Significance
Intracellular sterol distribution mediated by lipid transfer proteins (LTPs) is crucial for membrane function. LTPs anchored at membrane contact sites (LAMs), new members of LTPs in the StARkin superfamily, have sterol transport roles at contact sites between the endoplasmic reticulum (ER) and other membranes. The determinants for ligand specificity and protein targeting were elusive. Here, we determined the structures of the pleckstrin homology (PH)-like domain and the StARkin domains from LAM homologs. The Lam6 PH-like domain has a unique PH domain fold critical for targeting to ER–mitochondrial contacts. The LAM StARkin domains have a hydrophobic cavity that accommodates a sterol ligand. This work provides a structural explanation for the sterol recognition of LAMs, which can be extended to understand the sterol-binding mode of other LTPs in the StARkin superfamily.
Keywords: StARkin, sterol, nonvesicular transport, lipid transfer protein, membrane contact site
Abstract
Membrane contact sites (MCSs) in eukaryotic cells are hotspots for lipid exchange, which is essential for many biological functions, including regulation of membrane properties and protein trafficking. Lipid transfer proteins anchored at membrane contact sites (LAMs) contain sterol-specific lipid transfer domains [StARkin domain (SD)] and multiple targeting modules to specific membrane organelles. Elucidating the structural mechanisms of targeting and ligand recognition by LAMs is important for understanding the interorganelle communication and exchange at MCSs. Here, we determined the crystal structures of the yeast Lam6 pleckstrin homology (PH)-like domain and the SDs of Lam2 and Lam4 in the apo form and in complex with ergosterol. The Lam6 PH-like domain displays a unique PH domain fold with a conserved N-terminal α-helix. The Lam6 PH-like domain lacks the basic surface for phosphoinositide binding, but contains hydrophobic patches on its surface, which are critical for targeting to endoplasmic reticulum (ER)–mitochondrial contacts. Structures of the LAM SDs display a helix-grip fold with a hydrophobic cavity and a flexible Ω1-loop as a lid. Ergosterol is bound to the pocket in a head-down orientation, with its hydrophobic acyl group located in the tunnel entrance. The Ω1-loop in an open conformation is essential for ergosterol binding by direct hydrophobic interaction. Structural comparison suggested that the sterol binding mode of the Lam2 SD2 is likely conserved among the sterol transfer proteins of the StARkin superfamily. Structural models of full-length Lam2 correlated with the sterol transport function at the membrane contact sites.
The proper intracellular distribution of sterol is essential for many biological functions of mammalian cells, including regulation of membrane properties, signal transduction, and protein trafficking (1, 2). Although the endoplasmic reticulum (ER) is the site of cholesterol synthesis, it contains only about 0.5–1% of cellular cholesterol. Cholesterol composes 30–40% of the lipid molecules of the plasma membrane, which account for 60–80% of total cellular cholesterol (3). The intracellular distribution of cholesterol is mediated by multiple mechanisms including movement by transport vesicles and nonvesicular transport by lipid transfer proteins. Increasing evidence suggests that nonvesicular transport is the major transport route for cholesterol (4–6). In particular, transfer of lipid molecules over the narrow gap between lipid bilayers at membrane contact sites is a specific and efficient means of achieving lipid transport (7).
Nonvesicular lipid transport is mediated by lipid transfer proteins (LTPs), which contain one or more lipid-binding domains that can solubilize the hydrophobic ligands. The lipid-binding domain is characterized by the presence of an internal cavity often capped with a flexible lid, which allows the extraction of a water-insoluble lipid into the cavity and transfers it through the cytosol (8). So far, the number of discovered LTPs has expanded to 23 protein families displaying diverse ligand specificities (9). One of the widespread LTP folds is the StARkin (8) superfamily, which includes five family members: StART (steroidogenic acute regulatory protein-related lipid transfer), PITP (phosphatidylinositol/phosphatidylcholine transfer protein), Bet_v1 (major pollen allergen from birch Betula verrucosa), PRELI (proteins of relevant evolutionary and lymphoid interest), and LAMs (LTPs anchored at membrane contact sites) (9).
Membrane contact sites (MCSs) are closely apposed regions in which two organellar membranes are in close proximity, typically within a distance of 30 nm (7). The ER, a major site of lipid biosynthesis, makes contact with almost all types of subcellular organelles (10). Oxysterol-binding proteins, which are conserved from yeast to humans, are suggested to have a role in the directional transport of sterols at MCSs by coupling with a PI(4)P concentration gradient (11). The StART proteins play a major role in nonvesicular sterol transport in mammals (12). However, the StART proteins are absent from yeast and Archea (13).
LAM proteins were discovered recently as members of the StARkin superfamily and were suggested to have sterol transport and regulatory roles at contact sites between the ER and other organellar membranes (14–16). Humans have three LAM homologs (GramD1a–GramD1c), and budding yeast has six homologs: Lam1/Ysp1, Lam2/Ysp2/Ltc4, Lam3/Sip3, Lam4/Ltc3, Lam5/Ltc2, and Lam6/Ltc1. LAMs are integral membrane proteins with a C-terminal transmembrane helix that anchors the proteins to the ER membrane (8). They commonly contain a pleckstrin homology (PH)-like domain of the glucosyltransferases, rab-GTPase activating proteins, and myotubularins family in the N-terminal region, and one or two StARkin lipid transfer domains in the middle. Certain LAM homologs, such as Lam1 and Lam2, have membrane-targeting Bin/amphiphysin/RVS (BAR) domains in their N-termini. Lam1–Lam4 localize to ER–plasma membrane contacts and mediate sterol exchange between the membranes (14). Lam5 and Lam6 target to multiple sites at the ER–mitochondria and ER–vacuole contacts and play a role in lipid transport (14–16). Lam6 interacts with the ERMES (ER–mitochondrial encounter structure) complex, a protein complex involved in tethering mitochondria to the ER (15, 16). Lam6 localizes to the nuclear vacuole junction by interacting with the vacuolar protein Vac8 and plays a role in sterol-enriched membrane domain formation in response to stress (15). The targeting of Lam6 to ER–mitochondria contacts by its PH-like domain is Tom70/71-dependent (15). However, the molecular basis of the interaction between the Lam6 PH-like domain and Tom70/71 is not well-understood. In addition, the roles of the PH-like domains in other LAM homologs are unknown.
Determining the molecular and structural mechanisms of LAMs is important for understanding the interorganelle communication and exchange at the contact site. So far, many sterol-transfer proteins from the StART family have been characterized and their structures have been reported (9, 17). Despite the extensive biochemical and structural studies on the StARkin superfamily proteins in the last decade, the binding mode of the sterol ligand is elusive owing to the lack of known structures of the sterol-bound forms. It has yet to be determined how LAMs recognize sterols by the StARkin domains (SDs) and how LAMs are targeted to specific membrane contact sites. So far, there are no available structures showing sequence homologies to the LAM PH-like and SDs in the Protein Data Bank.
In this study, to elucidate the structural mechanisms of ligand recognition, transport, and MCS targeting of LAMs, we determined the structures of the PH-like domain and the SDs from several LAM homologs. The Lam6 PH-like domain, displaying a unique PH domain fold with a conserved N-terminal α-helix, contains exposed hydrophobic patches on the end of β-sheet, which are critical for targeting to ER–mitochondrial contacts. This study revealed the conserved binding mode of sterol ligand by StARkin superfamily proteins. Structures of the first and second SDs of Lam2 and Lam4 displayed a helix grip fold with a hydrophobic cavity and a flexible lid. Ergosterol bound to the pocket in a head-down orientation, with its hydrophobic acyl group located in the tunnel entrance. The partially open Ω1-loop is essential for ergosterol binding by direct hydrophobic interaction. In vitro sterol transfer assays suggested that hydrophobic interaction is a major determinant for sterol binding. Structural similarity implies the sterol-binding mode of the Lam2 SD2 is likely conserved in sterol transfer proteins of the StARkin superfamily.
