Significance
Abasic (apurinic/apyrimidinic, AP) sites in DNA result from spontaneous and repair-mediated base release. They may be processed by AP endonucleases or AP lyases, but the relative roles of both types of enzymes are poorly understood. Our study reveals that the model plant Arabidopsis uses an AP lyase-dependent pathway to repair AP sites generated by spontaneous loss of N7-methylguanine (N7-meG), a major lesion arising from DNA methylation damage. We further show that the main Arabidopsis AP endonuclease is active on AP sites generated by enzymatic excision of N7-meG, but not on those arising from N7-meG loss. Our findings identify an important role for AP lyase activity in plants and challenge the assumption that spontaneous and repair-generated AP sites have identical biochemical properties.
Keywords: base excision repair, AP endonucleases, AP lyases, abasic sites, N7-methylguanine
Abstract
Abasic (apurinic/apyrimidinic, AP) sites in DNA arise from spontaneous base loss or by enzymatic removal during base excision repair. It is commonly accepted that both classes of AP site have analogous biochemical properties and are equivalent substrates for AP endonucleases and AP lyases, although the relative roles of these two types of enzymes are not well understood. We provide here genetic and biochemical evidence that, in Arabidopsis, AP sites generated by spontaneous loss of N7-methylguanine (N7-meG) are exclusively repaired through an AP endonuclease-independent pathway initiated by FPG, a bifunctional DNA glycosylase with AP lyase activity. Abasic site incision catalyzed by FPG generates a single-nucleotide gap with a 3′-phosphate terminus that is processed by the DNA 3′-phosphatase ZDP before repair is completed. We further show that the major AP endonuclease in Arabidopsis (ARP) incises AP sites generated by enzymatic N7-meG excision but, unexpectedly, not those resulting from spontaneous N7-meG loss. These findings, which reveal previously undetected differences between products of enzymatic and nonenzymatic base release, may shed light on the evolution and biological roles of AP endonucleases and AP lyases.
Abasic (apurinic/apyrimidinic, AP) sites are ubiquitous DNA lesions generated by spontaneous hydrolysis of the N-glycosylic bond connecting the base with the deoxyribose moiety of the nucleotide (1). It has been estimated that 2,000–10,000 AP sites arise spontaneously per mammalian cell per generation (2). Abasic sites are also generated as intermediates during the base excision repair (BER) pathway, following excision of damaged bases by monofunctional DNA glycosylases (3, 4). In addition, they may be induced directly by oxygen radical species (5) and indirectly by spontaneous release of alkylated bases such as N7-methylguanine (N7-meG) (5–7). Under physiological conditions, AP sites exist in an equilibrium mixture of α- and β-hemiacetals of the closed furanose form, with ∼1% present in the opened aldehyde form (8). The latter is prone to spontaneous hydrolysis by β- and β, δ-elimination, generating single-strand breaks (SSB) (9). AP sites can block DNA replication and transcription and slowly decay to form SSB, therefore causing cytotoxic effects. They are also mutagenic due to erroneous bypass by translesion DNA synthesis (10).
Abasic sites are mainly repaired through BER initiated either by AP endonucleases or by AP lyase activities associated with bifunctional DNA glycosylases (4, 11, 12). AP endonucleases hydrolyze DNA at the 5′-side of the AP site, leaving 3′-hydroxyl (3′-OH) and 5′-deoxyribose phosphate (5′-dRP) termini (13). AP lyases cleave 3′ to the AP site by β-elimination, generating 3′-phosphor-α, β-unsaturated aldehyde (3′-PUA), and 5′-phosphate (5′-P) termini. A subset of AP lyases catalyze β, δ-elimination and generate 3′-phosphate (3′-P) termini (13). Therefore, AP endonucleases and AP lyases generate single-nucleotide gaps with 5′- and 3′-blocked ends, respectively. The processing of such noncanonical termini influences subsequent steps of the repair process, which may continue through insertion of either one (short-patch, SP-BER) or several (long-patch, LP-BER) nucleotides (14, 15).
In SP-BER, the 5′-dRP group generated by AP endonucleases is converted to 5′-P by a deoxyribophosphatase activity that in mammalian cells is associated to DNA polymerase β (16). However, such step is rate-limiting (17) and the 5′-dRP may be also removed as part of an oligonucleotide excised by FEN1 nuclease after strand displacement during LP-BER (14). Since AP lyases produce canonical 5′-P termini, it has been proposed that they usually initiate SP-BER (18). In this case, processing of 3′-PUA generated by β-elimination is carried out by a 3′-phosphodiesterase activity associated to AP endonucleases (19, 20) and 3′-P produced by β, δ-elimination is removed by a DNA 3′-phosphatase, such a mammalian PNK (21) or plant ZDP (22, 23). Therefore, AP site incision by either AP endonucleases or AP lyases determines downstream BER steps requiring different subsets of proteins. However, the factors influencing the choice between both types of enzymes are unknown.
BER has been extensively studied in bacterial, yeast, and mammalian systems, but knowledge about this crucial repair pathway has been gained in plants only recently. Results obtained so far, mostly in Arabidopsis, indicate that plants share many BER components with other organisms but possess some distinctive features and combinations. In Arabidopsis, repair of uracil is initiated by the monofunctional uracil DNA glycosylase UNG (24), and the ensuing AP sites may be processed by both AP lyase and AP endonuclease activities detectable in cell extracts (15, 25). Interestingly, and despite the lack of plant homologs of Pol β and LigIII, repair proceeds not only by LP-BER but also through SP-BER, and in both cases the final ligation step is catalyzed by LIGI (15, 25). Arabidopsis also possesses an MBD4-like protein active on U:G and T:G mismatches but, unlike its mammalian homolog, it lacks a methyl-CpG-binding domain (26). Removal of oxidized pyrimidines is carried out by an NTH1 homolog (27), whereas repair of oxidized guanine (8-oxoG) involves both FPG and OGG1 homologs (22, 28–30), a distinctive combination of bacterial-like and eukaryotic-like 8-oxoG DNA glycosylases characteristic of plants and some fungi (31). Repair intermediates generated by the bifunctional DNA glycosylases FPG and OGG1 are processed by the DNA 3′-phosphatase ZDP and ARP, the major AP endonuclease detectable in Arabidopsis cell extracts (22). In addition to removing damaged bases, plants use BER for epigenetic reprograming initiated by 5-meC DNA glycosylases/lyases of the ROS1/DME family (32, 33). These enzymes generate single-nucleotide gaps with either 3′-PUA or 3′-P ends (32, 33) that are processed by the 3′-phosphodiesterase activity of AP endonuclease APE1L (20) and the DNA 3′-phosphatase ZDP (23), respectively. Although Arabidopsis possesses several DNA glycosylases/lyases (27–29, 32, 33) and three different AP endonucleases (34), their relative roles in the repair of AP sites have not been established so far.
It is generally accepted that AP site repair in vivo is predominantly initiated by AP endonucleases, but there is evidence that AP lyases play a prominent role in yeast. Thus, AP sites in Schizosaccharomyces pombe are primarily incised by the DNA glycosylase/AP lyase Nth1, which generates 3′-PUA ends that are further processed by the phosphodiesterase activity of Apn2, the major AP endonuclease in fission yeast (35, 36). Rather than AP incision, the main function of Apn2 appears to be the removal of 3′-blocked ends generated by the AP lyase activity of Nth1. A similar mechanism operates in Saccharomyces cerevisiae (37).
The biological relevance of AP lyases in the processing of abasic sites is poorly understood, and it has been suggested that it may be a yeast-specific feature (38). In this paper we report biochemical and genetic evidence that the AP lyase activity of Arabidopsis DNA glycosylase FPG plays a major role in the repair of AP sites generated by spontaneous depurination of N7-meG. FPG incision generates a 3′-P terminus that is converted to 3′-OH by the DNA 3′-phosphatase ZDP, enabling DNA polymerase and ligase activities to complete repair in an AP endonuclease-independent pathway. Importantly, we found that ARP, the major AP endonuclease in Arabidopsis, is active on AP sites generated by enzymatic excision of N7-meG but not on products of spontaneous N7-meG depurination. Our results indicate that hitherto unknown differences between the products of enzymatic and nonenzymatic base release dictate AP site DNA repair choice in Arabidopsis.
