SUMMARY
Antigenic variation in malaria was discovered in Plasmodium knowlesi studies involving longitudinal infections of rhesus macaques (M. mulatta). The variant proteins, known as the P. knowlesi Schizont Infected Cell Agglutination (SICA) antigens and the P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1) antigens, expressed by the SICAvar and var multigene families, respectively, have been studied for over 30 years. Expression of the SICA antigens in P. knowlesi requires a splenic component, and specific antibodies are necessary for variant antigen switch events in vivo. Outstanding questions revolve around the role of the spleen and the mechanisms by which the expression of these variant antigen families are regulated. Importantly, the longitudinal dynamics and molecular mechanisms that govern variant antigen expression can be studied with P. knowlesi infection of its mammalian and vector hosts. Synchronous infections can be initiated with established clones and studied at multi-omic levels, with the benefit of computational tools from systems biology that permit the integration of datasets and the design of explanatory, predictive mathematical models. Here we provide an historical account of this topic, while highlighting the potential for maximizing the use of P. knowlesi – macaque model systems and summarizing exciting new progress in this area of research.
Key words: SICAvar genes, var genes, Plasmodium falciparum, epigenetics, mathematical models, systems biology, multi-omics, host–pathogen interactions, longitudinal infections, macaques
INTRODUCTION
Antigenic variation of parasite-encoded proteins expressed at the surface of Plasmodium infected RBCs (iRBCs) is critical to parasite survival. In turn, host immune responses against these proteins are important for host survival and the establishment and maintenance of chronic asymptomatic malaria infections (Howard, 1984; Galinski and Corredor, 2004; Arnot and Jensen, 2011). Upon repeated exposure to parasites with different variant antigen repertoires, individuals are less likely to exhibit severe clinical complications and will eventually cease having symptomatic infections, despite being parasitemic (Bull et al. 1998). On a population level, antigenic variation has explained observations of reduced incidence of severe disease in older individuals living in high transmission settings. Importantly, aside from being the basis, by definition, of an unhealthy state, chronic parasitemias may aid the propagation of the parasite via transmission (Zhou et al. 2016). Taking all these aspects into account, understanding antigenic variation mechanisms and chronicity may lead to new strategies to boost the host immune system for clearing parasitemias and in turn support malaria elimination strategies (Alonso et al. 2011).
Malaria antigenic variation has been most prominently associated with the Plasmodium falciparum Erythrocyte Membrane Protein-1 (EMP-1) variant antigens, which are encoded by the large var multigene family with about 60 members dispersed throughout all 14 chromosomes, with the majority located near the telomeres (Leech et al. 1984; Baruch et al. 1995; Smith et al. 1995; Su et al. 1995). However, the phenomenon of antigenic variation was discovered in Plasmodium knowlesi (Brown and Brown, 1965). In P. knowlesi, the Schizont Infected Cell Agglutination (SICA) variant antigens (Howard et al. 1983) are encoded by the related large SICAvar multigene family, which has at least 136 members that are likewise dispersed on all 14 chromosomes, yet more evenly distributed than the P. falciparum var genes and without as much concentration near the telomeres (Al-Khedery et al. 1999; Corredor et al. 2004; Pain et al. 2008; Lapp et al. 2017). The initial groundbreaking studies from 1965 involved experimental longitudinal infections of P. knowlesi in rhesus macaques, and this work raised a number of critical questions regarding the mechanisms of antigenic variation that remain unanswered today. In contrast to P. falciparum and P. knowlesi, the var gene family is not present in the human malaria species Plasmodium vivax, Plasmodium malariae and Plasmodium ovale (Carlton et al. 2008; Rutledge et al. 2017). Since P. knowlesi was recognized as a zoonotic parasite and a widespread public health threat in South East Asia (Singh et al. 2004; Cox-Singh and Singh, 2008; Cox-Singh et al. 2008), the relative importance of understanding antigenic variation in this species has been escalating.
This paper puts forth the overarching hypothesis that molecular mechanisms of Plasmodium antigenic variation in vivo, regulated by specific host responses and factors, can be discovered or computationally inferred with modern methods of systems biology and possibly result in groundbreaking strategies for treating malaria infections. Given the widespread implications of antigenic variation in malaria, with hundreds of millions of cases estimated annually in about 100 countries, we believe this research area should gain increased traction with translational goals in mind.
PERSPECTIVES
Importantly, the dynamics and molecular mechanisms of variant antigens in P. knowlesi can be studied in vivo using macaques, with highly synchronous blood-stage infections initiated by established (or new) SICA[+] and SICA[−] clones, which do or do not express the variant antigens, respectively (Barnwell et al. 1983a). The P. knowlesi rhesus macaque model system is the most advanced option for studying the regulatory mechanisms of antigenic variation based on in vivo investigations, with the use of in vivo derived, well-characterized parasite clones. Moreover, this nonhuman primate (NHP) model allows for experimental passage through the mosquito host, thereby offering the opportunity to observe possible reset patterns of SICAvar gene expression and to monitor the within-host expression dynamics for the first time during the course of longitudinal infections. In addition, the Anopheles dirus mosquito can be reared in the laboratory, providing an experimental model system for the study of regulatory mechanisms of antigenic variation that may occur in the vector. This vector has been implicated in natural P. knowlesi and P. falciparum transmission in the region (Nakazawa et al. 2009; Marchand et al. 2011). The value of investigations of the related simian species Plasmodium coatneyi and Plasmodium fragile, which also undergo antigenic variation and share biological features akin to P. falciparum, should also be stressed (Handunnetti et al. 1987; Galinski and Corredor, 2004; Chien et al. 2016). In-depth comparative investigations of these species may be worth pursuing in the future, as would be studies of P. knowlesi in the natural macaque hosts from South East Asia, Macaca fascicularis and Macaca nemestrina (Cox-Singh et al. 2008; Divis et al. 2015; Maeno et al. 2015). It has been predicted that the human and simian malaria species share regulatory mechanisms, and it is likely that similar molecular and immunobiological host factors govern their expression (Barnwell et al. 1983b; Howard, 1984; Howard and Barnwell, 1984b; Korir and Galinski, 2006; Arnot and Jensen, 2011).
Model systems and advanced technologies have reached a level of sophistication allowing for the realistic characterization of the in vivo dynamics of Plasmodium antigenic variation, as well as the identification of specific in vivo host-parasite factors that regulate antigenic variation. This can be done by analyzing infected host and parasite blood samples from longitudinal infections and conducting high-throughput studies with multi-omic analyses (e.g., genomics, epigenomics, transcriptomics, proteomics, lipidomics, immunomics and metabolomics). A refined Macaca mulatta genome (Zimin et al. 2014) (Version 7.8) has been available, but a refined P. knowlesi genome (Pain et al. 2008) assembly with fully annotated SICAvar sequences has been lacking. To fill this need, over the past year, a high-quality P. knowlesi de novo genome sequence has been generated in the Malaria Host–Pathogen Interaction Center (MaHPIC) by combining long-read sequences (PacBio) and genome-wide high-throughput chromosome conformation capture (Hi-C) data to produce accurate, chromosome-level scaffolds, followed by automated and manual annotation, including for all SICAvar genes. This genome sequence has been named the ‘MaHPIC Pk Genome Sequence’ and is being reported elsewhere in this Special Issue (Lapp et al. 2017). This sequence, combined with sophisticated mathematical and computational analyses to determine how the SICAvar expression is affected by passage through the Anopheles vector and NHP hosts, will aid future research toward understanding the molecular factors regulating antigenic variation in vivo throughout the parasite's life cycle.
HISTORICAL OVERVIEW
The P. knowlesi–rhesus macaque model system: discovery of malaria antigenic variation and a regulatory role of the spleen
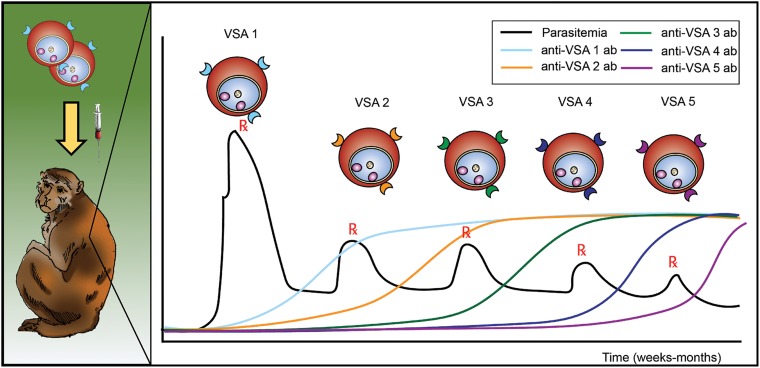
Antigenic variation in malaria was discovered in longitudinal experiments by inoculation of rhesus macaques with P. knowlesi blood-stage parasites (Brown and Brown, 1965). These studies used SICA agglutination assays (Eaton, 1938) to demonstrate that variant antigens were exposed at the surface of iRBCs and that the antigenicity changed during the course of an infection. The antigens were subsequently determined to be parasite-encoded proteins (Howard et al. 1983), and are often referred to, across species, as variable surface antigens or VSAs (Fig. 1). Immediately following this demonstration, efforts continued to understand the host responses to these antigens and to assess whether the changing of exposed antigens (alternatively referred to as ‘switching’) was a result of antibody induction or selection; the early studies concluded that both could be in play (Brown, 1973; Brown and Hills, 1974). At the time, it was unknown whether the host proteins were simply changing or if the parasite was producing different proteins at the surfaces of the host cells.
Fig. 1.
Schematic representing the longitudinal infection experiments performed with P. knowlesi in rhesus monkeys, demonstrating the phenomenon of malaria antigenic variation, and reported by K.N. Brown and I.N. Brown in 1965, in Nature (Brown and Brown, 1965). Different VSAs are expressed in the course of an infection, as antigenic variation occurs in response to the appearance of anti-VSA antibody (ab). VSA, variable surface antigens.