Results
Overall Structure of the LAM StARkin Domains.
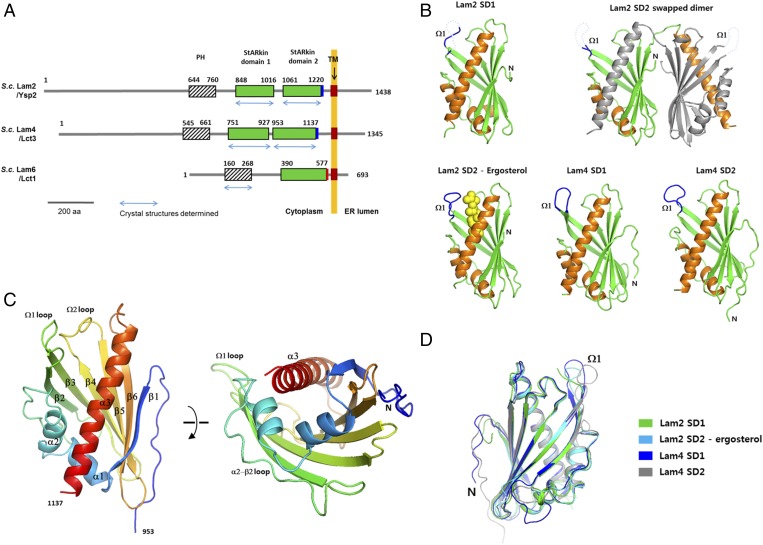
The six LAM homologs in yeast all contain a PH-like domain in their N-terminal region, one or two StARkin lipid transfer domains, and a transmembrane helix at the C-terminal region (Fig. 1A). The SDs are composed of 160–190 amino acids. LAMs often contain a short, charged sequence of fewer than 20 amino acids at the C-terminal end of their second SD (Fig. S1). To determine the structures of key domains of LAMs, we performed X-ray crystallographic studies of BAR domains, PH-like domains, and SDs from LAM homologs. We determined crystal structures of both the first and second SDs (hereafter referred to as SD1 and SD2, respectively) of Lam2 and Lam4 in apo forms at 1.5–1.9-Å resolutions (Table S1). The structure of the ergosterol-bound Lam2 SD2 was determined at 2.6-Å resolution (Fig. 1B). We solved the structure of the Lam4 SD1 by single anomalous dispersion using Se-methionine-labeled crystals. The structures of the other SDs were solved by molecular replacement, using the structure of the Lam4 SD1.
Fig. 1.
Overall structures of LAM homologs. (A) Schematic representation of the domain structures of Lam2, Lam4, and Lam6. The domains with the structures determined in this study are indicated with blue arrows. The polybasic regions and the acidic regions in the C-termini of the SDs are indicated with blue and red rectangles, respectively. The transmembrane regions of LAMs are indicated by brown rectangles. (B) The structures of the LAM SDs determined in this study. The disordered Ω1-loops are indicated with dashed lines. The bound ergosterol in the Lam2 SD2 is shown in yellow spheres. (C) The overall structure of the Lam2 SD1. The secondary structures are colored with blue to red from the N- to C- terminus. (D) Structural superposition of the LAM SDs.
The SDs of Lam2 and Lam4 contain three α-helices and a six-stranded antiparallel β-sheet, which fold into a half-barrel structure (Fig. 1C). The β-sheet wraps around the longest helix, α3, forming a helix-grip fold, which is commonly observed in StART and Bet_v1 family proteins. The β-sheet of the SD consists of six, rather than seven, β-strands. The residues connecting β6 and α2 form an extended loop, which is typically a β-strand in the related StART domains (17). The helix grip fold formed by the twisted β-sheet creates a large tunnel in the core of the protein. The C-terminal side of tunnel is plugged by α2 and α3 helices, and the other end is flanked by the β2–β3 and β4–β5 loops. The SD1 and SD2 from Lam2 and Lam4 display amino acid sequence identities of 34–56% between them (Fig. S1). The overall structures of four SDs from Lam2 and Lam4 are highly conserved. All LAM SDs are superimposable within a Cα rmsd of 0.7 Å (Fig. 1D).
The β2-β3 loop, which is referred to as the Ω1-loop, is the most flexible part of the SDs, displaying the highest B-factors in their structures. The Ω1-loops of all SDs of Lam2 and Lam4 reported in this study were in open conformation or disordered. The Ω1-loops of the Lam2 SD1 and Lam2 SD2 in apo forms were disordered in the crystal and were not visible. The Ω1-loops of the Lam4 SD1 and Lam4 SD2 were weakly visible owing to stabilization by lattice contact interactions in the crystals. The flexible Ω1-loop was suggested to serve as a gate to the ligand-binding cavity in StART domains (18, 19). A transient opening of the Ω1-loop was known to be sufficient for sterol uptake and release, as determined by molecular dynamics simulations (18).
The SDs of Lam2 and Lam4 are monomers in solution when analyzed by size-exclusion chromatography during protein purification. In the asymmetric unit of the apo Lam2 SD2, two molecules were found to form a domain-swapped dimer. The two Lam2 SD2 molecules exchanged the identical C-terminal helix (residues 1,191–1,223) between each other. However, size-exclusion chromatography analysis showed that the apparent oligomeric state of the Lam2 SD2 was a monomer before crystallization. The domain-swapped conformation likely originated from the flexible nature of SDs and seems to be a crystallographic artifact. This artifactual C-terminal helix swapping in the StART domain has also been observed in the Ups1-Mdm35 heterodimer of the StARkin superfamily (20).
Comparison of the LAM StARkin Domains to Other StARkin Family Proteins.
The LAM proteins share little sequence homology with other members of the StARkin superfamily. Structural comparison by the Dali search against the Protein Data Bank indicated that the LAM SDs show the highest structural similarity, with a Cα rmsd of 2.7 Å to the pathogenesis-related class 10 (PR10) protein of the Bet_v1 family (PDB id: 1IFV) with unknown function (21) (Fig. S2). Structural comparison of the LAM SD to a sterol transfer protein, STARD4 (PDB id: 1JSS) (22), revealed that the overall helix grip folds were conserved with the curved β-sheet with the central long α-helix (Fig. S2). However, the LAM SDs are more compact than StARD4 because they contain shorter connecting loops between secondary structure elements and they lack three β-strands and an N-terminal α-helix.
Sterol Recognition of the LAM StARkin Domains.
So far, many structures of the sterol transfer proteins in the StART family (STARD1, STARD3/MLN64, STARD4, STARD5, and STARD6) have been reported (17, 19, 22–24). However, the binding mode of the sterol ligand by StART domains has remained elusive owing to the lack of sterol-bound structures. In this study, we attempted to determine the structures of the LAM SDs in complex with ergosterol. We performed cocrystallization trials of the sterol ligand with all SDs of Lam2 and Lam4. However, the initial attempts to obtain a sterol complex by incubating the purified SDs with ergosterol in the crystallization setup were not successful. Therefore, we added ergosterol during protein expression and purification steps to increase the chance of ligand binding. The SDs from Lam2 and Lam4 treated with ergosterol were crystallized and the crystals were examined by X-ray analysis to determine the presence of a bound ligand. Only the Lam2 SD2 domain with a C2 space group displayed electron densities of sterol molecules (Fig. S3). The structure of the Lam2 SD2-ergosterol complex was determined at 2.6 Å resolution by molecular replacement, using the structure of the apo form. Although the crystals of all SDs in apo form diffracted to the resolutions higher than 1.9 Å, the best diffraction data for the sterol-bound form were 2.6 Å in resolution, perhaps because of the intrinsic disorder of sterol-bound crystals.
