Significance
Quantitative detection of protein biomarkers over a wide concentration range from minute amounts of blood is essential for clinical diagnostics. Proximity ligation assay combines antibody–oligo conjugates, enzymatic ligation, and PCR amplification into a sensitive method for quantitative protein detection from small volumes. This report describes a streamlined and more stringent assay format that takes advantage of DNA circle formation to remove unwanted DNA molecules. Kinetic analysis of antibody–antigen interactions shows that variation in assay performance between various biomarkers is an effect of antibody quality. We show that this assay format enables compatibility with low-affinity reagents, a major limitation for most protein quantitation methods, while improving sensitivity and reproducibility.
Keywords: proximity ligation assay, antibody affinity, kinetic analysis, immuno-PCR, qPCR
Abstract
Proximity ligation assay (PLA) is a powerful tool for quantitative detection of protein biomarkers in biological fluids and tissues. Here, we present the circular proximity ligation assay (c-PLA), a highly specific protein detection method that outperforms traditional PLA in stringency, ease of use, and compatibility with low-affinity reagents. In c-PLA, two proximity probes bind to an analyte, providing a scaffolding that positions two free oligonucleotides such that they can be ligated into a circular DNA molecule. This assay format stabilizes antigen proximity probe complexes and enhances stringency by reducing the probability of random background ligation events. Circle formation also increases selectivity, since the uncircularized DNA can be removed enzymatically. We compare this method with traditional PLA on several biomarkers and show that the higher stringency for c-PLA improves reproducibility and enhances sensitivity in both buffer and human plasma. The limit of detection ranges from femtomolar to nanomolar concentrations for both methods. Kinetic analyses using surface plasmon resonance (SPR) and biolayer interferometry (BLI) reveal that the variation in limit of detection is due to the variation in antibody affinity and that c-PLA outperforms traditional PLA for low-affinity antibodies. The lower background signal can be used to increase proximity probe concentration while maintaining a high signal-to-noise ratio, thereby enabling the use of low-affinity reagents in a homogeneous assay format. We anticipate that the advantages of c-PLA will be useful in a variety of clinical protein detection applications where high-affinity reagents are lacking.
Quantitative detection of protein biomarkers in biological fluids is essential for diagnosis, monitoring, and personalized treatment of disease. Despite considerable progress in recent years, the clinical use of validated proteomic biomarkers remains limited (1). The benchmark for affinity-based protein measurements is defined by the ELISA where affinity ligands (e.g., antibodies) are used in a sandwich format to detect and quantify the protein of interest (2, 3). ELISA generally involves several steps starting with sample incubation, where the target analyte is captured on a surface precoated with primary antibodies, followed by washing steps and recognition with a secondary antibody, which facilitates detection with a colorimetric, fluorescent, or luminescent label. ELISA offers reasonable sensitivity, but it requires a large sample volume, has limited dynamic range, and frequently suffers from false positives due to nonspecific binding (4, 5). These limitations interfere with the discovery and validation of novel biomarker candidates that have the potential to enable early diagnosis and regular molecular monitoring of disease.
Immunoassays combined with nucleic acid-based amplification and detection have facilitated new approaches and have extended the analytical sensitivity beyond that achievable with ELISA (6–9). One particularly promising approach is the proximity ligation assay (PLA) (10, 11). In PLA, pairs of affinity probes are individually conjugated to short ssDNA molecules to form proximity probes that carry either a phosphorylated 5′ end or a 3′-hydroxyl group. When the probe pairs subsequently bind to their cognate target analyte in solution, the associated DNA strands are brought into close proximity and aligned by hybridization to a third bridging oligonucleotide. The free DNA ends are ligated, forming a new DNA sequence that is amplified and quantified using qPCR (Fig. 1A). We have previously showed that PLA provides femtomolar sensitivity and a wide dynamic range over five orders of magnitude, consuming as little as 1 μL of sample (12). One key benefit of PLA is that it addresses the widespread problem of cross-reactivity in antibody-based protein detection. Potential signals from cross-reactive antibodies are eliminated by tailoring the DNA sequences, such that ligation only takes place when cognate proximity probes are bound to the target analyte. We have applied this technology to detect more than 20 different biomarkers in clinical plasma samples using panels of seven multiplex PLA reactions (13, 14). Others have expanded the multiplexing capability even further (15).
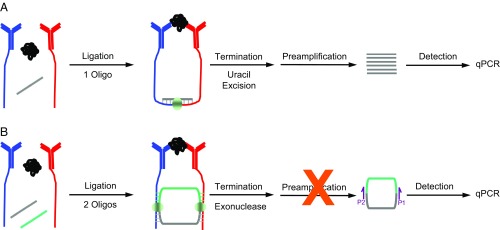
Fig. 1.
Schematic representation of t-PLA and c-PLA. (A) t-PLA detects proteins using pairs of antibody–DNA conjugates (red and blue), which are brought into close proximity on binding to target analyte. The addition of a bridge oligonucleotide and DNA ligase enables ligation of the antibody-tethered oligonucleotides to form a new DNA sequence. Ligation is terminated by selective degradation of the bridge oligonucleotide. The newly formed ligation product is subsequently preamplified followed by quantification using qPCR. (B) In c-PLA, the antibody-tethered oligonucleotides act as bridges for two ligation events between free oligonucleotides, resulting in the formation of a circular ligation product. The addition of an extra oligonucleotide increases stringency compared with t-PLA; it lowers the probability of random background ligation events, since four components must assemble in the absence of the target analyte to generate an independent circular ligation product. Circle formation also allows exonuclease treatment, which terminates ligation and reduces background by degrading all uncircularized DNA. The reduction in background also simplifies the workflow by eliminating the need for preamplification. Circular ligation products are quantified by qPCR using primer sites spanning the newly formed junctions (P1 and P2).
One of the limitations of affinity-based immunoassays, including PLA, is the so-called hook effect, in which the signal decreases at high antigen concentrations, resulting in incorrectly low signals or even false negatives (16). The hook effect becomes predominant when the analyte concentration exceeds the concentration of proximity probes (>>1 nM), and it effectively determines the upper quantification limit (17). The proximity probe concentration can potentially be increased but must be carefully balanced against a deteriorating signal-to-noise ratio from random background ligation events when probes and the bridge oligo come together by chance. The hook effect can ordinarily be avoided by sample dilution; however, dilution alters binding equilibrium. This does not present a problem for high-affinity interactions, but it may prevent low-affinity antibodies from binding to their target analytes, thereby limiting the effectiveness of PLA to high-affinity capture agents (18). While incompatibility with low-affinity antibodies is a major drawback for PLA, it is not restricted to this assay alone. The availability of high-affinity antibodies or other capture reagents is a general limitation for all sandwich immunoassays, because the sensitivity is ultimately determined by the quality of the reagents used (19).