In 1983, nearly 20 years later, in vivo derived parasite clones were developed by Barnwell, Howard and others and shown to switch variant types in animals in the presence of specific antibodies, whereas the clones did not switch phenotypes in naïve animals (Barnwell et al. 1983a). The original and other P. knowlesi related clones were developed by micromanipulation of single schizonts and expansion in naïve rhesus, followed by cryopreservation as ring-stage forms (Barnwell et al. 1983a, and unpublished data confirming additional switched clones). As one example, the Pk1(A+) clone switched when inoculated into a rhesus that had been previously inoculated with Pk1(A+) parasites, and the Pk1(B+)1+ clone was derived from the resulting switched population. The Pk1(A+) and Pk1(B+)1+ clones express dominant SICA proteins that were characterized as doublets of 190 and 210 kDa, and 200 and 205 kDa, respectively (Howard et al. 1983). These were identified with radioiodination surface labeling and immunoprecipitation studies based on detergent extracts and variant specific polyclonal sera, to be expressed at the surface of the iRBCs, predictably with a transmembrane domain and internal cytoplasmic domain (Howard et al. 1983; Howard and Barnwell, 1984a, b , c , 1985; Howard et al. 1984). These studies provided confirmatory data to support the theory that the parasites were expressing distinct parasite-encoded antigens that changed with each antigenic switch. Comparable biochemical methods used to identify the SICA proteins were subsequently applied by Leech and others to identify EMP-1 in P. falciparum. They showed that the proteins from both species shared basic characteristics and, in particular, basic common regulatory mechanisms (Leech et al. 1984).
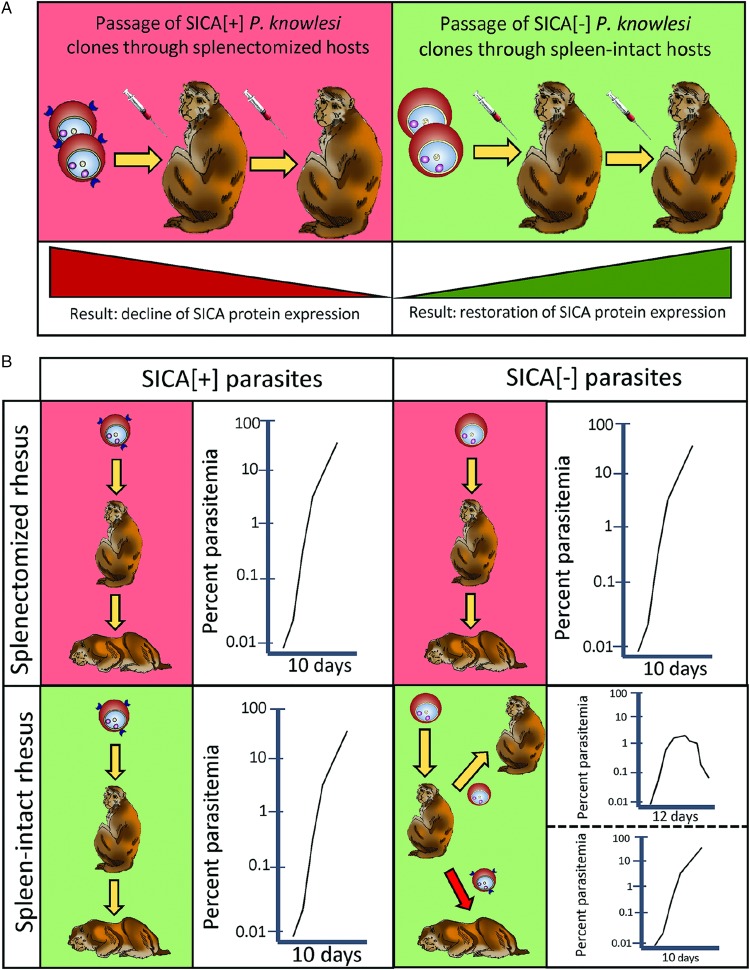
The spleen also proved to be important. An ongoing mechanistic mystery is the observation that the expression of variant antigens at the surface of the iRBCs is lost during passage in splenectomized monkeys, but that expression of variant antigens can be recovered with passage in intact monkeys (Barnwell et al. 1982, 1983a, b ). Specifically, immunofluorescence assays (IFAs) and labelling experiments by Barnwell et al. showed that the loss occurred in a matter of a few 24-h cycles and confirmed that the expression of SICA was associated with host factors and parasite virulence (Fig. 2). Intact rhesus monkeys almost universally succumbed to rapid, high rising parasitemias without antimalarial treatment, but they could control the virulent parasitemia if SICA proteins were not expressed. The loss of variant antigen expression in splenectomized hosts was subsequently shown to occur also with P. falciparum in squirrel monkeys (Hommel et al. 1983) and with P. fragile in toque monkeys (Handunnetti et al. 1987).
Fig. 2.
Depiction of the loss or gain of SICA protein expression in splenectomized and intact rhesus macaques (A) and of the association of virulence with the expression of SICA proteins (B), as described by Barnwell and colleagues in 1983 (Barnwell et al. 1983b). If SICA expression was regained by SICA[−] parasites in intact rhesus the parasites were highly virulent, but if SICA[−] parasites did not regain SICA expression, the infections were controlled. SICA, Schizont Infected Cell Agglutination.
The P. knowlesi SICAvar gene family: structure, expression, conservation and regulation
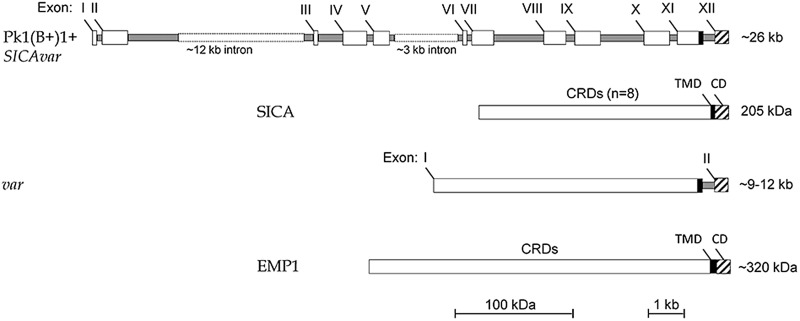
The large SICAvar multigene family was identified and characterized beginning in the mid-1990s, using traditional cloning, sequencing and whole genome hybridization methods (Al-Khedery et al. 1999). The initial report demonstrated the presence of the gene family and specifically characterized the SICAvar gene encoding the 205 kDa SICA protein that is expressed in the Pk1(B+)1+ clonal parasites. This gene was reported with a 10-exon structure (Al-Khedery et al. 1999) and later redefined as 12 exons when an approximated 12 kb intron and two additional exons were identified upstream (Lapp et al. 2009). An intriguing genomic rearrangement was also identified at the end of this SICAvar gene and shown to be associated with the in vivo switch from the Pk1(A+) to Pk1(B+)1+ phenotypes (Al-Khedery et al. 1999; Corredor et al. 2004; Lapp et al. 2009). Further research is required to determine if this observation was a unique mitotic rearrangement event or if it is a common mechanism associated with switching events. In any case, the initial 1999 report by Al-Khedery et al. (1999) suggested possible roles for transcriptional or post-transcriptional control of SICAvar expression. These early studies showed that a wide repertoire of transcripts was produced by the Pk1(B+)1+ parasites, with representation of various gene family members in a cDNA library but without particular dominant transcripts noted. Yet, northern blots demonstrated a dominant 205 kDa protein-encoding stage-specific full-length transcript in the ring stages of the Pk1(B+)1+ but not the Pk1(A+) parasites (Al-Khedery et al. 1999). One might predict that a majority of the iRBCs in an infection express those predominant transcripts known to make the characteristic SICA proteins in a clonal population, while a minority of iRBCs may be making other SICAvar transcripts. As discussed further below, single cell studies will be important to clarify whether all (or most) cells produce the repertoire of SICAvar transcripts observed in different SICA[+] clones.
Figure 3 shows that the structure of the Pk1(B+)1+ SICAvar gene encoding the 205 kDa SICA protein contrasts with the much simpler two-exon structure of var genes in P. falciparum (reviewed in Galinski and Corredor, 2004). Nonetheless, both protein structures have a series of variable cysteine-rich domains (CRDs) in the large externalized portion of the protein, and EMP1 and SICA proteins were shown by proteomics to share common peptides, suggesting their evolutionary relatedness (Korir and Galinski, 2006), which is supported in a recent evolutionary analysis of Plasmodium variant antigen gene families and their relationships (Frech and Chen, 2013). Despite the difference in exon structures (explained as loss or gain of intronic sequences in evolutionary time), both SICAvar and var gene families possess a transmembrane domain in the penultimate exon and a final conserved exon encoding a cytoplasmic domain. It is also interesting that the 205 kDA SICA CRDs were shown to be encoded by sequence beginning at the end of each exon, interrupted by the intron sequences, and then continued at the start of the next exon (Al-Khedery et al. 1999).
Fig. 3.
Schematic of the structure of the multi-exon SICAvar and var genes, along with the encoded proteins (SICA and EMP1, respectively). Open boxes are exons and grey rectangles represent introns; the two rectangular dotted open boxes denote particularly long intron sequences that range in size beyond the scale of the figure. CRD, Cysteine-rich domain; TMD, transmembrane domain; CD, cytoplasmic domain; SICA, Schizont Infected Cell Agglutination..
The SICA cytoplasmic domain has been hypothesized to be functionally critical and possibly involved in signalling processes induced by specific antibodies produced against the external variant domains (Al-Khedery et al. 1999). This hypothesis is consistent with the concept of antibody induced switching, which was proposed beginning in 1965 to explain waves of parasitemia with different iRBC cell-surface exposed antigens (Brown and Brown, 1965), with some subsequent experimental support for this notion (Brown and Hills, 1974). As expected, P. coatneyi, which is phylogenetically very closely related to P. knowlesi (Vargas-Serrato et al. 2003), has a comparable large SICAvar multigene family (Galinski and Corredor, 2004; Chien et al. 2016). Additionally, while the P. fragile genome sequence is not yet available, antigenic variation of iRBC surface-exposed antigens has been shown for this species in the toque monkey (Macaca sinica) (Handunnetti et al. 1987), and it is likely that P. fragile's genome is also endowed with SICAvar genes. In support of this prediction, experiments with P. fragile, reported by Handunnetti et al. demonstrated a sequential pattern of variant types upon switching in the course of blood-stage infections in multiple small cohorts of toque monkeys (Handunnetti et al. 1987). Clearly, complementary knowledge across different primate species can inform and guide the building of mechanistic models, and include comparisons of P. knowlesi infections in natural monkey hosts, such as M. fascicularis and M. nemestrina (Cox-Singh et al. 2008).