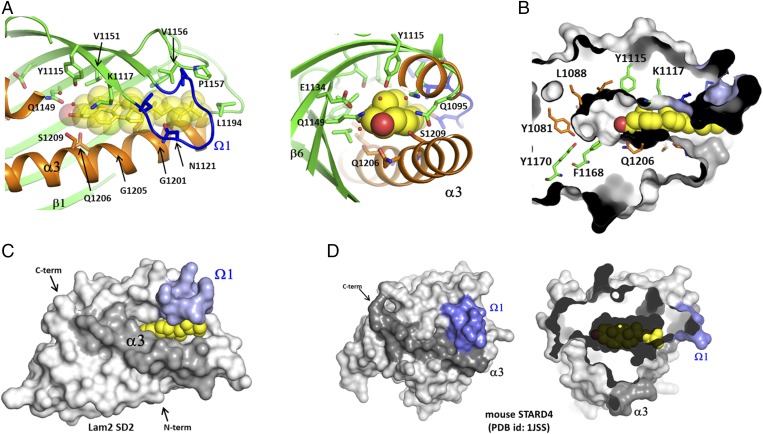
The crystals of the Lam2 SD2 domain complexed with ergosterol contained three molecules (chains A–C) in the asymmetric unit. Chain A and chain B of the Lam2 SD2 in the asymmetric unit showed electron densities of ergosterol in the ligand-binding cavity (Fig. S3). However, the third molecule completely lacked ergosterol electron densities in the pocket. Occupancy refinement using software Phenix suggested that the ergosterol molecules in chains A and B have 78% and 75% occupancies in the crystal structure, respectively. The Ω1-loop of chain C was invisible because of the disorder arising from the lack of ergosterol binding in the pocket. The LAM SDs have a large cavity with a typical volume of 650–800 Å3, when it is assumed that the hydrophobic tunnel is completely closed by the Ω1-loop. The cavity volumes are large enough to accommodate an ergosterol molecule with a molecular volume of 427.32 Å3. The tunnel bottom is relatively polar because of the presence of many hydrophilic residues (Gln1095, Lys1117, Glu1134, Gln1149, Gln1206, and Ser1209), which make hydrogen bond networks with water molecules in the cavity. The tunnel wall and the entrance are mainly composed of hydrophobic residues.
Ergosterol was bound to the hydrophobic pocket in a head-down orientation. The 3-hydroxyl group of ergosterol is located near the hydrophilic residues including Gln1206 and Gln1149. The ergosterol molecules in chain A and B make a hydrogen bond with Gln1149 and Gln1206, respectively, which are located in the two thirds of the hydrophobic tunnel (Fig. 2A). Ergosterol does not occupy the bottom of the hydrophobic cavity; rather, it binds closely to the tunnel entrance (Fig. 2B). The tunnel bottom is occupied by several water molecules forming a hydrogen bonding network in the apo form. The water molecules in the tunnel were not clearly visible in the ergosterol-bound form because of the resolution limit of the data. The hydrophobic rings of ergosterol interact with helix α3 and the walls formed by the β-sheet (β2, β3, β4, and β5). The acyl chain of ergosterol positioned in the tunnel entrance makes a hydrophobic interaction with the residues from the Ω1-loop (β2−β3) and the Ω2-loop (β4−β5). The Ω1-loop was disordered and invisible in the absence of ligand, suggesting that the flexible lid becomes ordered on sterol binding by tight hydrophobic interaction. Remarkably, the Ω1-loop in the sterol-bound form did not close the tunnel entrance. Therefore, the bound ergosterol in the Lam2 SD2 is not completely buried inside the cavity but, rather, partially exposes its hydrophobic acyl group to the solvent (Fig. 2C). The Ω1-loops of all SDs of Lam2 and Lam4, whether in a ligand-bound or ligand-free state, have a partially open conformation, which differs from the closed conformation of STARD4 and STARD3/MLN64 in their apo forms (22, 24). In the closed conformation of these StART domains, the Ω1-loop makes a hydrophobic contact with the residues on the central α-helix, covering the entrance of the hydrophobic cavity (Fig. 2D). In this closed form of STARD4, the small cavity cannot fully accommodate the cholesterol molecule (Fig. 2D), suggesting that conformational changes must accompany ligand binding. The Ω1-loops of the LAM SDs are too short to surround the acyl chain of the bound sterol, and therefore cannot close the tunnel entrance. Ergosterol binding induces a slight conformational change in the Lam2 SD2 around the tunnel entrance (Fig. 3A). The residues of the Ω1 and Ω2 loops are ordered to make hydrophobic contacts with ergosterol. However, there is no significant conformational difference in overall structures between apo and sterol-bound forms in the Lam2 SD2.
Fig. 2.
The sterol-binding site of the LAM SDs. (A) The ligand-binding site of the Lam2 SD2 with a bound ergosterol. (Right) View of the bound ergosterol from the tunnel bottom. Water molecules in the tunnel bottom are shown in small red spheres. (B) The surface representation of the ergosterol binding site. The side chains of the residues composing the tunnel walls are shown in sticks with the same color scheme as that used in A. (C) The surface representation of the Lam2 SD1 with a bound ergosterol. The Ω1-loop and the helix α3 are colored with light blue and dark gray, respectively. (D) Surface representation of mouse STARD4 (PDB id: 1JSS). The Ω1-loop and the helix α3 are colored with light blue and dark gray, respectively. (Right) Slap view of the sterol-binding pocket with a cholesterol molecule modeled into the pocket. Cholesterol is shown in yellow spheres. The closed form of STARD4 cannot accommodate cholesterol owing to the small dimensions of the binding pocket. The acyl chain of cholesterol clashes with the backbone of STARD4.
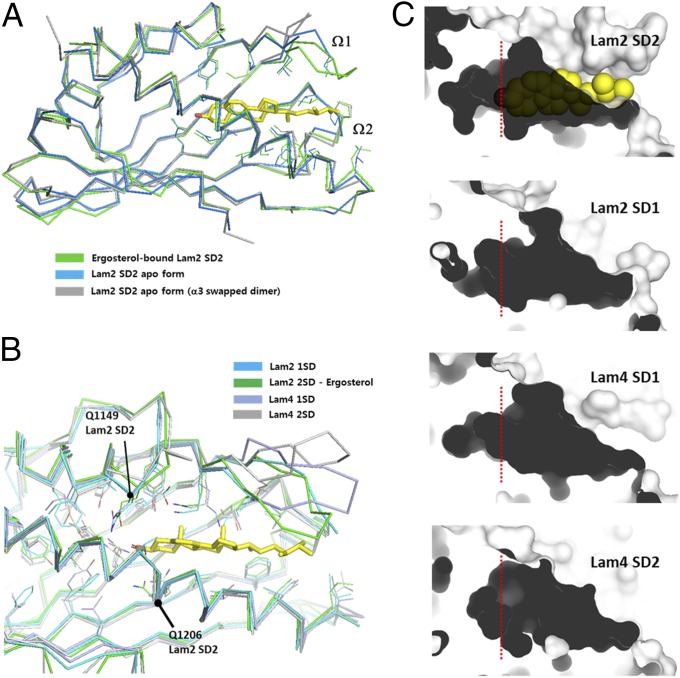
Fig. 3.
Structure comparison of the LAM SDs. (A) Structural comparison of apo forms and an ergosterol-bound form of the Lam2 SD2. (B) Structural comparison of the ligand-binding pockets of the LAM SDs. (C) Comparison of sterol-binding pockets of the LAM SDs by surface contours. Red dotted lines indicate the positions of the 3-hydroxyl group of ergosterol in the tunnel bottom.