Results
Preparation and Characterization of Substrates to Monitor Repair of DNA Methylation Damage in Arabidopsis Cell-Free Extracts.
ZDP is the major, if not the only, DNA 3′-phosphatase activity detected in Arabidopsis cells (23). We have previously reported that ZDP is required to process 3′-P termini generated by FPG during 8-oxoG repair (22) or by ROS1 during active DNA demethylation (23). Intriguingly, zdp−/− mutants are hypersensitive to methyl methanesulfonate (MMS) (23), suggesting that ZDP also plays an important role in the repair of alkylation DNA damage.
The most abundant lesion caused by MMS is N7-meG, which accounts for 80–85% of total DNA methylation (39). This lesion is neither cytotoxic nor mutagenic but under physiological conditions undergoes spontaneous depurination, exhibiting half-lives ranging from 69 to 192 h at neutral pH and 37 °C (7). In addition, N7-meG may suffer opening of its imidazole ring to yield 5-N-methyl-2,6-diamino-4-hydroxyformamidopyrimidine (me-FAPy-G), in a reaction favored by basic conditions (7).
To monitor repair of MMS-induced DNA damage in Arabidopsis cell extracts we prepared a DNA duplex containing a single N7-meG residue using an enzymatic method (40) (Fig. S1A). We also generated an analogous substrate with me-FAPy-G by incubating the N7-meG–containing DNA at 37 °C for 5 h at pH 11. N7-meG is known to be excised by the monofunctional human alkyladenine DNA glycosylase (hAAG) (41), whereas Escherichia coli Fpg excises me-FAPy-G very efficiently (42). As shown in Fig. S1B, the oligonucleotide containing N7-meG was completely cleaved upon incubation with hAAG and human AP endonuclease 1 (hAPE1), while no product was observed for the me-FAPy-G substrate. Conversely, the oligonucleotide containing me-FAPy-G was fully cleaved by E. coli Fpg but was resistant to treatment with hAAG and hAPE1. As expected, reaction products generated by hAPE1 and E. coli Fpg contained 3′-OH and 3′-P termini, respectively (Fig. S1B).
The DNA Phosphatase ZDP Functions Downstream of FPG During Repair of DNA Containing N7-meG.
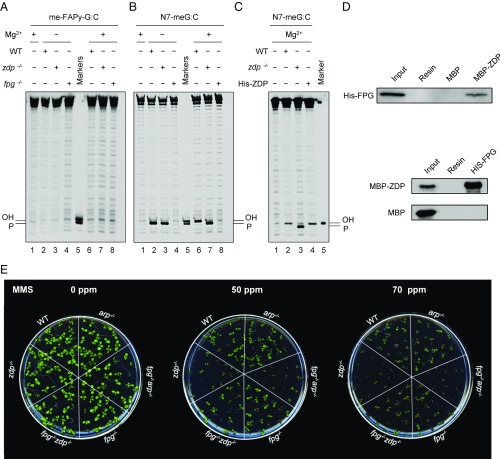
We next incubated cell extracts from WT and zdp−/− or fpg−/− mutant plants with DNA substrates containing either N7-meG or me-FAPy-G (Fig. 1 A–C). No incision products were detected in reactions with the DNA substrate containing me-FAPy-G (Fig. 1A). However, when incubated with the duplex containing N7-meG in the absence of Mg2+, WT extracts generated a product with a 3′-P terminus that was converted into a 3′-OH end upon Mg2+ addition (Fig. 1B, lanes 2 and 6). Such conversion was undetectable in zdp−/− mutant extracts (Fig. 1B, lanes 3 and 7), but was restored when purified recombinant ZDP protein was added to the repair reaction (Fig. 1C, lanes 3 and 4). These results indicate that ZDP phosphatase activity is required to process a 3′-P intermediate generated during the repair of DNA containing N7-meG.
Fig. 1.
ZDP 3′-phosphatase functions downstream of FPG DNA glycosylase/lyase during repair of DNA containing N7-meG. (A–C) Double-stranded oligonucleotide substrates (20 nM) containing either a single me-FAPy-G:C (A) or an N7-meG:C pair (B and C) were incubated with WT, fpg−/−, or zdp−/− Arabidopsis cell-free extracts for 16 h at 37 °C. Then, MgCl2 (2 mM) was added to reactions and incubation continued for 1 h. When indicated, His-ZDP protein (1.8 nM) was added to zdp−/− extracts. All reaction products were separated by denaturing PAGE and detected by fluorescence scanning. (D) Pull-down assays using either His-FPG (Upper) or MBP-ZDP protein (Lower) as baits. (E) MMS sensitivity assay. Arabidopsis seedlings were grown in MS nutrient agar containing increasing concentrations of MMS and photographs were taken after 14 d of growth.
We have previously shown that ZDP processes 3′-P termini generated by FPG during 8-oxoG repair (22). Therefore, we hypothesized that this DNA glycosylase/lyase might be also responsible for the generation of such intermediates during N7-meG repair. We found that cell extracts from mutant fpg−/− plants do not generate detectable repair incision products, either in the absence or the presence of Mg2+ (Fig. 1B, lanes 4 and 8). These results indicate that FPG functions in the repair of DNA containing N7-meG, performing a β, δ-elimination and generating a single-nucleotide gap with a 3′-P terminus that is converted to 3′-OH by the DNA 3′-phosphatase activity of ZDP.
Since ZDP is required to process FPG products, we tested for a direct interaction between both proteins using pull-down assays (Fig. 1D). We found His-FPG bound to MBP-ZDP, but not to MBP alone, immobilized in an amylose column (Fig. 1D, Upper). Conversely, MBP-ZDP, but not MBP alone, bound to His-FPG immobilized in a nickel-agarose column (Fig. 1D, Lower). These results suggest that FPG and ZDP directly interact in vitro.
The results described above suggest that FPG functions upstream ZDP during repair of DNA containing N7-meG. We therefore hypothesized that the hypersensitivity of zdp−/− mutants to MMS could be due to the accumulation of unprocessed SSB intermediates containing 3′-P ends generated by FPG. To test this idea, we generated a double fpg−/− zdp−/− mutant and assessed its resistance to MMS in comparison with WT and single fpg−/− or zdp−/− mutants (Fig. 1E). The results show that inactivation of FPG activity in zdp−/− mutant plants restores MMS resistance to nearly WT levels. Altogether, these results indicate that FPG functions upstream ZDP during repair of DNA containing N7-meG. We also found that, while single fpg−/− mutants are not sensitive to MMS, the combined deficiency of FPG and ARP, the major Arabidopsis AP endonuclease (25), causes an MMS sensibility similar to that of zdp−/− mutants. These results suggest that ARP also plays a role in the repair of MMS-induced DNA damage.
FPG Incises AP Sites Generated by Spontaneous Depurination of N7-meG.
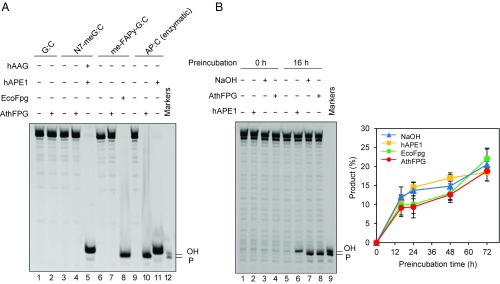
We next examined in detail the role of FPG during repair of DNA containing N7-meG. As indicated above, N7-meG may undergo either spontaneous depurination to generate an AP site or imidazole-ring opening to yield me-FAPy-G. We therefore tested the activity of Arabidopsis FPG protein against N7-meG and its two derivatives (Fig. 2). Control reactions confirmed that N7-meG was only incised by the simultaneous addition of hAAG and hAPE1, me-FAPy-G was processed by E. coli Fpg, and the AP site was incised by hAPE1 (Fig. 2A, lanes 5, 8, and 11, respectively). We found that Arabidopsis FPG did not display detectable incision activity against either N7-meG or me-FAPy-G (Fig. 2A, lanes 4 and 7). However, it efficiently incised the AP site, generating as a product a DNA repair intermediate with a 3′-P terminus (Fig. 2A, lane 10). The robust AP lyase activity of Arabidopsis FPG has been previously reported (31). It has been described that a truncated form of Arabidopsis FPG processes me-FAPy-G with low efficiency (43), but we could not detect such activity, either with the full-length enzyme or with cell extracts, at least under our experimental conditions.