Delving deeper into questions relating to the repertoires of SICAvar transcript vs protein expression in cloned parasites, quantitative RT-PCR and proteomics studies were performed. These studies identified the variant antigen repertoires of the Pk1(A+) and Pk1(B+)1+ parasite clones, and demonstrated a complete switch in SICA expression with dominant proteins and lesser proteins identified in each clone (Lapp et al. 2013). As shown for the first time in this study, the in vivo switch from the Pk1(A+) to the Pk1(B+)1+ phenotype resulted in the downregulation of one set of SICAvar genes and the upregulation of another set. The results, also reflected in northern blots, strongly suggested that both transcription and post-transcription processes were functioning to regulate the expression of these genes and proteins (Lapp et al. 2013).
Comparisons of SICA[+] and SICA[−] parasites furthermore revealed that the SICAvar gene family becomes downregulated when passaged in splenectomized animals. In the SICA[−] parasites, transcript detection is dramatically reduced, transcript signals are not detected by northern blot using specific or conserved probes and SICA products are not detected by proteomics after immunoprecipitation with an antisera that recognizes the conserved cytoplasmic domain (Lapp et al. 2013). These observations are consistent with possible transcriptional control mechanisms and specific post-transcriptional processing events (Galinski and Corredor, 2004); e.g. involving non-coding RNAs. The observed downregulation under these unnatural physiological conditions stresses the importance of host factors in the regulation of antigenic variation. Evidently, antigenic switch events at the genetic level that result in positive SICA expression require some unknown component or interactions provided by the presence of the spleen and specific antibodies to the expressed variant antigen, and possible other undefined regulatory factors. Of direct relevance to humans, several studies have reported the circulation of all P. falciparum RBC stages from splenectomized patients (Israeli et al. 1987; Ho et al. 1992; Bach et al. 2005; Bachmann et al. 2009). One of these studies specifically confirmed that the P. falciparum var gene family was not expressed in the iRBCs, thus connecting the lack of EMP1 expression with the loss of sequestration (Bachmann et al. 2009). Downregulation of the SICAvar family also occurs in P. knowlesi in vitro cultures (Lapp et al. 2015). Thus, long-term cultures are not particularly well suited for SICAvar expression studies, but ex vivo transfection experiments followed by in vivo growth can be useful to test hypotheses (van der Wel et al. 1997; Kocken et al. 2002; Kocken et al. 2009; Pasini et al. 2016). Curiously, our present culture-adapted line, derived from a line established by Kocken et al. (2002) did not grow when passaged back into rhesus, owl and squirrel monkeys, primate species normally highly susceptible to infection with P. knowlesi (unpublished data); this suggests that changes have occurred in this line over time upon in vitro propagation that now prohibit its successful infection and growth in vivo.
SICAvar gene expression may be regulated by non-coding RNAs that are antisense to those genes, supporting the possible involvement of post-transcriptional regulatory processes (Fig. 4 and unpublished results characterizing such sequences). Observations linking non-coding RNAs with SICAvar gene expression, such as the presence of antisense RNAs in SICA[−] parasites that lack full-length SICAvar mRNA, became evident at the turn of the century, before the broad discovery of such RNAs, and their importance as regulators of gene expression in nature (reviewed in Rinn and Chang, 2012). However, a P. knowlesi genome sequence was not available then, which precluded further research on this topic at that time. With the first P. knowlesi genome sequence (Pain et al. 2008), however, and since with RNA-Seq transcriptome analyses (unpublished results), non-coding RNA species have become evident as predominant species. Non-coding RNA species have also been observed in P. falciparum and are believed to be mechanistically important for the regulation of var gene expression at the transcriptional level, although their precise roles are not fully understood (Amit-Avraham et al. 2015). Interestingly, polysome profiling analyses throughout the P. falciparum erythrocytic cycle identified var gene introns associated with the polysome fraction at the ring stage (Bunnik et al. 2013). These recent data indicate that control of antigenic variation and translational repression of transcribed var genes can occur at the translational level and involve non-coding RNA species. The field of RNA regulatory control mechanisms is burdgeoning in general, and the same is true for Plasmodium research reviewied in Vembar et al. (2016).
Fig. 4.
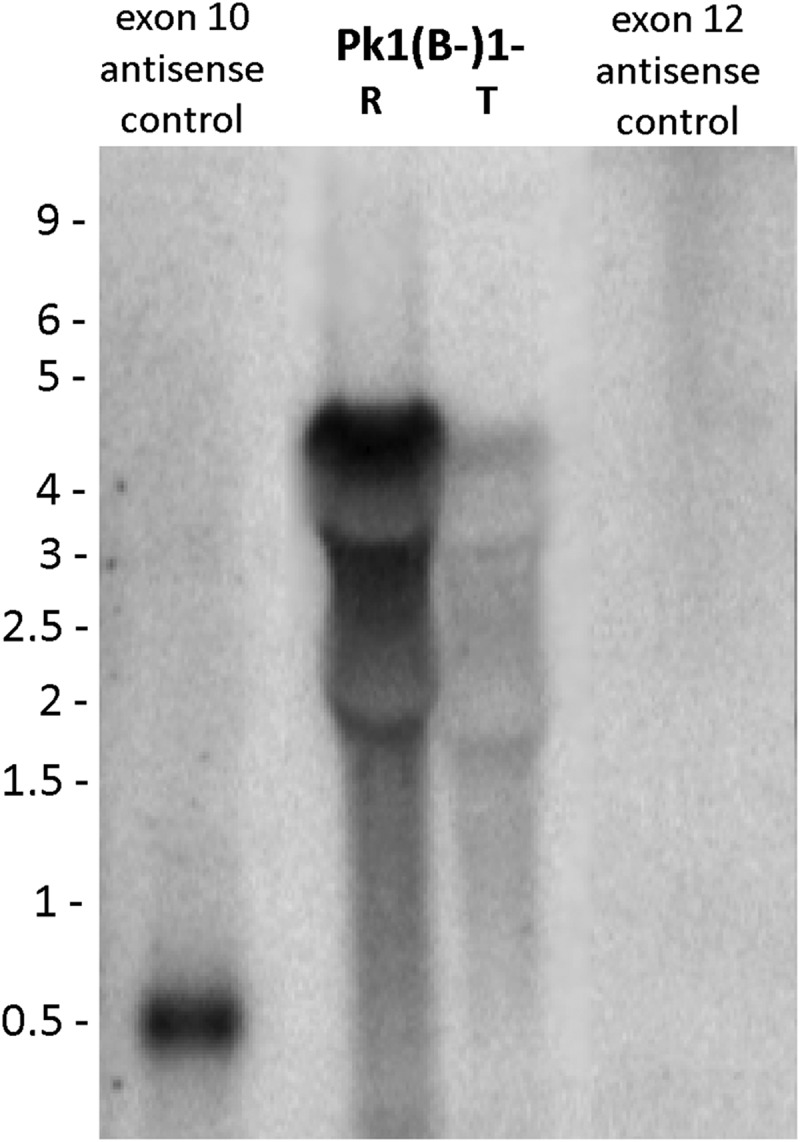
Northern blot experiment showing antisense transcripts to the SICAvar gene encoding the 205 kDa SICA protein are present in the SICA[-] ring (R), but not trophozoite (T) stages, of Pk1(B-)1- iRBCs. A gene-specific exon 10 sense riboprobe control hybridization, representing the probe used, is also shown. SICA, Schizont Infected Cell Agglutination.
THE QUEST FOR IDENTIFYING REGULATORY MECHANISMS OF ANTIGENIC VARIATION IN MALARIA
Plasmodium falciparum, 1995 onward
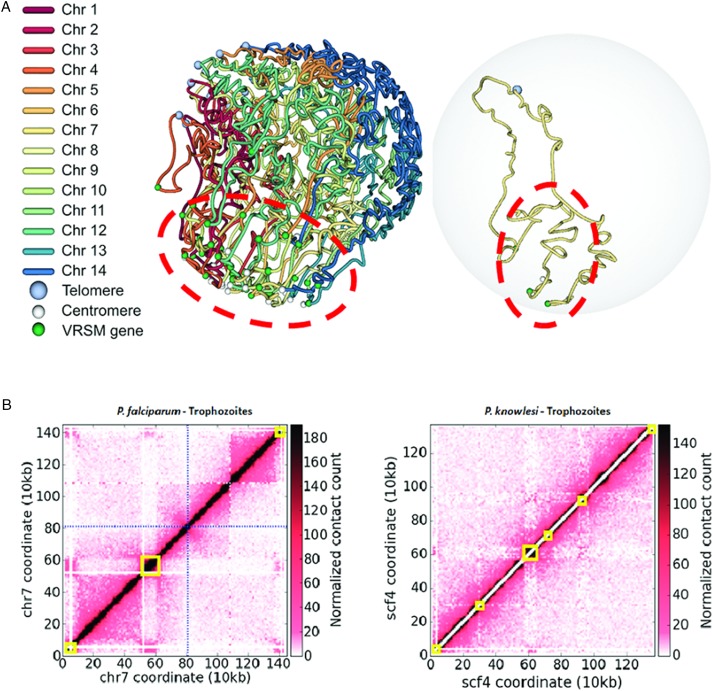
Numerous studies in laboratories and at field sites worldwide have been designed to better understand mechanisms regulating antigenic variation in the human malaria parasite P. falciparum since the discovery of the multigene var family in 1995 (Baruch et al. 1995; Smith et al. 1995; Su et al. 1995), and the publication of the first P. falciparum genome in 2002 (Gardner et al. 2002). Researchers have aimed to understand var gene expression patterns and switching mechanisms, and associations with immunity, illness severity and population dynamics (reviewed in Miller et al. 2002; Wellems et al. 2009; Arnot and Jensen, 2011; Guizetti and Scherf, 2013; Cortes and Deitsch, 2017). Plasmodium falciparum EMP1 is associated with pathogenicity mediated through the adherence of infected RBCs via CRDs to the endothelium of post-capillary venules and accumulated immunity to antigenic variants has been associated with the reduction of clinical severity in adults and older children (Smith et al. 1995; Bull et al. 1998; Smith et al. 2000; Warimwe et al. 2013). The ensuing body of epigenetic research has revealed processes involving histone modifications, subnuclear localization and movement of var genes, promoter/promoter interactions, and roles for long non-coding RNAs (Chen et al. 1998; Scherf et al. 1998; Deitsch et al. 2001; Duraisingh et al. 2005; Freitas-Junior et al. 2005; Chookajorn et al. 2007; Dzikowski et al. 2007; Frank et al. 2007; Dzikowski and Deitsch, 2008; Volz et al. 2012; Jiang et al. 2013). Silent var genes have been shown to co-localize with each other near the nuclear periphery, in heterochromatin regions, while a single active var gene was relocated elsewhere (Freitas-Junior et al. 2005; Lopez-Rubio et al. 2007; Lopez-Rubio et al. 2009). Recently, the development of next-generation sequencing combined with molecular assays that measure proximities of DNA (e.g., 4C-Seq and Hi-C, referring to one-vs-all and all-vs-all high-throughput chromosome conformation capture sequencing, respectively (Lieberman-Aiden et al. 2009; van de Werken et al. 2012) and histone modifications (e.g. ChIP-Seq; chromatin immunoprecipitation-sequencing) demonstrated that the var gene family adds a striking complexity to the genome organization and clusters of var genes act as structural elements that shape the genome architecture (Fig. 5A) (Lemieux et al. 2013; Ay et al. 2014; Ay et al. 2015); similar structural elements were also observed with SICAvar genes in P. knowlesi (Fig. 5B). The identification of specific players on the stage of complex interactions, such as PfSETvs-dependent H3K36me3 in var gene silencing, raised the prospect that the knowledge gained from these studies may lead to the development of translational, preventative or therapeutic tools (Jiang et al. 2013).