The sequence and conformation of the residues forming the hydrophobic walls of the cavities are well-conserved in LAM homologs (Fig. 3B and Fig. S1). However, Gln1149 and Gln1206, which hydrogen bond to the 3-hydroxyl group of sterol, are not well-conserved in LAMs (Fig. S1A). The equivalent residues of Gln1206 of Lam2 are Val999 in the Lam2 SD1 and Val911 in the Lam4 SD1. The tunnel bottoms of the LAM SDs are relatively variable in size and shape (Fig. 3C). For example, the Lam2 SD1 has a narrower tunnel bottom than the Lam2 SD2, and the tunnel bottom of the Lam4 SD2 is shallow compared with those of other SDs. However, all SDs of Lam2 and Lam4 contain a few water molecules in the tunnel bottom, which might form a hydrogen bonding network with the 3-hydroxyl group of the sterol molecule. Remarkably, the major cavities in the SDs contacting the sterol molecule are very similar in size and shape, and the residues on the cavity walls are well-conserved in LAM homologs. This observation suggests that the shape of the hydrophobic cavity is the key factor in ligand specificity. In conclusion, the high structural similarity of the SDs among LAM homologs indicates that the sterol binding mode observed in the Lam2 SD2 is likely to be conserved in other LAM homologs.
Sterol Transport of LAM StARkin Domains.
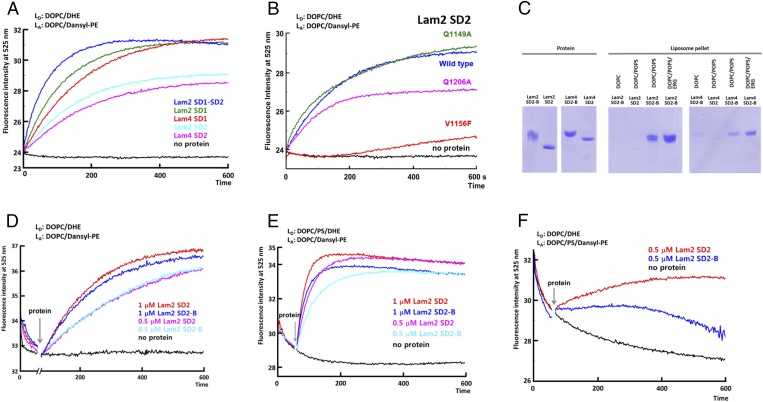
To identify the key residues for sterol recognition, we examined the sterol transport activities of the LAM SDs and their mutants. The transfer of sterol by the SDs was monitored by florescence resonance energy transfer of dehydroergosterol (DHE) to dansyl-phosphatidylethanolamine (dansyl-PE) in acceptor liposomes. The donor liposomes contained 10 mol % DHE and the acceptor liposomes contained 2.5% dansyl-PE. Each SDs of Lam2 and Lam4 showed DHE transfer activity between the liposomes (Fig. 4A). Of the seven LAM homologs, Lam2 and Lam4 contain two tandem SDs connected by flexible linkers of 45 and 26 residues, respectively. Although the first SD of Lam2 is dispensable for function, the C terminus and the second SD are essential (14). The tandem SDs of Lam2 showed a faster equilibrium of DHE transport than the single SDs, suggesting that an additional SD enhances the speed of sterol transport (Fig. 4A).
Fig. 4.
Sterol transport by the LAM SDs. (A) DHE transport assay between liposomes by the LAM SDs. DOPC/DHE liposomes (90/10 mol/mol, 13 μM total lipids) as donor were mixed with DOPC/Dansyl-PE (97.5/2.5 13 μM total lipids) liposomes as acceptor at 25 °C. After 1 min, LAM SD (1 μM) was added. FRET between DHE and Dansyl-PE was increased as DHE is transported to the acceptor liposomes. LD and LA denote donor and acceptor liposomes, respectively. (B) DHE transport assay of the Lam2 SD2 mutants. (C) Liposome pull-down assay. The liposomes were mixed with the various constructs of the LAM SDs (80 μg), and the mixture was incubated at 4 °C for 40 min. The protein in the supernatant and the liposome-bound protein were separated by centrifugation. The proteins in the liposome pellet were analyzed by SDS/PAGE. The liposomes in the pull-down assay were DOPC (100%), DOPC/POPS (molar ratio 80/20), and DOPC/POPS/ERG (molar ratio 70/20/10). (D) DHE transport assay of the LAM SDs with polybasic tails. (E) DHE transport assay of the LAM SDs with the donor liposomes containing PS. The donor liposomes contain DOPC/POPS/DHE (molar ratio 70/20/10). (F) DHE transport assay of the LAM SDs with the acceptor liposomes containing PS. The acceptor liposomes contain DOPC/POPS/Dansyl-PE (molar ratio 77.5/20/2.5).
Gln1206 and Gln1149 of the Lam2 SD2 are located within the hydrogen bond distance with the 3-hydroxyl group of ergosterol (ERG). However, both Gln1206 and Gln1149 are not conserved in the SDs of LAM homologs. The Gln1206Ala mutation in the Lam2 SD2 slightly reduced the DHE transfer activity, suggesting that a hydrogen bond between the protein and the sterol molecule is involved, but it is not essential for ligand binding and transfer activity (Fig. 4B). Similarly, the hydrogen bonding between sterol and the Gln residue of the binding pocket in oxysterol-binding protein, Osh4, is not essential for ligand binding (25). Another residue close to the 3-hydroxyl group of ergosterol is Gln1149 in the Lam2 SD2. The Gln1149Ala mutant has a similar sterol transfer activity to wild type, suggesting that Gln1149 does not contribute to sterol binding (Fig. 4B). The hydrogen bonds between ergosterol and the water molecules in the tunnel bottom are plausible in LAM homologs lacking the equivalent residue of Gln1206. However, hydrophobic interaction seems to be a dominant force for sterol binding in LAM proteins. The hydrophobic residues in the inner side of the Ω1-loop and Ω2-loop interact with the acyl chain of ergosterol. Mutating Val1156 in the Ω2-loop to a bulky phenylalanine residue significantly inhibited the sterol transfer activity, confirming that a tight hydrophobic interaction between the Ω-loops and ergosterol is essential for ligand binding (Fig. 4B).
The second SDs of most LAM homologs contain charged residue clusters as a C-terminal extension (8) (Fig. S1). Lam2 and Lam4 have poly basic residues in the C-terminal extension, whereas the SD of Lam6 contains an acidic tail. The basic residue extensions of Lam2 and Lam4 are expected to enhance the binding of the domains onto the acidic target membranes. The plasma membrane of budding yeast is composed of 17% phosphatidylcholine, 20% phosphatidylethanolamine (PE), and 34% phosphatidylserine (PS), whereas the ER membrane contains only 6.6% PS (26). We tested the membrane-binding properties of the Lam2 SD2 and Lam4 SD2 using DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) liposomes containing acidic POPS (1-palmitoyl-2- oleoyl-sn-glycero-3-phospho-l-serine). The Lam2 SD2 and Lam4 SD2 containing basic tails (Lam2 SD2-B and Lam4 SD2-B) associated with the DOPC/POPS (80/20 mol/mol) liposomes more strongly than with the DOPC liposomes (Fig. 4C). However, the SDs lacking the basic tail (Lam2 SD2 and Lam4 SD2) showed significantly weaker binding to the DOPC/POPS liposomes, confirming the role of charged residue extensions for membrane association.