Numerous approaches have been developed to improve PLA performance, including concentration of minute amounts of analyte on a solid phase before ligation (10, 18, 20), use of multivalent proximity probes (21), addition of multiple affinity probes (22, 23), special design of asymmetric bridge oligos (17), inclusion of protecting oligonucleotides prehybridized to proximity probes to reduce background ligation events (22, 24), and the use of novel amplification schemes (25–28). Some of the amplification schemes entail enzymatic manipulations, where the ligation products are released from antibodies and converted into circles used for isothermal rolling circle amplification (26, 29–31). Many of these approaches have improved assay reproducibility, although precision still remains a challenge for adaptation into clinical diagnostics. The relationship between affinity and assay sensitivity has been described by Gullberg et al. (11). They showed that the dose–response curve and limit of detection correlated with modeled equilibrium affinity constants, although detailed kinetic characterization of antibodies used in PLA and its relationship to assay performance remain unexplored. We suggest that a better understanding of how antibody–antigen affinity and kinetic parameters affect PLA performance has great value for assay development and optimization efforts.
Here, we describe a circular proximity ligation assay (c-PLA), where in contrast to traditional proximity ligation assay (t-PLA), the proximity probes are used as bridges that enable the connection of two free oligonucleotides via dual ligation events, resulting in the formation of a circle (Fig. 1B). The addition of an extra oligonucleotide decreases the probability of random background ligation events due to stringency in assay design. We define stringency as the minimization of background ligation, since four molecules (two proximity probes and two free connector oligonucleotides) must come together in the absence of target antigen to form a circle. In addition, circle formation has selective advantages, as uncircularized DNA can be removed by a simple exonuclease treatment (32), streamlining the workflow by eliminating preamplification before qPCR. As a result, our c-PLA is much more straightforward than not only PLA but also, most existing protein detection methods and can be performed in a single reaction tube with a tiny sample volume (2 μL). The assay format utilizes the same affinity probes as in t-PLA, which enable a direct performance comparison between the two methods. We apply both assays to six biomarkers and show equal or better performance for c-PLA with improvements in precision. We also provide kinetic binding analysis of the antibodies used and find a direct correlation between antibody affinity and assay performance. We show that kinetic information can be used to guide assay development: the enhanced stringency in c-PLA is utilized to increase affinity probe concentration while maintaining a low probability of background ligation events—thereby improving signal-to-background ratio. Finally, we show that c-PLA is compatible with complex mixtures, such as human plasma, and provides a path for adaptation with low-affinity antibodies as well as alternative capture agents (33, 34) without the need for preconcentration on solid phase.
Results and Discussion
c-PLA Workflow.
The workflows for both the t-PLA and the c-PLA are shown in Fig. 1. Both assays utilize the same set of proximity probes that are prepared by bioconjugation of polyclonal antibodies with amine-modified oligonucleotides through aromatic hydrazone chemistry (details are provided in SI Methods). Polyclonal antibodies are cost-efficient, as a single batch of antibodies is divided into two portions and coupled to oligonucleotides that are terminated by either a phosphorylated 5′ end or a 3′ end. This produces two heterogeneous mixtures of proximity probes against several different epitopes of the same target antigen. During sample incubation, the antibody portions of these proximity probes bind to distinct epitopes of the target analyte. This is followed by a ligation step, in which a solution containing DNA ligase is added to the incubation mixture. In t-PLA, this ligation mixture contains a third bridge oligonucleotide complementary to the ends of the proximity probes, thereby facilitating ligation to form a new DNA sequence. Ligation is terminated by the addition of a uracil excision mixture, which selectively degrades the bridge oligonucleotide. The ligation mixture is subsequently preamplified across the newly formed DNA junction to increase signal-to-background ratio and reproducibility (12, 35) before quantification by qPCR. In c-PLA, ligation products are not formed by a direct junction of proximity probes but rather, by the formation of a circle when two free connector oligonucleotides are joined in two distinct ligation events facilitated by target-bound proximity probes. The uracil excision step is replaced with exonuclease treatment, which has the selective advantage of enriching for circularized DNA (32). Consequently, the background is dramatically decreased as all uncircularized nucleic acids are degraded as opposed to only the bridge oligo in t-PLA. This allows for omission of the preamplification before qPCR analysis without any loss in signal-to-noise ratio or reproducibility.
Assay Characteristics.
We designed the c-PLA from existing t-PLA probes that have been optimized for minimization of heteroduplex formation (12). This allowed us to compare the two methods using the same set of proximity probes. For c-PLA, we relaxed the ligation conditions to account for the requirement of two ligation events and to accommodate the efficiency of Ampligase at higher temperature. Ligation was consequently performed for 30 min at 45 °C instead of 15 min at 30 °C. A comparable modification of t-PLA did not yield any improvements.
A direct comparison between t-PLA and c-PLA for six biomarkers is shown in Fig. 2, and the results are summarized in Table 1. These experiments were performed with three biological replicates and quantified in triplicate qPCR experiments, generating nine data points for each concentration. For VEGF, a dose-dependent response across nearly five orders of magnitude is shown for both methods (Fig. 2A). The difference of almost four orders of magnitude in counts (estimated number of ligated molecules) generated by the two methods is due to both the elimination of preamplification and the enhanced stringency for c-PLA. For comparison, we performed preamplification in c-PLA and found that the omission of this step accounts for approximately three orders of magnitude difference in signal, whereas stringency in assay design accounts for the remainder (Fig. S2). Preamplification was originally introduced to improve signal-to-background ratio and precision, although t-PLA is still impeded by relatively high coefficients of variation (CVs), an obstacle for standard use in clinical diagnostics. Variation in t-PLA has traditionally been addressed by addition of internal controls and normalization of data, allowing for relative biomarker profiling (13, 15). Background signals for PLA are generated from random ligation events, nonspecific binding of two cognate proximity probes, or nucleic acid amplification artifacts. In c-PLA, random ligation events are minimized by the stringency in assay design, as four molecules and two ligation events are required for signal generation. Nonspecific binding effects are addressed by the addition of excess bulk IgG molecules from the same species as the proximity probes (12). Nucleic acid amplification artifacts are minimized in c-PLA, as dual ligation events result in two new DNA sequences at the junction sites. We tested multiple primers targeting different sites on the newly formed circular DNA and found that a primer pair spanning both junctions produced the lowest background. Exonuclease treatment in c-PLA also reduces background, which enhances signal-to-noise ratio and assay performance. This improves precision within the linear dynamic range as shown by a decrease in the average CVs from 29% for t-PLA to below 16% for c-PLA (Table 1). PLA precision in general is limited by the qPCR readout, and variation at low counts can be decreased further with digital quantification methods (31, 36).