Fig. 5.
(A) 3D model of the P. falciparum genome – var genes co-localize in repressive center(s) within the red dashed ellipse (left). Co-localization of the var genes shapes chromosome conformation (right) in chromosome 7. (B) Hi-C contact maps of trophozoite stage parasites for P. falciparum chromosome 7 (left) and P. knowlesi scaffold 4 (Lapp et al. 2017). Hi-C data were generated and analysed as described (Ay et al. 2014), and heatmaps for normalized contact counts were created at 10 kb resolution. Yellow boxes indicate var and SICAvar gene loci for P. falciparum and P. knowlesi, respectively. P. falciparum gene annotations were accessed from PlasmoDB (v9·0). SICAvar gene annotations were curated manually (Lapp et al. 2017). SICA, Schizont Infected Cell Agglutination.
Single cell expression
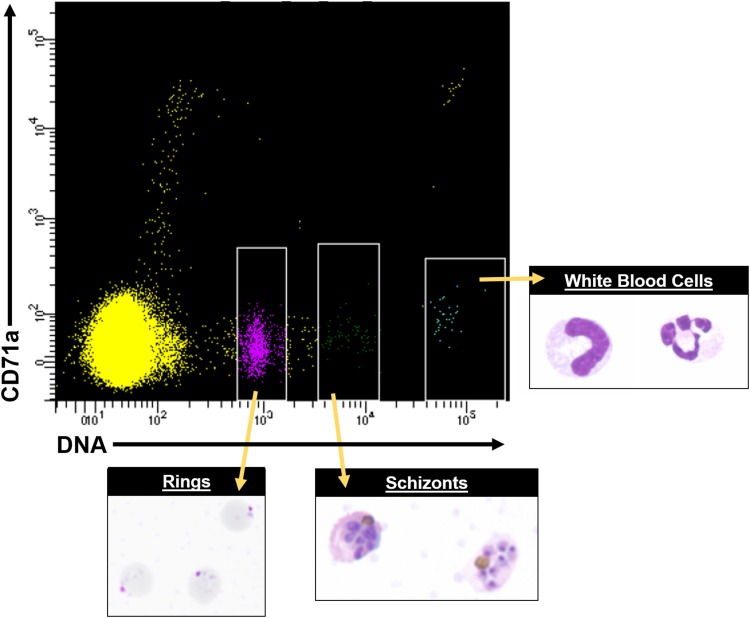
Most mechanistic studies on variant antigens have been performed on populations of cells, due to the historical limitations of single-cell technologies, and these populations were often derived with receptor binding/panning selection methods (Biggs et al. 1992; Roberts et al. 1992). However, recent advances in single-cell sorting and single-cell transcriptomics suggest powerful potential for studying the variant antigen gene expression profiles of single cells, or defined subpopulations, and how they relate to the larger populations from which they were derived, and possible proteomic profiles. Figure 6 shows an example of how fluorescence activated cell sorting (FACS) can be leveraged to obtain purified asexual stages from P. knowlesi infected blood. In vitro fluorescence in situ hybridization (FISH) and other techniques of modern molecular biology have indicated that multiple P. falciparum var transcripts can be present at the same time in single cells (Chen et al. 1998; Brolin et al. 2009; Joergensen et al. 2010), and the same has been determined for P. knowlesi (Lapp et al. unpublished data).
Fig. 6.
FACS data showing distinct cell populations, including different parasite stages from P. knowlesi infected blood, using antibodies conjugated to fluorophores and Hoechst 33342 DNA dye. A representative FACS plot from multiple experiments is shown. White rimmed boxes indicate specific cell populations that were sorted based on the single-cell expression of CD71a and DNA content, demonstrating the ability to resolve infected RBCs from white blood cells and uninfected RBCs. A representative image of each sorted population is provided. FACS, fluorescent activated cell sorting.
Single cell analyses will become important to address questions regarding the dynamics of antigenic variation in vivo, to clarify to what degree parasites maintain clonality with regard to SICAvar/SICA expression after short-term passage in naïve hosts, and how this clonality may change as a chronic infection is established. Answering such questions is key to understanding variant antigen regulation on a population level and addressing possible implications for cell–cell communication mechanisms relating to switching of variant antigen phenotypes among iRBC (reviewed in Rivkin et al. 2017) and possible intracellular signalling pathways as suggested in response to host antibodies (Al-Khedery et al. 1999). Moreover, the combination of population and single-cell transcriptomic studies along with mathematical modelling can quite readily advance understanding of mechanisms of transcriptional and post-transcriptional control, including the possible role(s) of non-coding RNAs, as on/off or antigenic switch changes are occurring over time in SICA[+] and SICA[−] infections, in intact or splenectomized animals.
THE HOST MATTERS: THE IN VIVO ENVIRONMENT INFLUENCES REGULATORY PROCESSES
Over the past 10 years, evidence has been mounting that regulatory controls of the host can influence the expression of variant antigen genes, for both P. knowlesi and P. falciparum. Thus, according to our current understanding, the basic mechanisms governing antigenic variation must be evaluated from in vivo-derived data to capture a complete picture and the dynamics that include such influences. This point has been stressed by many researchers, taking the spleen and immune responses into account (Galinski and Corredor, 2004; Daily et al. 2005; Bachmann et al. 2009; Arnot and Jensen, 2011; Bachmann et al. 2011; Lapp et al. 2015), and recent studies involving humans and mice have shown a ‘reset’ of the expression of the variant antigen gene repertoire after passage through the mosquito vector (Lavstsen et al. 2005; Wang et al. 2009; Spence et al. 2013; Bachmann et al. 2016). However, investigations with humans or rodent models have caveats. First, longitudinal infection discovery is limited in human investigations due to ethical restrictions and the need of immediate blood-stage treatment. Second, genetic counterparts to the var gene family do not exist in rodent malaria species. Consequently, a deeper understanding of the mechanisms regulating antigenic variation of the var or SICAvar families – before, during and after mosquito passage – will enormously benefit from in vivo investigations using appropriate animal models, and there is little doubt that the host–parasite interactions at the molecular and immunobiological levels in humans will be most similar to those in NHPs.
So far, an NHP model has not been available to conduct comparable research with P. falciparum akin to what is possible with P. knowlesi or other simian malaria parasites in macaques, which is due to various factors including the limited number of P. falciparum isolates adapted to New World monkey hosts and the limitations of those parasite–host combinations (reviewed in Galinski and Barnwell, 2012). Work on antigenic variation of P. falciparum in NHPs to date has been limited to the Falciparum Uganda Palo Alto (FUP) strain and Saimiri sciureus monkeys (Hommel et al. 1983). The blood stages of this strain have been adapted to produce robust infections in these animals, but transmission through a mosquito vector has not been possible. Moreover, the animals weigh only about 1 kg, which imposes severe limitations on experimental blood volume draws in comparison with macaques. Fortuitously, as shown in Table 1, biological features of P. falciparum EMP1 and P. knowlesi SICA are quite similar, and one might legitimately expect the regulatory mechanisms to reflect corresponding similarities.
Table 1.
Features shared by P. knowlesi SICA and P. falciparum EMP1
| Variant antigen characteristic | SICA protein | PfEMP1 |
|---|---|---|
| Large parasite-encoded proteins | ✓ | ✓ |
| Exposed at infected RBC surface | ✓ | ✓ |
| Extractable only by ionic detergents | ✓ | ✓ |
| Undergo antigenic variation | ✓ | ✓ |
| Associated with virulence | ✓ | ✓ |
| Encoded by large multigene family | ✓ | ✓ |
| Contain cysteine-rich domains | ✓ | ✓ |
| Subject to much diversity | ✓ | ✓ |
| Genes on all chromosomes | ✓ | ✓ |
| In situ transcription | ✓ | ✓ |
| Abundant transcription in ring stage | ✓ | ✓ |
| Translation in trophozoite stage | ✓ | ✓ |
| Antisense long non-coding RNAs in gene regulation | ✓ | ✓ |
| Epigenetic gene control | ✓ | ✓ |
| Role for spleen in expression control | ✓ | ✓ |
| Associated with typical cytoadherence/sequestration | × a | ✓ |
Comparative studies in macaques
Rhesus monkeys have been used for decades as well-established experimental hosts of P. knowlesi, however, they are not known to be infected with this species in the wild. Macaca fascicularis and M. nemestrina are natural hosts and disease reservoirs for human infections in South East Asia (Cox-Singh et al. 2008; Divis et al. 2015). These species have been studied to a lesser degree with Plasmodium infections, but should be utilized as well to develop within-host knowledge of antigenic variation. In contrast to rhesus, these species do not exhibit overwhelming parasitemia with P. knowlesi infections, nor a comparable high risk of mortality (Knowles and Das Gupta, 1932; Anderios et al. 2010). Clinical manifestations of P. knowlesi in these species more closely parallel human cases (Daneshvar et al. 2009), although lower parasitemias and controlled infections make them less suitable for studying and modeling switch events in longitudinal infections. Ultimately understanding the differences in the dynamics of SICA protein expression in these distinct host species may be enlightening with regard to the dramatic differences in virulence, the immune reponses and their clinical manifestations.
