repair. Dev Neurobiol. 2011;71:1073–1089. doi: 10.1002/dneu.20974. [DOI] [PubMed] [Google Scholar]
- 48.Callaway EM. Structure and function of parallel pathways in the primate early visual system. J Physiol. 2005;566:13–19. doi: 10.1113/jphysiol.2005.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruz-Martín A, et al. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature. 2014;507:358–361. doi: 10.1038/nature12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng G, et al. Monocular visual deprivation in macaque monkeys: A profile in the gene expression of lateral geniculate nucleus by laser capture microdissection. Mol Vis. 2008;14:1401–1413. [PMC free article] [PubMed] [Google Scholar]
- 51.Klochendler A, et al. A transgenic mouse marking live replicating cells reveals in vivo transcriptional program of proliferation. Dev Cell. 2012;23:681–690. doi: 10.1016/j.devcel.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Brockschnieder D, Sabanay H, Riethmacher D, Peles E. Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J Neurosci. 2006;26:757–762. doi: 10.1523/JNEUROSCI.4317-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T, et al. Human Ermin (hErmin), a new oligodendrocyte-specific cytoskeletal protein related to epileptic seizure. Brain Res. 2011;1367:77–84. doi: 10.1016/j.brainres.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Li W, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collinson JM, Marshall D, Gillespie CS, Brophy PJ. Transient expression of neurofascin by oligodendrocytes at the onset of myelinogenesis: Implications for mechanisms of axon-glial interaction. Glia. 1998;23:11–23. doi: 10.1002/(sici)1098-1136(199805)23:1<11::aid-glia2>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 56.Werner HB, et al. A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia. 2013;61:567–586. doi: 10.1002/glia.22456. [DOI] [PubMed] [Google Scholar]
- 57.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 58.Gomes WA, Mehler MF, Kessler JA. Transgenic overexpression of BMP4 increases astroglial and decreases oligodendroglial lineage commitment. Dev Biol. 2003;255:164–177. doi: 10.1016/s0012-1606(02)00037-4. [DOI] [PubMed] [Google Scholar]
- 59.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 60.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung WS, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheridan GK, Murphy KJ. Neuron-glia crosstalk in health and disease: Fractalkine and CX3CR1 take centre stage. Open Biol. 2013;3:130181. doi: 10.1098/rsob.130181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson AD, Picard N, Min L, Fagiolini M, Chen C. Cortical feedback regulates feedforward retinogeniculate refinement. Neuron. 2016;91:1021–1033. doi: 10.1016/j.neuron.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seabrook TA, El-Danaf RN, Krahe TE, Fox MA, Guido W. Retinal input regulates the timing of corticogeniculate innervation. J Neurosci. 2013;33:10085–10097. doi: 10.1523/JNEUROSCI.5271-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohan H, et al. Transcript profiling of different types of multiple sclerosis lesions yields FGF1 as a promoter of remyelination. Acta Neuropathol Commun. 2014;2:168. doi: 10.1186/s40478-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou YX, Pannu R, Le TQ, Armstrong RC. Fibroblast growth factor 1 (FGFR1) modulation regulates repair capacity of oligodendrocyte progenitor cells following chronic demyelination. Neurobiol Dis. 2012;45:196–205. doi: 10.1016/j.nbd.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miron VE. Microglia-driven regulation of oligodendrocyte lineage cells, myelination, and remyelination. J Leukoc Biol. 2017;101:1103–1108. doi: 10.1189/jlb.3RI1116-494R. [DOI] [PubMed] [Google Scholar]
- 68.Pang Y, et al. Differential roles of astrocyte and microglia in supporting oligodendrocyte development and myelination in vitro. Brain Behav. 2013;3:503–514. doi: 10.1002/brb3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuelling LM, Fuss B. Autotaxin (ATX): A multi-functional and multi-modular protein possessing enzymatic lysoPLD activity and matricellular properties. Biochim Biophys Acta. 2008;1781:525–530. doi: 10.1016/j.bbalip.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anliker B, et al. Lysophosphatidic acid (LPA) and its receptor, LPA1, influence embryonic Schwann cell migration, myelination, and cell-to-axon segregation. Glia. 2013;61:2009–2022. doi: 10.1002/glia.