Fig. 2.
Dose–response curves of t-PLA and c-PLA for detection of VEGF (A), GDNF (B), IL-6 (C), MIF (D), TNF-α (E), and IGF-II (F). The x axis displays antigen (Ag) concentration, and the y axis displays an estimated number of ligated molecules. The enhanced stringency for c-PLA is shown by a lower number of counts because of the rigor imposed by circle formation, background reduction through exonuclease treatment, elimination of preamplification, and tailored qPCR primer sites. Error bars denote 1 SD (n = 9), and the dashed lines denote limit of detection, defined as the mean signal of a blank sample +3 SD.
Table 1.
Comparison of limit of detection and dynamic range for six biomarkers measured by c-PLA and t-PLA
| Analyte | Function | Data in figure | c-PLA | t-PLA | ||||
| Limit of detection | Dynamic range | CVs, % | Limit of detection | Dynamic range | CVs, % | |||
| VEGF | Growth factor | Fig. 2A | <10 fM | <5 | 15.6 | 10 fM | <5 | 29.3 |
| GDNF | Cell survival | Fig. 2B | 15 fM | <5 | 12.4 | 10 fM | 5 | 30.8 |
| IL-6 | Inflammation | Fig. 2C | 100 fM | <4 | 28.2 | 150 fM | <4 | 48.5 |
| MIF | Innate immunity | Fig. 2D | 3 pM | 3 | 10.4 | 5 pM | 3 | 39.9 |
| TNF-α | Cell signaling | Fig. 2E | 20 pM | 3 | 19.4 | 70 pM | <3 | 44.2 |
| IGF-II | Inflammation | Fig. 2F | 1 nM | 2 | 17.0 | 3 nM | <2 | 24.3 |
We compared c-PLA with t-PLA for five additional biomarkers: glial cell line-derived neurotrophic factor (GDNF), IL-6, macrophage migration inhibitory factor (MIF), TNF-α, and insulin-like growth factor II (IGF-II) (Fig. 2 B–F and raw data provided in SI Methods). We found that the higher stringency for c-PLA yielded improvements in precision and signal-to-background ratios and equal or better assay performance in terms of limit of detection and dynamic range for all analytes except GDNF. Average CVs across the linear range were all below 20% for c-PLA, with the exception of IL-6 (28%), while CVs for t-PLA varied from 24 to 48%. Reproducibility is generally better at higher concentrations, while precision deteriorates at low concentrations. This represents a common problem for PLA and other highly sensitive methods for protein quantitation (37–39).
Notable observations are that the limit of detection for the analytes varies more than five orders of magnitude from low femtomolar to nanomolar concentration and that the dynamic range is confined by the limit of detection on the lower end and the proximity probe concentration on the upper end, where the hook effect starts to interfere. Consequently, IGF-II, which exhibits the worst limit of detection (1 nM) among the analytes that we tested, also displays a restricted dynamic range of about two orders of magnitude. The assay performance for the same biomarkers in t-PLA generally follows the same trend as found for c-PLA, albeit with a slightly higher limit of detection. We note that previous proximity ligation studies of the same analytes have reported limit of detection using various definitions (12, 15). Here, we strictly define limit of detection as the mean signal corresponding to a blank sample plus three SDs (40). Discrepancies in reported limit of detection can be affected by the use of alternative definitions, differences in protocol, or batch-to-batch variation in proximity probe conjugation, as affinity probes without conjugated oligonucleotides may bind to target analytes and interfere with the ability to facilitate ligation. One benefit of using the same batch of proximity probes for both traditional and circular proximity ligation is that such effects are eliminated, making comparison between the two methods more straightforward. Furthermore, keeping the oligonucleotide sequences constant for all analytes eliminates any variation that may arise from sequence specificity, albeit that this limits the possibility of multiplexing for the purpose of this study. The similarities in performance between the two methods indicate that the variation in limit of detection between analytes is inherently an effect of the quality of the proximity probes and their ability to enable antibody–antigen interactions. This motivated us to further investigate the kinetic properties of these interactions.
Kinetic Analysis of Affinity Reagents.
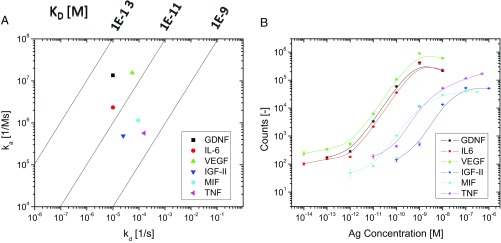
Knowledge about the kinetics of binding between antibodies and antigens is important for all immunoassays (3, 19). The wide range in limit of detection among the analytes studied in this report led us to further explore the relationship between kinetics and assay performance. A previous report showed good correlation in t-PLA between limit of detection and theoretical affinity but did not determine equilibrium dissociation constants (Kd) for the antibodies used (11). Furthermore, no analyses have been reported on either association rate constants, ka (on rates), or dissociation rate constants, kd (off rates) and their significance in PLAs. To address this, we used surface plasmon resonance (SPR) (41) to determine Kd values as well as on and off rates for the antibody–antigen interactions investigated. Kinetic parameters are listed in Table 2, and SPR sensorgrams for individual analytes fitted to a 1:1 binding model are provided in SI Methods. The results are also summarized in an isoaffinity graph (Fig. 3A), where the two rate constants are plotted against each other to provide Kd values along diagonal isoaffinity lines (42). The on rates varied 30-fold from 4.9 × 105 M−1 s−1 for IGF-II to 1.6 × 107 M−1 s−1 for VEGF. Off rates varied from 1.6 × 10−4 s−1 for TNF-α to exceptionally slow off rates below the sensitivity range of the Biacore T200 instrument (1.0 × 10−5 s−1) for both GDNF and IL-6. The software used to fit the data still provided off rates beyond the limit of the instrument (results are in SI Methods), but because of uncertainty in the accuracy, we limited off rates to no lower than 1.0 × 10−5 s−1 in the calculation of Kd. The resulting Kd values spanned more than two orders of magnitude, ranging from 2.8 × 10−10 M for TNF-α to below 7.1 × 10−13 M for GDNF.