Then, we compare the sterol transport activities of the SDs with or without the basic tails. There was no significant difference of transport activities for the SDs when DOPC liposomes were used (Fig. 4D). However, when either acceptor or donor liposomes contained POPS, the sterol transport activities of the Lam2 SD2-B and Lam4 SD2-B were reduced compared with those of constructs without basic tails (Fig. 4 E and F). The reduction of sterol transport by the polybasic region in our experimental conditions seemed to be caused by the stable membrane binding of the SDs, which might reduce the dissociation of the protein after sterol release from the acceptor membranes. A theoretical analysis of nonvesicular sterol transport suggested that the extraction of a sterol from a membrane rather than diffusion through the cytoplasm might be the rate-limiting step in physiological conditions (27). So far, the determinants for the plasma membrane targeting of Lam2 and Lam4 are unknown. We speculate that the enhanced association of the SDs by the basic tail to the acidic membranes would be advantageous for ligand extraction and proper targeting of proteins in physiological situations.
The cellular localizations of Lam5/6 are ER–mitochondria and ER–vacuole contact sites (14–16). The composition of acidic PS in the mitochondrial and vacuolar membranes is very low. The lack of basic extensions in Lam5/6 correlates with the properties required for the binding to their target membranes. In conclusion, although the C-terminal transmembrane helix anchors LAMs to the ER, the charged extensions of the SDs might enhance the association of ligand-binding domains to the membrane of the opposite side.
Structure of the Lam6 PH-like Domain.
All LAM homologs in yeast and humans contain a PH-like domain that is related to the PH domains found in glucosyltransferases, rab-GTPase activating proteins, and myotubularins family proteins (8). The PH-like domain is present upstream of the StARkin lipid transfer domain in LAM homologs. Proteomics studies identified many binding partners of Lam6, including Tom70/71 and Vac8 (15, 16). The PH-like domain contributes to the targeting of Lam6 to the ER–mitochondrial contacts in a Tom70/71-dependent manner (15). However, the molecular basis of the PH-like domain–Tom70/71 interaction is unknown. The functional role of the PH-like domains of other LAM homologs has not been defined, with the exception of Lam5/Lam6. The LAM PH-like domains show very low sequence homologies to canonical PH domains, although the overall PH domain fold is expected to be conserved. The LAM PH-like domains are characterized by the presence of a short amphipathic helix in addition to the canonical PH domain topology (8).
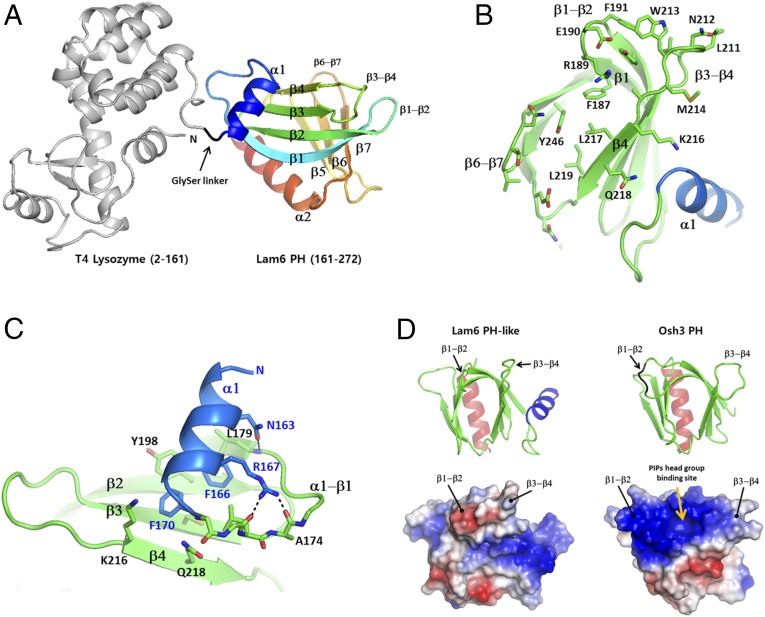
To reveal the structural features of the LAM PH-like domains, we performed crystallographic studies on the PH-like domains of all yeast LAM homologs. We obtained a soluble construct for the PH-like domain from the Lam6 homolog. We determined the crystal structure of the PH-like domain in fusion with an N-terminal T4 lysozyme (T4L) at 2.4 Å resolution. The Lam6 PH-like domain (residue 161–272) was connected to the C terminus of T4L, using GlySer as a dipeptide linker (Fig. 5A). The structure of the T4L-Lam6 PH was determined by molecular replacement, using the T4L structure as a search model. The Lam6 PH-like domain with 112 amino acids consists of two seven-stranded antiparallel β sheets in the middle and two α-helices in the N and C termini. The overall structure of the Lam6 PH-like domain displays a typical PH domain fold with an additional N-terminal short α helix (Fig. 5 A and B). The amphipathic helix α1 is conserved in LAM homologs with the consensus sequence of NxxFxxxF. The helix α1 covers the surface of the β-sheet formed by the strands β1–β4. The two phenyl groups (Phe166 and Phe170) of α1 make hydrophobic contacts with the hydrophobic surface of the β-sheet (Fig. 5C). The conserved Asn168 at the beginning of α1 hydrogen bonds with the carbonyl oxygen of Leu179 of β1. The typical PH domains often contain strong positive charges on the ends of their β-sheets, which serve as a binding site for the acidic head group of phosphoinositides (28). For example, Osh3 PH domain has a phosphoinositide binding site on the basic concave surface (29). However, the Lam6 PH-like domain has only one arginine in the β1-β2 loop (Fig. 5D). Instead, many hydrophobic residues such as Trp213, Phe191, Leu217, Leu219, and Met214 are exposed at the ends of the β-sheets, forming hydrophobic patches on the surface. This feature explains the lack of direct membrane binding on the Lam6 PH-like domain. The sequence for the core of the PH-like domains is well-conserved in LAM homologs. However, the loop residues on the ends of the β-sheets are relatively variable, suggesting that the binding partners of the PH-like domains might be specific to each LAM homolog (Fig. S1B).
Fig. 5.
Structure of the Lam6 PH-like domain. (A) The overall structure of the T4 lysozyme-fused Lam6 PH-like domain. The T4L is shown in gray, and the PH-like domain is colored from blue to red based on the secondary structure succession. (B) The surface residues on the end of the β-sheet. (C) Structure of the unique N-terminal conserved helix of the PH-like domain. (D) Structural comparison of Lam6 PH-like domain and Osh3 PH domain (PDB id: 4IAP). (Bottom) Electrostatic surface representations of the domains.
Targeting of Lam6 by the PH-like Domain.