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wheeler NA, Lister JA, Fuss B. The autotaxin-lysophosphatidic acid axis modulates histone acetylation and gene expression during oligodendrocyte differentiation. J Neurosci. 2015;35:11399–11414. doi: 10.1523/JNEUROSCI.0345-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kucukdereli H, et al. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci USA. 2011;108:E440–E449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh SK, et al. Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via hevin. Cell. 2016;164:183–196. doi: 10.1016/j.cell.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eroglu C. The role of astrocyte-secreted matricellular proteins in central nervous system development and function. J Cell Commun Signal. 2009;3:167–176. doi: 10.1007/s12079-009-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huberman AD, Murray KD, Warland DK, Feldheim DA, Chapman B. Ephrin-As mediate targeting of eye-specific projections to the lateral geniculate nucleus. Nat Neurosci. 2005;8:1013–1021. doi: 10.1038/nn1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfeiffenberger C, et al. Ephrin-As and neural activity are required for eye-specific patterning during retinogeniculate mapping. Nat Neurosci. 2005;8:1022–1027. doi: 10.1038/nn1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfeiffenberger C, Yamada J, Feldheim DA. Ephrin-As and patterned retinal activity act together in the development of topographic maps in the primary visual system. J Neurosci. 2006;26:12873–12884. doi: 10.1523/JNEUROSCI.3595-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waimey KE, Cheng HJ. Axon pruning and synaptic development: How are they per-plexin? Neuroscientist. 2006;12:398–409. doi: 10.1177/1073858406292631. [DOI] [PubMed] [Google Scholar]
- 79.Fiore R, Püschel AW. The function of semaphorins during nervous system development. Front Biosci. 2003;8:s484–s499. doi: 10.2741/1080. [DOI] [PubMed] [Google Scholar]
- 80.Byström B, Carracedo S, Behndig A, Gullberg D, Pedrosa-Domellöf F. Alpha11 integrin in the human cornea: Importance in development and disease. Invest Ophthalmol Vis Sci. 2009;50:5044–5053. doi: 10.1167/iovs.08-3261. [DOI] [PubMed] [Google Scholar]
- 81.Peavey M, Salleh N, Leppert P. Collagen-binding α11 integrin expression in human myometrium and fibroids utilizing a novel RNA in situ probe. Reprod Sci. 2014;21:1139–1144. doi: 10.1177/1933719114522548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 83.López-Murcia FJ, Terni B, Llobet A. SPARC triggers a cell-autonomous program of synapse elimination. Proc Natl Acad Sci USA. 2015;112:13366–13371. doi: 10.1073/pnas.1512202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ben-Zvi A, et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keaney J, Campbell M. The dynamic blood-brain barrier. FEBS J. 2015;282:4067–4079. doi: 10.1111/febs.13412. [DOI] [PubMed] [Google Scholar]
- 86.Schnepp A, et al. Mouse testican-2. Expression, glycosylation, and effects on neurite outgrowth. J Biol Chem. 2005;280:11274–11280. doi: 10.1074/jbc.M414276200. [DOI] [PubMed] [Google Scholar]
- 87.Vannahme C, et al. Molecular cloning of testican-2: Defining a novel calcium-binding proteoglycan family expressed in brain. J Neurochem. 1999;73:12–20. doi: 10.1046/j.1471-4159.1999.0730012.x. [DOI] [PubMed] [Google Scholar]
- 88.Duong T, Lopez IA, Ishiyama A, Ishiyama G. Immunocytochemical distribution of WARP (von Willebrand A domain-related protein) in the inner ear. Brain Res. 2011;1367:50–61. doi: 10.1016/j.brainres.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fitzgerald J, Tay Ting S, Bateman JF. WARP is a new member of the von Willebrand factor A-domain superfamily of extracellular matrix proteins. FEBS Lett. 2002;517:61–66. doi: 10.1016/s0014-5793(02)02579-6. [DOI] [PubMed] [Google Scholar]
- 90.Haubensak W, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hooks BM, Chen C. Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron. 2006;52:281–291. doi: 10.1016/j.neuron.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 92.Hooks BM, Chen C. Vision triggers an experience-dependent sensitive period at the retinogeniculate synapse. J Neurosci. 2008;28:4807–4817. doi: 10.1523/JNEUROSCI.4667-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Allen Institute for Brain Science 2017 Allen Brain Atlas. Available at celltypes.brain-map.org/rnaseq. Accessed November 27, 2017.
- 94.Abrahams BS, et al. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.