Table 2.
Summary of kinetic analysis data for six biomarkers using SPR
| Analyte | Association rate constant ka, M−1 s−1 | Dissociation rate constant kd, s−1 | Equilibrium dissociation constant Kd, M |
| GDNF | 1.4 E+07 | <1.0 E−05 | <7.1 E−13 |
| IL-6 | 2.3 E+06 | <1.0 E−05 | <4.3 E−12 |
| VEGF | 1.6 E+07 | 5.6 E−05 | 3.6 E−12 |
| IGF-II | 4.9 E+05 | 2.5 E−05 | 5.1 E−11 |
| MIF | 1.1 E+06 | 9.4 E−05 | 8.3 E−11 |
| TNF-α | 5.7 E+05 | 1.6 E−04 | 2.8 E−10 |
Fig. 3.
Isoaffinity analysis and corresponding c-PLA performance for six biomarkers. (A) Kinetic analysis reveals two distinct groups for Kd values: one group with high affinity (single-digit picomolar) and another group with affinity above 50 pM. (B) The differences in antibody affinities are directly reflected in the c-PLA dose–response curves, where analytes with low Kd values display limits of detection in the subpicomolar range, while the other group exhibits limits of detection in the midpicomolar range or higher.
Because some of the off rates determined by SPR were found to be below the sensitivity range of the Biacore instrument, we performed complementary analysis using biolayer interferometry (BLI) (43). These measurements corroborated the SPR results; off rates for GDNF and IL-6 were also beyond the sensitivity of the Fortebio Octet RED instrument. Kinetic data for BLI measurements, binding curves for individual analytes fitted to a 1:1 binding model, and an isoaffinity chart for all analytes are provided in SI Methods. Kd values determined by BLI varied more than three orders of magnitude, ranging from 6.4 × 10−9 M for TNF-α to below 2.5 × 10−12 M for GDNF and IL-6. These values are approximately an order of magnitude higher than the corresponding SPR data, largely due to differences in the determination of on rates. SPR is a flow cell-based method with a 3D dextran matrix, whereas BLI utilizes planar fiberoptic sensors in a well-plate format. It has previously been reported that the BLI system underestimates fast on rates due to mass transfer limitations, which is consistent with our findings (44). The numeric values of kinetic constants determined by surface-based methods are likely to differ from solution-based values, which are presumably more applicable to homogenous PLA. Nonetheless, the relative rankings between the two methods display good agreement and are important indicators of comparative antibody quality.
All proximity probes described in this work originate from affinity-purified polyclonal antibodies. These antibodies are mixtures derived from different cell lineages, producing antibodies that recognize distinct epitopes of the antigen each with their own individual kinetic characteristics. Due to this heterogeneity, kinetic properties of polyclonal antibody are inherently difficult to characterize with precision, and the derived kinetic constants and affinities are considered an average for the different subpopulations existing within a batch of antisera. One prospective effect is that extended incubation time between sample and proximity probes will allow continued exchanges that progress toward higher-affinity interactions.
Using Kinetics Data to Improve c-PLA Performance.
The isoaffinity graph in Fig. 3A includes the kinetic information for all antibody–antigen interactions investigated, revealing a clear differentiation in affinities. The antibodies with the highest affinity are located in the upper left corner, as they are characterized by low off rates and high on rates that result in low Kd values. GDNF, VEGF, and IL-6 antibodies have high affinity, with Kd values below 5 pM. Accordingly, these analytes provide subpicomolar limits of detection in c-PLA and wide dynamic ranges of at least four orders of magnitude as seen in dose–response curves (Fig. 3B: dose–response curves are derived from the same batches of antibodies and antigens as those used in Fig. 3A but are not completely the same batches as those used in Fig. 2). The remaining analytes, MIF, TNF-α, and IGF-II, are clustered together, with Kd values above 50 pM. Consequently, c-PLA for these analytes does not perform nearly as well, exhibiting limits of detection in the picomolar range (or higher) and narrower dynamic ranges that are constrained by the lower affinity of the antibodies. This comparison shows that the inherent difference in sensitivity and dynamic range among the six analytes is not entirely dependent on the assay design but rather, is caused by the quality of the affinity reagents used. We note that the only biomarker for which c-PLA did not improve assay performance compared with t-PLA is GDNF, the analyte with the highest affinity. The improvements in limit of detection for c-PLA over t-PLA are also larger for the lowest-affinity interactions (Tables 1 and 2), indicating a trend toward increased benefits of c-PLA when only low-affinity reagents are available.
The detailed information obtained by kinetic analysis is beneficial for assay development, as it suggests modifications that may result in improved signals. We used the kinetic constants to predict the number of complexes formed at equilibrium during sample incubation and the new equilibrium that is established when the volume is increased by addition of the ligation mixture. Details are provided in SI Methods. The calculations suggest that ∼50% of the antigens present in solution are captured in a complex at equilibrium during sample incubation when the Kd value of an interaction equals 1 × 10−10 M (comparable with MIF and TNF-α; 8.3 × 10−11 and 2.8 × 10−10 M, respectively) (Fig. S6B). However, the subsequent addition of ligation mixture triggers complex dissociation until a new equilibrium is established. Modeling using Kd = 1 × 10−10 M indicates that, at the new equilibrium, essentially no antigens would remain in a complex, resulting in no circle formation and consequently, no detectable qPCR signal (Fig. S6C). However, the assay still functions for analytes with these Kd values, and the discrepancy is explained by suppressed dissociation as predicted by the low off rates. The complex half-life (t1/2) is defined as ln2/kd, indicating that, for both MIF and TNF-α (which have off rates of ∼1 × 10−4 s−1), it would take roughly 2 h for one-half of the complexes to dissociate. This is a sufficient duration to avoid significant losses during a 30-min ligation step (or 15 min in the case of t-PLA). In addition, these calculations do not consider any added advantages that the two circle-forming connector oligos have over t-PLA in retarding proximity probe diffusion away from the analyte, thereby facilitating antibody rebinding. DNA duplexes can be remarkably stable as exemplified by our SPR measurements, which use DNA-directed immobilization of the capture agents. Previous studies have determined the off rates for short oligonucleotides to range from 10−4 to below 10−5 s−1, values that are comparable with or lower than the off rates for the antibody–antigen interactions described in this work (45, 46). The calculations above highlight not only the benefits of high-affinity probes in PLAs but also, the importance of slow off rates or other means to demote dissociation, especially when it entails large-volume additions for ligation.