Fig. 2.
FPG incises AP sites generated by spontaneous depurination of N7-meG. (A) Double-stranded oligonucleotide substrates (20 nM) containing a single lesion opposite C (N7-meG, me-FAPy-G, or AP site generated by uracil excision) were incubated for 2 h at 37 °C with Arabidopsis FPG (10 nM), hAPE1 (1 U), hAAG (2 U), or E. coli Fpg (8 U). (B) A double-stranded oligonucleotide substrate containing a single N7-meG:C pair was preincubated in DNA incision assay buffer (Methods) at 37 °C for the indicated times and then treated with human APE1 (1 U), E. coli Fpg (8 U), or Arabidopsis FPG (10 nM) for 1 h at 37 °C, or with NaOH (15 mM) for 10 min. at 70 °C. After stabilization with NaBH4, reaction products were separated by denaturing PAGE and detected by fluorescence scanning. Values are means with SEs from three independent experiments.
We therefore hypothesized that, during DNA repair reactions with cell extracts, spontaneous depurination of N7-meG generates AP sites that are substrates for FPG. To test this idea, we preincubated a DNA duplex with a single N7-meG residue during different time periods in the absence of cell extract and then submitted DNA either to an alkali treatment at 70 °C or to incubation with hAPE1, E. coli Fpg, or Arabidopsis FPG. We found that alkali/heat-labile sites sensitive to all three enzymes accumulated in DNA at position 28 (Fig. 2B). The observed accumulation rate is compatible with that reported for depurination of N7-meG in dsDNA at neutral pH and 37 °C (7). As expected for AP sites, incision by hAPE1 generated 3′-OH termini whereas incision by Arabidopsis FPG or heat/alkali treatment generated 3′-P termini (Fig. 2B). Altogether, these results suggest that FPG enzyme present in Arabidopsis cell extracts efficiently processes AP sites generated by spontaneous depurination of N7-meG, but it is not active either on N7-meG itself or its me-FAPy-G derivative.
ARP, the Major Arabidopsis AP Endonuclease, Plays a Negligible Role in the Repair of Depurinated N7-meG.
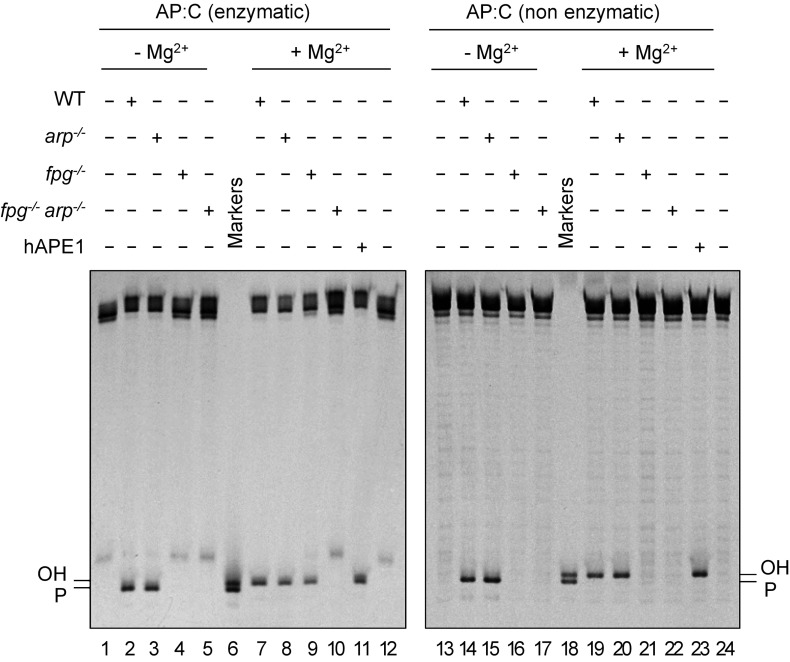
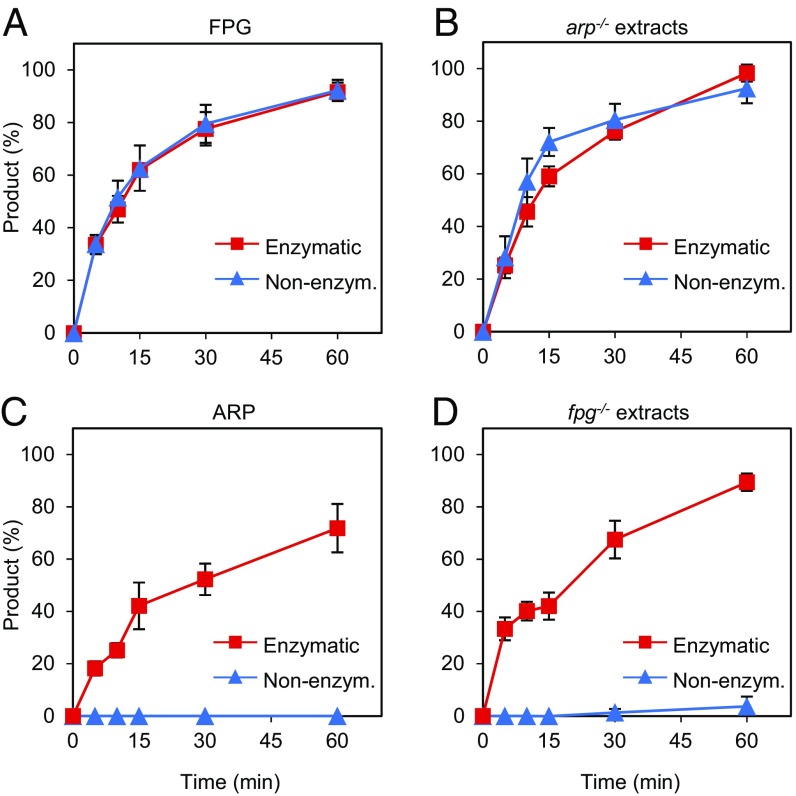
We next examined the relative roles of FPG and ARP in the repair of AP sites generated by spontaneous depurination of N7-meG. We first analyzed the level of AP endonuclease and AP lyase activity in cell extracts from WT, fpg−/−, arp−/−, and double fpg−/− arp−/− mutant plants. Equivalent cell extract quality and DNA repair competence were previously verified by measuring UDG activity on a DNA duplex containing a U:C mismatch (Fig. S2). We then incubated cell extracts with a DNA substrate containing an AP site opposite C generated by uracil excision (Fig. 3, Left). To mimic the partial depurination of DNA containing N7-meG (discussed below), heteroduplex DNA with the enzymatically generated AP:C was mixed with homoduplex G:C at a 1:9 ratio before initiating repair reactions. We found that arp−/− extracts catalyzed AP incision with efficiency similar to WT extracts, either in the absence or the presence of Mg2+ (Fig. 3, lanes 2 and 3 and 7 and 8). This result indicates that arp−/− extracts only exhibit AP lyase activity, which is Mg2+-independent. However, fpg−/− extracts also catalyzed AP incision at levels comparable to those of WT extracts, but only in the presence of Mg2+ (Fig. 3, lanes 4 and 9). This result indicates that they only exhibit AP endonuclease activity, which is Mg2+-dependent. No AP incision activity was detected in fpg−/− arp−/− mutant extracts, although limited spontaneous hydrolysis was detected in the form of β-elimination products (Fig. 3, lanes 5 and 10). These results indicate that the only AP lyase and AP endonuclease activities detectable in Arabidopsis cell extracts under the experimental conditions used are FPG and ARP, respectively.