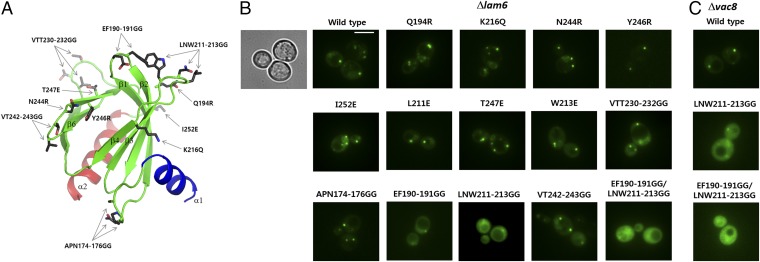
Lam6 is localized to several contact sites including ERMES, vacuole and mitochondria patch, and nuclear vacuolar junction (15, 16). The targeting of Lam6 to ERMES is dependent on Tom70/71, the paralogous mitochondrial preprotein import receptors. Deleting the PH-like domain (residues 145–360) in Lam6 relocalizes Lam6 to ER–vacuole contact sites, suggesting an essential role of the PH-like domain in ER–mitochondrial targeting (15). To understand the molecular basis of ER–mitochondrial targeting by the Lam6 PH-like domain, we examined the cellular localization of the Lam6 PH-like domain and its mutants. Expression of the wild-type Lam6 PH-like domain in lam6 knockout cells showed a strong punctate pattern, which is consistent with the reported localization of intact Lam6 in the ERMES structures (16). This observation indicates that the PH-like domain alone is sufficient for targeting Lam6 to ERMES. Then, to identify the key regions in the PH-like domain for Lam6 targeting, we generated a series of various surface mutations in the Lam6 PH-like domain based on the crystal structure and examined the localization of the mutants in yeast cells (Fig. 6A). The mutants, Q194R (β2), K216Q (β4), N244R (β6−β7 loop), I252E (β7−α2 loop), T247E (β7), L211E (β4−β5 loop), W213E (β3−β4 loop), Y246R (β7), APN174-176GG (α1−β1 loop), VTT230-232GG (β2), and VT242-243GG (β5−β6 loop), all showed a similar punctate localization to that of wild type. However, EF190-191GG (β1-β2 loop), and LNW211-213GG (β3-β4 loop) mutants exhibited reduced ERMES localization and showed more cytosolic distribution compared with wild type (Fig. 6B). The combination of EF190-191GG and LNW211-213GG completely abolished the ERMES localization of the Lam6 PH to cytosolic distribution. Then, we tested the localization of the Lam6 PH-like domain in vac8 knockout cells. The wild type and mutants of the GFP-Lam6 PH-like domains showed a similar localization pattern to that observed in lam6 knockout cells, suggesting that Vac8 does not contribute to the ER–mitochondrial targeting of the Lam6 PH-like domain (Fig. 6C). EF190-191 and LNW211-213 are located next to each other in the same face of the β-sheet in the Lam6 PH-like domain (Fig. 6A), which is consistent with the synergistic effect of double mutations (EF190-191GG and LNW211-213GG). Typical PH domains have poly charged residues in the end of β-sheets for the interaction with hydrophilic head groups of lipids (Fig. 5D) (28, 29). However, these two hair-pin loops critical for proper targeting of Lam6 expose three hydrophobic residues, F190, L211, and W213, suggesting that hydrophobic interaction is important for specific protein-targeting. This feature supports the idea that the Lam6 PH-like domain does not bind membrane lipids, but targets Lam6 to the ERMES complex at the MCSs, perhaps through protein–protein interaction.
Fig. 6.
Cellular localization of the GFP-Lam6 PH-like domain constructs. (A) The mutated residues in the Lam6 PH-like domain are indicated by stick models. (B) Cellular localization of the GFP-Lam6 PH-like domains in lam6 knockout cells. The differential interference contrast image is shown only for the wild type. (Scale bar, 3 μm.) (C) Cellular localization of the GFP-Lam6 PH-like domains in vac8 knockout cells.
Discussion
MCSs in eukaryotic cells are hotspots for lipid exchange and cell signaling, allowing a short transport distance and close tethering of the proteins involved in these processes (7). In addition, MCSs are advantageous for regulatory processes by compartmentalizing lipid synthesis or transport machinery (27). The gaps of certain MCSs are narrow (10–30 nm) enough to be spanned by individual proteins between organellar membranes. LAMs are known to have a major role in nonvesicular lipid transport at the specific MCSs, which is important for sterol distribution in cells (14).
The structures of the LAM SDs display an α-helix/β-grip fold, which is reminiscent of the overall fold of the StART domains in the StARkin superfamily (17). The StART domains of STARD family proteins have been studied extensively. The structures of all members of the mammalian sterol-binding StART domains were determined in an open form for STARD1 (17), and in closed forms for STARD3/MNL64 (24, 30), STARD4 (22), STARD5 (17), and STARD6 (19). The backbone of these StART domains are highly conserved with the Cα rmsd of less than 2.0 Å (19). However, understanding the mechanism of sterol binding and dissociation has been complicated by the lack of structure of a sterol-bound form and by the variation in cavity size and shape among the homologs. The cavity volumes of StART domains are 308–535 Å3, which is similar to or slightly smaller than the volume of sterol ligands (19). For example, cholesterol-binding STARD6 with a closed lid has a cavity volume smaller than that of its ligand (19). Still, the key determinants for the ligand specificity remained elusive, and they could not be defined by sequence conservation of hydrophilic residues.
This structural study revealed key determinants of ligand recognition by LAM proteins, which can be extended to the understanding of the sterol-binding mode of StARkin superfamily proteins. Sterols bind to the upper portion of the pocket near the tunnel entrance, which is relatively conserved in shape for all LAM homologs. Structural conservation and sterol transfer assays suggested that the major force of sterol binding is hydrophobic interaction, which correlates with the lack of conserved hydrophilic residues in the binding cavity. A sterol binds close to the tunnel entrance and makes hydrophobic contacts with the Ω-loops. The tunnel bottom of LAM SDs is not directly involved in sterol binding. We observed that the tunnel bottoms of LAM SDs are occupied by several water molecules, not by the ligand. The presence of water clusters in the tunnel bottom was as also observed in the sterol-bound states of oxysterol-binding proteins, Osh1 and Osh4 (25, 31). Therefore, if we exclude the space for the water clusters in the tunnel bottom of StART domains, the cavity volumes in the closed conformation of the Ω1-loop are too small to accommodate the sterol molecules. Therefore, conformational changes of the binding pockets or opening of the lid seem to be essential for the sterol-binding of STARD domains. This feature, if conserved in the StART subfamily as well, seems to explain the common sterol-binding mode of STARD homologs.
The StART family proteins, owing to their variation in binding cavities, recognize various lipid types such as phosphatidylcholine, PE, sterols, and ceramides (17). At this time, sterol is the only ligand discovered to be transported by LAM homologs. However, considering the variations in the tunnel bottom, the dual lipid selectivity toward another lipid type in a certain LAM homolog cannot be excluded completely. The direction of sterol transport between the ER and other organellar membranes, whether forward or reverse, could not be determined in this study. Considering the structural simplicity of the LAM SDs for ligand-binding, a possible role of LAMs might be sterol transport along the concentration gradient, counter balancing the directional sterol distribution by oxysterol-binding proteins. In addition to the major role of LAMs in nonvesicular lipid transport, other regulatory roles of LAMs seem to be also plausible (8). LAMs might affect the relative size of sterol pools in the plasma membrane. Lam6 seems to have structural roles in maintaining the contact sites by tethering the two membranes (8). Another role of LAMs in sterol sensing, by conveying either the unbound or bound state to downstream targets, has been proposed (8). For LTPs to act as lipid sensors, ligand binding must produce unique conformational changes that can be transformed to signaling events. For example, the yeast oxysterol-binding protein, Osh4, undergoes significant conformational changes around the tunnel entrance on sterol binding (25). However, there is no unique conformational difference between the apo and sterol-bound forms of the Lam2 SD2 except the slight change in the Ω1−loop, suggesting sterol sensing is unlikely to be a function of the SDs.
LAMs contain multiple targeting modules that enable localization at the contact sites of two different organellar membranes. All LAMs are basically anchored to the ER membrane by their C-terminal transmembrane helix at contact sites that are more than 15 nm in gap distance (8, 32). The other common targeting module is the LAM PH-like domain. The LAM PH-like domains have an N-terminal conserved helix in addition to the canonical PH domain fold. The typical PH domains in many oxysterol-binding proteins, which bind to acidic head groups of phosphoinositides, contain more than eight basic residues in the β1-β2 and β3-β4 loops (29). However, the PH-like domains of all LAM homologs in yeast and humans do not contain more than two basic residues in these regions and expose hydrophobic residues, which are shown to be critical for protein targeting of Lam6. The LAM PH-like domains are well-conserved in amino acid sequence with variations in the loops between secondary structure elements, suggesting that other LAM homologs might use their PH-like domains for protein targeting with different target selectivity.