Aside from having a clear role in parasite virulence, the biological function of SICA proteins and their precise localizations on the host cell membrane are still unknown. As both SICA[−] and SICA[+] expand rapidly in splenectomized hosts, the function of SICA proteins is not thought to be metabolic (Barnwell et al. 1983a). Plasmodium knowlesi exhibits sequestration behaviour, which however is limited compared with P. falciparum, P. coatneyi or P. fragile, for which only the ring-stage forms and gametocytes typically circulate. In synchronous P. knowlesi infections, fewer than the expected number of schizonts are often observed on a morning blood smear compared with the number of ring-stage parasites observed the afternoon prior. Miller et al. described extensive P. knowlesi schizogony in several tissues in rhesus monkeys (Miller et al. 1971), and more recent studies showed P. knowlesi iRBC distribution in baboon tissues (Ozwara et al. 2003; Onditi et al. 2015). Additionally, unlike P. falciparum and P. coatneyi, the P. knowlesi variant antigens are not concentrated at parasite-induced protrusions at the RBC surface (reviewed in Galinski and Corredor, 2004). The much-simplified P. knowlesi infected RBC surface, without knobby protrusions also introduces questions regarding the minimal requirements of parasitism of the primate species and the fundamental biological function of the variant antigens across the species. Plasmodium knowlesi manages with a shorter asexual blood-stage life cycle of ~24 h, vs ~48, and simply by comparison with producing a number of sparse internal caveolae structures at the iRBC surface. The function(s) of these structures has yet to be defined, though they are of high interest, as they likely perform critical physiological functions, and could become targets of intervention against P. knowlesi, as well as other species.
SYSTEMS BIOLOGY: THE WAY FORWARD
Enormous advances in modern biology, including the omics revolution, and in mathematical and computational modeling are aligning and current research suggests that methods of systems biology have great potential for elucidating various aspects of antigenic variation in P. knowlesi during experimental infections of NHPs. For instance, the combination of targeted experimental and modelling methods could focus on the parasite's growth and development ex vivo, as well as in both NHP and mosquito hosts, and ultimately reveal molecular mechanisms that govern antigenic variation in vivo and by definition contribute to successful parasitism, transmission and pathology. This emerging holistic research strategy will demand novel mathematical models and computational approaches, customized for the P. knowlesi rhesus-macaque model systems, to characterize gene and protein expression of variant antigens, and immune responses, as well as the principles of the switching dynamics and epigenetic players.
Through the integration of multiple data sources, careful interpretation of epigenetic changes, and the identification of external stimuli that activate these changes, novel signalling pathways may be discovered that might become the target of future vaccine or antimalarial intervention strategies. An improved understanding of the multilayer regulation of the SICAvar/var gene expression including epigenetics is within reach, and such knowledge will lead to a greater understanding of parasite immune evasion mechanism and host survival.
Antigenic variation is such a complex process that one can easily imagine diverse modeling strategies toward investigating different aspects. These could mathematically describe anything from the molecular interactions of SICA proteins with other intracellular proteins, to the sequence of expressed genes, as well as the overall competitive advantage that different SICAvar genes might confer to clonal populations. Here we briefly sketch two such strategies.
Stochastic modelling of switching patterns
Several mathematical models have been proposed to describe and predict mechanistic features of antigenic variation in a number of pathogens, including Plasmodium (Recker et al. 2004; Blyuss and Gupta, 2009; Recker et al. 2011; Buckee and Recker, 2012; Noble and Recker, 2012; Severins et al. 2012), Streptococcus, Neisseria (Lipsitch and O'Hagan, 2007), trypanosomes (Agur et al. 1989; Antia et al. 1996), and HIV (Nowak and May, 1991). The main targets of these studies have been switching patterns, infection duration and chronicity. As an example, switching patterns of var gene repertoires were modelled based on in vitro derived data from P. falciparum blood-stage cultures. In one of these studies, Recker's group observed a non-random, highly structured switching pathway where an initially dominant transcript switched via a set of intermediates either to a new dominant transcript, or back to the original (Noble et al. 2013). It was furthermore proposed that var genes have intrinsic probabilities of being activated or silenced and that switching patterns can be inferred from transcription time courses of several parasite populations from the same isolate, each starting with different variant distributions (Noble and Recker, 2012).
As an alternative to this strategy, and capitalizing on P. knowlesi model systems, it is in principle possible to design and test stochastic models that permit a dynamic characterization of switching patterns in vivo. For instance, one could develop Markov chain models to shed light on antigen switching during longitudinal infection of NHPs. Such models contain states, which here are combinations of expressed SICAvar genes, epigenomic profiles, or protein-based phenotypes and transitions, which correspond to switches among states and can be estimated from RNA-Seq studies of expressed genes before and after switches. Interestingly, this type of model automatically subsumes two null hypotheses as special cases, namely, that the switching is entirely deterministic, or that it is entirely random. The truth is naturally bounded by these two scenarios. Specifically, it will be of interest to study the ‘next set’ of expressed SICAvar genes, if some particular set is observed on different occasions. It is clear that such a strategy cannot be pursued in a human host, but it is possible to study these transitions in NHP infections. A disadvantage of the Markov approach is that it has no memory. That is, the probability of switching from one state to another is implicitly assumed to depend only on this state, but not on the history of the process before entering this state. With sufficient data, however, Markov chain models with memory can be established (Ross, 2006). If memory remains a problem, it is also possible to formulate differential equations whose state variables are the probabilities of expressing a certain gene set. This method accounts for some memory and is vaguely reminiscent of the Chemical Master Equation approach, but here leads to a homogeneous Poisson process that has an explicit solution (Stumpf et al. 2017). One could furthermore reformulate the process as a stochastic process with ‘short memory approximation,’ expressed as a stochastic differential equation (Zhabin, 2004; Goldwyn et al. 2011).
Kinetic modeling of infections
Rather than attempting to predict which variant might be expressed next, given a certain present state, one could ask if the expression of certain variants confers a dynamic advantage over other alternatives. Such an advantage could be due to an intrinsic growth advantage, enhanced immune evasion, or suppression of other variants. So far, there have been only a few attempts to model the kinetics of malarial antigenic variation and its impact on the outcome of the infection (Childs and Buckee, 2015). For example, Childs and Buckee analysed P. falciparum infections but suggested that antigenic variation does not explain the length of chronic infections (Childs and Buckee, 2015). Antia's group (Johnson et al. 2012) proposed that persistent infections could be explained by a high level of immunodominance coupled with either killing saturation or immune exhaustion. They also suggested that immune exhaustion plays an important role in the determination of when the primary infection should be treated in order to allow development of protection against a secondary infection.
As an alternative to these documented approaches, and with the aim of characterizing the immunogenicity, growth rates and competition among different variants, one could pursue a combination of an SIR (Susceptibile, Infected, Recovered) type of model with a Lotka-Volterra model of interspecies competition. The SIR formulation could be used to describe how different variants infect susceptible RBCs and thus produce different SICA-expressing iRBCs. The Lotka-Volterra formulation could add a variant-specific and cross-reactive (Johnson et al. 2012) immune response that could control the growth of each variant, and a set of interaction terms between parasite variants would characterize the suppression or promotion of one variant over another. This approach could just as well be applied to coinfections with different strains or species.
Key experimental considerations
As a first step towards these goals, a high-quality P. knowlesi nuclear genome assembly with manual annotation was generated recently and called the ‘MaHPIC Pk Genome Sequence’ (see Lapp et al. 2017 in this Special Issue). This nuclear genome sequence can serve as the basis for studies of gene regulation as a whole, for capturing regulatory gene networks, and specifically for propelling the field forward toward understanding SICAvar expression and switching dynamics in vivo. The combination of long reads Pacific Biosciences (PacBio) with Hi-C technology (Kaplan and Dekker, 2013), which generates genomic distance proxies to accurately position contigs without requiring sequence overlap, has greatly improved de novo scaffolds in P. knowlesi; this is contrasted in Table 2 with the basic information on the original nuclear genome sequence (Pain et al. 2008). Similar to the original genome sequence, the MaHPIC Pk Genome Sequence is based on genomic DNA from the Pk1(A+) clone of the Malayan strain of P. knowlesi. The 2008 assembly provided a glimpse into the SICAvar family on all 14 chromosomes, but had 190 gaps, many SICAvar fragments and misplaced genes. In total 29 full-length SICAvar genes were identified in 2008, and we have now confirmed at least 136 SICAvar genes (117 Type I and 19 Type II) and 22 ‘SICAvar gene segments’ that mainly (and curiously, as they may in fact have a functional purpose) contain the first two or last 3 exons (Lapp et al. 2017). The current MaHPIC assembly is advanced in terms of the overall confirmed 24·6 M genome size, with high sequencing coverage (151X with PacBio and 68X with Hi-C), 15 contigs and only 25 gaps (Table 2). Another P. knowlesi de novo genome assembly, based on the use of PacBio and culture adapted lines, was also reported recently (Moon et al. 2016): a thorough manual assessment of the SICAvar sequences in this assembly would be of interest as performed for the MaHPIC Pk Genome Sequence (Lapp et al. 2017).
Table 2.
MaHPIC Pk Nuclear Genome Sequence: basic information comparisons with previously reported P. knowlesi nuclear genome sequences from the Malayan Strain and two in vitro culture adapted lines
| Malayan strain a | Technology | Size | CDS | Coverage | Chr/ scaffolds | Contigs | Gaps |
|---|---|---|---|---|---|---|---|
| PKNOH/MaHPIC (Lapp et al. 2017) | PacBio+HiC | 24·8 M | 5217 | 151X (PacBio) 68X (Hi-C) | 14 | 14 | 25 |
| PKNH/Sanger v2 PlasmoDB b 2017 | Illumina | 24·4 M | 5282 | UA | 14 | 148 | 77 |
| PKNH/Sanger v1 (Pain et al. 2008) | WGS 4 & 40 kb insert libraries | 24·4 M | 5188 | 8X | NA | 204 | 190 |
| In vitro culture adapted lines | |||||||
| PKNA1-C.2 (Moon et al. 2016) | PacBio | 24·4 M | 5172 | 121X | 14 | 45 | NA |
| PKNA1-H.1 (Moon et al. 2016) | PacBio | 24·0 M | 5138 | 126X | 14 | 37 | NA |
To advance the field of systems biology of antigenic variation processes and mechanisms, P. knowlesi – macaque longitudinal infections can be monitored daily and and blood samples collected strategically with the goal of capturing and defining molecular host-parasite events and immune reponses related to the switching dynamics. Using methods reported recently for P. coatneyi and P. cynomolgi (Fonseca et al. 2016; Joyner et al. 2016), time series data can be generated for clinical parameters and all cell types available in blood count measurements and integrated with omics information to build and calibrate cellular models of antigenic variation. Such studies can be performed in spleen-intact and splenectomized macaque hosts. Chromatin remodelling and histone modifications are dynamic epigenetic changes responsive to external stimuli and, hence, can be integrated into these dynamic models, at least in principle.