The large-volume addition associated with ligation was originally introduced to minimize background ligation in t-PLA (12). Background ligation events in t-PLA occur when affinity probes and bridge oligo are randomly brought into close proximity. For c-PLA, the likelihood of background ligation is lower, as this complex requires an additional connector oligo and two ligation events before producing a detectable signal. This results in less background and a better signal-to-noise ratio. Additional modeling studies provided in SI Methods (Fig. S6E) also indicate that higher proximity probe concentrations result in more complex formation, which may increase signal-to-noise ratios even further. This is analogous to experimental findings on solid phase, where higher densities of surface-bound capture antibodies have been shown to improve both limit of detection and dynamic range (47). Consequently, the combination of using a more stringent assay format, like c-PLA, with increased proximity probe concentrations should improve assay performance with low-affinity antibodies.
Stringency of c-PLA Improves Signal-to-Background Ratio and Limit of Detection.
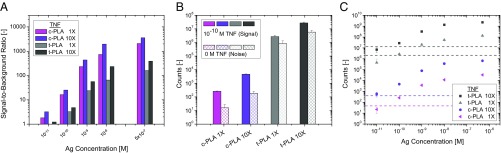
Based on simulations described above, we tested the impact of increased proximity probe concentration for both t-PLA and c-PLA. We chose to study this using the TNF-α assay, as it displayed the highest Kd and kd, providing a good example of low-affinity reagents with fast off rates. We performed both assay formats with the standard affinity probe concentration (1× = 0.25 nM) as well as a 10-fold increase. The results are shown in Fig. 4A and show a consistently higher signal-to-background ratio for c-PLA compared with t-PLA. The increase in signal-to-background ratio between the 10× and 1× affinity probe cases for the two methods is an expansion of our previous findings that probe concentration can be increased for low-affinity interactions (12), although it is a complex subject that must be carefully balanced between antibody affinity and a higher probability of random background ligation (48). The greater signal-to-noise ratio for c-PLA is explained by a reduction in background noise as seen in Fig. 4B. This is a consequence of rigorous assay design in combination with exonuclease treatment, which effectively eliminates the background noise. Note that the higher stringency of c-PLA also lowers the signal but that the stability of circle-forming complexes combined with even lower background noise more than compensates for this decline. The higher signal-to-background ratio for c-PLA 10× improves limit of detection about one order of magnitude compared with the original t-PLA (Fig. 4C). Additional experiments suggest that the probe concentration can be increased even further for low-affinity interactions, although it must be accompanied by an increase in the amount of connector oligonucleotides to ensure efficient circle formation.
Fig. 4.
Comparison of PLA methods at different probe concentrations for TNF-α. (A) c-PLA results offer a larger signal-to-background ratio than t-PLA. (B) Individual components for 100 pM signal-to-noise ratio. The greater signal-to-noise ratio for c-PLA is a consequence of higher stringency in c-PLA, which produces lower overall signals and larger differences between positive signal (100 pM) and negative background noise. (C) Dose–response curves showing that the higher signal-to-background ratios result in a more than 10-fold improvement in limit of detection (7 pM) for c-PLA when the probe concentrations are increased 10-fold compared with t-PLA (1×). Error bars denote 1 SD (n = 9), and the dashed lines denote limit of detection, defined as the mean signal of a blank sample +3 SD. Ag, antigen.
c-PLA Effectively Detects Proteins in Human Plasma.
The results described thus far were all from buffer solutions, as we sought to investigate the relationship between affinity and assay performance. Affinity analysis using SPR is not amenable to complex mixtures, like plasma, and although BLI is less sensitive to matrix effects (43), both methods require pure ligands for accurate determination of kinetic constants. The correlation between kinetic constants and assay performance is established in Fig. 3 and provides valuable information for assay development. However, assay performance is known to differ greatly between different matrices, and assay compatibility with complex mixtures is essential for diagnostic and clinical relevance (1).
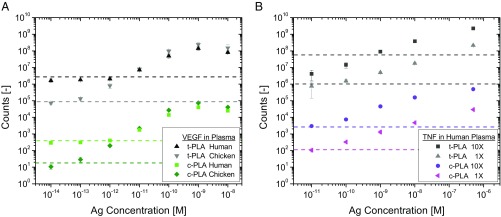
We have previously shown that multiplexed t-PLA enables quantitation of biomarkers in plasma samples (12–14). To determine the performance of c-PLA in a complex matrix, we tested two analytes: VEGF as an example of a high-affinity interaction (low Kd and slow off rates) and TNF-α as an example of a low-affinity interaction (high Kd and fast off rates). VEGF was effectively detected in human plasma by c-PLA down to physiological single-digit picomolar concentrations (12, 49) (Fig. 5A). Limit of detection is increased 100-fold from femtomolar concentration in buffer to 1 pM for c-PLA and 2 pM for t-PLA in human plasma. Dynamic range is reduced accordingly by two orders of magnitude for both methods. Average CVs were lower for c-PLA than t-PLA (at 11 and 22%, respectively), which is consistent with our findings in buffer solutions. These results confirm that the c-PLA provides less variation in a simplified assay format without the need for preamplification. We performed additional experiments with VEGF in chicken plasma (in which human VEGF is absent) to verify that the changes in limit of detection and dynamic range are not impeded by the assay format but are rather an effect of naturally occurring background levels of VEGF in human plasma (Fig. 5A). In chicken plasma, we achieved femtomolar limit of detection and more than four orders of magnitude dynamic range, which more closely reflect our original measurements in buffer solution. Hence, both PLA methods are compatible with complex mixtures, like human plasma, with additional benefits in assay performance for c-PLA.
Fig. 5.
Performance of PLAs in human plasma. (A) Dose–response curves for VEGF detection show assay compatibility for both PLA methods in human and chicken plasma. The difference in limit of detection between the two matrices is attributed to endogenous VEGF levels in human plasma that are absent in chicken plasma. (B) Dose–response curves for TNF-α detection in human plasma showing improvement in assay performance for c-PLA over t-PLA. A 10-fold increase in probe concentration (10× = 2.5 nM) improves reproducibility for c-PLA, while there is no improvement for t-PLA. Error bars denote 1 SD (n = 9), and the dashed lines denote limit of detection, defined as the mean signal of a blank sample +3 SD. Ag, antigen.