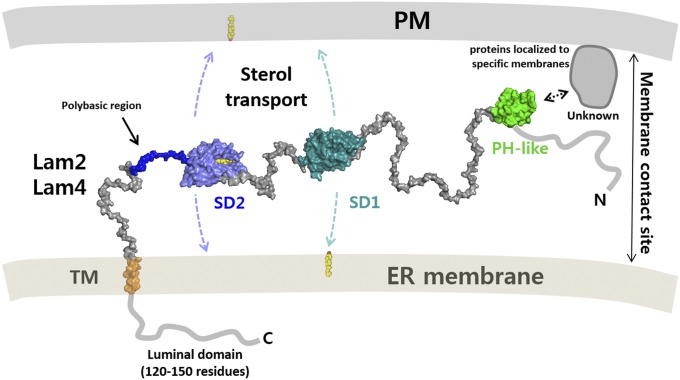
To perform lipid transport at the membrane contact sites, the linker between the anchor and the ligand-binding domain should be long enough to cover the gap so that the SD is able to reach to the other membrane. This requirement for Lam2 was previously demonstrated by examining the phenotype of mutants with different linker lengths, suggesting that the SDs reach out to the plasma membrane to function (14). Based on the findings from this study and those of previous reports (8, 14), we made a schematic model of LAM function at the MCSs (Fig. 7). The SDs are connected by flexible linkers between targeting modules. Owing to the limited length of linkers and the anchoring of LAMs to the ER, direct sterol transport seems to occur in the narrow membrane contacts within 15-nm gap distances. Several LAM homologs contain polybasic regions at the end of their second SD, which might assist direct membrane binding of the domain for sterol uptake or release.
Fig. 7.
Schematic model of the LAM function at the membrane contact sites. LAMs are anchored to the ER membrane by their C-terminal transmembrane helix. The two SDs transport ergosterol between the membranes. The domains are connected by flexible linkers that allow the movement of the sterol transfer domains between the membranes. The PH-like domains of Lam6 contribute to protein targeting by interacting with specific membrane-localized proteins. The binding partner of the PH-like domain of Lam2 and Lam4 are unknown.
In conclusion, we have identified the structural features of the key domains and the determinants of sterol recognition by the LAM SDs. The unique features of the LAM PH-like domains suggested a protein-targeting role by protein–protein interaction. The sterol-binding mode observed in the LAM SD2 is likely to be conserved in other LAM homologs, as well as in sterol-binding StART family proteins. The structural features of the LAM homologs observed in this study well correlate with the lipid transfer function at membrane contact sites.
Materials and Methods
Cloning of the LAM PH-Like and StARkin Domains.
DNA encoding the first SD (residues 848–1,016) and the second SD (residues 1,060–1,223) of Lam2 (UniProt ID: Q06681) were amplified by PCR using yeast genomic DNA as a template. The PCR products of the Lam2 SD1 and Lam2 SD2 were subcloned into the NcoI/XhoI and BamHI/XhoI site of a modified pHIS-2 vector, respectively. The SDs were tagged with an N-terminal hexa-histidine followed by a thrombin protease cleavage site (LVPR/GS). The first and second StART-like domains of Lam4 (UniProt ID: P38800) were cloned into the BamHI/XhoI of the pHIS-2 vector. The PH-like domain (residues 161–272) of LAM6 (UniProt ID: Q08001) was subcloned into a BamHI/XhoI site of the modified pHIS-T4L vector providing an N-terminal T4 lysozyme fusion. The T4 lysozyme inactivated by a Glu20Gln mutation was tagged with a cleavable N-terminal hexa-histidine by thrombin protease. The Lam6 PH-like domain was fused to the C terminus of a T4 lysozyme (residues 2–160), with GlySer as a dipeptide linker.
Protein Purification.
Escherichia coli BL21(DE3) cells transformed with the plasmid encoding the LAM StARkin or PH-like domain were grown to an OD600 of 0.8 at 37 °C. Cells were induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.25 mM and were incubated for 12 h at 20 °C before harvesting. Cells were resuspended in 2× PBS containing 30 mM imidazole and were lysed by sonication. After centrifugation at 23,108 × g for 45 min, the supernatant containing the his-tagged protein was applied to a Ni-NTA affinity column. The protein was eluted from the column using 0.1 M Tris⋅HCl at pH 7.0, 0.3 M NaCl, 0.3 M imidazole. The protein eluate was concentrated to 10 mg/mL, and the His-tag was cleaved by addition of thrombin protease. The target protein was subjected to size exclusion chromatography on a Superdex 200 column (GE healthcare) equilibrated with 20 mM Tris⋅HCl at pH 7.5, 150 mM NaCl. The peak fractions containing the Lam2 SD1, Lam2 SD2, and T4L-Lam6 PH were concentrated to 10 mg/mL for crystallization. The Lam4 SD1 and Lam4 SD2 were concentrated to 60 mg/mL and 20 mg/mL for crystallization studies, respectively.
Crystallization and Crystallographic Analysis.
Preliminary crystallization experiments were carried out in 96-well crystallization plates using a multichannel pipette and customized crystallization screening solutions by dispensing 0.8 μL protein solution and 0.8 μL precipitant solution. Protein crystals were grown by the hanging-drop vapor-diffusion method at 22 °C. Crystals of Lam2 SD1 were grown in 0.1 M Mes·NaOH at pH 6.0, 30% PEG 3350, 0.1 M KNO3. Crystals of the Lam2 SD2 were obtained using a crystallization condition containing 0.1 M Tris⋅HCl at pH 8.0, 30% PEG 8000. To obtain sterol-bound complex for the Lam2 SD2, ergosterol dissolved in ethanol was added during purification steps and to the purified protein at a final concentration of 1.5 mM. The Lam4 SD1 crystals were grown in 0.1 M Na3 Citrate·HCl at pH 5.0, 0.1 M Li2SO4, and 30% PEG 1500. Crystals of the selenomethionine-labeled Lam4 SD1 were grown using same conditions. The Lam4 SD2 was crystallized in 0.1 M Bicine at pH 9.0, 0.1 M MgCl2, and 30% PEG 1500. Crystals of The T4L-LAM6 PH were grown in 0.1 M Hepes·HCl at pH 7.0, 10% PEG 8000, and 0.1 M Na3Citrate.
The crystals were cryoprotected in a reservoir solution supplemented with 10% glycerol and were flash-cooled by immersion in liquid nitrogen. Diffraction data were collected at a fixed wavelength of 0.97949 Å using an ADSC Q270 CCD detector on the 7A and 5C beamlines at Pohang Light Source (PLS), Pohang Accelerator Laboratory, Republic of Korea. All data were processed and scaled using HKL-2000. The structure of the Lam4 SD1 was determined by the single anomalous dispersion method using selenomethionine-labeled crystals. Diffraction data were collected at a wavelength of 0.97934–1.6 Å resolution, and single anomalous dispersion phasing was performed using the software Phaser (33). The structures of the Lam6 PH-like domain and other SDs were solved by molecular replacement, using the structures of T4 lysozyme and the Lam4 SD1, respectively. Modeling was performed using the software COOT (34), and the structures were refined using Phenix (35). The statistics for the X-ray crystallographic studies are shown in Table S1.
Preparation of Liposomes.