Concluding remarks/future directions
Many hypotheses regarding the molecular and immunobiological basis of antigenic variation in malaria, can effectively be addressed with in vivo NHP infection models, beyond what is possible with humans, and advance quickly with the use of modern methods of multi-omic data generation and analysis, and sophisticated strategies of integrating heterogeneous information with tools from systems biology. In the past 20 years, many breakthroughs in our understanding of variant antigen gene expression have involved P. falciparum and in vitro culture systems, but it has become abundantly evident that gene and protein expression patterns are influenced by host factors. This truth is problematic for fully understanding antigenic variation in humans, because clinical studies with humans require treatment of blood-stage parasitemias. The well-established in vivo NHP – P. knowlesi infection model system provides a superb alternative for investigating longitudinal infections. With this host-pathogen system, the dynamics of antigenic variation and switching processes can be explored, and the full lifecyle interplay can be studied between the primate and vector hosts, including epigenetic regulation, cell–cell communication mechanisms and uncharted aspects of host-factor involvement; including the elusive role of the spleen. The potential of mechanistic studies is now opportune with the release of the MaHPIC Pk Genome Sequence, with the correct placement and annotation of 136 members of the SICAvar gene family (Lapp et al. 2017). Researchers can now test whether and how (i) Plasmodium variant antigen expression and switching patterns undergo predictable dynamics in the primate and mosquito hosts; (ii) epigenetic mechanisms within the parasite can drive in vivo switching of variant antigen expression; (iii) host factors can drive epigenetic control of antigenic variation in the parasite; and (iv) variant antigen switches and the switching rate correlate with antibody response patterns. As the scientific community takes on these challenges, it may also become viable to effectively explore selected question using P. falciparum – NHP models. One day, interventions may come to light that can interrupt switching mechanisms and thus accelerate the elimination of parasites, which would otherwise remain disguised and survive the host's immune response.
ACKNOWLEDGEMENTS
The authors would like to thank Jeremy D. DeBarry and Jessica C. Kissinger for review of the P. knowlesi nuclear genome information provided in Table 2, the Wellcome Trust Sanger Institute and GeneDB for making their P. knowlesi genome sequence (version 2) and annotation available to the community, PlasmoDB for analysis tools, and John W. Barnwell for his enthusiastic review of the manuscript.
FINANCIAL SUPPORT
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases; National Institutes of Health, Department of Health and Human Services (M.R.G., contract number HHSN272201200031C), (M.R.G., grant number R01AI065961), (K.G.L.R., grant number R01AI06775); The National Center for Research Resources (Y.N.P.R.C., grant number ORIP/OD P51OD011132); The University of California, Riverside (K.G.L.R., NIFA-Hatch-225935); and Institute Leadership Funds from La Jolla Institute for Allergy and Immunology (F.A.).
REFERENCES
- Agur Z., Abiri D. and Van Der Ploeg L. H. (1989). Ordered appearance of antigenic variants of African trypanosomes explained in a mathematical model based on a stochastic switch process and immune-selection against putative switch intermediates. Proceedings of the National Academy of Sciences of the United States of America 86, 9626–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khedery B., Barnwell J. W. and Galinski M. R. (1999). Antigenic variation in malaria: a 3′ genomic alteration associated with the expression of a P. knowlesi variant antigen. Molecular Cell 3, 131–141. [DOI] [PubMed] [Google Scholar]
- Alonso P. L., Brown G., Arevalo-Herrera M., Binka F., Chitnis C., Collins F., Doumbo O. K., Greenwood B., Hall B. F., Levine M. M., Mendis K., Newman R. D., Plowe C. V., Rodriguez M. H., Sinden R., Slutsker L. and Tanner M. (2011). A research agenda to underpin malaria eradication. PLoS Medicine 8, e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit-Avraham I., Pozner G., Eshar S., Fastman Y., Kolevzon N., Yavin E. and Dzikowski R. (2015). Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America 112, E982–E991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderios F., Noorrain A. and Vythilingam I. (2010). In vivo study of human Plasmodium knowlesi in Macaca fascicularis. Experimental Parasitology 124, 181–189. [DOI] [PubMed] [Google Scholar]
- Antia R., Nowak M. A. and Anderson R. M. (1996). Antigenic variation and the within-host dynamics of parasites. Proceedings of the National Academy of Sciences of the United States of America 93, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnot D. E. and Jensen A. T. (2011). Antigenic variation and the genetics and epigenetics of the PfEMP1 Erythrocyte Surface Antigens in Plasmodium falciparum Malaria. Advances in Applied Microbiology 74, 77–96. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J. Jr., Treatman C. and Wang H. (2009). PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Research 37, D539–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F., Bunnik E. M., Varoquaux N., Bol S. M., Prudhomme J., Vert J. P., Noble W. S. and Le Roch K. G. (2014). Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Research 24, 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F., Bunnik E. M., Varoquaux N., Vert J. P., Noble W. S. and Le Roch K. G. (2015). Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum: gene regulation via histone modifications, nucleosome positioning and nuclear architecture in P. falciparum. Bioessays 37, 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach O., Baier M., Pullwitt A., Fosiko N., Chagaluka G., Kalima M., Pfister W., Straube E. and Molyneux M. (2005). Falciparum malaria after splenectomy: a prospective controlled study of 33 previously splenectomized Malawian adults. Transactions of the Royal Society of Tropical Medicine and Hygiene 99, 861–867. [DOI] [PubMed] [Google Scholar]
- Bachmann A., Esser C., Petter M., Predehl S., Von Kalckreuth V., Schmiedel S., Bruchhaus I. and Tannich E. (2009). Absence of erythrocyte sequestration and lack of multicopy gene family expression in Plasmodium falciparum from a splenectomized malaria patient. PLoS ONE 4, e7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A., Predehl S., May J., Harder S., Burchard G. D., Gilberger T. W., Tannich E. and Bruchhaus I. (2011). Highly co-ordinated var gene expression and switching in clinical Plasmodium falciparum isolates from non-immune malaria patients. Cell Microbiology 13, 1397–1409. [DOI] [PubMed] [Google Scholar]
- Bachmann A., Petter M., Krumkamp R., Esen M., Held J., Scholz J. A., Li T., Sim B. K., Hoffman S. L., Kremsner P. G., Mordmuller B., Duffy M. F. and Tannich E. (2016). Mosquito passage dramatically changes var gene expression in controlled human plasmodium falciparum Infections. PLoS Pathogens 12, e1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell J., Howard R., Coon H. and Miller L. (1983a). Splenic requirement for antigenic variation and expression of the variant antigen on the erythrocyte membrane in cloned Plasmodium knowlesi malaria. Infection and Immunity 40, 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell J. W., Howard R. J. and Miller L. H. (1982). Altered expression of Plasmodium knowlesi variant antigen on the erythrocyte membrane in splenectomized rhesus monkeys. Journal of Immunology 128, 224–226. [PubMed] [Google Scholar]
- Barnwell J. W., Howard R. J. and Miller L. H. (1983b). Influence of the spleen on the expression of surface antigens on parasitized erythrocytes. Ciba Foundation Symposium 94, 117–136. [DOI] [PubMed] [Google Scholar]
- Baruch D. I., Pasloske B. L., Singh H. B., Bi X., Ma X. C., Feldman M., Taraschi T. F. and Howard R. J. (1995). Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82, 77–87. [DOI] [PubMed] [Google Scholar]
- Biggs B. A., Anders R. F., Dillon H. E., Davern K. M., Martin M., Petersen C. and Brown G. V. (1992). Adherence of infected erythrocytes to venular endothelium selects for antigenic variants of Plasmodium falciparum. Journal of Immunology 149, 2047–2054. [PubMed] [Google Scholar]
- Blyuss K. B. and Gupta S. (2009). Stability and bifurcations in a model of antigenic variation in malaria. Journal of Mathematical Biology 58, 923–937. [DOI] [PubMed] [Google Scholar]
- Brolin K. J., Ribacke U., Nilsson S., Ankarklev J., Moll K., Wahlgren M. and Chen Q. (2009). Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biology 10, R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. and Hills L. (1974). Antigenic variation and immunity to Plasmodium knowlesi: antibodies which induce antigenic variation and antibodies which destroy parasites. Transactions of the Royal Society of Tropical Medicine and Hygiene 68, 139–142. [DOI] [PubMed] [Google Scholar]
- Brown K. N. (1973). Antibody induced variation in malaria parasites. Nature 242, 49–50. [DOI] [PubMed] [Google Scholar]
- Brown K. N. and Brown I. N. (1965). Immunity to Malaria – Antigenic variation in chronic infections of Plasmodium Knowlesi. Nature 208, 1286–1288. [DOI] [PubMed] [Google Scholar]
- Buckee C. O. and Recker M. (2012). Evolution of the multi-domain structures of virulence genes in the human malaria parasite, Plasmodium falciparum. PLoS Computational Biology 8, e1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull P. C., Lowe B. S., Kortok M., Molyneux C. S., Newbold C. I. and Marsh K. (1998). Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nature Medicine 4, 358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik E. M., Chung D. W., Hamilton M., Ponts N., Saraf A., Prudhomme J., Florens L. and Le Roch K. G. (2013). Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biology 14, R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., Caler E., Crabtree J., Angiuoli S. V., Merino E. F., Amedeo P., Cheng Q., Coulson R. M., Crabb B. S., Del Portillo H. A., Essien K., Feldblyum T. V., Fernandez-Becerra C., Gilson P. R., Gueye A. H., Guo X., Kang'a S., Kooij T. W., Korsinczky M., Meyer E. V., Nene V., Paulsen I., White O., Ralph S. A., Ren Q., Sargeant T. J. et al. (2008). Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Fernandez V., Sundstrom A., Schlichtherle M., Datta S., Hagblom P. and Wahlgren M. (1998). Developmental selection of var gene expression in Plasmodium falciparum. Nature 394, 392–395. [DOI] [PubMed] [Google Scholar]