Fig. 3.
ARP plays a negligible role in repair of depurinated N7-meG. DNA substrates (20 nM) were a 9:1 mixture of homoduplex G:C and heteroduplex AP:C generated either by uracil excision (enzymatic AP:C, Left) or by spontaneous N7-meG depurination (nonenzymatic AP:C, Right). Similar concentrations of each type of AP site were verified by incision with hAPE1 (10 U) (lanes 11 and 23). Substrates were incubated with Arabidopsis cell-free extracts (8 µg) for 1 h at 37 °C. After stabilization with NaBH4, reaction products were separated by denaturing PAGE and detected by fluorescence scanning.
We next performed analogous repair reactions with a DNA substrate containing depurinated N7-meG (Fig. 3, Right). Depurination was achieved by preincubating a DNA duplex containing N7-meG in the absence of cell extracts for 16 h (Fig. 2B and Methods). As previously observed (Fig. 1B), we found that in the absence of Mg2+, WT extracts generated a product with a 3′-P terminus that was converted into a 3′-OH end upon Mg2+ addition (Fig. 3, lanes 14 and 19), whereas fpg−/− extracts did not catalyze any incision, either in the absence or the presence of Mg2+ (Fig. 3, lanes 16 and 21). In contrast, arp−/− mutant extracts displayed an incision pattern very similar to that of WT plants (Fig. 3, lanes 15 and 20). Extracts from double fpg−/− arp−/− mutant plants did not display any detectable incision activity (Fig. 3, lanes 17 and 22). Altogether, these results indicate that incision of depurinated N7-meG in Arabidopsis cell extracts is ARP-independent and it is carried out exclusively by FPG.
Depurinated N7-meG Is Repaired Through FPG-Dependent SP-BER.
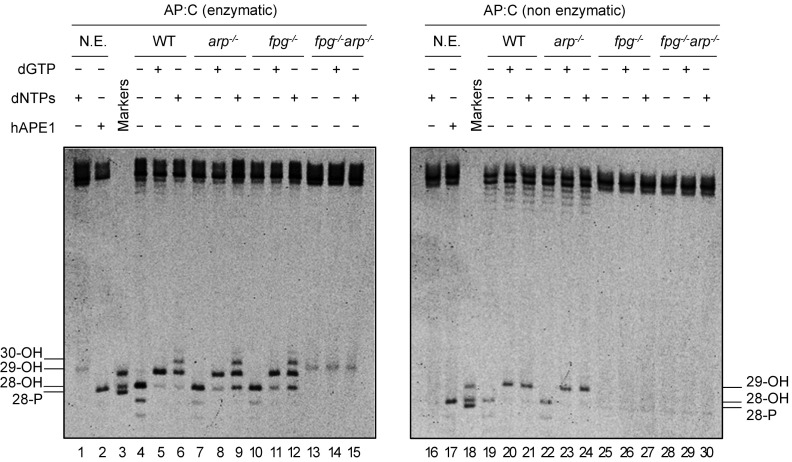
It has been previously suggested that BER initiated by monofunctional DNA glycosylases continues via both SP- and LP-BER, whereas that initiated by bifunctional DNA glycosylases/lyases continues primarily via SP-BER (18). Since repair of depurinated N7-meG is AP lyase-dependent and AP endonuclease-independent we hypothesized that it should mainly involve SP-BER. To test this idea, we performed gap-filling DNA repair reactions either in the presence of dGTP or all four dNTPs (Fig. 4).
Fig. 4.
Depurinated N7-meG is repaired through FPG-dependent SP-BER. DNA substrates (20 nM) were a 9:1 mixture of homoduplex G:C and heteroduplex AP:C generated either by uracil excision (enzymatic AP:C, Left) or by spontaneous N7-meG depurination (nonenzymatic AP:C, Right). Similar concentrations of each type of AP site were verified by incision with hAPE1 (10 U) (lanes 2 and 17). Substrates were incubated with Arabidopsis cell-free extracts (8 µg) for 1 h at 37 °C with or without either dGTP or all four dNTPs. After stabilization with NaBH4, reaction products were separated by denaturing PAGE and detected by fluorescence scanning. N.E., nonextract.
When gap-filling DNA repair reactions were performed with DNA containing an AP site generated by uracil excision, WT, arp−/−, and fpg−/− extracts catalyzed the insertion of up to three deoxynucleotides when all four dNTPs were present in the repair reaction (Fig. 4, lanes 6, 9, and 12), suggesting the operation of an LP-BER. As expected, no DNA repair intermediates were detected with double mutant fpg−/− arp−/− extracts (Fig. 4, lanes 13–15). Since only FPG and ARP activities are detectable in arp−/− and fpg−/− extracts, respectively (Fig. 3), these results suggest that the nature of the enzyme performing AP incision is not the only factor influencing the choice between SP- and LP-BER.
When reactions catalyzed by WT extracts were performed with DNA containing depurinated N7-meG the insertion of just one deoxynucleotide was detected, regardless of the presence of only dGTP or all four dNTPs in the repair reaction (Fig. 4, lanes 20 and 21). The same insertion pattern was observed with arp−/− mutant cell extracts, which only exhibit FPG incision activity (Fig. 4, lanes 23 and 24). As expected, no DNA repair intermediates were detected with either single mutant fpg−/− or double mutant fpg−/− arp−/− extracts (Fig. 4, lanes 25–30). These results suggest that depurinated N7-meG is repaired through FPG-dependent SP-BER.
ARP Discriminates Between AP Sites Generated by Enzymatic and Nonenzymatic Release of N7-meG.
The results described above suggested that nonenzymatic hydrolysis of N7-meG channels repair into an AP endonuclease-independent pathway. To test this idea, we compared the incision activity of recombinant and native FPG and ARP on DNA substrates containing AP sites generated either by enzymatic or nonenzymatic release of N7-meG (Fig. 5). We found that recombinant FPG incised both types of DNA substrates with similar efficiency (Fig. 5A). Native FPG enzyme present in arp−/− mutant extracts, which, as previously shown (Fig. 3) only display FPG-dependent AP incision activity, was also active on both types of AP sites (Fig. 5B). When tested on the same DNA substrates recombinant ARP-incised AP sites generated by enzymatic release of N7-meG but, unexpectedly, did not display detectable activity on AP sites generated by spontaneous depurination of N7-meG (Fig. 5C). Mutant fpg−/− extracts, which only show ARP-dependent AP incision activity (Fig. 3), were also active on enzymatic AP sites but lacked activity on nonenzymatic AP sites (Fig. 5D). Importantly, recombinant hAPE1 did not exhibit such differential activity (Fig. S3). Altogether, these results indicate that ARP discriminates between AP sites generated by enzymatic and nonenzymatic release of N7-meG.
Fig. 5.
ARP discriminates between AP sites generated by enzymatic and nonenzymatic release of N7-meG. DNA substrates (20 nM) were a 9:1 mixture of homoduplex G:C and heteroduplex AP:C generated either by spontaneous N7-meG depurination (nonenzymatic, blue triangles), or N7-meG excision by hAAG (enzymatic, red squares). Substrates were incubated either with purified proteins [(A) FPG: 0.5 nM; (C) ARP: 10 nM] or Arabidopsis cell-free extracts [(B) arp−/−: 8 µg; (D) fpg−/−: 8 µg]. Reactions for detection of AP endonuclease activity (C and D) were supplemented with 2 mM MgCl2. After stabilization with NaBH4, reaction products were separated by denaturing PAGE and detected by fluorescence scanning. Values are means with SEs from three independent experiments.