The advantages in assay performance of c-PLA over t-PLA for TNF-α (a low-affinity interaction) also persist in human plasma (Fig. 5B). The limit of detection for c-PLA is about 10 pM for both 1× and 10× proximity probe concentrations, with lower CVs for the higher probe concentration (8% for 10× vs. 16% for 1×), presumably due to more efficient capture of antigens at the higher concentration. The limit of detection for t-PLA is approximately an order of magnitude higher compared with c-PLA, which reduces the dynamic range accordingly. CVs remain at around 30% across the linear range for both proximity probe concentrations. In contrast to c-PLA, a 10-fold increase in proximity probe concentration for t-PLA did not improve sensitivity (limit of detection about 700 pM), underscoring the challenges when using this method with low-affinity antibodies. The ability to increase probe concentration in c-PLA also partially addresses the hook effect, as it results in a later onset of proximity probe saturation. Furthermore, statistical analysis of the CVs for TNF-α in plasma reveals that the lower CVs for c-PLA 10× are significantly different from CVs for c-PLA 1× as well as t-PLA 1× and 10× (P < 0.001). The difference in CVs between c-PLA 1× and t-PLA 1× is also significant (P < 0.001). This is encouraging, as the generally accepted precision requirement for clinical immunoassays is below 20% (40, 50). These findings indicate that the increased stringency of c-PLA provides advantages over t-PLA in terms of ease of use and sensitivity, while reducing variation. Most importantly, the opportunity to improve the performance of low-affinity antibodies by simply increasing their concentration in c-PLA is maintained in complex matrices, like plasma and serum.
Conclusions
In this report, we present a highly specific, sensitive, and convenient assay for quantitative protein detection. Our method builds on the existing PLA but also benefits from the formation of circular DNA molecules. PLAs combined with circle formation have traditionally been used for rolling circle amplification and have shown impressive results for in situ detection and digital quantification. We show that the circle formation increases reproducibility by minimizing noise across the linear dynamic range. This is facilitated by the inclusion of exonuclease treatment, which hydrolyzes noncircular DNA. This design also simplifies the workflow, as it eliminates the need for preamplification before qPCR quantification without any loss in assay performance. We compare both methods using six commonly used biomarkers and show equal or better performance for c-PLA.
Our kinetic analysis of the required antibody–antigen interactions, an essential precaution rarely taken during assay development for protein detection, shows a direct correlation between kinetic constants and assay performance. We establish the value of kinetic information to screen for suitable antibodies before proximity probe conjugation and to optimize proximity probe concentrations as well as assay procedures. Our work thus indicates that the regular use of kinetic analysis will be highly beneficial in improving assay performance across a wide variety of analytes.
The advantages of c-PLA over t-PLA for low-affinity interactions are aided by suppressed dissociation provided by longer DNA duplexes. We show that the additional proofreading step facilitated by the extra oligo in c-PLA can be exploited to increase proximity probe concentration without a concomitant increase in background ligation. This improves the signal-to-noise ratio and limit of detection compared with t-PLA and enables the use of low-affinity reagents. This is a major benefit, as low-affinity antibodies and variation in antibody specificity often contribute to systematic errors in research and clinical diagnostics (51). We also show that the benefits of c-PLA over t-PLA persist in complex media, such as human plasma. This will facilitate applications when high-affinity reagents are not available, allowing not only more analytes to be detected but also, greater sensitivity for protein detection. These advantages are strengthened by the ease of application of our assay as well as its amenability to automation and a variety of quantification methods (digital, fluorescent, electrical, and more). The formation of circular DNA also supports detection using rolling circle amplification and other isothermal amplification methods that can be accomplished on low-cost point-of-care devices (52, 53). We anticipate that an assay of this versatility and performance will help pave the way for much broader adoption of protein quantification into clinical and even resource-poor settings.
Methods
Materials.
Affinity-purified polyclonal antibodies and antigen targets were from R&D Systems. Product numbers are provided in SI Methods. All oligonucleotides were from Integrated DNA Technologies. Sequences are listed in SI Methods and were kept the same for all analytes to allow for an unbiased comparison. Oligonucleotides used for proximity probe conjugation were HPLC purified and designed to minimize probe–probe heteroduplex formation. All other reagents were from Sigma-Aldrich unless otherwise indicated.
Proximity Probe Conjugation.
Antibody–oligonucleotide conjugation was performed using hydrazone chemistry (Solulink) followed by purification according to the manufacturer’s protocol (Fig. S1). One polyclonal antibody batch was divided into two portions and coupled to amine-modified oligonucleotides containing either a free phosphorylated 5′ end or a free 3′ end. Antibody–oligonucleotide conjugates were analyzed on a 2100 Agilent Bioanalyzer using the protein 80 kit under reducing conditions following the manufacturer’s protocol. The estimated yield for all conjugates varied between 0.65 and 1.35 oligonucleotides per antibody. Final antibody–oligonucleotide concentration was determined using a Bradford protein assay (Bio-Rad) according to the manufacturer’s specification. Antibody–oligo probes were diluted to 5 nM in 1× PBS supplemented with 2 mM EDTA (Thermo Fisher Scientific), 0.10% BSA, and 0.02% NaN3 and were stored at 4 °C.
Proximity Probe Target Incubation.
Two microliters of sample [diluted in either 1× PBS with 0.1% BSA or neat plasma from chicken or human (Sigma-Aldrich)] was added to 2 μL of proximity probe mix and incubated for 2 h at 37 °C to establish complex formation between the target analyte and antibodies in proximity probes. The combined 4 μL incubation mixture contained a final concentration of 250 pM for each proximity probe and was supplemented with 0.35 mg/mL polyadenylic acid potassium salt, 1% BSA, 0.1% Triton X-100, 0.05% IgG, 0.01% aprotinin, 1 mM PMSF, and 4 mM EDTA (Thermo Fisher Scientific) in 0.375× PBS.
Ligation Step for c-PLA.
One hundred twenty microliters of ligation mixture was added to each 4-μL sample and incubated for 30 min at 45 °C. The ligation mixture contained 100 nM each of the two circle-forming connector oligos (SI Methods), 0.025 U/μL Ampligase (Epicentre), 0.5 mM NAD, 1 mM DTT (Sigma-Aldrich), 0.01% Triton X-100, and 0.01% BSA in 20 mM Tris, pH 8.4, 10 mM MgCl2, and 50 mM KCl (Thermo Fisher Scientific). Ligation was terminated by adding 10 μL of exonuclease mixture and incubated for 30 min at 37 °C followed by heat inactivation of exonuclease enzymes for 20 min at 80 °C. Exonuclease mixture contained 2 U/μL exonuclease I (New England Biolabs) and 2 U/μL exonuclease III (New England Biolabs) in 1× NEBuffer 1 containing 10 mM Bis⋅Tris⋅Propane⋅HCl, pH 7.0, 10 mM MgCl2, and 1 mM DTT (New England Biolabs).