- Chien J. T., Pakala S. B., Geraldo J. A., Lapp S. A., Humphrey J. C., Barnwell J. W., Kissinger J. C. and Galinski M. R. (2016). High-quality genome assembly and annotation for Plasmodium coatneyi, generated using single-molecule real-time PacBio technology. Genome Announcements 4. doi: 10.1128/genomeA.00883-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs L. M. and Buckee C. O. (2015). Dissecting the determinants of malaria chronicity: why within-host models struggle to reproduce infection dynamics. Journal of Royal Society Interface 12, 20141379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A. Z., Hartl D. L. and Deitsch K. W. (2007). Epigenetic memory at malaria virulence genes. Proceedings of the National Academy of Sciences of the United States of America 104, 899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor V., Meyer E. V., Lapp S., Corredor-Medina C., Huber C. S., Evans A. G., Barnwell J. W. and Galinski M. R. (2004). A SICAvar switching event in Plasmodium knowlesi is associated with the DNA rearrangement of conserved 3′ non-coding sequences. Molecular and Biochemical Parasitology 138, 37–49. [DOI] [PubMed] [Google Scholar]
- Cortes A. and Deitsch K. W. (2017). Malaria epigenetics. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J. and Singh B. (2008). Knowlesi malaria: newly emergent and of public health importance? Trends in Parasitology 24, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J., Davis T. M. E., Lee K. S., Shamsul S. S. G., Matusop A., Ratnam S., Rahman H. A., Conway D. J. and Singh B. (2008). Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clinical Infectious Diseases 46, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily J. P., Le Roch K. G., Sarr O., Ndiaye D., Lukens A., Zhou Y., Ndir O., Mboup S., Sultan A., Winzeler E. A. and Wirth D. F. (2005). In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. Journal of Infectious Diseases 191, 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar C., Davis T. M. E., Cox-Singh J., Rafa'ee M. Z., Zakaria S. K., Divis P. C. S. and Singh B. (2009). Clinical and laboratory features of human Plasmodium knowlesi infection. Clinical Infectious Diseases 49, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K. W., Calderwood M. S. and Wellems T. E. (2001). Malaria. Cooperative silencing elements in var genes. Nature 412, 875–876. [DOI] [PubMed] [Google Scholar]
- Divis P. C., Singh B., Anderios F., Hisam S., Matusop A., Kocken C. H., Assefa S. A., Duffy C. W. and Conway D. J. (2015). Admixture in humans of two divergent Plasmodium knowlesi populations associated with different macaque host species. PLoS Pathogens 11, e1004888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh M. T., Voss T. S., Marty A. J., Duffy M. F., Good R. T., Thompson J. K., Freitas-Junior L. H., Scherf A., Crabb B. S. and Cowman A. F. (2005). Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121, 13–24. [DOI] [PubMed] [Google Scholar]
- Dzikowski R. and Deitsch K. W. (2008). Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. Journal of Molecular Biology 382, 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S., Wellems T. E. and Deitsch K. W. (2007). Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Reports 8, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton M. D. (1938). The agglutination of Plasmodium Knowlesi by immune serum. Journal of Experimental Medicine 67, 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca L. L., Alezi H. S., Moreno A., Barnwell J. W., Galinski M. R. and Voit E. O. (2016). Quantifying the removal of red blood cells in Macaca mulatta during a Plasmodium coatneyi infection. Malaria Journal 15, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Dzikowski R., Amulic B. and Deitsch K. (2007). Variable switching rates of malaria virulence genes are associated with chromosomal position. Molecular Microbiology 64, 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech C. and Chen N. (2013). Variant surface antigens of malaria parasites: functional and evolutionary insights from comparative gene family classification and analysis. BMC Genomics 14, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., Mancio-Silva L., Leal-Silvestre R. J., Gontijo A. M., Shorte S. and Scherf A. (2005). Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36. [DOI] [PubMed] [Google Scholar]
- Galinski M. R. and Barnwell J. W. (2012). Chapter 5 – Nonhuman Primate Models for Human Malaria Research A2 – Abee, Christian R In Nonhuman Primates in Biomedical Research, 2nd Edn. (ed. Mansfield K., Tardif S. and Morris T.), pp. 299–323. Academic Press, Boston. [Google Scholar]
- Galinski M. R. and Corredor V. (2004). Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology. Molecular Biochemistry Parasitology 134, 17–25. [DOI] [PubMed] [Google Scholar]
- Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M. et al. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwyn J. H., Imennov N. S., Famulare M. and Shea-Brown E. (2011). Stochastic differential equation models for ion channel noise in Hodgkin-Huxley neurons. Physical Review E 83, 041908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J. and Scherf A. (2013). Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiology 15, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handunnetti S. M., Mendis K. N. and David P. H. (1987). Antigenic variation of cloned Plasmodium fragile in its natural host Macaca sinica. Sequential appearance of successive variant antigenic types. Journal of Experimental Medicine 165, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M., Bannister L. H., Looareesuwan S. and Suntharasamai P. (1992). Cytoadherence and ultrastructure of Plasmodium falciparum-infected erythrocytes from a splenectomized patient. Infection and Immunity 60, 2225–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel M., David P. H. and Oligino L. D. (1983). Surface alterations of erythrocytes in Plasmodium falciparum malaria. Antigenic variation, antigenic diversity, and the role of the spleen. Journal of Experimental Medicine 157, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R. J. (1984). Antigenic variation of bloodstage malaria parasites. Philosophical Transactions of Royal Society of London B Biological Sciences 307, 141–158. [DOI] [PubMed] [Google Scholar]
- Howard R. J. and Barnwell J. W. (1984a). The detergent solubility properties of a malarial (Plasmodium knowlesi) variant antigen expressed on the surface of infected erythrocytes. Journal of Cell Biochemistry 24, 297–306. [DOI] [PubMed] [Google Scholar]
- Howard R. J. and Barnwell J. W. (1984b). Roles of surface antigens on malaria-infected red blood cells in evasion of immunity. Contemporary Topocs in Immunobiology 12, 127–200. [DOI] [PubMed] [Google Scholar]
- Howard R. J. and Barnwell J. W. (1984c). Solubilization and immunoprecipitation of 125I-labelled antigens from Plasmodium knowlesi schizont-infected erythrocytes using non-ionic, anionic and zwitterionic detergents. Parasitology 88 (Pt 1), 27–36. [DOI] [PubMed] [Google Scholar]
- Howard R. J. and Barnwell J. W. (1985). Immunochemical analysis of surface membrane antigens on erythrocytes infected with non-cloned SICA[+] or cloned SICA[−] Plasmodium knowlesi. Parasitology 91 (Pt 2), 245–261. [DOI] [PubMed] [Google Scholar]
- Howard R. J., Barnwell J. W. and Kao V. (1983). Antigenic variation in Plasmodium knowlesi Malaria – identification of the variant antigen on infected erythrocytes. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences 80, 4129–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R. J., Kao V. and Barnwell J. W. (1984). Protein antigens of Plasmodium knowlesi clones of different variant antigen phenotype. Parasitology 88(Pt 2), 221–237. [PubMed] [Google Scholar]
- Israeli A., Shapiro M. and Ephros M. A. (1987). Plasmodium falciparum malaria in an asplenic man. Transactions of Royal Society of Tropical Medicine and Hygiene 81, 233–234. [DOI] [PubMed] [Google Scholar]
- Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., Yang W., Turner L., Lavstsen T., Theander T. G., Peng W., Wei G., Jing Q., Wakabayashi Y., Bansal A., Luo Y., Ribeiro J. M., Scherf A., Aravind L., Zhu J., Zhao K. and Miller L. H. (2013). PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joergensen L., Bengtsson D. C., Bengtsson A., Ronander E., Berger S. S., Turner L., Dalgaard M. B., Cham G. K., Victor M. E., Lavstsen T., Theander T. G., Arnot D. E. and Jensen A. T. (2010). Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathogens 6, e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. L., Kochin B. F., Ahmed R. and Antia R. (2012). How do antigenically varying pathogens avoid cross-reactive responses to invariant antigens? Proceedings Biological Sciences 279, 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner C., Moreno A., Meyer E. V., Cabrera-Mora M., Ma H. C., Kissinger J. C., Barnwell J. W. and Galinski M. R. (2016). Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malaria Journal 15, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. and Dekker J. (2013). High-throughput genome scaffolding from in vivo DNA interaction frequency. Nature Biotechnology 31, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R. and Das Gupta B. (1932). A study of monkey-malaria and its experimental transmission to man. Indian Medical Gazette 67, 301–320. [PMC free article] [PubMed] [Google Scholar]
- Kocken C. H., Ozwara H., Van Der Wel A., Beetsma A. L., Mwenda J. M. and Thomas A. W. (2002). Plasmodium knowlesi provides a rapid in vitro and in vivo transfection system that enables double-crossover gene knockout studies. Infection and Immunity 70, 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocken C. H., Zeeman A. M., Voorberg-Van Der Wel A. and Thomas A. W. (2009). Transgenic Plasmodium knowlesi: relieving a bottleneck in malaria research? Trends in Parasitology 25, 370–374. [DOI] [PubMed] [Google Scholar]
- Korir C. C. and Galinski M. R. (2006). Proteomic studies of Plasmodium knowlesi SICA variant antigens demonstrate their relationship with P. falciparum EMP1. Infection Genetics and Evolution 6, 75–79. [DOI] [PubMed] [Google Scholar]
- Lapp S. A., Korir C. C. and Galinski M. R. (2009). Redefining the expressed prototype SICAvar gene involved in Plasmodium knowlesi antigenic variation. Malaria Journal 8, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp S. A., Korir-Morrison C., Jiang J., Bai Y., Corredor V. and Galinski M. R. (2013). Spleen-dependent regulation of antigenic variation in malaria parasites: Plasmodium knowlesi SICAvar expression profiles in splenic and asplenic hosts. PLoS ONE 8, e78014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp S. A., Mok S., Zhu L., Wu H., Preiser P. R., Bozdech Z. and Galinski M. R. (2015). Plasmodium knowlesi gene expression differs in ex vivo compared to in vitro blood-stage cultures. Malaria Journal 14, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp S. A., Geraldo J. A., Chien J. T., Ay F., Pakala S. B., Batugedara G., Humphrey J., The MaHPIC Consortium, Debarry J. D., Le Roche K. G., Galinski M. R. and Kissinger J. C. (2017). PacBio assembly of a Plasmodium knowlesi genome sequence with Hi-C correction and manual annotatin of the SICAvar gene family. Parasitology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T., Magistrado P., Hermsen C. C., Salanti A., Jensen A. T., Sauerwein R., Hviid L., Theander T. G. and Staalsoe T. (2005). Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malaria Journal 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech J. H., Barnwell J. W., Aikawa M., Miller L. H. and Howard R. J. (1984). Plasmodium falciparum malaria: association of knobs on the surface of infected erythrocytes with a histidine-rich protein and the erythrocyte skeleton. Journal of Cell Biology 98, 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J. E., Kyes S. A., Otto T. D., Feller A. I., Eastman R. T., Pinches R. A., Berriman M., Su X. Z. and Newbold C. I. (2013). Genome-wide profiling of chromosome interactions in Plasmodium falciparum characterizes nuclear architecture and reconfigurations associated with antigenic variation. Molecular Microbiology 90, 519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., Van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S. and Dekker J. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M. and O'hagan J. J. (2007). Patterns of antigenic diversity and the mechanisms that maintain them. Journal of Royal Society Interface 4, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio J. J., Riviere L. and Scherf A. (2007). Shared epigenetic mechanisms control virulence factors in protozoan parasites. Current Opinion in Microbiology 10, 560–568. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio J. J., Mancio-Silva L. and Scherf A. (2009). Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host & Microbe 5, 179–190. [DOI] [PubMed] [Google Scholar]