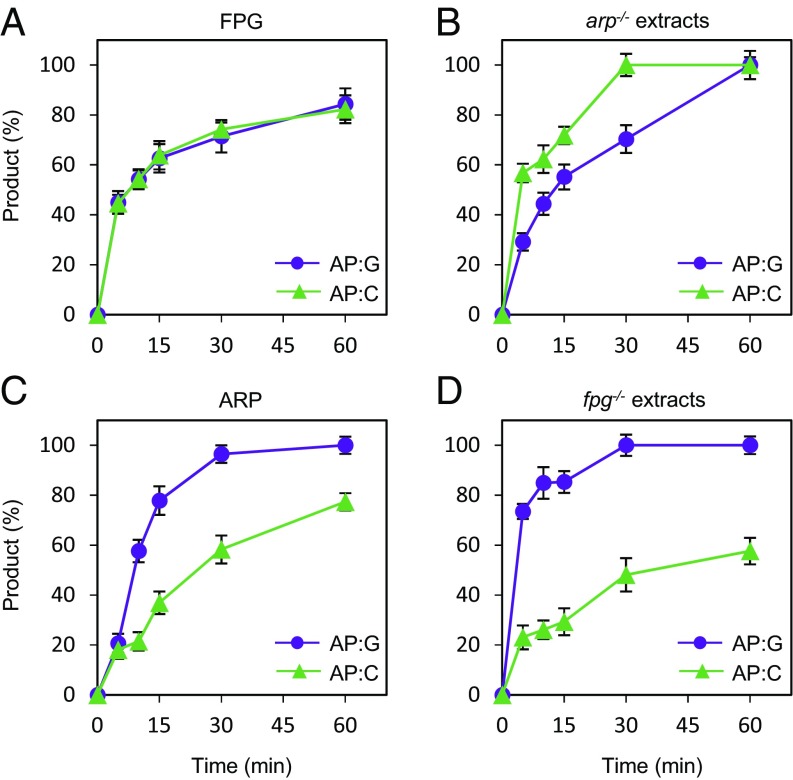
The Identity of the Base Opposite an Enzymatically Generated AP Site Influences Both AP Endonuclease and AP Lyase Activities.
The results described above indicate that both ARP and FPG are active on enzymatically generated AP sites, regardless of whether the excised base is either N7-meG or uracil. It is important to emphasize that all previous experiments were performed using C as the opposite base in the complementary strand. However, the relevant in vivo lesions arising from enzymatic excision of N7-meG and uracil (deaminated cytosine) are expected to be AP sites opposite C and G, respectively. To further explore the substrate preferences of the major AP incision activities in Arabidopsis we tested the effect of the base identity, either C or G, opposite an enzymatically generated AP site. We compared the incision activity of purified recombinant ARP and FPG proteins on DNA substrates containing AP sites generated by excision of uracil opposite either C or G (Fig. 6). Whereas FPG displayed similar activity on both DNA substrates (Fig. 6A), ARP activity was higher on AP sites opposite G (Fig. 6C). Native ARP in fpg−/− extracts (Fig. 6D) and recombinant hAPE1 (Fig. S4) also exhibited a similar preference for G as the orphan base. Interestingly, and unlike recombinant FPG, native FPG activity detected in arp−/− extracts was higher on AP sites opposite C (Fig. 6B). Altogether, these results suggest that the identity of the base opposite an enzymatically generated AP site influences the choice between AP endonuclease- and AP lyase-dependent repair.
Fig. 6.
Identity of the base opposite the abasic site influences the choice between AP endonuclease- and AP lyase-mediated repair. DNA substrates (2 nM) contained either a single AP:G (purple circles) or AP:C (green triangles), both generated by uracil excision. Substrates were incubated either with purified proteins [(A) FPG: 0.5 nM; (C) ARP: 10 nM] or Arabidopsis cell-free extracts [(B) arp−/−: 8 µg; (D) fpg−/−: 8 µg]. Reactions for detection of AP endonuclease activity (C and D) were supplemented with 2 mM MgCl2. After stabilization with NaBH4, reaction products were separated by denaturing PAGE and detected by fluorescence scanning. Values are means with SEs from two independent experiments.
Discussion
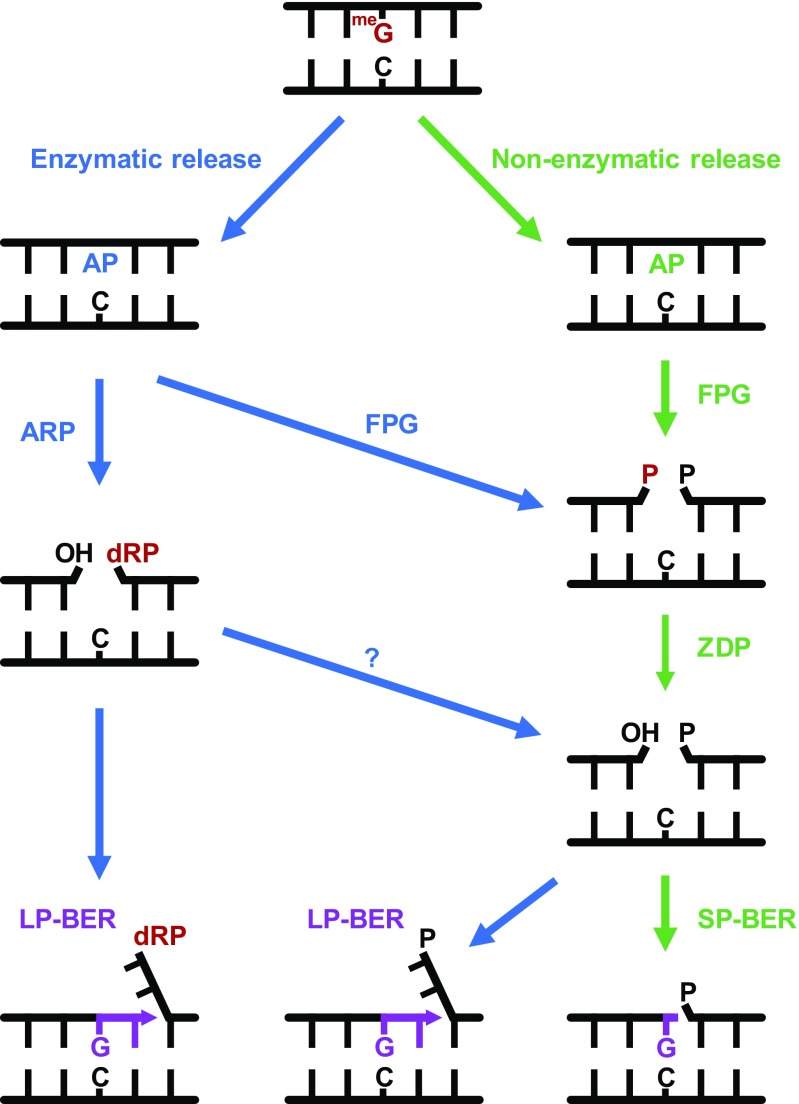
The initial motivation of this study was to understand why zdp−/− mutant plants are hypersensitive to MMS. Our biochemical and genetic analysis strongly suggests that the nonenzymatic release of MMS-induced N7-meG channels repair into an AP endonuclease-independent pathway in which FPG and ZDP perform consecutive steps (Fig. 7). The fact that FPG inactivation in zdp−/− mutants partially restores MMS resistance (Fig. 1E) suggests that the SSB intermediates with blocked 3′-P ends generated by FPG are more cytotoxic than AP sites. A similar observation has been previously reported in S. cerevisiae, where deletion of its two Nth homologs (NTG1 and NTG2) partially rescues the MMS hypersensitivity of a double apn1 apn2 mutant, presumably by avoiding accumulation of 3′-PUA ends (37). In S. pombe, the deletion of nth1 also relieves the MMS sensitivity of the single apn2 mutant (35). Our results suggest that it would be interesting to examine the relative activities of yeast AP lyases and AP endonucleases on AP sites of enzymatic and nonenzymatic origin.
Fig. 7.