Ligation Step for t-PLA.
One hundred twenty microliters of ligation mixture was added to each 4-μL sample and incubated for 15 min at 30 °C. The ligation mixture contained 100 nM t-PLA bridge oligo, 0.025 U/μL Ampligase (Epicentre), 0.25 mM NAD, 10 mM DTT, 0.02% Triton X-100, and 0.01% BSA in 20 mM Tris, pH 8.4, 1.5 mM MgCl2, and 50 mM KCl (Thermo Fisher Scientific). Ligation was terminated by adding 2 μL of stop ligation mixture followed by a 5-min incubation at room temperature. The stop ligation mixture contained 0.125 U/μL Uracil-DNA Excision Mix (Epicentre) in 20 mM Tris, pH 8.4, and 50 mM KCl (Thermo Fisher Scientific).
Preamplification Step for t-PLA.
Twenty-five microliters of the terminated ligation mixture was added to 25 μL preamplification mixture and cycled with the following conditions: 95 °C for 3 min (1 cycle), 95 °C for 30 s and 60 °C for 4 min (13 cycles), and a final hold at 4 °C. The preamplification mixture contained 20 nM preamplification primers, 0.06 U/μL Platinum Taq DNA Polymerase (Thermo Fisher Scientific), and 1.6 mM dNTP Mix (Agilent Technologies) in 20 mM Tris, pH 8.4, 6 mM MgCl2, and 50 mM KCl (Thermo Fisher Scientific). After preamplification, the product was diluted 10-fold in 10 mM Tris, pH 8, 0.1 mM EDTA buffer and stored at 4 °C until qPCR quantification.
qPCR Quantification and Data Analysis.
Two microliters of either exonuclease-treated ligation product (for c-PLA) or diluted preamplification product (for t-PLA) was added to qPCR master mix to a final volume of 10 μL containing 400 nM primers and 1× Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). Samples were quantified using real-time qPCR (ABI 7900 HT) with the following thermal cycling conditions: 95 °C for 10 min (1 cycle) and 95 °C for 15 s and 60 °C for 60 s (40 cycles). Cycle threshold (Ct) values were converted to an estimated number of ligated molecules using the formula 10−0.301 × Ct + 11.439 as previously described (13). Experiments were performed with three biological replicates that were quantified in triplicate qPCR experiments, generating nine data points per concentration analyzed. Limit of detection was derived relative to the blank sample, where the dose–response curve and the dashed line (mean signal of a blank sample + 3 SDs) intersect (40). Statistical analysis was performed using bootstrapping (random sampling with replacement on the independent measurements of biological replicates 1,000 times).
Kinetic Analysis Using SPR.
SPR analysis was performed at 25 °C using a Biacore T200 instrument and a Biotin CAPture Kit (GE Healthcare) (Fig. S3A). All antibodies were biotinylated using the EZ-Link NHS-PEO4-Biotin kit (Thermo Fisher Scientific) according to the manufacturer’s recommendation using a 1:1 mol ratio of biotin to antibody. Excess biotin was removed using Zeba spin columns to avoid interference with the binding of biotinylated antibodies to the surface of the sensor chip. Antibody immobilization was tailored to a level that resulted in a maximum analyte binding capacity (Rmax) of 25–50 RU/s. Analytes were diluted in 0.01 M Hepes, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% vol/vol Surfactant P20 (GE Healthcare) buffer and passed over the chip at a rate of 30 μL/min. Association was measured for 120 s, and dissociation was measured for 1,800 s. Sensors were regenerated using a solution containing 6 M Guanidine-HCl and 0.25 M NaOH. Rate constants (ka and kd) were determined using BIAevaluation software, version 4.1 (Biacore) with a 1:1 model and global fitting of at least five concentrations in twofold dilution series ranging from 200 to 0 nM (Fig. S4 and Table S1). Affinity constants (Kd) were subsequently derived from the ratio of ka and kd.
Kinetic Analysis Using BLI.
BLI analysis was performed on a Fortebio Octet RED instrument using protein G biosensors (Fortebio) (Fig. S3B). All antibodies and analytes were diluted in assay buffer (0.1 mg/mL BSA and 0.13% Triton X-100 in 10 mM Tris, pH 7.5, 1 mM CaCl2, and 150 mM NaCl). Antibody immobilization was tailored to a binding level of ∼3.6 nm by varying the antibody loading time. All measurements were conducted at 30 °C in 200 μL total with constant agitation. Association was measured for 120 s, and dissociation was measured for 1,200 s. Sensors were regenerated using a 10 mM glycine solution at pH 1.7. For each analyte, three replicates covering full kinetics cycles were performed and resulted in identical binding curves across all cycles. Rate constants (ka and kd) were determined using data analysis software, version 8.2 (Fortebio) with a 1:1 model and global fitting of at least five concentrations in threefold dilution series ranging from 500 to 0 nM. Affinity constants (Kd) were subsequently derived from the ratio of ka and kd. The results of the kinetic analysis by BLI are provided in Fig. S5 and Table S2.
Supplementary Material
Acknowledgments
We thank Andrew Hill, Terence Hui, Raeka Aiyar, and Wenzhong Xiao for valuable discussions. This project was supported by NIH Grant HG000205.
Footnotes
Conflict of interest statement: S.F. owns stock in a company (Olink AB) with patents on the core technology described, "Proximity Ligation Assay."
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718283115/-/DCSupplemental.