- Maeno Y., Quang N. T., Culleton R., Kawai S., Masuda G., Nakazawa S. and Marchand R. P. (2015). Humans frequently exposed to a range of non-human primate malaria parasite species through the bites of Anopheles dirus mosquitoes in South-central Vietnam. Parasites & Vectors 8, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand R. P., Culleton R., Maeno Y., Quang N. T. and Nakazawa S. (2011). Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among Humans and Anopheles dirus Mosquitoes, Southern Vietnam. Emerging Infectious Diseases 17, 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. H., Fremount H. N. and Luse S. A. (1971). Deep vascular schizogony of Plasmodium knowlesi in Macaca mulatta. Distribution in organs and ultrastructure of parasitized red cells. American Journal of Tropical Medicine and Hygiene 20, 816–824. [DOI] [PubMed] [Google Scholar]
- Miller L. H., Baruch D. I., Marsh K. and Doumbo O. K. (2002). The pathogenic basis of malaria. Nature 415, 673–679. [DOI] [PubMed] [Google Scholar]
- Moon R. W., Sharaf H., Hastings C. H., Ho Y. S., Nair M. B., Rchiad Z., Knuepfer E., Ramaprasad A., Mohring F., Amir A., Yusuf N. A., Hall J., Almond N., Lau Y. L., Pain A., Blackman M. J. and Holder A. A. (2016). Normocyte-binding protein required for human erythrocyte invasion by the zoonotic malaria parasite Plasmodium knowlesi. Proceedings of the National Academy of Sciences of the United States of America 113, 7231–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa S., Marchand R. P., Quang N. T., Culleton R., Manh N. D. and Maeno Y. (2009). Anopheles dirus co-infection with human and monkey malaria parasites in Vietnam. International Journal of Parasitology 39, 1533–1537. [DOI] [PubMed] [Google Scholar]
- Noble R. and Recker M. (2012). A statistically rigorous method for determining antigenic switching networks. PLoS ONE 7, e39335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble R., Christodoulou Z., Kyes S., Pinches R., Newbold C. I. and Recker M. (2013). The antigenic switching network of Plasmodium falciparum and its implications for the immuno-epidemiology of malaria. Elife 2, e01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. A. and May R. M. (1991). Mathematical biology of HIV infections: antigenic variation and diversity threshold. Mathematical Biosciences 106, 1–21. [DOI] [PubMed] [Google Scholar]
- Onditi F. I., Nyamongo O. W., Omwandho C. O., Maina N. W., Maloba F., Farah I. O., King C. L., Moore J. M. and Ozwara H. S. (2015). Parasite accumulation in placenta of non-immune baboons during Plasmodium knowlesi infection. Malaria Journal 14, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozwara H., Langermans J. A., Maamun J., Farah I. O., Yole D. S., Mwenda J. M., Weiler H. and Thomas A. W. (2003). Experimental infection of the olive baboon (Paplio anubis) with Plasmodium knowlesi: severe disease accompanied by cerebral involvement. American Journal of Tropical Medicine and Hygiene 69, 188–194. [PubMed] [Google Scholar]
- Pain A., Bohme U., Berry A. E., Mungall K., Finn R. D., Jackson A. P., Mourier T., Mistry J., Pasini E. M., Aslett M. A., Balasubrammaniam S., Borgwardt K., Brooks K., Carret C., Carver T. J., Cherevach I., Chillingworth T., Clark T. G., Galinski M. R., Hall N., Harper D., Harris D., Hauser H., Ivens A., Janssen C. S., Keane T., Larke N., Lapp S., Marti M., Moule S. et al. (2008). The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 455, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini E. M., Zeeman A. M., Voorberg V. A. N. D. E. R. W. A. and Kocken C. H. (2016). Plasmodium knowlesi: a relevant, versatile experimental malaria model. Parasitology, 1–15. doi: 10.1017/S0031182016002286. [DOI] [Google Scholar]
- Recker M., Nee S., Bull P. C., Kinyanjui S., Marsh K., Newbold C. and Gupta S. (2004). Transient cross-reactive immune responses can orchestrate antigenic variation in malaria. Nature 429, 555–558. [DOI] [PubMed] [Google Scholar]
- Recker M., Buckee C. O., Serazin A., Kyes S., Pinches R., Christodoulou Z., Springer A. L., Gupta S. and Newbold C. I. (2011). Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathogens 7, e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L. and Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annual Review of Biochemistry 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin A., Ben-Hur S. and Regev-Rudzki N. (2017). Malaria parasites distribute subversive messages across enemy lines. Trends in Parasitology 33, 2–4. [DOI] [PubMed] [Google Scholar]
- Roberts D. J., Craig A. G., Berendt A. R., Pinches R., Nash G., Marsh K. and Newbold C. I. (1992). Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature 357, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S. M. (2006). Introduction to Probability Models, 9th Edn, Academic Press, Inc. [Google Scholar]
- Rutledge G. G., Bohme U., Sanders M., Reid A. J., Cotton J. A., Maiga-Ascofare O., Djimde A. A., Apinjoh T. O., Amenga-Etego L., Manske M., Barnwell J. W., Renaud F., Ollomo B., Prugnolle F., Anstey N. M., Auburn S., Price R. N., Mccarthy J. S., Kwiatkowski D. P., Newbold C. I., Berriman M. and Otto T. D. (2017). Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature 542, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., Gysin J. and Lanzer M. (1998). Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO Journal 17, 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severins M., Klinkenberg D. and Heesterbeek H. (2012). How selection forces dictate the variant surface antigens used by malaria parasites. Journal of the Royal Society Interface 9, 246–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Kim Sung L., Matusop A., Radhakrishnan A., Shamsul S. S., Cox-Singh J., Thomas A. and Conway D. J. (2004). A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I. and Miller L. H. (1995). Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. D., Subramanian G., Gamain B., Baruch D. I. and Miller L. H. (2000). Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Molecular and Biochemical Parasitology 110, 293–310. [DOI] [PubMed] [Google Scholar]
- Spence P. J., Jarra W., Levy P., Reid A. J., Chappell L., Brugat T., Sanders M., Berriman M. and Langhorne J. (2013). Vector transmission regulates immune control of Plasmodium virulence. Nature 498, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf P. S., Smith R. C. G., Lenz M., Schuppert A., Müller F.-J., Babtie A., Chan T. E., Stumpf M. P. H., Please C. P., Howison S. D., Arai F. and Macarthur B. D. (2017). Stem cell differentiation is a stochastic process with memory. bioRxiv doi: 10.1101/101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A. and Wellems T. E. (1995). The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100. [DOI] [PubMed] [Google Scholar]
- Van De Werken H. J., Landan G., Holwerda S. J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B. A., Verstegen M. J., De Wit E., Tanay A. and De Laat W. (2012). Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature Methods 9, 969–972. [DOI] [PubMed] [Google Scholar]
- Van Der Wel A. M., Tomas A. M., Kocken C. H., Malhotra P., Janse C. J., Waters A. P. and Thomas A. W. (1997). Transfection of the primate malaria parasite Plasmodium knowlesi using entirely heterologous constructs. Journal of Experimental Medicine 185, 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Serrato E., Corredor V. and Galinski M. R. (2003). Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi to Plasmodium knowlesi. Infection Genetics and Evolution 3, 67–73. [DOI] [PubMed] [Google Scholar]
- Vembar S. S., Droll D. and Scherf A. (2016). Translational regulation in blood stages of the malaria parasite Plasmodium spp.: systems-wide studies pave the way. Wiley Interdisciplinary Reviews RNA 7, 772–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz J. C., Bartfai R., Petter M., Langer C., Josling G. A., Tsuboi T., Schwach F., Baum J., Rayner J. C., Stunnenberg H. G., Duffy M. F. and Cowman A. F. (2012). PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host & Microbe 11, 7–18. [DOI] [PubMed] [Google Scholar]
- Wang C. W., Hermsen C. C., Sauerwein R. W., Arnot D. E., Theander T. G. and Lavstsen T. (2009). The Plasmodium falciparum var gene transcription strategy at the onset of blood stage infection in a human volunteer. Parasitology International 58, 478–480. [DOI] [PubMed] [Google Scholar]
- Warimwe G. M., Recker M., Kiragu E. W., Buckee C. O., Wambua J., Musyoki J. N., Marsh K. and Bull P. C. (2013). Plasmodium falciparum var gene expression homogeneity as a marker of the host-parasite relationship under different levels of naturally acquired immunity to malaria. PLoS ONE 8, e70467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T. E., Hayton K. and Fairhurst R. M. (2009). The impact of malaria parasitism: from corpuscles to communities. Journal of Clinical Investigation 119, 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabin D. N. (2004). Stochastic processes with short memory. ARXIV arXiv:math/0401144v0401141.
- Zhou Z., Mitchell R. M., Kariuki S., Odero C., Otieno P., Otieno K., Onyona P., Were V., Wiegand R. E., Gimnig J. E., Walker E. D., Desai M. and Shi Y. P. (2016). Assessment of submicroscopic infections and gametocyte carriage of Plasmodium falciparum during peak malaria transmission season in a community-based cross-sectional survey in western Kenya, 2012. Malaria Journal 15, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A. V., Cornish A. S., Maudhoo M. D., Gibbs R. M., Zhang X., Pandey S., Meehan D. T., Wipfler K., Bosinger S. E., Johnson Z. P., Tharp G. K., Marcais G., Roberts M., Ferguson B., Fox H. S., Treangen T., Salzberg S. L., Yorke J. A. and Norgren R. B. Jr. (2014). A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biology Direct 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]