A model for repair of AP sites arising from enzymatic and nonenzymatic release of N7-meG. FPG incises AP sites generated by spontaneous release of N7-meG, generating a single-nucleotide gap with a 3′-P terminus that is processed by ZDP to generate a 3′-OH terminus. Repair is continued through SP-BER. AP sites generated by enzymatic release of N7-meG may be incised by both ARP and FPG, and repair may be continued by either SP- or LP-BER. See text for details.
Unlike the yeast pathway, where AP endonucleases are required to process the 3′-PUA blocking ends generated by Nth homologs, the Arabidopsis pathway described here is AP endonuclease-independent, since 3′-P ends generated by FPG are processed by ZDP DNA 3′-phosphatase. This scenario is reminiscent of the AP endonuclease-independent pathway reported for oxidatively damaged bases in mammalian cells (21), in which NEIL1 and NEIL2 DNA glycosylases generate DNA strand breaks with 3′-P termini that are processed by the ZDP homolog polynucleotide kinase (PNK). Interestingly, PNK-deficient cells are sensitive to MMS (44), thus raising the possibility that the mammalian NEIL/PNK pathway may also operate in the repair of depurinated N7-meG. It has also been suggested that a PNK-dependent pathway functions as a backup mechanism for AP site repair in S. pombe (45, 46).
An unexpected observation arising from our work is that, unlike its mammalian homolog APE1, Arabidopsis ARP endonuclease does not exhibit detectable activity on abasic sites arising from N7-meG depurination (Fig. 5 C and D). We propose that such incapacity underlies the critical function of the FPG/ZDP pathway in protection of Arabidopsis against MMS. However, it is likely that ARP plays a role in the repair of AP sites generated by enzymatic excision of alkylated bases. In fact, fpg−/− mutant extracts, which only display ARP-dependent AP incision activity, are active on AP sites generated by N7-meG excision (Fig. 5D). Moreover, the simultaneous deficiency of FPG and ARP causes a hypersensitivity to MMS similar to that of single zdp−/− mutants (Fig. 1E). Although we could not detect N7-meG excision in cell extracts, the Arabidopsis genome encodes homologs of enzymes that excise N7-meG, such as E. coli AlkA and human AAG (47, 48). In any case, AP sites generated by such alkylpurine DNA glycosylases would also be substrates for the potent AP lyase activity of FPG (Figs. 5 and 7).
The absolute preference of ARP for enzymatically generated AP sites in comparison with those generated by nonenzymatic release (Fig. 5 C and D) deserves further investigation, particularly since its mammalian homolog does not exhibit such discrimination (Fig. S3). ARP and APE1 are members of the exonuclease III (ExoIII) family of AP endonucleases (49) and share a 57% sequence identity in their respective catalytic DNA repair domains (50). Both proteins have N-terminal extensions absent from E. coli ExoIII, with a length of 61 extra residues in APE1 and 270 residues in ARP (50). The N-terminal domain of APE1 is required for a redox activity that is independent of the DNA repair function (51). Although there is no sequence conservation between the N-terminal extensions of both proteins, it has been reported that ARP possesses a redox function similar to that of APE1 (52). However, the redox domain of APE1 is less well-defined than the DNA repair domain, and they overlap in a region that contains some residues conserved in both ARP and APE1 (53).
The negligible activity of ARP against N7-meG depurination products strongly suggests that AP sites generated by spontaneous hydrolysis and those generated by monofunctional DNA glycosylases have differential features not yet identified. Observed kinetic isotope effects, pH dependencies, and structure–reactivity comparisons suggest that enzymatic and nonenzymatic glycosidic bond cleavage follow the same reaction coordinate, with an oxocarbenium ion-like transition state (54–56). However, detailed transition-state analysis of nonenzymatic N-glycoside hydrolysis of deoxynucleosides has been reported only for dAMP (reviewed in ref. 56). The chemical instability of abasic sites poses a challenge for biochemical and structural studies. AP sites exist in an equilibrium mixture of four species, with ∼99% as two hemiacetal anomers (α- and β-2-deoxy-d-ribofuranose) and 1% as ring-opened aldehyde and hydrated aldehyde forms (57, 58). Since the aldehyde forms are prone to spontaneous cleavage by β-elimination, most biochemical, thermodynamic, and structural studies have been performed using an stable analog containing a tetrahydrofuran (THF) moiety lacking the hydroxyl group at C-1 of 2-deoxyribose (59). A derivative retaining such group but with the 3′-phosphate replaced by methylene (3CAPS) has also been suggested as a useful AP-site analog (60). However, both THF and 3CAPS are refractory to incision by AP lyases (60). Structural models of THF in naked DNA reveal an intrahelical conformation of the deoxyribose moiety (reviewed in ref. 9). In contrast, studies with genuine AP sites generated by UDG treatment of uracil-containing DNA have reported extrahelical conformations either for the β-anomer (61) or for both anomeric forms (62). Interestingly, it has been suggested that the ratio of the anomeric forms may depend upon the identity of the base in the complementary strand (8, 57). AP sites have been also generated in oligonucleotides by thermal depurination of either normal (63) or alkylated purines (64), but unfortunately there are no reports on their structure. Our results suggest that direct structural comparison between enzymatically and nonenzymatically generated AP sites, in the same sequence context and conditions, will be essential to fully understand the biological consequences of this ubiquitous lesion.
Methods
Plant Material and Growth Conditions.
The Arabidopsis mutant line SALK_076932 harboring a T-DNA insertion in the FPG gene was obtained from the Arabidopsis Biological Resource Center. Homozygous plants for the T-DNA insertion were identified by PCR using primers FPG_F1, FPG_R5, and LBa_3 (Table S1). Arabidopsis arp−/− (SALK_021478) and zdp−/− (SAIL_60_C08) lines were previously described (23, 25). All T-DNA insertion mutants were in the Col-0 background. Double fpg−/− zdp−/− and fpg−/− arp−/− mutant lines were obtained by crossing homozygous fpg−/−, zdp−/− and arp−/− plants and self-crossing the progeny. F2 plants were genotyped by PCR using specific primers for FPG (FPG_F1, FPG_F5, and LBa1), ARP (ARP_F1, ARP_R1, and LBa1), and ZDP (ZDP_F1, ZDP_R1, and LB3) loci (Table S1).
Arabidopsis WT (Col-0) and mutant plants were grown in pots in a growth chamber at 23 °C under long-day conditions (16 h light, 8 h darkness). For in vitro culture, sterilized seeds were cold-treated at 4 °C for 2 d and plated on 10-cm Petri dishes containing 25 mL of 0.44% (wt/vol) MS medium (Sigma) supplemented with 3% (wt/vol) sucrose and 0.8% (wt/vol) agar, pH 5.8. Plates were transferred to the growth chamber and incubated under long-day conditions at 23 °C. For genotoxic treatments, plants were grown in MS nutrient agar containing increasing concentrations of MMS (Sigma).
Arabidopsis Cell Extract Preparation.
Whole-cell extracts were prepared from snap-frozen 15-d-old seedlings grown in Petri dishes as described above. All steps were performed at 0–4 °C. Frozen plant material was ground in a handle mortar with liquid N2, and the resulting powder was resuspended in two to three volumes (wt/vol) of homogenization buffer containing 25 mM Hepes-KOH, pH 7.8, 100 mM KCl, 5 mM MgCl2, 250 mM sucrose, 10% glycerol, 1 mM DTT, and 1 µL⋅mL−1 protease inhibitor mixture (Sigma-Aldrich). The homogenate was incubated for 30 min at 4 °C and centrifuged at 11,400 × g for 1 h. The supernatant was filtered through a 20-µm nylon mesh and dialyzed overnight against 25 mM Hepes-KOH, pH 7.8, 100 mM KCl, 17% glycerol, and 2 mM DTT. Protein concentration was determined by the Bradford assay, and the extract was stored in small aliquots at −80 °C.
DNA Substrates.