References
- 1.Landegren U, et al. Opportunities for sensitive plasma proteome analysis. Anal Chem. 2012;84:1824–1830. doi: 10.1021/ac2032222. [DOI] [PubMed] [Google Scholar]
- 2.Kingsmore SF. Multiplexed protein measurement: Technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wild D, editor. The Immunoassay Handbook. Elsevier; Oxford: 2013. [Google Scholar]
- 4.Zhang H, Zhao Q, Li XF, Le XC. Ultrasensitive assays for proteins. Analyst (Lond) 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 5.Spengler M, Adler M, Niemeyer CM. Highly sensitive ligand-binding assays in pre-clinical and clinical applications: Immuno-PCR and other emerging techniques. Analyst (Lond) 2015;140:6175–6194. doi: 10.1039/c5an00822k. [DOI] [PubMed] [Google Scholar]
- 6.Sano T, Smith CL, Cantor CR. Immuno-PCR: Very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 7.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 8.Schweitzer B, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie S, Walton SP. Development of a dual-aptamer-based multiplex protein biosensor. Biosens Bioelectron. 2010;25:2663–2668. doi: 10.1016/j.bios.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 11.Gullberg M, et al. Cytokine detection by antibody-based proximity ligation. Proc Natl Acad Sci USA. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredriksson S, et al. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 13.Fredriksson S, et al. Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin Chem. 2008;54:582–589. doi: 10.1373/clinchem.2007.093195. [DOI] [PubMed] [Google Scholar]
- 14.Chang ST, et al. Identification of a biomarker panel using a multiplex proximity ligation assay improves accuracy of pancreatic cancer diagnosis. J Transl Med. 2009;7:105. doi: 10.1186/1479-5876-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg M, et al. Multiplexed homogeneous proximity ligation assays for high-throughput protein biomarker research in serological material. Mol Cell Proteomics. 2011;10:M110.004978. doi: 10.1074/mcp.M110.004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tate J, Ward G. Interferences in immunoassay. Clin Biochem Rev. 2004;25:105–120. [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Hu J, Sollie RS, Easley CJ. Improvement of sensitivity and dynamic range in proximity ligation assays by asymmetric connector hybridization. Anal Chem. 2010;82:6976–6982. doi: 10.1021/ac101762m. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, et al. A sensitive proximity ligation assay for active PSA. Biol Chem. 2006;387:769–772. doi: 10.1515/BC.2006.096. [DOI] [PubMed] [Google Scholar]
- 19.Xie S, Moya C, Bilgin B, Jayaraman A, Walton SP. Emerging affinity-based techniques in proteomics. Expert Rev Proteomics. 2009;6:573–583. doi: 10.1586/epr.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro-López V, Elizalde J, Pacek M, Hijona E, Bujanda L. A simple and portable device for the quantification of TNF-α in human plasma by means of on-chip magnetic bead-based proximity ligation assay. Biosens Bioelectron. 2014;54:499–505. doi: 10.1016/j.bios.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Pai S, Ellington AD, Levy M. Proximity ligation assays with peptide conjugate ‘burrs’ for the sensitive detection of spores. Nucleic Acids Res. 2005;33:e162. doi: 10.1093/nar/gni150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schallmeiner E, et al. Sensitive protein detection via triple-binder proximity ligation assays. Nat Methods. 2007;4:135–137. doi: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- 23.Tavoosidana G, et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc Natl Acad Sci USA. 2011;108:8809–8814. doi: 10.1073/pnas.1019330108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Li X-F, Le XC. Binding-induced DNA assembly and its application to yoctomole detection of proteins. Anal Chem. 2012;84:877–884. doi: 10.1021/ac203207g. [DOI] [PubMed] [Google Scholar]
- 25.Di Giusto DA, Wlassoff WA, Gooding JJ, Messerle BA, King GC. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvius J, et al. Digital quantification using amplified single-molecule detection. Nat Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 27.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 28.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ericsson O, et al. A dual-tag microarray platform for high-performance nucleic acid and protein analyses. Nucleic Acids Res. 2008;36:e45. doi: 10.1093/nar/gkn106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez de la Torre TZ, et al. Sensitive detection of spores using volume-amplified magnetic nanobeads. Small. 2012;8:2174–2177. doi: 10.1002/smll.201102632. [DOI] [PubMed] [Google Scholar]
- 31.Ke R, Nong RY, Fredriksson S, Landegren U, Nilsson M. Improving precision of proximity ligation assay by amplified single molecule detection. PLoS One. 2013;8:e69813. doi: 10.1371/journal.pone.0069813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 33.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S, et al. A general synthetic approach for designing epitope targeted macrocyclic peptide ligands. Angew Chem Int Ed Engl. 2015;54:13219–13224. doi: 10.1002/anie.201505243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai CT, Robinson PV, Spencer CA, Bertozzi CR. Ultrasensitive antibody detection by Agglutination-PCR (ADAP) ACS Cent Sci. 2016;2:139–147. doi: 10.1021/acscentsci.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albayrak C, et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol Cell. 2016;61:914–924. doi: 10.1016/j.molcel.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 37.Darmanis S, et al. Sensitive plasma protein analysis by microparticle-based proximity ligation assays. Mol Cell Proteomics. 2010;9:327–335. doi: 10.1074/mcp.M900248-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd J, et al. Ultrasensitive flow-based immunoassays using single-molecule counting. Clin Chem. 2007;53:1990–1995. doi: 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- 40.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29:S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper MA. Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002;1:515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 42.Markgren P-O, et al. Relationships between structure and interaction kinetics for HIV-1 protease inhibitors. J Med Chem. 2002;45:5430–5439. doi: 10.1021/jm0208370. [DOI] [PubMed] [Google Scholar]
- 43.Concepcion J, et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb Chem High Throughput Screen. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 44.Estep P, et al. High throughput solution-based measurement of antibody-antigen affinity and epitope binning. MAbs. 2013;5:270–278. doi: 10.4161/mabs.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gotoh M, Hasegawa Y, Shinohara Y, Shimizu M, Tosu M. A new approach to determine the effect of mismatches on kinetic parameters in DNA hybridization using an optical biosensor. DNA Res. 1995;2:285–293. doi: 10.1093/dnares/2.6.285. [DOI] [PubMed] [Google Scholar]
- 46.Liebermann T, Knoll W, Sluka P, Herrmann R. Complement hybridization from solution to surface-attached probe-oligonucleotides observed by surface-plasmon-field-enhanced fluorescence spectroscopy. Colloids Surf A Physicochem Eng Asp. 2000;169:337–350. [Google Scholar]
- 47.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen MC, Nederby L, Henriksen MOB, Hansen M, Nyvold CG. Sensitive ligand-based protein quantification using immuno-PCR: A critical review of single-probe and proximity ligation assays. Biotechniques. 2014;56:217–228. doi: 10.2144/000114164. [DOI] [PubMed] [Google Scholar]
- 49.Larsson A, Sköldenberg E, Ericson H. Serum and plasma levels of FGF-2 and VEGF in healthy blood donors. Angiogenesis. 2002;5:107–110. doi: 10.1023/a:1021588227705. [DOI] [PubMed] [Google Scholar]
- 50.Andreasson U, et al. A practical guide to immunoassay method validation. Front Neurol. 2015;6:179. doi: 10.3389/fneur.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 52.Shen F, et al. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal Chem. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh E-C, et al. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci Adv. 2017;3:e1501645. doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.