DOPC and POPS were obtained from Avanti Polar Lipids Inc. Ergosterol and DHE were obtained from Sigma-Aldrich. To prepare the liposomes for lipid transport assays, the various lipids dissolved in chloroform or ethanol were mixed at the desired molar ratio, incubated at 37 °C for 5 min, and then dried to a thin layer of film using nitrogen gas stream. The dried lipids were resuspended in 1 mL buffer in 20 mM Hepes⋅NaOH at pH 7.2 and 150 mM NaCl by vortexing. The liposomes were prepared at a total lipid concentration of 260 μM in 1 mL. The hydrated lipid mixture was frozen and thawed five times using a water bath and ethanol at −70 °C. The lipid mixture was extruded 10 times through a polycarbonate filter with a pore size of 0.1 mm. The liposomes were stored at 4 °C and used within 24 h.
Sterol Transfer Assays.
The liposomes for sterol transfer assays were prepared as described earlier with desired lipid compositions. The donor and acceptor liposomes contained 10 mol % DHE and 2.5 mol % dansyl-PE, respectively. For each transfer assay, 100 μL donor liposomes and 100 μL acceptor liposomes were added to the buffer in a quartz cuvette to a final volume of 2 mL. The final lipid concentration of each donor and acceptor liposomes was 13 μM. The purified LAM SDs were added to a final concentration of 1 μM. The fluorescence measurement was initiated immediately after adding the purified protein to the liposome mixture in the cuvette with gentle stirring at 25 °C. The fluorescence at 525 nm with an excitation wavelength of 324 nm was monitored for 10 min, using an FP-6200 spectrofluorometer (JASCO).
Liposome Pull-Down Assay.
The various lipids dissolved in methanol or in chloroform were mixed in desired molar ratio. The dried lipids were swollen by adding 150 μL 0.3 M sucrose and sonicated for 30 min. Subsequently, 1 mL deionized water was added and centrifuged at 18,945 × g for 30 min, and the supernatant was removed. The lipid pellet was resuspended in 20 mM Tris⋅HCl at pH 8.0, 0.1 M NaCl and was extruded 10 times through a polycarbonate filter with a pore size of 0.1 mm. The liposomes (100 μg of lipids) were mixed with the various constructs of the LAM SDs (80 μg), and the mixture was incubated at 4 °C for 40 min. The protein in the supernatant and the liposome-bound protein were separated by centrifugation at 17,592 × g for 1 h. The pellet was washed with 200 μL of the buffer, and the proteins in the supernatant and in the liposome pellet were analyzed by SDS/PAGE.
Yeast Cell Imaging.
The various constructs of the Lam6 PH-like domains were cloned into the yeast vector pRS416Met, providing an N-terminal GFP fusion. Yeast strains (Δlam6, BY4741 and Δvac8, BY4742) transformed with the plasmids containing the various GFP-Lam6 PH-like domains were grown to an OD600 of 0.4–0.6 in Ura− selection media. The cells were washed two times with water and resuspended in selection media for microscopic observation. Visualization of cells was performed on an Eclipse Ti fluorescence microscope (Nikon) equipped with a fluorescein isothiocyanate filter, and the images were captured with an Andor ixon EMCCD camera.
Supplementary Material
Acknowledgments
We thank the beamline staff at PLS for their technical assistance. This project was supported by the National Research Foundation of Korea grant funded by the Ministry of Education, Science and Technology (Grant NRF-2017R1A2B4004914).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID code 5YQI (Lam2 SD1), 5YQQ (Lam2 SD2 swapped dimer), 5YS0 (Lam2 SD2 complexed with ergosterol), 5YQJ (Lam4 SD1), 5YQP (Lam4 SD2), and 5YQR (Lam6 PH-like domain)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719709115/-/DCSupplemental.
References
- 1.Mesmin B, Maxfield FR. Intracellular sterol dynamics. Biochim Biophys Acta. 2009;1791:636–645. doi: 10.1016/j.bbalip.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soffientini U, Graham A. Intracellular cholesterol transport proteins: Roles in health and disease. Clin Sci (Lond) 2016;130:1843–1859. doi: 10.1042/CS20160339. [DOI] [PubMed] [Google Scholar]
- 3.Maxfield FR, Wüstner D. Intracellular cholesterol transport. J Clin Invest. 2002;110:891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann NA, et al. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- 5.Hao M, et al. Vesicular and non-vesicular sterol transport in living cells. The endocytic recycling compartment is a major sterol storage organelle. J Biol Chem. 2002;277:609–617. doi: 10.1074/jbc.M108861200. [DOI] [PubMed] [Google Scholar]
- 6.Mesmin B, et al. STARD4 abundance regulates sterol transport and sensing. Mol Biol Cell. 2011;22:4004–4015. doi: 10.1091/mbc.E11-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 8.Wong LH, Levine TP. Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem Soc Trans. 2016;44:517–527. doi: 10.1042/BST20150275. [DOI] [PubMed] [Google Scholar]
- 9.Wong LH, Čopič A, Levine TP. Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem Sci. 2017;42:516–530. doi: 10.1016/j.tibs.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser von Filseck J, Drin G. Running up that hill: How to create cellular lipid gradients by lipid counter-flows. Biochimie. 2016;130:115–121. doi: 10.1016/j.biochi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Alpy F, Tomasetto C. START ships lipids across interorganelle space. Biochimie. 2014;96:85–95. doi: 10.1016/j.biochi.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Alpy F, Tomasetto C. Give lipids a START: The StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 14.Gatta AT, et al. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015;4:e07253. doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murley A, et al. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol. 2015;209:539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbaz-Alon Y, et al. Lam6 regulates the extent of contacts between organelles. Cell Rep. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorsell AG, et al. Comparative structural analysis of lipid binding START domains. PLoS One. 2011;6:e19521. doi: 10.1371/journal.pone.0019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murcia M, Faráldo-Gómez JD, Maxfield FR, Roux B. Modeling the structure of the StART domains of MLN64 and StAR proteins in complex with cholesterol. J Lipid Res. 2006;47:2614–2630. doi: 10.1194/jlr.M600232-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Létourneau D, et al. STARD6 on steroids: Solution structure, multiple timescale backbone dynamics and ligand binding mechanism. Sci Rep. 2016;6:28486. doi: 10.1038/srep28486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe Y, Tamura Y, Kawano S, Endo T. Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat Commun. 2015;6:7922. doi: 10.1038/ncomms8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biesiadka J, Bujacz G, Sikorski MM, Jaskolski M. Crystal structures of two homologous pathogenesis-related proteins from yellow lupine. J Mol Biol. 2002;319:1223–1234. doi: 10.1016/S0022-2836(02)00385-6. [DOI] [PubMed] [Google Scholar]
- 22.Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iaea DB, Dikiy I, Kiburu I, Eliezer D, Maxfield FR. STARD4 membrane interactions and sterol binding. Biochemistry. 2015;54:4623–4636. doi: 10.1021/acs.biochem.5b00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 25.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinser E, et al. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dittman JS, Menon AK. Speed limits for nonvesicular intracellular sterol transport. Trends Biochem Sci. 2017;42:90–97. doi: 10.1016/j.tibs.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JW, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 29.Tong J, Yang H, Yang H, Eom SH, Im YJ. Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 2013;21:1203–1213. doi: 10.1016/j.str.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Horvath MP, et al. Structure of the lutein-binding domain of human StARD3 at 1.74 Å resolution and model of a complex with lutein. Acta Crystallogr F Struct Biol Commun. 2016;72:609–618. doi: 10.1107/S2053230X16010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manik MK, Yang H, Tong J, Im YJ. Structure of yeast OSBP-related protein Osh1 reveals key determinants for lipid transport and protein targeting at the nucleus-vacuole junction. Structure. 2017;25:617–629. doi: 10.1016/j.str.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 32.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.