Oligonucleotides used as DNA substrates (Table S2) were synthesized by IDT and purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5 µM solution of a 5′-fluorescein (Fl)–labeled oligonucleotide with a 10 µM solution of an unlabeled complementary oligonucleotide. Annealing reactions were carried out by heating at 95 °C for 5 min followed by slowly cooling to room temperature.
DNA duplex containing a single N7-meG was synthesized by the primer extension method (40) using a 5′-fluorescein–labeled 28-nt oligonucleotide annealed to a 51-nt oligonucleotide (Table S2). DNA synthesis was performed in a reaction mixture containing 0.1 U⋅µL−1 Klenow Fragment (3′→5′ exo−, NEB), 20 µM dCTP, 20 µM dATP, 20 µM dTTP, and 200 µM 7-methyl-dGTP (Jena Bioscience) at 37 °C for 1 h in NEBuffer 2. A control DNA substrate was synthetized in presence of 20 μM dGTP instead of 7-methyl-dGTP. Reactions were stopped by adding 10 mM EDTA, and DNA was ethanol-precipitated at −20 °C in the presence of 0.3 mM NaCl and 16 mg⋅mL−1 glycogen. Samples were resuspended in deionized water and stored at −20 °C. The N7-meG was converted into its ring-opened me-FAPy-G form by incubating in 50 mM phosphate buffer (pH 11) at 37 °C for 5 h (65). DNA was ethanol-precipitated as described above.
DNA substrates containing an enzymatic AP site were generated by incubating a DNA duplex containing either a U:G or a U:C mismatch, prepared as described above, with E. coli UDG (1.5 U; New England BioLabs, NEB) at 37 °C for 1 h. Enzymatic AP sites were also generated by incubating a DNA duplex containing a N7-meG:C pair with hAAG (2 U; NEB), at 37 °C for 8 h in NEBuffer 1. DNA substrates containing an AP site generated by spontaneous depurination were generated by incubating a DNA duplex with a single N7-meG:C pair for 16 h at 37 °C in DNA incision assay buffer (45 mM Hepes-KOH, pH 7.8, 70 mM KCl, 1 mM DTT, 0.4 mM EDTA, 36 µg BSA, and 0.2% glycerol).
Validation of DNA Substrates Containing N7-meG and Me-FAPy-G.
DNA substrates (20 nM) containing N7-meG or me-FAPy-G were validated in reactions (50 µL) with hAAG (2 U; NEB), hAPE1 (1 U; NEB), or E. coli Fpg (EcoFpg, 8 U; NEB). Incubations were performed at 37 °C for 8 h in NEBuffer 1 (hAAG and hAPE1) or ThermoPol Reaction Buffer (EcoFpg). DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol-precipitated as described above. Samples were resuspended in 10 μL of 90% formamide and heated at 50 °C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide gel containing 7 M urea. Labeled DNA was visualized using FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
DNA Incision Assay.
Reactions (50 µL) contained 45 mM Hepes-KOH, pH 7.8, 70 mM KCl, 1 mM DTT, 0.4 mM EDTA, 36 µg BSA, 0.2% glycerol, DNA substrate (2 or 20 nM), the indicated amount of cell extract or protein, and, when specified, MgCl2 (2 mM). After incubation at 37 °C during the indicated time, reactions were stopped by adding 20 mM EDTA, 0.6% SDS, and 0.5 mg⋅mL−1 proteinase K, and mixtures were incubated at 37 °C for 30 min. When indicated, reaction products were stabilized by the addition of freshly prepared sodium borohydride (NaBH4; Sigma-Aldrich) to a final concentration of 230 mM and incubated at 0 °C for 30 min. Finally, samples were buffered with 100 mM Tris, pH 8, and DNA was extracted as described above. Reaction products were separated and visualized as previously described.
For assays comparing enzymatic and nonenzymatic AP sites, DNA substrates (20 nM) were a 9:1 mixture of homoduplex G:C and heteroduplex AP:C. Nonenzymatic AP:C was generated by spontaneous N7-meG depurination for 16 h, whereas enzymatic AP:C was generated either by N7-meG excision by hAAG or uracil excision by E. coli UDG (discussed above). Product (percent) was calculated as the incision fragments detected relative to those generated by hAPE1 (10 U).
Gap-Filling Assay.
Reactions (50 µL) contained 45 mM Hepes-KOH, pH 7.8, 70 mM KCl, 1 mM DTT, 0.4 mM EDTA, 36 µg BSA, 1 mM NAD, 0.2% glycerol, 10 mM ATP, 110 mM phosphocreatine, 0.25 µg⋅mL−1 creatine phosphokinase, 2 mM MgCl2, and 20 µM of the indicated deoxynucleotides. After incubation at 37 °C for 3 h, reactions were stopped, DNA was extracted, and samples were processed as described above.
Protein Expression and Purification.
Arabidopsis FPG cDNA, a gift from Scott Kathe and Susan Wallace, University of Vermont, Burlington, VT (31), was subcloned into pET30b expression vector (Novagen) using XhoI and XbaI sites. Expression was carried out in E. coli BL21 (DE3) dcm− Codon Plus cells (Stratagene) induced during 2 h by adding 1 mM isopropyl-1-thio-β-d-galactopyranoside. His-FPG was purified by affinity chromatography on a Ni2+-Sepharose column (HisTrap HP; GE Healthcare). Protein was eluted with a 60 mM to 1 M gradient of imidazole and analyzed by SDS/PAGE (10%) using broad-range molecular weight standards (Bio-Rad). Protein concentration was determined by the Bradford assay. His-ZDP, MBP-ZDP, and His-ARP were expressed and purified as previously described (23, 25).
Pull-Down Assays.
For His-FPG pull-down, 30 pmol of purified MBP or MBP-ZDP in 30 µL of column buffer (20 mM Tris⋅HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 10 mM β-mercaptoethanol) was added to 100 µL of amylose resin (NEB) and incubated for 1 h at 4 °C. The resin was washed three times with 1 mL of binding buffer (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 10 mM β-mercaptoethanol). Purified His-FPG (5 pmol) was incubated at 25 °C for 30 min at 450 rpm with either MBP or MBP-ZDP bound to resin. The resin was washed three times with 1 mL of washing buffer (20 mM Tris, pH 7.4, 1 mM EDTA, 10 mM β-mercaptoethanol, 1.5% Triton X-100, and 250 mM NaCl). Bound proteins were analyzed by Western blot using antibodies against His6 tag (Santa Cruz).
For MBP-ZDP pull-down, 30 pmol of purified His-FPG in 30 µL of dialysis buffer (50 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, and 1 mM DTT) was added to 100 µL of Ni2+ Sepharose resin (NEB) and incubated for 1 h at 4 °C. The resin was washed three times with 1 mL of binding buffer 2 (10 mM Tris, pH 8, 1 mM DTT, 0.01 mg⋅mL−1 BSA, and 60 mM Imidazol). Purified MBP or MBP-ZDP (5 pmol) was incubated at 25 °C for 30 min at 450 rpm with His-FPG bound to resin. The resin was washed three times with 1 mL of washing buffer 2 (20 mM Tris, pH 7.4, 1 mM EDTA, 10 mM β-mercaptoethanol, 1% Triton X-100, 100 mM NaCl, and 60 mM imidazol). Bound proteins were analyzed by Western blot using antibodies against MBP tag (Santa Cruz).
Supplementary Material
Acknowledgments
We thank S. D. Kathe and S. S. Wallace (University of Vermont) for the kind gift of Arabidopsis FPG cDNA, Ana M. Maldonado and Jesús V. Jorrín (University of Córdoba) for generously sharing plant-growing facilities, and Jara Teresa Parrilla-Doblas (University of Córdoba) for assistance with FPG purification. This study was supported by the Spanish Ministry of Economy, Industry and Competitivity and the European Regional Development Fund under Grant BFU2016-80728-P and by the Junta de Andalucía and the European Regional Development Fund under Grant P11-CVI-7576. C.B. was the recipient of a PhD FPI (Formación de Personal Investigador) Fellowship from the Junta de Andalucía, Spain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719497115/-/DCSupplemental.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- 4.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talpaert-Borlé M. Formation, detection and repair of AP sites. Mutat Res. 1987;181:45–56. doi: 10.1016/0027-5107(87)90286-7. [DOI] [PubMed] [Google Scholar]
- 6.Strauss B, Hill T. The intermediate in the degradation of DNA alkylated with a monofunctional alkylating agent. Biochim Biophys Acta. 1970;213:14–25. doi: 10.1016/0005-2787(70)90003-1. [DOI] [PubMed] [Google Scholar]
- 7.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 8.Beger RD, Bolton PH. Structures of apurinic and apyrimidinic sites in duplex DNAs. J Biol Chem. 1998;273:15565–15573. doi: 10.1074/jbc.273.25.15565. [DOI] [PubMed] [Google Scholar]
- 9.Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S, Guillet M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Demple B, Harrison L. Repair of oxidative damage to DNA: Enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 12.Seeberg E, et al. Base removers and strand scissors: Different strategies employed in base excision and strand incision at modified base residues in DNA. Cold Spring Harb Symp Quant Biol. 2000;65:135–142. doi: 10.1101/sqb.2000.65.135. [DOI] [PubMed] [Google Scholar]
- 13.Levin JD, Demple B. Analysis of class II (hydrolytic) and class I (beta-lyase) apurinic/apyrimidinic endonucleases with a synthetic DNA substrate. Nucleic Acids Res. 1990;18:5069–5075. doi: 10.1093/nar/18.17.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortini P, Dogliotti E. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Córdoba-Cañero D, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009;60:716–728. doi: 10.1111/j.1365-313X.2009.03994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava DK, et al. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 18.Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 19.Pascucci B, et al. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, et al. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis [corrected] PLoS Genet. 2015;11:e1004905, and erratum (2015) 11:e1005198. doi: 10.1371/journal.pgen.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiederhold L, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Córdoba-Cañero D, Roldán-Arjona T, Ariza RR. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014;79:824–834. doi: 10.1111/tpj.12588. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Macías MI, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Córdoba-Cañero D, Dubois E, Ariza RR, Doutriaux MP, Roldán-Arjona T. Arabidopsis uracil DNA glycosylase (UNG) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J Biol Chem. 2010;285:7475–7483. doi: 10.1074/jbc.M109.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Córdoba-Cañero D, Roldán-Arjona T, Ariza RR. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011;68:693–702. doi: 10.1111/j.1365-313X.2011.04720.x. [DOI] [PubMed] [Google Scholar]
- 26.Ramiro-Merina Á, Ariza RR, Roldán-Arjona T. Molecular characterization of a putative plant homolog of MBD4 DNA glycosylase. DNA Repair (Amst) 2013;12:890–898. doi: 10.1016/j.dnarep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Roldán-Arjona T, García-Ortiz MV, Ruiz-Rubio M, Ariza RR. cDNA cloning, expression and functional characterization of an Arabidopsis thaliana homologue of the Escherichia coli DNA repair enzyme endonuclease III. Plant Mol Biol. 2000;44:43–52. doi: 10.1023/a:1006429114451. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsubo T, et al. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol Gen Genet. 1998;259:577–590. doi: 10.1007/s004380050851. [DOI] [PubMed] [Google Scholar]
- 29.García-Ortiz MV, Ariza RR, Roldán-Arjona T. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol Biol. 2001;47:795–804. doi: 10.1023/a:1013644026132. [DOI] [PubMed] [Google Scholar]
- 30.Dany AL, Tissier A. A functional OGG1 homologue from Arabidopsis thaliana. Mol Genet Genomics. 2001;265:293–301. doi: 10.1007/s004380000414. [DOI] [PubMed] [Google Scholar]
- 31.Kathe SD, et al. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst) 2009;8:643–653. doi: 10.1016/j.dnarep.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 34.Murphy TM, Belmonte M, Shu S, Britt AB, Hatteroth J. Requirement for abasic endonuclease gene homologues in Arabidopsis seed development. PLoS One. 2009;4:e4297. doi: 10.1371/journal.pone.0004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alseth I, Korvald H, Osman F, Seeberg E, Bjørås M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto T, et al. Roles of base excision repair enzymes Nth1p and Apn2p from Schizosaccharomyces pombe in processing alkylation and oxidative DNA damage. DNA Repair (Amst) 2005;4:1270–1280. doi: 10.1016/j.dnarep.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Hanna M, et al. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Meadows KL, Song B, Doetsch PW. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: Implications for biological function. Nucleic Acids Res. 2003;31:5560–5567. doi: 10.1093/nar/gkg749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 40.Asaeda A, et al. Substrate specificity of human methylpurine DNA N-glycosylase. Biochemistry. 2000;39:1959–1965. doi: 10.1021/bi9917075. [DOI] [PubMed] [Google Scholar]
- 41.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 42.Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 43.Duclos S, et al. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012;11:714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc Natl Acad Sci USA. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsen L, Forstrøm RJ, Bjørås M, Alseth I. AP endonuclease independent repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2000–2009. doi: 10.1093/nar/gkr933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kashkina E, Qi T, Weinfeld M, Young D. Polynucleotide kinase/phosphatase, Pnk1, is involved in base excision repair in Schizosaccharomyces pombe. DNA Repair (Amst) 2012;11:676–683. doi: 10.1016/j.dnarep.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Britt AB. Repair of damaged bases. In: Somerville C, Meyerowitz EM, editors. The Arabidopsis Book. Am Soc Plant Biol; Rockville, MD: 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santerre A, Britt AB. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:2240–2244. doi: 10.1073/pnas.91.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: Definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorman MA, et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babiychuk E, Kushnir S, Van Montagu M, Inzé D. The Arabidopsis thaliana apurinic endonuclease Arp reduces human transcription factors Fos and Jun. Proc Natl Acad Sci USA. 1994;91:3299–3303. doi: 10.1073/pnas.91.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 54.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 55.O’Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 56.Drohat AC, Maiti A. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. Org Biomol Chem. 2014;12:8367–8378. doi: 10.1039/c4ob01063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manoharan M, et al. The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon-13. J Am Chem Soc. 1988;110:1620–1622. [Google Scholar]
- 58.Wilde JA, Bolton PH, Mazumder A, Manoharan M, Gerlt JA. Characterization of the equilibrating forms of the aldehydic abasic site in duplex DNA by oxygen-17 NMR. J Am Chem Soc. 1989;111:1894–1896. [Google Scholar]
- 59.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 60.Schuermann D, et al. 3CAPS–A structural AP-site analogue as a tool to investigate DNA base excision repair. Nucleic Acids Res. 2016;44:2187–2198. doi: 10.1093/nar/gkv1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goljer I, Kumar S, Bolton PH. Refined solution structure of a DNA heteroduplex containing an aldehydic abasic site. J Biol Chem. 1995;270:22980–22987. doi: 10.1074/jbc.270.39.22980. [DOI] [PubMed] [Google Scholar]
- 62.Hoehn ST, Turner CJ, Stubbe J. Solution structure of an oligonucleotide containing an abasic site: Evidence for an unusual deoxyribose conformation. Nucleic Acids Res. 2001;29:3413–3423. doi: 10.1093/nar/29.16.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vasseur JJ, Peoc’h D, Rayner B, Imbach JL. Derivatization of oligonucleotides through abasic site formation. Nucleosides Nucleotides. 1991;10:107–117. [Google Scholar]
- 64.Coleman RS, Pires RM. Site-specific formation of abasic lesions in DNA. Nucleosides Nucleotides. 1999;18:2141–2146. doi: 10.1080/07328319908044871. [DOI] [PubMed] [Google Scholar]
- 65.Alekseyev YO, Hamm ML, Essigmann JM. Aflatoxin B1 formamidopyrimidine adducts are preferentially repaired by the nucleotide excision repair pathway in vivo. Carcinogenesis. 2004;25:1045–1051. doi: 10.1093/carcin/bgh098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.